Panel A in Fig 4 and panels A and F in S4 Fig are not presented correctly. The lanes in the corrected figures are separated by tracks for other mutants that are not relevant for the current work and can be seen in the original blots, provided here as S7 and S8 Figs. The authors apologise for the mistake and have provided corrected versions, along with the original blots that were used to create the figures. These errors do not affect the conclusions of this article.
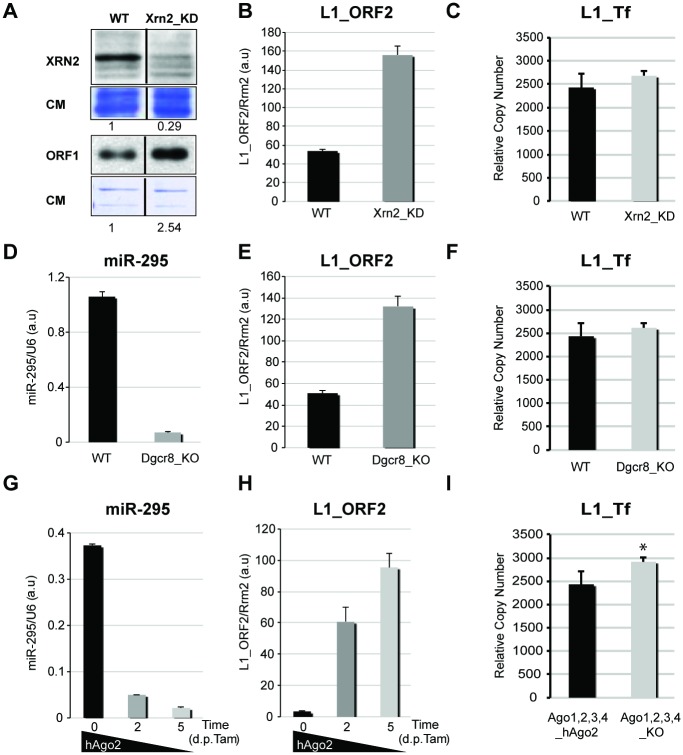
Fig 4. L1 mRNA levels and genomic copy-number in various knock-out and knock-down mESC lines.
A. Western analysis of XRN2 and L1_ORF1 accumulation in WT and Xrn2_KD mESCs; CM: Coomassie staining of total protein. B. qRT-PCR analysis of L1_ORF2 mRNA levels in WT and Xrn2_KD mESCs. C. qPCR analysis of L1_Tf copy-number in WT and Xrn2_KD mESCs. D–E. qRT-PCR analysis of miR-295 (D) and L1_ORF2 mRNA (E) levels in WT and Dgcr8_KO mESCs. F. qPCR analysis of L1_Tf copy-number in WT and Dgcr8_KO mESCs. G–H. qRT-PCR analysis of miR-295 (G) and L1_ORF2 mRNA (H) levels upon hAgo2 deletion in Tamoxifen-treated Ago1,2,3,4_KO mESCs. I. qPCR analysis of L1_Tf copy-number in Ago1,2,3,4_KO_hAgo2 mESCs before and after hAgo2 deletion. *: p-value<0.1.
Supporting Information
A. Western analysis of RRP6 and L1_ORF1 accumulation in WT and Rrp6_KD mESCs; CM: Coomassie staining of total protein. B. Accumulation of Tf_5′-UTR (+) and (−) sRNAs detected by qRT-PCR in WT and Xrn2_KD mESCs. C. qPCR analysis of L1_Tf copy-number in WT and Rrp6_KD mESCs. D. L1_ORF2, Tf, Gf and A sub-type mRNAs accumulation detected by qRT-PCR in Xrn2_KD and Rrp6_KD mESCs. E. Accumulation of miR-320 detected by qRT-PCR in WT and Dgcr8_KO mESCs. F. Western analysis of AGO2 accumulation in WT and Ago1,2,3,4_KO_hAgo2 mESCs before and after hAgo2 deletion induced by tamoxifen; CM: Coomassie staining of total protein. G. Accumulation of the Hmga2 and Btg2 mRNAs, respectively targeted by mmu-miR-196a and mmu-let-7a/mmu-miR-132, analyzed by qRT-PCR before and after deletion of hAgo2. H. mRNA accumulation of L1_Tf, _Gf and _A sub-types detected by qRT-PCR before and after hAgo2 deletion. I. mRNA accumulation of a single Tf_L1 subtype located on chromosome 17, analyzed by semi-quantitative RT-PCR before and after hAgo2 deletion.
(TIF)
The red rectangles highlight the part of the gel presented in Fig 4A. CM: Coomassie staining of total protein.
(TIF)
A. Original blot for the S4A Fig. The red rectangles highlight the part of the gel presented in S4A Fig. B. The original blot for the S4F Fig is presented. CM: Coomassie staining of total protein.
(TIF)
Reference
- 1. Ciaudo C, Jay F, Okamoto I, Chen C-J, Sarazin A, et al. (2013) RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells. PLoS Genet 9(11): e1003791 doi: 10.1371/journal.pgen.1003791 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Western analysis of RRP6 and L1_ORF1 accumulation in WT and Rrp6_KD mESCs; CM: Coomassie staining of total protein. B. Accumulation of Tf_5′-UTR (+) and (−) sRNAs detected by qRT-PCR in WT and Xrn2_KD mESCs. C. qPCR analysis of L1_Tf copy-number in WT and Rrp6_KD mESCs. D. L1_ORF2, Tf, Gf and A sub-type mRNAs accumulation detected by qRT-PCR in Xrn2_KD and Rrp6_KD mESCs. E. Accumulation of miR-320 detected by qRT-PCR in WT and Dgcr8_KO mESCs. F. Western analysis of AGO2 accumulation in WT and Ago1,2,3,4_KO_hAgo2 mESCs before and after hAgo2 deletion induced by tamoxifen; CM: Coomassie staining of total protein. G. Accumulation of the Hmga2 and Btg2 mRNAs, respectively targeted by mmu-miR-196a and mmu-let-7a/mmu-miR-132, analyzed by qRT-PCR before and after deletion of hAgo2. H. mRNA accumulation of L1_Tf, _Gf and _A sub-types detected by qRT-PCR before and after hAgo2 deletion. I. mRNA accumulation of a single Tf_L1 subtype located on chromosome 17, analyzed by semi-quantitative RT-PCR before and after hAgo2 deletion.
(TIF)
The red rectangles highlight the part of the gel presented in Fig 4A. CM: Coomassie staining of total protein.
(TIF)
A. Original blot for the S4A Fig. The red rectangles highlight the part of the gel presented in S4A Fig. B. The original blot for the S4F Fig is presented. CM: Coomassie staining of total protein.
(TIF)