Abstract
Eggshell damages lead to economic losses in the egg production industry and are a threat to human health. We examined 49-wk-old Rhode Island White hens (Gallus gallus) that laid eggs having shells with significantly different strengths and thicknesses. We used HiSeq 2000 (Illumina) sequencing to characterize the chicken transcriptome and whole genome to identify the key genes and genetic mutations associated with eggshell calcification. We identified a total of 14,234 genes expressed in the chicken uterus, representing 89% of all annotated chicken genes. A total of 889 differentially expressed genes were identified by comparing low eggshell strength (LES) and normal eggshell strength (NES) genomes. The DEGs are enriched in calcification-related processes, including calcium ion transport and calcium signaling pathways as reveled by gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis. Some important matrix proteins, such as OC-116, LTF and SPP1, were also expressed differentially between two groups. A total of 3,671,919 single-nucleotide polymorphisms (SNPs) and 508,035 Indels were detected in protein coding genes by whole-genome re-sequencing, including 1775 non-synonymous variations and 19 frame-shift Indels in DEGs. SNPs and Indels found in this study could be further investigated for eggshell traits. This is the first report to integrate the transcriptome and genome re-sequencing to target the genetic variations which decreased the eggshell qualities. These findings further advance our understanding of eggshell calcification in the chicken uterus.
Introduction
Calcified eggshells provide protection to egg contents, prevent contamination by microorganisms, and protect the embryo [1]. Eggshells also regulate the exchange of metabolic gases and water, and provide calcium to the embryo. Eggshell damage results in large economic losses each year [2]. Eggshells are composed of two shell membranes, a calcium carbonate layer, and a foamy layer of cuticle [3]. Mineralization of the avian eggshell is a complex process involving the precipitation of calcium carbonate that results from interaction between this mineral and the organic matrix during the nucleation and growth phases [4–7]. Calcification occurs on the shell membrane in the cellular uterine fluid, which contains the inorganic minerals and organic matrix precursors of the eggshell [8]. Calcification is the lengthiest phase of egg formation and occurs in three stages over a 6–22h period following ovulation.
Eggshells contain a proteinaceous matrix of 95% calcium carbonate and 3.5% organic material. Calcium (Ca2+) and bicarbonate (HCO3 -) comprise approximately 37.5% and 58% of the eggshell weight, respectively; other minerals are present in trace amounts. Calcium is not stored in the uterus before formation of the shell [5, 9]. Eggshell calcification requires the interaction of numerous processes, including transcellular and/or paracellular transport of Ca2+ and secretion of matrix proteins. Many researchers have noted increased uterine Ca2+ content during eggshell calcification [10–16]. This supports the hypothesis that a major portion of the Ca2+ secreted into the uterus occurs against the electrochemical potential gradient and, therefore, involves active transport. Both Ca2+ and HCO3 - are continuously provided during eggshell formation via blood plasma [17]. Successful interaction of calcium deposition and organic matrix proteins is critically important in the formation of eggshells. Uncovering the key genes that regulate calcium deposition and their mutations would provide an important basis for understanding the eggshell formation process and for altering eggshell qualities.
A farm-reared Rhode Island White population showed variable eggshell strength. Approximately 15% of individuals laid eggs with very weak eggshells in the same rearing environment as the remainder of the population. This is an excellent model for studying the genetic basis of eggshell formation. In this study, we used systematic whole transcriptome and genome re-sequencing approaches to examine the gene expression and mutations in Rhode Island White hens that lay eggs with extremely different eggshell qualities (Fig 1).
Fig 1. Graphical representation of the experimental strategy.
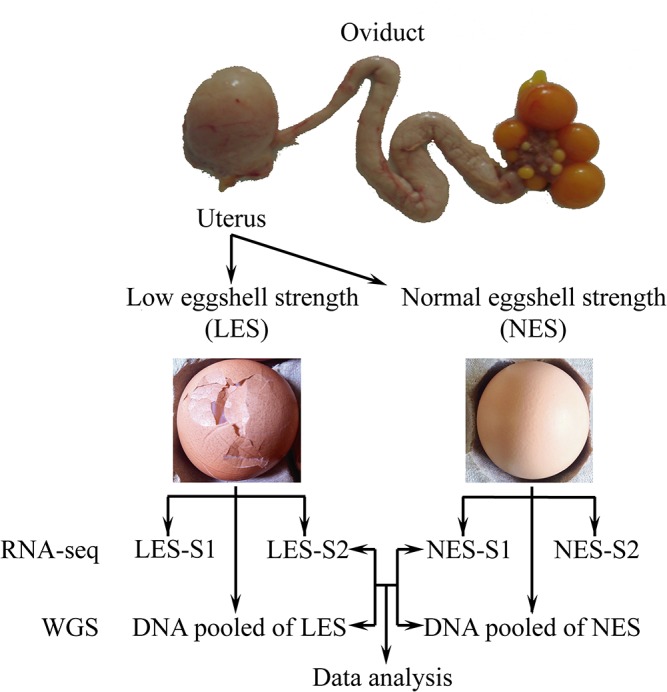
The oviposition cycle of all birds was measured daily, and birds were slaughtered 18 h after ovulation. This stage generally represents the fast growth stage during the eggshell calcification. Individuals from the same sire family that laid eggs with one of two extremely different eggshell qualities were selected. We selected eight birds with normal eggshell strength (NES) and eight birds with very low eggshell strength (LES) for further study. Two biological replicate samples from each group were selected for transcriptome sequencing, and one pooled DNA library for each group was made for whole genome re-sequencing. Finally, we integrated transcriptome and whole genome re-sequencing data to discover the genetic information related to eggshell quality.
Materials and Methods
Ethics statement
Animal experiments were approved by the Animal Care and Use Committee of China Agricultural University (permit number: SYXK 2007–0023). Euthanasia was performed by cervical dislocation in order to quickly obtain the tissue samples to minimize any effect on gene expression changes. All experiments were performed according to regulations and guidelines established by this committee.
Animals and samples preparation
We observed an exceptionally high proportion of broken eggs in Rhode Island White hens (broken eggshell rate in the laying house >15%). Then, we decided to use this population to study the genetic basis of this phenomenon. All birds were individually reared in cages located in a windowless, air-conditioned poultry house. They were subjected to a cycle of 14 h light: 10 h darkness and were fed ad libitum on a layer diet. All the birds used in this study are healthy. Hens that laid broken eggs were generally evenly distributed across the house.
This population was one of the breeding lines in a breeding farm. The egg production of each bird was recorded daily. The selected birds were representative of the normal production ratio of differing shell quality in this species based on the 25–48 weeks daily records. When the birds were 49 weeks of age, we measured the eggshell qualities of the eggs laid by each bird for 3 days. We measured the eggshell strength and thickness. We carefully checked each hen cage to ensure there was no physical cause for the broken eggs. The experimental design is shown in Fig 1.
Measurements of eggshell qualities
Hens from which 50% or more of the eggs were broken during a 10-day recording period were considered to lay low eggshell strength (LES) eggs. After the first observation stage, the eggshell qualities were measured for each hen in the population. Three eggs were measured for each hen and the average values were considered to represent the eggshell qualities. Eggshell breaking strength and eggshell thickness were measured using the Egg Multi Tester EMT-5200 (Robotmation Co. Ltd). Measurement methods were as previously published [18]. After measurement, descriptive statistics were calculated for all individuals (Fig 2). We chose the individuals that had significantly different eggshell breaking strength and thickness. Finally, we chose eight dam families, each dam family include one normal eggshell strength (NES) bird and one low eggshell strength (LES) bird, for further study (Table 1). The oviposition cycle of all 16 birds was measured daily, and they were slaughtered 18 h after the previous oviposition. This stage generally represents the fast growth stage during eggshell calcification [6].
Fig 2. The eggshell strength (A) and eggshell thickness (B) in the normal eggshell strength (NES) group and the low eggshell strength (LES) group.
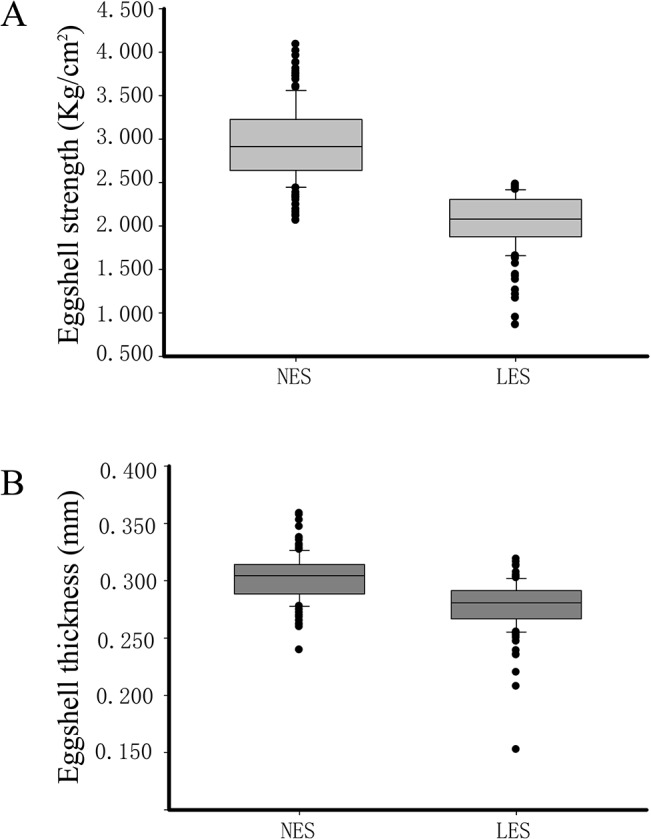
There are 154 and 159 individuals for the NES group and LES group, respectively.
Table 1. Eggshell quality in Rhode Island White hens.
| F. | Low eggshell quality | Normal eggshell quality | ||||
|---|---|---|---|---|---|---|
| Samples | EST (mm) | ESS (Kg/cm2) | Samples | EST (mm) | ESS (Kg/cm2) | |
| 51 | L1 | 0.263 | 2.080 | N1 | 0.240 | 2.722 |
| 67 A | L2 | 0.247 | 1.776 | N2 | 0.353 | 3.776 |
| 30 | L3 | 0.282 | 1.757 | N3 | 0.291 | 3.072 |
| 48 | L4 | 0.250 | - | N4 | 0.283 | 2.383 |
| 43 A | L5 | 0.235 | - | N5 | 0.306 | 3.132 |
| 73 | L6 | 0.279 | - | N6 | 0.307 | 3.188 |
| 42 | L7 | 0.255 | 1.420 | N7 | 0.302 | 2.890 |
| 18 | L8 | 0.275 | - | N8 | 0.313 | 3.199 |
| Mean+SD | 0.261±0.017** | - | Mean+SD | 0.299±0.030** | 3.045±0.380 | |
F.: Families. EST: Eggshell thickness. ESS: Eggshell-breaking strength. “-”: Eggshell broken prior to collection.
**: P<0.01 (one way ANOVA analysis).
A Four chickens from two dam families were randomly selected for RNA-Seq analysis. Eight equal quantities of DNA samples were mixed to form a pooled DNA for each group.
Uterus tissues were collected from the hens at 18 h after previous oviposition and were snap-frozen in liquid nitrogen and stored at −80°C prior to total RNA extraction. Two individuals were randomly selected from each group for RNA-Seq analysis (Table 1). Blood samples were collected in acid citrate dextrose (ACD) anticoagulant and stored at −20°C prior to DNA extraction. Equal-quantity DNA samples from all eight hens in each group were mixed to form pooled DNA samples.
RNA-Seq and whole genome re-sequencing
Total mRNA was extracted for sequencing using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. The total mRNA was treated with DNase I and purified with the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The quality and concentration of RNA was checked with an Agilent 2100 Bioanalyzer (Agilent) and Nanodrop ND-2000 spectrophotometer (Thermo Scientific). Genomic DNA for whole genome re-sequencing was prepared with a TIANamp Blood DNA Kit (Tiangen) according to the manufacturer’s instructions.
Two biological replicates were generally used in the RNA-Seq related studies [19, 20] as the estimate biological variation highly reliably even when the number of replicates is very small [21]. We used an Illumina HiSeq2000 [22] to perform genome re-sequencing and RNA-Seq. Libraries were constructed following the Illumina Paired-end Sequencing Library Preparation Protocol. A 100-bp paired-end library was constructed for each sample according to the manufacturer’s instructions (Illumina). Library quality and concentration were determined using an Agilent 2100 Bioanalyzer. A standard Illumina base-calling pipeline was used to process the raw fluorescent images and the called sequences. Read qualities were evaluated using the FastQC package (www.bioinformatics.babraham.ac.uk/projects/fastqc/). For genome re-sequencing data, short-reads were trimmed 15-bp from the 3′-end, according the base quality distributions. For RNA-Seq data, short-reads were trimmed 20-bp from the 3′-end according the base quality distributions.
Reads alignment and mutation analysis for whole genome re-sequencing data
Paired-end short reads were aligned to the G. gallus reference genome (Ensembl v72, Galgal 4) using the Burrows-Wheeler Aligner (BWA, version 0.6.2) algorithm with default parameters [23]. SAMTools [24] was used to remove duplicate reads that might have been caused by PCR. To enhance the accuracy of read alignment, aligned reads were realigned at putative SNPs and Indel positions using the Genome Analysis Toolkit (GATK) realigner algorithm (version 2.5.2) [25]. Base quality scores were recalibrated using the GATK recalibration algorithm. The options used for SNP and Indel calling were a minimum 10-read mapping depth, consensus quality of 30, and prior likelihood for heterozygosity value of 0.001.
Transcriptomic data analysis
Reads were mapped to the G. gallus genome assembly (Ensembl v72, Galgal 4) using TopHat version 2.0.8 [26]. TopHat RNA-Seq analysis was used with a maximum of 5 mismatches per read. Expression levels of the transcripts were quantified as reads per kilobase per million reads (RPKM) [27]. Genes with RPKM < 0.04 were not included in the analysis. The DESeq [28] algorithm was used to analyze DEGs between LES and NES samples with a P-value of 0.05 as a threshold. Ensembl gene IDs from each group were uploaded to the DAVID Functional Annotation Tool and analyzed for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment [29]. The classification stringency was set to the default parameters. Novel isoform detection and differential expression analysis was carried out using the Cufflinks pipeline [30]. Cufflinks transcripts marked as "u" (intergenic transcript), were treated as novel transcripts.
Genetic mutation annotation
We used SnpEff [31] to annotate mutations and classify mutations into different categories from whole genome re-sequencing and RNA-Seq data. Ensembl (version 72) and dbSNP (ftp://ftp.ncbi.nih.gov/snp/organisms/chicken_9031/VCF/, updated in June 11, 2013) were the source databases used for annotation.
Quantitative real-time PCR (qPCR) and direct PCR sequencing
Expression of mRNA was verified by qPCR with cDNA from 16 uterine tissue samples. Chicken β-actin (GenBank Accession ID: NM_204305) served as a housekeeping gene. Primer 3 Input (v. 0.4.0) was used with default parameters to generate primer pairs for selected genes (Table A in S1 File). Total RNA was extracted from uterine tissue using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Total RNA (50 ng/μl) was reverse transcribed using M-MLV reverse transcriptase (Promega Corporation) as recommended by the supplier. The qPCR was performed using the SYBR Green Master Mix (Life Technologies) on an ABI 7500 Real Time system (Applied Biosystems, USA). The experiments were carried out in triplicate. The cycling conditions were 95°C for 5 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. A melting curve was obtained at 60 to 95°C for each sample amplified. We used the 2−ΔΔCT method [32] to analyze the relative changes in gene expression from qPCR experiments.
DNA was extracted by TIANamp Blood DNA Kit (Tiangen) according to the manufacturer’s instructions for isolation of genomic DNA from 16 individual blood samples. Direct PCR sequencing is the method of choice for detecting mutations in the target samples. The primers used for PCR amplification are listed in Table B in S1 File. PCR amplifications were performed with 50ng genomic DNA, 10μl PCR Mix (Yeasen, USA), and add to 20 μl with water. Amplification conditions were: 5 min at 95°C, 35 cycles of 30 s at 95°C, 30 s at the T m of the primers, and 1 min per kb to be amplified at 72°C, followed by 10 min at 72°C. The PCR products were run on a 1% agarose gel (Gene tech, Spain) in 1×TAE buffer. Sequencing was performed with the ABI 3730XL sequencing.
Results
Identification of differentially expressed genes and novel isoforms
The total number of reads varied from 37 to 71 million, and 78.09–80.72% of filtered reads were mapped on the reference chicken genome (Table C in S1 File). Among these, 99.4% of the reads mapped to protein-coding genes. Approximately 68% of the reads were mapped to exons (Fig. A in S2 File). Some differences in read counts were detected between biological replicates, especially between NES-S1 and NES-S2. The quantity of total mRNA analyzed was identical in these samples, indicating that the different read counts may have arisen during processing. However, this discrepancy did not significantly affect gene expression analysis because the RPKM values were corrected for the total number of read counts for each sample.
To identify the DEGs between the two groups, we used DESeq [28] with adjusted P < 0.05 for all comparisons. A total of 889 DEGs were detected between the samples (Fig 3, S1 Dataset). Of these, 255 DEGs were significantly up-regulated in NES hens and 634 were significantly up-regulated in LES hens, indicating that uterine genes are differentially expressed in these groups during the calcification of eggshells.
Fig 3. Differentially expressed genes (DEGs) between the low eggshell strength (LES) group and the normal eggshell strength (NES) group.
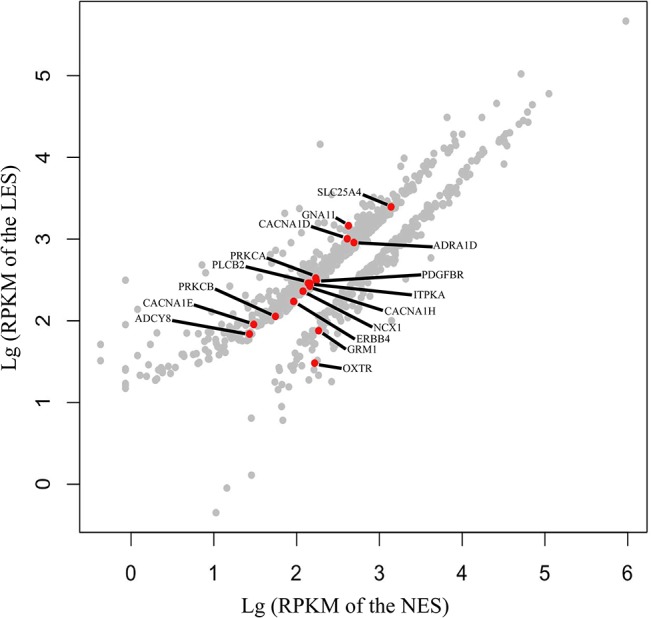
The red spots represent DEGs belonging to a calcium signaling pathway (KO: 04020) (P < 0.01).
Different isoforms of the gene and novel transcripts were also measured in this study (S2 Dataset). On average 11,092 genes, 21,696 known isoforms and 5,048 novel isoforms were expressed in each sample group (RPKM > 1). There were 118 different promoters and 124 different transcription start sites (P < 0.01). However, these promoter and transcription start sites were not significantly different between the two sample groups.
Functional annotation and pathway enrichment analysis of the DEGs
To gain insight into the biological processes regulated during eggshell formation and to determine which processes were coded by DEGs, the DEGs were subjected to GO term enrichment analysis using the DAVID system [29]. We set the threshold P value <0.01 for the GO terms and enrichment score value >1. The GO terms were then classified according to biological process (Fig 4, S3 Dataset). The majority of DEGs were involved in ion transport in the uterus during formation of the eggshell. The GO terms included metal ion transport (GO: 0030001), di- or tri- valent inorganic cation transport (GO: 0015674), ion transport (GO: 0006811), calcium ion transport (GO: 0006816), and cation transport (GO: 0006812). Another important group of DEGs encoded extracellular matrix proteins. Two GO terms were related to the extracellular matrix and were involved in extracellular matrix organization (GO: 0030198) and extracellular structure organization (GO: 0043062). GO terms for muscle contraction (GO: 0006936) and muscle system processes (GO: 0003012) corresponded to seven DEGs for binding actin, ATP, and Ca2+. Finally, the GO term for positive regulation of cell-substrate adhesion (GO: 0010811) was related to Ca2+ and extracellular matrix binding and cytokine activity.
Fig 4. Enriched biological process GO term among differentially expressed genes.
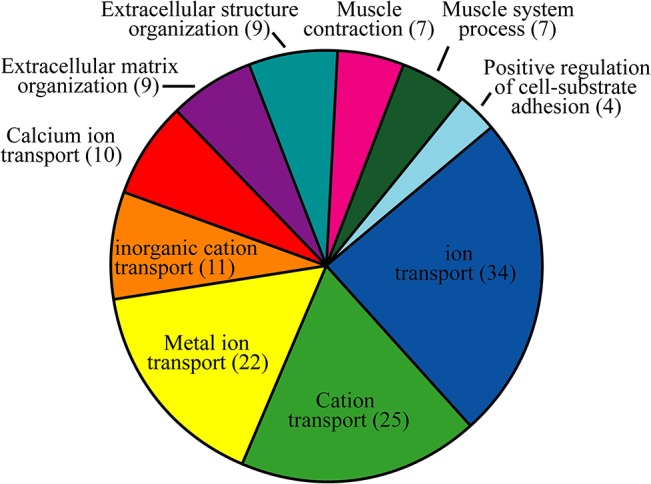
Numbers in parentheses indicate the number of differentially expressed genes.
KEGG is a knowledge base for systematic analysis of gene functions in terms of the networks of genes and molecules [33]. In this study, KEGG pathway analysis of the DEGs revealed several enriched pathways (P < 0.01), including a calcium signaling pathway (KO: 04020), focal adhesion (KO: 04510), extracellular matrix (ECM)-receptor interaction (KO: 04512), and vascular smooth muscle contraction (KO: 04270) (Table 2). The 16 DEGs significantly enriched in Ca2+ signaling pathway, and these DEGs belong to G proteins, voltage-dependent Ca2+ channels, phosphatidylinositol phospholipase C, Na+/Ca2+ exchangers, and Na+ channels (Fig. B in S2 File). We also confirmed the expression patterns by qPCR which of 16 DEGs in Ca2+ signaling pathways (Fig 5), the results of which correlated to the RPKM values estimated by RNA sequencing (r = 0.71).
Table 2. KEGG pathway enriched in differentially expressed genes.
| Pathway | Pathway definition | P value | Genes |
|---|---|---|---|
| KO: 04510 | Focal adhesion | 3.88E-05 | TNR, MAPK10, MYL2, TNC, ITGA8, COL6A3, COL3A1, PARVA, PRKCB, PRKCA, COL5A1, ITGA9, THBS2, MBSP, PDGFRB, MAPK9, VCL, ITGAV, ACTN2, COL11A1, VIN, SPP1, MET |
| KO: 04512 | ECM-receptor interaction | 2.04E-04 | TNR, SV2B, TNC, ITGA8, COL6A3, COL3A1, COL5A1, ITGA9, THBS2, ITGAV, CD36, COL11A1, VTN, SPP1 |
| KO: 04270 | Vascular smooth muscle contraction | 1.20E-03 | GNA11, ADRA1D, CACNA1D, CO6, PLCB2, MRV11, PRKCB, PRKCA, AVTG2, ADCY8, MBSP, KCNMB4 |
| KO: 04020 | Calcium signaling pathway | 2.32E-03 | GNA11, CACNA1E, ADRA1D, CACNA1D, PLCB2, PRKCB, ITPKA, PRKCA, NCX1, ERBB4, ADCY8, CACNA1H, SLC25A4, PDGFRB, GRM1, OXTR |
Fig 5. Validation of gene expression in the calcium signaling pathway.
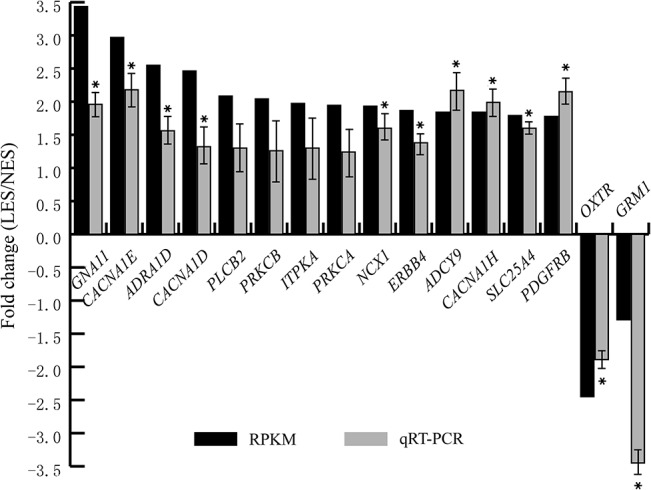
The qPCR was performed to quantify gene expression level. The 2−ΔΔCT method was used analyze changes in gene expression relative to chicken β-actin. Fold changes between the low eggshell strength (LES) group and the normal eggshell strength (NES) group were calculated for each gene. * represent the significant differentially expressed genes by the qPCR (P < 0.05, one way ANOVA analysis). Fold-changes based on RNA-Seq and qPCR were highly correlated (r = 0.71).
Whole genome re-sequencing to identify mutations related to eggshell qualities
Whole genome re-sequencing of chickens produced 32.2 and 36.8 Gbp, with coverage depths of 29.82X and 33.77X, for the LES and NES groups, respectively. There were 7,450,661 SNPs in the LES group and 7,588,813 SNPs in the NES group, which corresponded to approximately 8.6 and 7.6 SNP/kb, respectively. Of the SNPs identified in the LES and NES groups, 35.98% and 36.82%, respectively, were novel in comparison to the dbSNP database [34] (Table D in S1 File). SNP density varied and was lowest in sex chromosomes (Z and W chromosomes; avg. 3.78 and 3.90 SNP/kb, respectively) and highest in chromosomes 16 and LEG64 (avg. 12.63 and 13.00 SNP/kb, respectively). SNP density in chromosomes varied from 7.19 to 9.27 SNP/kb (Fig. C in S2 File). We detected 3,671,519 SNPs (484,857 and 563,437 unique SNPs in the LES and NES groups, respectively) for protein-coding genes in the two pooled samples. The number of SNPs and SNP density is similar with previous studies [35–37]. The majority of SNPs were detected in the intronic regions (38.82% for NES group, 38.82% for LES group) and intergenic regions (42.04% for NES group, 42.03% for LES group).
A total of 809,833 and 829,107 Indels were found, corresponding to approximately 0.79 and 0.81 Indels/kb for LES and NES hens, respectively. Of these Indels, 88.83% and 88.90% in the LES and NES groups, respectively, were novel (Table D in S1 File) compared with dbSNP. We detected 508,035 Indels (67,925 and 76,319 unique Indels in the LES and NES groups, respectively) for protein-coding genes in the two pooled samples. Number of detected Indels is also similar with previous report [35, 38]. The majority of Indels were detected in the intronic regions (40.21% for NES group, 40.26% for LES group) and intergenic regions (42.74% for NES group, 42.56% for LES group). Indels found in the exons were deserved in the further analysis.
Non-synonymous or frame-shift mutations may cause an amino acid substitution in the corresponding protein product, thus affecting the phenotype of the host organism. In this study, 1775 instances of non-synonymous and 19 frame-shift mutations were detected in 427 DEGs. The GO terms of the 427 DEGs included di- or tri- valent inorganic cation transport, calcium ion transport, ion transport, metal ion transport, and cation transport. Seven genes were assigned to calcium ion transport biological processing of gene ontology (GO-BP) terms. In the LES group, two common non-synonymous variations were detected in NCX1, two frame-shift mutations and one common non-synonymous variation were detected in PRKCB, and one non-synonymous variation and one unique non-synonymous variation was detected in TRPC6. In the NES group, one non-synonymous variation and one unique non-synonymous variation was detected in CACNA1H. Ovocleidin-116 (OC-116) is an important matrix protein that is involved in eggshell calcification. We conducted further validation one non-synonymous mutation of CACNA1H gene to test relationship of the SNP and eggshell strength in larger population (Table 3). In our experimental population, we observed that the genotype (AG) has a strong impact on the eggshell strength and thickness (P value<0.001, Table 3). According to the RNA-seq and whole genome re-sequencing, 10 non-synonymous variations occurred in OC-116, including one and two unique variations in the LES and NES groups, respectively. The SNP mutations in the DEGs of the calcium signaling pathway and matrix proteins are presented in Table 4. The Indel mutations in the DEGs are presented in Table E in S1 File.
Table 3. Eggshell qualities of different genotypes for CACNA1H gene.
| Genotypes | n | Eggshell strength (Kg/cm2) | Eggshell thickness (mm) |
|---|---|---|---|
| AG | 68 | 2.572±0.390*** | 0.300±0.022*** |
| AA | 160 | 2.216±0.513 | 0.287±0.024 |
| GG | 10 | 2.225±0.591 | 0.282±0.030 |
*** represent significant difference in line (P<0.001) used one-way analysis of variance.
Table 4. Non-synonymous mutations of the differentially expressed genes in calcium signaling pathways and matrix proteins.
| Genes | Depth | Position | Ref/new coding | Common | PULES. | PUNES. |
|---|---|---|---|---|---|---|
| NCX1 | 16 | 3:15742695 R | Atc/Ttc | * | ||
| NCX1 | 19 | 3:15742743 R | Gac/Aac | * | ||
| CACNA1E | 20 | 8:5693839 R | gaT/gaA | * | ||
| CACNA1E | 16 | 8:5715790 R | Acg/Ccg | * | ||
| CACNA1H | 33 | 14:5238069 R | cGc/cAc | * | ||
| CACNA1H | 19 | 14:5306181 R | tTg/tCg | * | ||
| OXTR | 14 | 12:19266364 R | Gtg/Atg | * | ||
| PRKCB | 17 | 14:6644303 R | Tcg/Ccg | * | ||
| DGKE | 38 | 18:6239253 R | Aac/Gac | * | ||
| ITGA8 | 33 | 2:20316822 R | Gca/Aca | * | ||
| ADCY8 | 29 | 2:140544236 R | gCg/gTg | * | ||
| GRM1 | 26 | 3:46087815 R | Gct/Act | * | ||
| GRM1 | 27 | 3:46087816 R | gCt/gAt | * | ||
| ADRA1D | 38 | 4:87899647 R | aCt/aAt | * | ||
| PLCB2 | 20 | 5:665344 R | Atc/Gtc | * | ||
| OC-116 | 19 | 4:45085851 R | cAt/cGt | * | ||
| OC-116 | 19 | 4:45086211 R | cCc/cTc | * | ||
| OC-116 | 29 | 4:45086662 R | Gct/Tct | * | ||
| OC-116 | 28 | 4:45086718 R | aGt/aCt | * | ||
| OC-116 | 26 | 4:45086821 R | Atg/Gtg | * | ||
| OC-116 | 22 | 4:45087409 R | Cct/Act | * | ||
| OC-116 | 14 | 4:45087496 R | Ggt/Agt | * | ||
| OC-116 | 34 | 4:45087542 R | caA/caC | * | ||
| OC-116 | 24 | 4:45087709 R | Acc/Ccc | * | ||
| OC-116 | 21 | 4:45087750 R | gCt/gTt | * | ||
| LTF | 27 | 9:4098717 R | Gtt/Att | * | ||
| LTF | 19 | 9:4099069 R | aGg/aAg | * | ||
| LTF | 23 | 9:4105617 R | Att/Gtt | * | ||
| LTF | 26 | 9:4106072 R | Tca/Gca | * | ||
| LTF | 22 | 9:4103017 R | aGc/aCc | * | ||
| SPP1 | 20 | 4:45075410 R | Gtg/Atg | * |
Depth: the number of reads to support the variation. Uppercase letter represent single-nucleotide polymorphism (SNP).
R SNP sites were also detected using RNA-Seq.
PULES: Potential unique low eggshell strength. PUNES: Potential unique normal eggshell strength.
Discussion
In chickens, eggshell calcification occurs in the hen uterus and is one of the most rapid biological mineralization processes known [5].
We compared gene expression and genetic variation between two different phenotypic groups to facilitate identification of candidate genes associated with eggshell calcification in chickens. Previous global gene expression profiling studies of hen oviducts during sexual maturation and eggshell formation revealed large numbers of DEGs [1, 39]. GO analysis and KEGG prediction showed that these DEGs are involved in several major pathways. The Ca2+ signaling system includes a very large set of components, including receptors, channels, and Ca2+ pumps and exchangers that can be mixed and matched to create a diverse array of signaling units for delivery of Ca2+ signals that have very different spatial and temporal properties [40]. The free intracellular Ca2+ concentration is an important link in the excitation-contraction coupling mechanism of vascular smooth muscle [41]. In the present study, 16 DEGs (Table 2, Fig. B in S2 File) were included in predicted Ca2+ signaling pathways and 14 DEGs (Table 2) were involved in extracellular matrix (ECM)-receptor interaction, suggesting that these are candidate genes for eggshell calcification in chickens. Previous studies have shown that the most highly represented GO terms are related to genes encoding ion-transport proteins [1, 5, 42]. The chicken uterine model provides a large list of ion-transport proteins that supply Ca2+ and HCO3 - and maintain cellular ionic homeostasis [43].
Five genes (CACNA1D, CACNA1E, CACNA1H, PRKCB, and NCX1) (Table 2, Fig. B in S2 File, S3 Dataset) are involved in the Ca2+ transport and the Ca2+ signaling pathway. Expression of the five Ca2+ transport genes was up-regulated in the LES group relative to that in the NES group. The NCX1 gene, encoding a calcium pump and exchanger, is predicted to be present only in the apical membranes of uterine glandular cells [43] and to participate in uterine Ca2+ secretion [44]. The role of the transporter is clearly established in mammalian intestines and kidneys [45]. Influx of Ca2+ through voltage-dependent Ca2+ channels plays a critical role in various biological functions [46]. Seven non-synonymous SNP variations were found in the CACNA1E, CACNA1H, PRKCB, and NCX1 genes.
Calcium transport in the uterus is associated with the counter-transport of Na+ and Cl- [5, 47] and with the secretion of minute masses of shell matrix proteins. In this study, three subunits (SCNN1A, SCNN1B, and SCNN1G) of the Na+ channel and the CLCN2 gene were down-regulated in the LES group as compared to the NES group, and one non-synonymous SNP variation was found in the SCNN1A gene. Earlier observations indicate that the SCNN1 gene family is over-expressed in the uterus; expression of SCNN1A is higher in older hens compared to younger hens and SCNN1B variations are associated with eggshell strength [48]. The SCNN1G gene is over-expressed during shell calcification relative to SCNN1A and SCNN1B, suggesting its involvement in shell calcification in the uterus [43]. The CLCN2 channel, a member of the CLCN (Cl- channel) family, is thought to participate in various functions such as regulation of cardiac activity [49, 50]. Thus, our study suggests that those ion exchangers and channels may lead to changes in calcium carbonate deposition during eggshell calcification in the uterus.
Lots of QTL regions affecting eggshell thickness have been detected by previous linkage studies and they distribute on GGA1, GGA2, GGA5, and GGA7 [51–54]. Some candidate genes for eggshell thickness were also identified on GGA2, GGA4, GGA8 and GGA9[55, 56]. About 64 DEGs were located in the 15 QTL regions that are related with eggshell strength, and 36 DEGs were located in the 3 QTL regions which are related with eggshell thickness (S4 Dataset). Among those DEGs that had been confirmed in the related QTL regions, CACNA1D, GNA11 and OXTR were found in calcium signaling pathway.
Eggshells consist of minerals associated with an organic matrix that is composed of proteins, glycoproteins, and proteoglycans. Proteins specifically produced in the uterus are more likely to play a role in eggshell formation than those produced elsewhere [43]. Different organic matrices participate in the three stages of eggshell calcification [17].
Recently, a high-throughput tandem mass spectrometry approach allowed the identification of >500 eggshell matrix proteins [57]. In our study, expression of OC-116, osteopontin (SPP1), and ovalbumin was down-regulated in the LES group as compared to the NES group. Ovalbumin presence in uterine fluid is predominant at the initial stage of eggshell formation [17] and ovocleidin-116 is a major component of the chicken eggshell matrix observed throughout the palisade layer and most abundant in uterine fluid during the intense eggshell calcification phase [58, 59]. These proteins are likely to play a fundamental role in eggshell formation since it potentially modifies calcite crystal growth in vitro [5, 6, 60]. Our results indicated that OC-116 and ovalbumin gene expressions positively correlated eggshell calcification during the intense eggshell calcification phase. We also found that expression of ovotransferrin (LTF) was up-regulated in the LES group as compared to the NES group. The LTF protein is observed in the eggshell [61] and in the uterine fluid, especially at the initial stage of eggshell formation. Calcium carbonate crystals grown in vitro in the presence of purified LTF showed large modifications of the calcite morphology [62]. The mechanism of the LTF on the calcite morphology and final eggshell ultrastructure needs to be further studies. These observations indicated that these differential expression genes might be one of the reasons for the eggshell quality variations. These matrix proteins are, therefore, good choice for the study of any relationship between eggshell matrix proteins and shell quality. Ten, one, and five non-synonymous mutations were detected in OC-116, SPP1, and LTF, respectively. Organic matrix proteins involved in eggshell formation have been identified [6] and polymorphisms in eggshell organic matrix genes have been associated with eggshell quality [55]. The extracellular matrix genes, ACAD (carbohydrate binding), ITGA8 (metal ion binding), LGALS3 (carbohydrate binding), and COL11A1 (extracellular matrix binding) were differentially expressed between the two groups of the present study.
We sequenced eight predicted group-specific mutations using Sanger-sequencing for each individual to verify the results of whole genome re-sequencing. However, the results are not fully supported by Sanger sequencing. Inconsistence of the RNA-Seq, whole genome re-sequencing and direct PCR sequencing results were found in this study. All eight mutations for further examined, which discovered by whole-genome resequencing, were also detected by Sanger sequencing. However, examined eight predicted group-specific mutations are not the real group-specific mutations after further validating. Predicted group-specific mutations were found at low frequencies in the control group. We filtered out low frequency mutations when analyzing group-specific mutations, however, this method introduced false-positive mutations which were found in these predicted sites (Fig. D in S2 File). Pooled whole genome resequencing results are good at finding mutations, but not good at discovering the group-specific mutations if resequencing coverage is not high enough, especially in difficulty to locate rare mutations. This needs to be further studied in the whole genome resequencing.
In summary, we analyzed DEGs and mutations from Rhode Island White hens with different eggshell qualities. The identified DEGs are involved in calcium ion transport and calcium signaling pathways. Some important matrix proteins, such as OC-116, LTF and SPP1, were also differentially expressed in the uterus between two groups during eggshell formation stage. Thirty-one non-synonymous mutations (as identified using RNA-Seq and WGS) were found in the DEGs. Genes which involved in ion transport and matrix proteins might be the major genetic factors affecting the eggshell strength in the studied Rhode Island White hen population. The genotype (rs312834462) of CACNA1H gene has a strong impact on the eggshell strength and thickness. This is the first report to integrate the transcriptic and genomic re-sequencing to target the genetic variations which decreased the eggshell qualities. Our study enhances our understanding of genetic variation in eggshell quality in relation to eggshell formation.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Primers used for qPCR of calcium signaling pathway genes. Table B. Primers used for direct PCR sequencing of the potential group-specific mutations. Table C. RNA sequencing of eggshell quality in Rhode Island White hens. Table D. Whole genome re-sequencing of eggshell quality in Rhode Island White hen. Table E. Frameshift mutations in the differentially expressed genes in KEGG pathway.
(PDF)
Distribution of mapped reads to different transcript types and gene regions. Top graph (A and B) indicates the proportion of transcripts belonging to different RNA species. Numbers within pie chart indicate number of reads while numbers in parenthesis indicate number of transcripts. Bottom graph (C) shows the read distribution within protein coding genes. Fig. B. Differentially expressed genes (DEGs) in calcium signal pathway. The red markers represent the DEGs in LES compared with NES. NCX was a NCX1 gene; ADCY was a ADCY8 gene; CaV1 was a CACNA1D gene; CaV2 was a CACNA1E gene; CaV3 was a CACNA1H gene; GPCR included ADRA1D, GRM1 and OXTR genes; PTK included PDGFRB and ERBB4 genes; Gq was a GNA11 gene; PLC β was a PLCB2 gene; ANT was a SLC25A4 gene; IP3 was a ITPKA gene and PKC included PRKCA and PRKCB genes. Red marker circle of Genes in KEGG pathway also involved in biological process (BP) term of calcium ion transport. Fig. C. Graphical representation of two pooled samples genomes in a Circos plots. Form outside to inside of circles represent chromosome, density of total genes within the chromosome that from red to blue represent for gene density which is from high to low, mapping depth of NES and LES, SNP mutations in the chromosome of NES and LES, Indel mutations in the chromosome of NES and LES. A’s chromosmes are from 1 to 15 based on Mb and B’s chromosomes are from 16 to 32 based on 10 Kb, including sex chromosomes. Fig. D. Direct PCR sequencing detected mutations of CACNA1H and LTF gene from the RNA-seq and whole genome re-sequencing. The results indicated that potential unique SNPs for each group by RNA-seq and genome re-sequencing were not unique SNPs, but the SNP frequencies have difference for each group. The SNP of CACNA1H gene is rs312834462, and LTF gene’s SNP is rs10724671.
(PDF)
Acknowledgments
The authors thank the Beijing Agricultural University Group (BAU) for maintaining and providing birds.
Data Availability
The data sets supporting the results presented here are available in the Sequence Read Archive (SRA) repository under accession numbers SRP034444, SRP034445, and SRP034511.
Funding Statement
This work was supported by the Chinese Agricultural Research System (CARS-41), Program for Changjiang Scholar and Innovation Research Team in University (IRT1191), the Fundamental Research Funds for the Central Universities (2011JS003) and the National Scientific Supporting Projects of China (2011BAD28B03). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jonchère V, Réhault-Godbert S, Hennequet-Antier C, Cabau C, Sibut V, Cogburn LA, et al. Gene expression profiling to identify eggshell proteins involved in physical defense of the chicken egg. BMC genomics. 2010;11(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nys Y, Bain M, Van Immerseel F. Improving the Safety and Quality of Eggs and Egg Products: Egg chemistry, production and consumption: Elsevier; 2011. [Google Scholar]
- 3. Solomon SE. Egg & eggshell quality: Iowa State University Press; 1997. [Google Scholar]
- 4. Arias JL, Fink DJ, Xiao SQ, Heuer AH, Caplan AI. Biomineralization and eggshells-cell-mediated acellular compartments of mineralized extracellular-matrix. International Review of Cytology—a Survey of Cell Biology, Vol 145. 1993;145:217–50. [DOI] [PubMed] [Google Scholar]
- 5. Nys Y, Hincke M, Arias J, Garcia-Ruiz J, Solomon S. Avian eggshell mineralization. poultry and avian biology reviews. 1999;10(3):143–66. [Google Scholar]
- 6. Nys Y, Gautron J, Garcia-Ruiz JM, Hincke MT. Avian eggshell mineralization: biochemical and functional characterization of matrix proteins. Comptes Rendus Palevol. 2004;3(6):549–62. [Google Scholar]
- 7. Hincke MT, Chien Y-C, Gerstenfeld LC, McKee MD. Colloidal-gold immunocytochemical localization of osteopontin in avian eggshell gland and eggshell. Journal of Histochemistry & Cytochemistry. 2008;56(5):467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gautron J, Hincke MT, Mann K, Panhéleux M, Bain M, McKee MD, et al. Ovocalyxin-32, a Novel Chicken Eggshell Matrix Protein isolation, amino acid sequencing, cloning, and immunocytochemical localization. Journal of Biological Chemistry. 2001;276(42):39243–52. [DOI] [PubMed] [Google Scholar]
- 9.Sauveur B. Poultry reproduction and egg production. 1988; XXI, 449P.
- 10. El Jack M, Lake P. The content of the principal inorganic ions and carbon dioxide in uterine fluids of the domestic hen. Journal of Reproduction and Fertility. 1967;13(1):127–32. [DOI] [PubMed] [Google Scholar]
- 11. Mongin P, Sauveur B. Composition of the uterine fluid and albumin during the stay of ovum in the uterus in domestic hens]. Comptes rendus hebdomadaires des séances de l'Académie des sciences Série D: Sciences naturelles. 1970;270(13):1715 [PubMed] [Google Scholar]
- 12. Edwards N. The secretion of glucose into the egg of Gallus domesticus and observations on the uterine secretion of cations. British Poultry Science. 1977;18(6):641–9. [DOI] [PubMed] [Google Scholar]
- 13. Arad Z, Eylath U, Ginsburg M, Eyal-Giladi H. Changes in uterine fluid composition and acid-base status during shell formation in the chicken. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 1989;257(4):R732–R7. [DOI] [PubMed] [Google Scholar]
- 14. Nakada T, Koga O. Stimulation of secretion of shell gland fluid and calcium by the presence of ovum or ovum-like mass containing artificial yolk in the oviduct uterus of the hen. Japanese Poultry Science. 1990;27. [Google Scholar]
- 15. Nys Y, Zawadzki J, Gautron J, Mills A. Whitening of brown-shelled eggs: mineral composition of uterine fluid and rate of protoporphyrin deposition. Poultry Science. 1991;70(5):1236–45. [DOI] [PubMed] [Google Scholar]
- 16. Panheleux M, Kalin O, Gautron J, Nys Y. Features of eggshell formation in guinea fowl: kinetics of shell deposition, uterine protein secretion and uterine histology. British Poultry Science. 1999;40(5):632–43. [DOI] [PubMed] [Google Scholar]
- 17. Gautron J, Hincke M, Nys Y. Precursor matrix proteins in the uterine fluid change with stages of eggshell formation in hens. Connective tissue research. 1997;36(3):195–210. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y, Hou Z, Chen Z, Zheng J, Chen S, Qu L, et al. Low-density lipoprotein receptor-related protein 2 gene is associated with egg-quality traits in dwarf layers. Poultry science. 2011;90(12):2718–22. 10.3382/ps.2011-01751 [DOI] [PubMed] [Google Scholar]
- 19. Chitwood JL, Rincon G, Kaiser GG, Medrano JF, Ross PJ. RNA-seq analysis of single bovine blastocysts. BMC genomics. 2013;14(1):350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laiho A, Kotaja N, Gyenesei A, Sironen A. Transcriptome profiling of the murine testis during the first wave of spermatogenesis. PloS One. 2013;8(4):e61558 10.1371/journal.pone.0061558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9(2):321–32. [DOI] [PubMed] [Google Scholar]
- 22. Shendure J, Ji H. Next-generation DNA sequencing. Nature Biotechnology. 2008;26(10):1135–45. 10.1038/nbt1486 [DOI] [PubMed] [Google Scholar]
- 23. Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–95. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20(9):1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5(7):621–8. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 28. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Da Wei Huang BTS, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. nature protocols. 2008;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 30. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28(5):511–5. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cingolani P, Platts A, Coon M, Nguyen T, Wang L, Land SJ, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 33. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29(1):308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fan WL, Ng CS, Chen CF, Lu MYJ, Chen YH, Liu CJ, et al. Genome-wide patterns of genetic variation in two domestic chickens. Genome biology and evolution. 2013;5(7):1376–92. 10.1093/gbe/evt097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J, He X, Ruan J, Dai M, Chen J, Zhang Y, et al. ChickVD: a sequence variation database for the chicken genome. Nucleic acids research. 2005;33(suppl 1):D438–D41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Daetwyler HD, Capitan A, Pausch H, Stothard P, Van Binsbergen R, Brøndum RF, et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nature genetics. 2014;46(8):858–65. 10.1038/ng.3034 [DOI] [PubMed] [Google Scholar]
- 38. Yan Y, Yi G, Sun C, Qu L, Yang N. Genome-Wide Characterization of Insertion and Deletion Variation in Chicken Using Next Generation Sequencing. PloS One. 2014;9(8):e104652 10.1371/journal.pone.0104652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dunn I, Wilson P, Lu Z, Bain M, Crossan C, Talbot R, et al. New hypotheses on the function of the avian shell gland derived from microarray analysis comparing tissue from juvenile and sexually mature hens. General and comparative endocrinology. 2009;163(1):225–32. 10.1016/j.ygcen.2009.03.006 [DOI] [PubMed] [Google Scholar]
- 40. Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nature Reviews Molecular Cell Biology. 2003;4(7):517–29. [DOI] [PubMed] [Google Scholar]
- 41. Himpens B, Missiaen L, Casteels R. Ca2+ homeostasis in vascular smooth muscle. Journal of vascular research. 1995;32(4):207–19. [DOI] [PubMed] [Google Scholar]
- 42. Bar A. Differential regulation of calbindin in the calcium-transporting organs of birds with high calcium requirements. The journal of poultry science. 2009;46(4):267–85. [Google Scholar]
- 43. Jonchère V, Brionne A, Gautron J, Nys Y. Identification of uterine ion transporters for mineralisation precursors of the avian eggshell. BMC physiology. 2012;12(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sauveur B, Mongin P. Comparative study of uterine fluid and egg albumen in shell gland of hen. Annales De Biologie Animale Biochimie Biophysique. 1971;11(2):213. [Google Scholar]
- 45. Philipson KD, Nicoll DA. Sodium-calcium exchange: a molecular perspective. Annual Review of Physiology. 2000;62(1):111–33. [DOI] [PubMed] [Google Scholar]
- 46. Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annual review of cell and developmental biology. 2000;16(1):521–55. [DOI] [PubMed] [Google Scholar]
- 47. Vetter AE, O'Grady SM. Sodium and anion transport across the avian uterine (shell gland) epithelium. The Journal of Experimental Biology. 2005;208(3):479–86. [DOI] [PubMed] [Google Scholar]
- 48. Fan Y, Hou Z, Yi G, Xu G, Yang N. The sodium channel gene family is specifically expressed in hen uterus and associated with eggshell quality traits. BMC Genetics. 2013;14(90). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Britton FC, Wang G- L, Huang ZM, Ye L, Horowitz B, Hume JR, et al. Functional characterization of novel alternatively spliced ClC-2 chloride channel variants in the heart. Journal of Biological Chemistry. 2005;280(27):25871–80. [DOI] [PubMed] [Google Scholar]
- 50. Huang ZM, Prasad C, Britton FC, Ye LL, Hatton WJ, Duan D. Functional role of CLC-2 chloride inward rectifier channels in cardiac sinoatrial nodal pacemaker cells. Journal of molecular and cellular cardiology. 2009;47(1):121–32. 10.1016/j.yjmcc.2009.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu W, Li D, Liu J, Chen S, Qu L, Zheng J, et al. A genome-wide SNP scan reveals novel loci for egg production and quality traits in white leghorn and brown-egg dwarf layers. PloS One. 2011;6(12):e28600 10.1371/journal.pone.0028600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sasaki O, Odawara S, Takahashi H, Nirasawa K, Oyamada Y, Yamamoto R, et al. Genetic mapping of quantitative trait loci affecting body weight, egg character and egg production in F2 intercross chickens. Animal Genetics. 2004;35(3):188–94. [DOI] [PubMed] [Google Scholar]
- 53. Wardêcka B, Olszewski R, Jaszczak K, Ziêba G, Pierzchala M, Wiciñska K. Relationship between microsatellite marker alleles on chromosomes 1–5 originating from the Rhode Island Red and Green-legged Partrigenous breeds and egg production and quality traits in F~ 2 mapping population. Journal of applied genetics. 2002;43(3):319–30. [PubMed] [Google Scholar]
- 54. Wright D, Kerje S, Lundström K, Babol J, Schütz K, Jensen P, et al. Quantitative trait loci analysis of egg and meat production traits in a red junglefowl× White Leghorn cross. Animal Genetics. 2006;37(6):529–34. [DOI] [PubMed] [Google Scholar]
- 55. Dunn I, Joseph N, Bain M, Edmond A, Wilson P, Milona P, et al. Polymorphisms in eggshell organic matrix genes are associated with eggshell quality measurements in pedigree Rhode Island Red hens. Animal Genetics. 2009;40(1):110–4. 10.1111/j.1365-2052.2008.01794.x [DOI] [PubMed] [Google Scholar]
- 56. Yao J, Chen Z, Xu G, Wang X, Ning Z, Zheng J, et al. Low-density lipoprotein receptor-related protein 8 gene association with egg traits in dwarf chickens. Poultry Science. 2010;89(5):883–6. 10.3382/ps.2009-00613 [DOI] [PubMed] [Google Scholar]
- 57. Mann K, Maček B, Olsen JV. Proteomic analysis of the acid‐soluble organic matrix of the chicken calcified eggshell layer. Proteomics. 2006;6(13):3801–10. [DOI] [PubMed] [Google Scholar]
- 58. Hincke MT, Gautron J, Tsang CP, McKee MD, Nys Y. Molecular cloning and ultrastructural localization of the core protein of an eggshell matrix proteoglycan, ovocleidin-116. Journal of Biological Chemistry. 1999;274(46):32915–23. [DOI] [PubMed] [Google Scholar]
- 59. Mann K, Hincke MT, Nys Y. Isolation of ovocleidin-116 from chicken eggshells, correction of its amino acid sequence and identification of disulfide bonds and glycosylated Asn. Matrix biology. 2002;21(5):383–7. [DOI] [PubMed] [Google Scholar]
- 60. Wang X, Wu C, Tao K, Zhao K, Wang J, Xu H, et al. Influence of ovalbumin on caco3 precipitation during in vitro biomineralization. The Journal of Physical Chemistry B. 2010;114(16):5301–8. 10.1021/jp1008237 [DOI] [PubMed] [Google Scholar]
- 61. Panheleux M, Bain M, Fernandez M, Morales I, Gautron J, Arias J, et al. Organic matrix composition and ultrastructure of eggshell: a comparative study. British Poultry Science. 1999;40(2):240–52. [DOI] [PubMed] [Google Scholar]
- 62. Gautron J, Hincke M, Panheleux M, Garcia-Ruiz J, Boldicke T, Nys Y. Ovotransferrin is a matrix protein of the hen eggshell membranes and basal calcified layer. Connective tissue research. 2001;42(4):255–67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Primers used for qPCR of calcium signaling pathway genes. Table B. Primers used for direct PCR sequencing of the potential group-specific mutations. Table C. RNA sequencing of eggshell quality in Rhode Island White hens. Table D. Whole genome re-sequencing of eggshell quality in Rhode Island White hen. Table E. Frameshift mutations in the differentially expressed genes in KEGG pathway.
(PDF)
Distribution of mapped reads to different transcript types and gene regions. Top graph (A and B) indicates the proportion of transcripts belonging to different RNA species. Numbers within pie chart indicate number of reads while numbers in parenthesis indicate number of transcripts. Bottom graph (C) shows the read distribution within protein coding genes. Fig. B. Differentially expressed genes (DEGs) in calcium signal pathway. The red markers represent the DEGs in LES compared with NES. NCX was a NCX1 gene; ADCY was a ADCY8 gene; CaV1 was a CACNA1D gene; CaV2 was a CACNA1E gene; CaV3 was a CACNA1H gene; GPCR included ADRA1D, GRM1 and OXTR genes; PTK included PDGFRB and ERBB4 genes; Gq was a GNA11 gene; PLC β was a PLCB2 gene; ANT was a SLC25A4 gene; IP3 was a ITPKA gene and PKC included PRKCA and PRKCB genes. Red marker circle of Genes in KEGG pathway also involved in biological process (BP) term of calcium ion transport. Fig. C. Graphical representation of two pooled samples genomes in a Circos plots. Form outside to inside of circles represent chromosome, density of total genes within the chromosome that from red to blue represent for gene density which is from high to low, mapping depth of NES and LES, SNP mutations in the chromosome of NES and LES, Indel mutations in the chromosome of NES and LES. A’s chromosmes are from 1 to 15 based on Mb and B’s chromosomes are from 16 to 32 based on 10 Kb, including sex chromosomes. Fig. D. Direct PCR sequencing detected mutations of CACNA1H and LTF gene from the RNA-seq and whole genome re-sequencing. The results indicated that potential unique SNPs for each group by RNA-seq and genome re-sequencing were not unique SNPs, but the SNP frequencies have difference for each group. The SNP of CACNA1H gene is rs312834462, and LTF gene’s SNP is rs10724671.
(PDF)
Data Availability Statement
The data sets supporting the results presented here are available in the Sequence Read Archive (SRA) repository under accession numbers SRP034444, SRP034445, and SRP034511.


