Abstract
Copper is universally toxic in excess, a feature exploited by the human immune system to facilitate bacterial clearance. The mechanism of copper intoxication remains unknown for many bacterial species. Here, we demonstrate that copper toxicity in Streptococcus pneumoniae is independent from oxidative stress but, rather, is the result of copper inhibiting the aerobic dNTP biosynthetic pathway. Furthermore, we show that copper-intoxicated S. pneumoniae is rescued by manganese, which is an essential metal in the aerobic nucleotide synthesis pathway. These data provide insight into new targets to enhance copper-mediated toxicity during bacterial clearance.
1. INTRODUCTION
It is becoming increasingly apparent that manipulating the levels of transition metals during infection is a powerful weapon wielded by vertebrates to combat invading pathogens. These elements are essential for life but are toxic when present in excess. Highlighting the importance of maintaining appropriate intracellular metal levels is the observation that numerous acquisition and efflux systems contribute to bacterial pathogenesis.1–3 Vertebrates take advantage of both the essential and toxic nature of these elements to combat infection.4, 5 During infection, the host restricts the availability of iron, manganese, and zinc to pathogens in an attempt to starve them of these metals.6–8 The host also uses the toxic properties of copper, which has antimicrobial and antiviral properties that have been appreciated since antiquity, to combat invading microbes.9, 10,7, 11 Genes under the control of copper-responsive regulation contribute to bacterial pathogenesis in the mammalian host.12 Understanding, how bacterial species adapt to fluctuations in metal availability is critical to understanding bacterial physiology and virulence.
S. pneumoniae, or pneumococcus, is a significant public health concern in pediatric and elderly populations and is a major cause of pneumonia, otitis media, and meningitis. Recent evidence indicates that S. pneumoniae experiences copper toxicity during infection.2, 13 A widely used mechanism to counter copper toxicity is the expression of dedicated copper efflux systems.2 These systems are functionally conserved amongst diverse bacterial species and typically mirror the pneumococcal system, which consists of a regulator to control expression (CopY), an ATPase to mediate efflux (CopA), and a copper chaperone (CupA).14 Loss of CopA reduces the virulence of pneumococcus in multiple infection models.2 This observation underscores the importance of evading copper-mediated intoxication within the mammalian host. In addition to export, it seems likely that bacteria use other mechanisms to cope with metal toxicity because they can alter their metabolic profile in response to intoxicating concentrations of transition metals by altering their expression of amino acid biosynthesis and transport genes.15 Alterations in multiple cellular pathways are not entirely unexpected given the involvement of metals in approximately 30% of cellular proteins.16 This is especially true of copper because pneumococcus does not contain any known copper-utilizing proteins, yet cellular levels of copper are still approximately 10% of those of the widely utilized metals manganese and zinc, underscoring the potential for mismetallation.17
Although the antibacterial properties of copper are well established, the specific mechanism underlying copper toxicity remains unknown for many bacterial species, including pneumococcus. Copper can produce free radicals when oxidized via Fenton reactions. This process occurs when copper (I) is oxidized in the presence of hydrogen peroxide, producing copper (II) + ·OH + OH−. The accumulation of free radicals can irreversibly damage bacterial cellular components, including proteins, lipids, and DNA.18 This reaction is of particular concern in pneumococcus, which not only lacks catalase and other widely distributed antioxidant defenses but also produces millimolar amounts of hydrogen peroxide, a precursor to Fenton chemistry.19 Oxidative damage can itself result in the mismetallation of proteins.20, 21 However, copper-induced oxidative stress is not sufficient to kill E. coli, leading to the suggestion that additional mechanisms of copper-mediated toxicity may exist.5, 22–24 Other proposed mechanisms of copper toxicity include disrupting the photosystem oxidase HemN in Rubrivivax gelatinosus, inactivating solvent-exposed iron-sulfur clusters in dehydratases such as fumarase A and displacing manganese from the active site of superoxide dismutase.25–27 Because of the universal nature of copper toxicity in bacteria, identification of both conserved and species-specific copper targets will enhance our understanding of how metals affect bacterial physiology and could provide potential targets for the development of novel therapeutics.
In this study, we elucidated a mechanism of copper toxicity in S. pneumoniae by examining the cellular consequences of copper stress. Copper toxicity in pneumococcus occurs independently of oxidative damage, as copper toxicity was readily observed under strict anaerobic conditions. Subsequent transcriptional profiling and mutagenesis studies revealed that the primary mechanism of copper toxicity in pneumococcus is inhibition of the essential manganese-dependent nucleotide synthesis pathway. This pathway has not been identified previously as a target of copper intoxication; thus, this work significantly expands our understanding of the underlying mechanisms of copper toxicity.
2. MATERIALS AND METHODS
2.1 Bacterial constructs
Mutations of SP_0202 (nrdD), SP_0727 (copY), SP_0728 (cupA), SP_0729 (copA), and SP_0766 (sodA) were created via the splicing by overhang extension method of PCR (SOE-PCR). Fragments approximately 1 kb upstream and downstream of the target gene were amplified and spliced to an erythromycin- (copA) or spectinomycin- (nrdD and sodA) resistance cassette. SOE-PCR products were subsequently used to transform pneumococcus (TIGR4), and the expected size of each knockout was verified by PCR to confirm insertion of the SOE-PCR product and deletion of the target gene.28
2.2 Growth curves and colony-forming units
Each S. pneumoniae strain was grown at 37°C in ThyB medium (30 g of Todd Hewitt Broth [Sigma], 2 g of yeast extract [Sigma], 1 L dH2O, pH 6.5) under aerobic conditions, with varying amounts of CuSO4 (hereafter, copper), other metals, and hydroxyurea (HU) as indicated. Bacteria was back-diluted 1:50 at optical density [O.D.] λ620 (unless otherwise noted) = 0.1 in experimental ThyB medium for overnight growth assessments. For colony-forming unit experiments, 10 μL of culture was serially diluted and plated on TSA (Sigma) sheep's blood (I-Tek) agar 8 hours after inoculation. All O.D. measurements used to construct growth curves were read at λ620 wavelength on a Turner Model 340 spectrophotometer. Because of HU's apparent instability at 37°C for extended time periods, 8-hour time points were reported for the HU studies.
2.3 Inductively coupled plasma mass spectrometry
Bacteria were grown in ThyB to an O.D. of 0.40, diluted by 37.5%, and grown for another hour. Then, 100 μM copper was added to the cultures, and cells were grown for an additional hour before being pelleted and washed with 10 mL PBS + 10 mM EDTA. Pelleted cells were resuspended in 1 mL PBS + 10 mM EDTA and transferred to an Eppendorf tube. Cells were then dried overnight at 70°C, collected in 10 mL of 10% nitric acid, vortexed vigorously for 1 minute, and incubated for 1 hour at 55°C. Samples were then vortexed and filtered for spectrophotometry (Varian 820 ICPMS System).
2.4 Macrophage intracellular growth assay
J774.1 murine macrophages (ATCC) were maintained in a 37°C, 5% CO2 incubator with Dulbecco's modified Eagle's medium (DMEM, Sigma) containing 10% (vol/vol) fetal bovine serum (FBS, Sigma), glutamine (2 mM, Sigma), penicillin (50 units/mL, Sigma), streptomycin (50 μg/mL, Sigma), and 0.015% sodium bicarbonate. Cells were grown to 80% confluence in 12-well tissue culture plates, washed twice with PBS, and resuspended in growth medium without antibiotics or FBS. Macrophages were then infected with 50 μL of S. pneumoniae at an O.D. of 0.1 for 90 minutes with or without 250 μM manganese supplementation. Wells were then washed twice: each wash was followed by a 1-minute incubation with DMEM containing gentamycin (50 μg/mL). Macrophages were lysed and serially diluted to determine intracellular bacterial content. Each value shown was normalized to the bacterial content of wild-type TIGR4 with no manganese supplementation.
2.5 Protein oxidation
S. pneumoniae were grown to an O.D. of 0.1 and stressed with 0 μM, 25 μM, 50 μM, or 100 μM copper for 1 hour. Each sample was serially diluted and plated to determine CFU titers used to ensure equivalent loading. Samples were centrifuged at 6000 × g for 5 minutes. Bacterial samples were lysed and processed by using the OxyBlot Protein Oxidization Detection Kit (Millipore), with 2% deoxycholate added to each sample to promote pneumococcal lysis. Samples added to the polyacrylamide gel were normalized by O.D. to the 0 μM copper inoculum of each individual sample after copper stress. Transfer, blocking, incubation with primary and secondary antibodies, and exposure were carried out according to manufacturer suggestions.
2.6 Hydrogen peroxide stress assay
Bacteria were grown to an O.D. of 0.1 in ThyB. Then, 500 μL of bacteria was combined with 0.1% (33 mM) hydrogen peroxide for 60 minutes. Samples were serially diluted in PBS and plated on TSA blood agar plates. Bar values represent percent survival compared to that of the respective strain grown without hydrogen peroxide added. Experiments were performed in triplicate.
2.7 RNA extraction
S. pneumoniae were grown to an O.D. of 0.1 in ThyB. Cultures were then diluted 50-fold in fresh ThyB and grown to an O.D. of 0.3. Indicated metals were added to each culture, and the mixtures were incubated for 15 minutes at 37°C. Culture samples (5 mL) were incubated with RNAprotect Bacterial Reagent (10 mL, Qiagen) for 20 minutes before the bacteria were isolated via centrifugation. RNA was extracted from the cell pellets by using an RNeasy Mini Kit (Qiagen).
2.8 Quantitative real-time PCR assays
SuperScript III First-Strand Synthesis SuperMix (Invitrogen) was used to synthesize cDNA from the isolated RNA (50 ng/μL). SYBR Green (Invitrogen) was used to monitor dsDNA synthesis on an ABI-Prism 7300 Real-Time PCR machine (Applied Biosystems). Samples were normalized relative to gyrA expression. Fold-change was calculated by using the ΔΔCt method.
2.9 Zone-of-inhibition assays
Bacteria were grown in C+Y to an O.D. of 0.2 at 620 nm. Then, 100 μL of culture was spread onto a blood agar plate. A disc of filter paper with 10 μL of 1M copper was placed in the middle of the plate, and bacteria were grown aerobically in 5% CO2 or anaerobically in a GasPak jar. The zone of inhibition was measured as the distance between the outer edge of the disc and that of bacterial growth.
2.10 Microarray analysis
The Qiagen RNeasy mini kit was used to harvest bacterial RNA from mid-log phase cultures (O.D. of 0.4 at 620 nm) grown in ThyB with or without 200 μM copper supplementation for 30 minutes. Microarray experiments were performed as described previously.29 Briefly, whole-genome S. pneumoniae version 8.0 cDNA microarrays were obtained from the Pathogen Functional Genomics Resource Center (PFGRC) at the J. Craig Venter Institute. Microarray experiments were performed by the Functional Genomics laboratory of the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital by using standard protocols provided by the PFGRC (http://pfgrc.tigr.org/protocols.shtml) as previously described.1
3. RESULTS
3.1 Copper stress does not affect accumulation of other metals
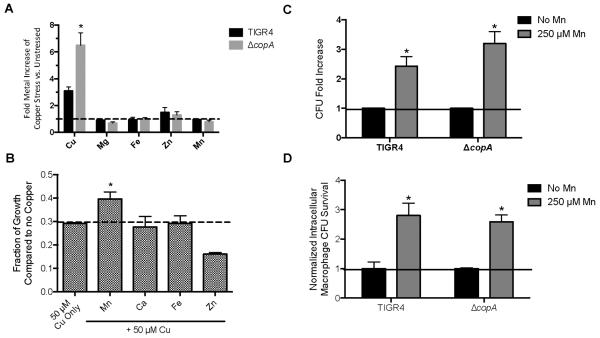
Although it is known that pneumococcus experience copper stress during infection,5 the precise mechanism of copper-mediated toxicity has not been determined. Thus, we sought to elucidate the underlying mechanism of copper toxicity in pneumococcus. Previous studies in pneumococcus have revealed that elevated concentrations of zinc can interfere with the uptake of manganese by PsaA.30 To evaluate whether copper can interfere with the uptake of other metals, the intracellular elemental composition was measured by performing inductively coupled plasma mass spectrometry on bacteria exposed to toxic levels of copper.2 These experiments revealed that both wild-type and ΔcopA mutant bacteria accumulate excess intracellular copper when exposed to high levels of this metal (Figure 1A). Consistent with the role of CopA in the copper efflux system, the mutant accumulated significantly more intracellular copper than did the wild type. While intracellular copper rapidly increased, the concentrations of the other metals examined (i.e., zinc, iron, manganese, and magnesium) were unaffected in wild-type and ΔcopA mutant bacteria (Fig. 1A). This result indicates that intracellular concentrations of other metals are not perturbed under copper stress. Furthermore, this result emphasizes the specificity of the CopA transporter for copper.
Figure 1. Manganese partially rescues S. pneumoniae from copper stress.
(A) Inductively coupled plasma mass spectrometry of copper-stressed wild-type TIGR4 and the ΔcopA mutant. Fold-increase is based on comparison to the strain without copper stress relative to bacterial CFU. *Student's t-test indicates p<.01 compared to copper-stressed, wild-type TIGR4. Error bars represent standard deviation (SD) n = 3. (B) Rescue of ΔcopA under copper stress by manganese supplementation. Indicated metals were added (250 μM). Values represent optical density at 620 nm as a fraction of bacteria without copper. * Student's t-test indicates p<.01 compared to 50 μM copper alone for the ΔcopA mutant. Error bars represent SD n = 5. (C) Replication under copper stress increases in the presence of manganese. The amount of copper used for each strain is relative to the estimated IC50 of copper stress respective to the strain's endpoint growth (500 μM for TIGR4 and 50 μM for the ΔcopA mutant). Error bars represent SEM n = 3. * Student's t-test indicates p<.01. (D) Manganese rescues bacterial survival from macrophage-mediated killing. Survival is normalized to the amount of the individual strain (TIGR4 and ΔcopA) without manganese added because of the heightened sensitivity of ΔcopA to macrophage-mediated killing. * Student's t-test indicates p<.01. Error bars represent SEM n = 3.
3.2 Manganese Rescues Copper Toxicity
The interactions between copper and proteins are very stable. As such, elevated levels of copper could result in mismetallation of proteins that use metals with weaker affinities. Although there is no discernable intracellular perturbation of other metals during short-term copper stress, we hypothesized that supplementation with different metals could overcome copper toxicity by preventing mismetallation. The ΔcopA mutant was used in these studies because of its increased sensitivity to copper toxicity and heightened intracellular accumulation of copper. The addition of calcium or iron did not rescue the growth of the ΔcopA mutant in the presence of toxic levels of copper. The addition of manganese partially alleviated the effects of copper toxicity in the ΔcopA mutant, increasing both the overall optical density and colony forming units respective to the values observed after adding copper alone (Fig. 1B, C). Interestingly, supplementation with zinc, which also has a high affinity for proteins, exacerbated copper toxicity (Fig. 1B). These data indicate that excess manganese can partially rescue the inhibitory effects of copper. Because copper stress did not reduce intracellular manganese levels (Fig. 1A), manganese rescue is unlikely to be due to a competition for uptake by the manganese importer PsaA.31 Manganese supplementation at concentrations used in this study have previously been shown to confer minimal alterations in the intracellular levels of other transition metals, indicating this effect is likely specific to manganese1.
To determine whether the manganese rescue observed in vitro could be recapitulated in a more biologically relevant assay, we investigated whether the addition of manganese would enhance the survival of S. pneumoniae in J774 murine lung macrophages. During infection, macrophages are a primary source of copper toxicity encountered by pathogens.7, 32, 33 Manganese supplementation enhanced the survival of TIGR4 and the ΔcopA mutant by roughly 3-fold (Fig. 1D). Levels of manganese utilized were non-toxic and within range of those previously utilized to investigate the role of manganese on macrophage function34. Thus, manganese supplementation partially rescues pneumococci under copper stress in vitro and during macrophage-mediated killing.
3.3 Oxidative Stress is not the Primary Cause of Copper Toxicity
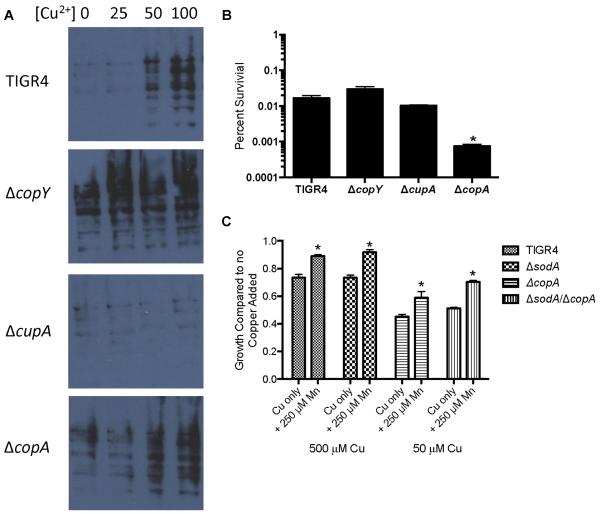
Manganese is a vital aspect of the pneumococcal defense against oxidative stress.35 Copper can generate free radicals via the Fenton reaction in vivo;36 hence, we next investigated the effect of copper on oxidative stress in pneumococcus. First, we assessed the effect of copper toxicity on protein oxidation by using 2, 4-dinitrophenylhydrazine to measure oxidized carbonyl groups in the individual cop operon mutants and the wild-type bacteria grown in standard media with or without copper stress. Increasing concentrations of copper led to progressively greater levels of protein oxidation in wild-type TIGR4 (Fig. 2A). The ΔcopA mutant showed a similar trend with increasing copper concentrations but, overall, had more protein oxidation than did the wild-type. This result suggested that protein oxidation could partially explain some of the copper-mediated toxicity in the ΔcopA mutant. However, ΔcopY, which overexpresses copA and maintains wild-type virulence and sensitivity to copper, displayed even higher levels of protein oxidation. Additionally, the ΔcupA mutant experienced less protein oxidation than did the wild-type bacteria across all concentrations of copper yet is as sensitive to copper toxicity as ΔcopA is (Fig. 2A).2 These observations indicate that copper sensitivity does not correlate with protein oxidation in pneumococcus.
Figure 2. Oxidative damage in S. pneumoniae does not correlate with copper stress.
(A) Protein oxidation in response to copper. TIGR4 and the cop operon mutants at an O.D. of 0.1 were exposed to varying amounts of copper for 1 hour and probed for protein oxidation. (B) Sensitivity to hydrogen peroxide stress. TIGR4 and the cop operon mutants at an O.D. of 0.1 were exposed to 0.1% hydrogen peroxide for 60 minutes and plated to determine the number of colony-forming units (CFU) present. * Student's t-test indicates p<.01 compared to wild-type. Error bars represent SD n = 5. (C) ΔsodA mutants are equally susceptible to copper stress and equally rescued by manganese. End-point growth of wild-type, ΔcopA, ΔsodA, and ΔcopAΔsodA TIGR4 pneumococci. * Student's t-test indicates p<.01 compared to strains grown without manganese. Error bars represent SEM, n = 4.
We also assessed pneumococcal susceptibility to exogenous hydrogen peroxide in the presence of toxic levels of copper. The ΔcopA mutant had the greatest sensitivity to hydrogen peroxide stress, and the ΔcopY and ΔcupA mutants had sensitivities similar to those of wild-type TIGR4 (Fig. 2B). Although the ΔcopA mutant is sensitive to hydrogen peroxide, the ΔcupA mutant, which is equally sensitive to copper toxicity, is not.2 These data indicate that copper sensitivity and hydrogen peroxide sensitivity do not correlate.
To further evaluate whether oxidative stress underlies copper toxicity, we investigated the contribution of superoxide dismutase (SodA) to resisting this stress. In other bacteria, SodA promotes resistance to transition metal stress.37 Additionally, exposure to high levels of copper have been shown to interfere with the activity of manganese-dependent SodA.38 We found that deletion of sodA in either the TIGR4 or the ΔcopA background did not significantly alter sensitivity to copper stress. (Fig. 2C). Of note, manganese supplementation partially protected wild-type bacteria, the ΔsodA mutant, and ΔsodA/ΔcopA double mutants equally from the toxic effects of copper (Fig. 2C). These results indicate that the manganese-mediated rescue of copper toxicity occurs independently of SodA activity. In total, these results suggest that oxidative damage is not the major factor in copper intoxication.
3.4 Intracellular Copper Alters Expression of Nucleotide Synthesis Pathways
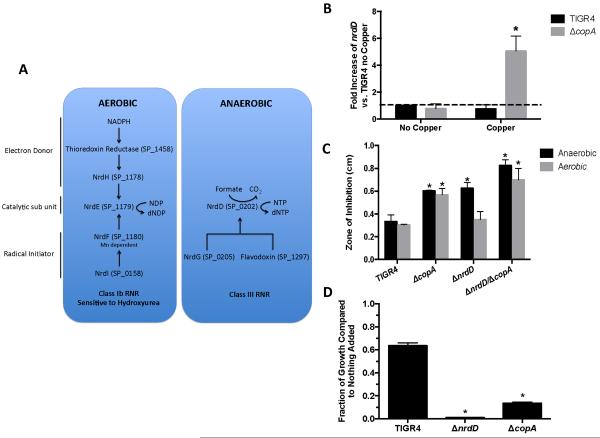
To ascertain the potential targets of copper toxicity in pneumococci, we performed a microarray comparing the mRNA expression of wild-type TIGR4 and that of the ΔcopA mutant under copper stress. In addition to the expected changes in the cop operon, a rather limited transcriptional response to intracellular copper accumulation was observed (Table 1). The czcD zinc efflux system was upregulated, potentially indicating some degree of cross-talk between zinc and copper. The chaperones clpP and clpE were also upregulated, indicative of a cellular stress response. Additionally, genes encoding the anaerobic nucleotide (dNTP) synthesis pathway were upregulated. Pneumococci encode two functional dNTP biosynthesis pathways, one aerobic (SP_1458, SP_1178, SP_1179, SP_1180, SP_0158) and one anaerobic (SP_0202, SP_0205, SP_1297) (Fig. 3A). By using real-time PCR assays, upregulation of the anaerobic nucleotide synthesis pathway (SP_0202, NrdD) was observed in the ΔcopA mutant during copper stress (Fig. 3B). Additionally, essential members of the aerobic nucleotide synthesis pathway (SP_1179 NrdH [electron transport] and SP_1713 NrdR [the Nrd regulator]) were more highly expressed by the ΔcopA mutant during copper stress than during unstressed conditions (Fig. S1A). While it was surprising to observe both dNTP biosynthetic pathways being expressed concurrently, previous reports have shown detectable levels of both pathways under aerobic culture39. Taken together, these results indicate that pneumococci increase expression of the dNTP biosynthesis pathways in response to intracellular copper accumulation.
Table 1. Microarray analysis comparing transcription in wild-type and ΔcopA bacteria during intracellular copper accumulation.
Data show the log(2) change in each transcript from 3 independent experiments comparing TIGR4 and ΔcopA bacteria under copper-stress conditions. Experimental conditions are detailed in the Methods.
| TIGR4 Gene Number | 2^X change | SD | Gene Name | Predicted Function |
|---|---|---|---|---|
| SP0202 | 2.2 | ±0.21 | nrdD | anaerobic ribonucleoside-triphosphate reductase |
| SP0203 | 1.8 | ±0.26 | hypothetical protein | |
| SP0204 | 2.2 | ±0.17 | acetyltransferase, GNAT family | |
| SP0205 | 2.2 | ±0.26 | nrdG | anaerobic ribonucleoside-triphosphate reductase activating protein |
| SP0206 | 2.1 | ±0.18 | hypothetical protein | |
| SP0207 | 2.1 | ±0.25 | conserved domain protein | |
| SP0338 | 5.4 | ±0.47 | clpP | ATP-dependent Clp protease, ATP-binding subunit, putative |
| SP0474 | 1.8 | ±0.18 | licC | PTS system, cellobiose-specific IIC component |
| SP0478 | 1.6 | ±0.31 | lacE | PTS system, lactose-specific IIBC components |
| SP0727 | 4 | ±0.11 | copY | Copper repressor |
| SP0728 | 3.8 | ±0.24 | cupA | Copper chaperone |
| SP0729 | −6 | ±0.45 | copA | Copper exporter |
| SP0820 | 1.7 | ±0.24 | clpE | ATP-dependent Clp protease, ATP-binding subunit CIpE |
| SP1774 | 1.7 | ±0.15 | transcriptional regulator, putative | |
| SP1775 | 2.5 | ±0.31 | hypothetical protein | |
| SP1776 | 1.8 | ±0.08 | trxA | Thioredoxin reductase |
| SP1857 | 3.4 | ±0.16 | czcD | Zinc efflux protein |
| SP2026 | 1.5 | ±0.21 | Iron containing alcohol dehydrogenase |
Figure 3. Copper downregulates aerobic nucleotide synthesis.
(A) Pathways for aerobic and anaerobic nucleotide synthesis in S. pneumoniae. (B) The anaerobic nucleotide synthesis pathway gene nrdD (SP_0202) is upregulated in the ΔcopA mutant after a sub-lethal dose of copper. Error bar represents SEM n = 3. * Student's t-test indicates p<.01 compared to real-time data of parental strain with no copper added. (C) The ΔnrdD mutant has increased copper toxicity in anaerobic conditions. Error bar represents SD n = 5. * Student's t-test indicates p<.01 compared to similarly treated TIGR4 strain in the copper zone-of-inhibition assay. (D) The ΔcopA mutant is more susceptible to 10 mM HU than is wild-type TIGR4. Error bar represents SD, n = 4. * Student's t-test indicates p<.01 compared to parental strain with nothing added.
3.5 Copper Toxicity Reduces Aerobic Ribonucleotide Synthesis
The aerobic pathway is predicted to require a manganese cofactor and to be essential.13, 40 The increased expression of the anaerobic dNTP synthesis pathway leads to the hypothesis that copper inhibits the aerobic pathway. This hypothesis predicts that if the anaerobic pathway were disrupted, then the pneumococcus would display greater sensitivity to copper-mediated growth inhibition because it would lack a backup pathway. To test this hypothesis, we created a mutation in nrdD in both the wild-type and ΔcopA backgrounds to disrupt the anaerobic pathway. Deletion of nrdD was chosen as this gene encodes the enzymatic catalytic subunit and NrdD alone has been previously shown to catalyze reduction of dNTPs 41. The ΔnrdD mutants grew both aerobically and anaerobically, indicating that the aerobic nucleotide synthesis pathway retains some activity in anaerobic conditions (Fig. S1B). The zone of clearance around a copper-treated disk was assessed to test the sensitivity of the ΔnrdD and ΔcopA/ΔnrdD mutants to copper toxicity. The sensitivity of the mutants to copper was assessed in both aerobic and anaerobic conditions. Consistent with previous data, the ΔcopA mutant had larger zones of inhibition than did wild-type under both aerobic and anaerobic conditions (Fig. 3C).2 The ΔnrdD single mutant had wild-type sensitivity under aerobic conditions. However, the mutant had a larger zone of clearance when grown anaerobically, indicating that copper toxicity prevented the aerobic pathway from supporting growth under these conditions. The ΔcopAΔnrdD double mutant had the largest zone of inhibition in both aerobic and anaerobic conditions (Fig. 3C). Similar experiments performed with other metals revealed no differences between wild-type TIGR4 and the mutants (Figure S1C). These data indicate that pneumococcus is more sensitive to copper intoxication when the metal-independent anaerobic pathway is deleted, suggesting that the aerobic pathway is not functional.
If intracellular copper toxicity inactivates the aerobic dNTP biosynthetic pathway, then further perturbation of this pathway should lead to an even greater sensitivity to copper. Attempts to delete genes in the aerobic dNTP biosynthetic pathway were unsuccessful, likely because these genes are essential.13, 42 Therefore, we used a chemical approach exploiting the ability of hydroxyurea (HU) to inhibit aerobic dNTP synthesis, which destroys a tyrosine free radical essential for enzymatic function in ribonucleotide reductase (NrdF).43 To confirm that HU inhibited the aerobic pathway, the sensitivity of the ΔnrdD mutant to HU was assessed. The ΔnrdD mutant was significantly more sensitive to HU inhibition than was the parental TIGR4 (Fig. 3D, Fig. S1D). Consistent with copper targeting the aerobic nucleotide synthesis pathway, the growth of ΔcopA was significantly more impaired by HU than was that of the wild-type TIGR4 (Fig. 3D), as might be expected if both copper and HU are targeting distinct steps of the same pathway. The addition of manganese partially reversed the growth defect of the ΔnrdD and ΔcopAΔnrdD mutants exposed to copper. However, the addition of manganese did not rescue the mutants when only HU was present (Figures S1D, S1E). These data suggest that copper toxicity and HU disrupt the aerobic dNTP synthesis via different mechanisms. 43 In total, these data indicate that intracellular copper may compete with manganese for a metal-dependent step in the aerobic dNTP biosynthetic pathway to facilitate bacterial killing.
4. Discussion
4.1 Oxidative stress is not the critical target of copper toxicity
In these investigations, we sought to clarify the mechanism by which copper is toxic to pneumococcus. We found that pneumococci do not alter the uptake of magnesium, iron, zinc, or manganese to combat the entrance of copper into the cell. In fact, manganese partially rescues the growth of wild-type TIGR4 and the ΔcopA mutant under copper-stress conditions. This rescue by manganese may partially explain the enhanced sensitivity to zinc under copper-stress conditions, as zinc can effectively compete for manganese uptake when present at sufficiently high ratios. Theoretically, the manganese rescue of copper-stressed pneumococci could occur via reduction of oxidative damage, independent of the manganese-dependent SodA.44 However, previous studies and data shown here indicate that copper stress does not cause significant oxidative stress in E. coli 22 or streptococci. Finally, the observations that culturing under strict anaerobic conditions did not alleviate copper sensitivity and that copper sensitivity did not correlate with oxidative damage further support a mechanism of copper toxicity that is independent of reactive oxygen species.
4.2 Copper inhibits the aerobic nucleotide synthesis pathway through potential mismetallation
Under copper stress conditions, the anaerobic nucleotide synthesis pathway is upregulated, implying that the anaerobic pathway may be partially complementing the aerobic pathway. Previous transcriptional data have also shown altered regulation of the anaerobic pathway in response to intracellular zinc stress in pneumococcus, indicating that this potential mismetallation mechanism of intoxication may extend to other transition metals.45 Because the anaerobic nucleotide synthesis pathway mutant (ΔnrdD) could grow in an anaerobic environment, it is likely that the aerobic nucleotide synthesis pathway can complement the anaerobic pathway in pneumococcus. Even so, in the ΔnrdD mutant, we observed heightened sensitivity to copper stress when only the aerobic pathway was functional, likely due to its still-intact copper export system. Copper sensitivity was further exacerbated in the absence of functional copper export. When the aerobic pathway was inhibited by hydroxyurea (as determined by no growth of the ΔnrdD mutant in the presence of 10 mM HU), the ΔcopA mutant, which has increased intracellular copper, underwent less growth than did wild-type TIGR4. Therefore, we propose that copper disrupts aerobic nucleotide synthesis, which occurs through a manganese-dependent pathway.
Although the precise mechanism by which copper could be interfering with the aerobic dNTP pathway is unknown, proteomics data and structural predictions provide some insight. Interestingly, pneumococcal proteins in the aerobic dNTP pathway have been predicted to have copper-binding properties via proteomics approaches.46 One hypothesis is that copper blocks aerobic nucleotide synthesis in S. pneumoniae by binding to and inhibiting the function of NrdF, a protein that is homologous (50% identity) to the manganese-dependent ribonucleotide reductase found in E. coli (Fig. S2A).40, 47 This hypothesis is based on the promiscuous binding nature of metals toward ribonucleotide reductases such as NrdF, which has residues in its metallocenter that classically coordinate manganese or copper and typically have a higher affinity for the latter (Fig. S2B).46, 48–50
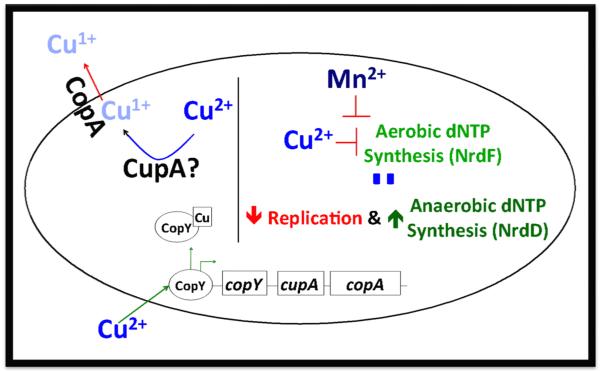
Our proposed model (Figure 4) is that when copper is added to the media, it enters the bacteria and triggers transcription of the cop operon. While the bacteria is attempting to alleviate the copper-induced stress by exporting copper, copper itself can inhibit aerobic nucleotide synthesis by binding to and inhibiting NrdF activity; this can lead to decreased replication and increased transcription of the anaerobic nucleotide synthesis pathway in an attempt to supplement the dNTP pool, which we observed transcriptionally. Addition of excess manganese, which binds to NrdF, can then potentially alleviate the copper stress by successfully competing for NrdF (Fig. S2A).47 There is diversity in the strategies utilized by bacterial to synthesize nucleotides, with bacterial species encoding between one and three classes of ribonucleotide reductases 51. Even amongst the streptococci there is inherent diversity, with S. pyogenes encoding a secondary functional nrdEF locus52. Hence, this mechanism of intoxication likely only extends to subset of bacterial species with similar classes of ribonucleotide reductases as the pneumococcus. Although these results indicate that copper toxicity can inhibit nucleotide biosynthesis, they do not preclude the possibility that other cellular pathways are inhibited by mismetallation with copper. Additional studies in pneumococcus and other bacteria are needed to elucidate the full effect of copper toxicity and will likely reveal other cellular pathways perturbed by accumulation of this metal.
Figure 4.
Graphical model of copper export and toxicity in S. pneumoniae
4.3 Synergism between metal intoxication and starvation in host immunity
During infection, metal concentrations in various organs can be dramatically altered, underscoring the dynamic nature of the environments that bacteria encounter.53 Furthermore, it is evident that both the absolute concentrations of various metals and the appropriate balance of these metals are vital for optimal cellular function. A prime example of this is manganese import by PsaABC, which is thought to be inhibited by elevated zinc levels during infection.5 The ability of this system to import manganese is influenced not by the absolute concentration of extracellular zinc but by the ratio of zinc to manganese.53 The observation that manganese can mitigate the effects of copper toxicity further highlights the importance of metal homeostasis to infection. This result also suggests that the nutrient-withholding response and metal intoxication may function synergistically. The phagolysosome is a prime example of where this synergy may occur: In this organelle, copper is actively pumped into the lumen while manganese is removed. The reduced availability of manganese would enhance the ability of copper to inhibit nucleotide biosynthesis via the aerobic pathway, thus increasing the efficacy of macrophage-mediated killing. Manganese starvation induced by calprotectin, which restricts extracellular metal availability, may have a similar effect.8 These findings and our data highlight the importance of understanding mechanisms of metal homeostasis under a dynamic range of metal bioavailability as well as understanding transporter-independent mechanisms by which bacteria circumvent metal toxicity. Although it is widely recognized that excess metals can be detrimental, the precise mechanisms by which metals intoxicate cells remain unknown for many bacterial pathogens whose sole environmental niche is the human host.
5. Conclusion
Although the antimicrobial properties of copper have been appreciated since antiquity, the mechanism underlying this activity in many bacterial species has remained poorly understood. The data presented here suggest that the main mechanism of copper intoxication in pneumococcus is independent of oxidative damage and likely occurs via inhibition of dNTP biosynthesis. Knowing the precise cellular targets of metal intoxication may provide subjects for future investigations aimed at targeting bacterial pathogens with novel therapeutic strategies.
Supplementary Material
Acknowledgements
Microarrays were kindly provided by the Pathogen Functional Genomics Resource Center at the J. Craig Venter Institute. Work in the laboratory of T.E.K is supported by NIH K22AI104805-01 from the NIH. Work in the laboratory of J.W.R. is supported by NIAID R01AI110618 and the American Lebanese Syrian Associated Charities (ALSAC).
References
- 1.Rosch JW, Gao G, Ridout G, Wang YD, Tuomanen EI. Molecular microbiology. 2009;72:12–25. doi: 10.1111/j.1365-2958.2009.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Wandel M, Andrew PW, Kuipers OP, Morrissey JA. Molecular microbiology. 2011;81:1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x. [DOI] [PubMed] [Google Scholar]
- 3.Rowland JL, Niederweis M. Tuberculosis (Edinb) 2012;92:202–210. doi: 10.1016/j.tube.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Festa RA, Thiele DJ. PLoS Pathog. 2012;8:e1002887. doi: 10.1371/journal.ppat.1002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honsa ES, Johnson MD, Rosch JW. Front Cell Infect Microbiol. 2013;3:92. doi: 10.3389/fcimb.2013.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hood MI, Skaar EP. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White C, Lee J, Kambe T, Fritsche K, Petris MJ. J Biol Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salgado CDMD, Sepkowitz KAMD, John JFMD, Cantey JRMD, Attaway HHMS, Freeman KDD, Sharpe PAMBA, Michels HTP, Schmidt MGP. Infection Control and Hospital Epidemiology. 2013;34:479–486. doi: 10.1086/670207. [DOI] [PubMed] [Google Scholar]
- 10.Fujimori Y, Sato T, Hayata T, Nagao T, Nakayama M, Nakayama T, Sugamata R, Suzuki K. Appl Environ Microb. 2012;78:951–955. doi: 10.1128/AEM.06284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forbes JR, Gros P. Trends Microbiol. 2001;9:397–403. doi: 10.1016/s0966-842x(01)02098-4. [DOI] [PubMed] [Google Scholar]
- 12.Shi X, Festa RA, Ioerger TR, Butler-Wu S, Sacchettini JC, Darwin KH, Samanovic MI. mBio. 2014;5 doi: 10.1128/mBio.00876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Opijnen T, Camilli A. Genome Res. 2012;22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banci L, Bertini I, Ciofi-Baffoni S, Del Conte R, Gonnelli L. Biochemistry. 2003;42:1939–1949. doi: 10.1021/bi027096p. [DOI] [PubMed] [Google Scholar]
- 15.Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD. Molecular microbiology. 2005;57:27–40. doi: 10.1111/j.1365-2958.2005.04642.x. [DOI] [PubMed] [Google Scholar]
- 16.Thomson AJ, Gray HB. Curr Opin Chem Biol. 1998;2:155–158. doi: 10.1016/s1367-5931(98)80056-2. [DOI] [PubMed] [Google Scholar]
- 17.Jacobsen FE, Kazmierczak KM, Lisher JP, Winkler ME, Giedroc DP. Metallomics. 2011;3:38–41. doi: 10.1039/c0mt00050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halliwell B. Nutr Rev. 2012;70:257–265. doi: 10.1111/j.1753-4887.2012.00476.x. [DOI] [PubMed] [Google Scholar]
- 19.Pericone CD, Park S, Imlay JA, Weiser JN. J Bacteriol. 2003;185:6815–6825. doi: 10.1128/JB.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imlay JA. J Biol Chem. 2014 DOI: 10.1074/jbc.R114.588814. [Google Scholar]
- 21.Gu M, Imlay JA. Molecular microbiology. 2013;89:123–134. doi: 10.1111/mmi.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macomber L, Rensing C, Imlay JA. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nandakumar R, Espirito Santo C, Madayiputhiya N, Grass G. Biometals. 2011;24:429–444. doi: 10.1007/s10534-011-9434-5. [DOI] [PubMed] [Google Scholar]
- 24.Braymer JJ, Giedroc DP. Current opinion in chemical biology. 2014;19:59–66. doi: 10.1016/j.cbpa.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macomber L, Imlay JA. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azzouzi A, Steunou AS, Durand A, Khalfaoui-Hassani B, Bourbon ML, Astier C, Bollivar DW, Ouchane S. Molecular microbiology. 2013;88:339–351. doi: 10.1111/mmi.12188. [DOI] [PubMed] [Google Scholar]
- 27.Batinic-Haberle I, Liochev SI, Spasojevic I, Fridovich I. Arch Biochem Biophys. 1997;343:225–233. doi: 10.1006/abbi.1997.0157. [DOI] [PubMed] [Google Scholar]
- 28.Horton RM, Cai ZL, Ho SN, Pease LR. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 29.Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, Tuomanen EI. Infection and immunity. 2004;72:5582–5596. doi: 10.1128/IAI.72.10.5582-5596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Counago RM, Ween MP, Begg SL, Bajaj M, Zuegg J, O'Mara ML, Cooper MA, McEwan AG, Paton JC, Kobe B, McDevitt CA. Nat Chem Biol. 2014;10:35–41. doi: 10.1038/nchembio.1382. [DOI] [PubMed] [Google Scholar]
- 31.Ogunniyi AD, Mahdi LK, Jennings MP, McEwan AG, McDevitt CA, Van der Hoek MB, Bagley CJ, Hoffmann P, Gould KA, Paton JC. Journal of bacteriology. 2010;192:4489–4497. doi: 10.1128/JB.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Festa RA, Helsel ME, Franz KJ, Thiele DJ. Chemistry & biology. 2014;21:977–987. doi: 10.1016/j.chembiol.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.German N, Doyscher D, Rensing C. Future microbiology. 2013;8:1257–1264. doi: 10.2217/fmb.13.100. [DOI] [PubMed] [Google Scholar]
- 34.Smialowicz RJ, Rogers RR, Riddle MM, Rowe DG, Luebke RW. Journal of toxicology and environmental health. 1986;19:243–254. doi: 10.1080/15287398609530924. [DOI] [PubMed] [Google Scholar]
- 35.Yesilkaya H, Kadioglu A, Gingles N, Alexander JE, Mitchell TJ, Andrew PW. Infection and immunity. 2000;68:2819–2826. doi: 10.1128/iai.68.5.2819-2826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadiiska MB, Mason RP. Spectrochim Acta A. 2002;58:1227–1239. doi: 10.1016/s1386-1425(01)00713-2. [DOI] [PubMed] [Google Scholar]
- 37.Geslin C, Llanos J, Prieur D, Jeanthon C. Research in microbiology. 2001;152:901–905. doi: 10.1016/s0923-2508(01)01273-6. [DOI] [PubMed] [Google Scholar]
- 38.Behera M, Dandapat J, Rath CC. Journal of basic microbiology. 2014;54:1201–1209. doi: 10.1002/jobm.201300805. [DOI] [PubMed] [Google Scholar]
- 39.Torrents E, Grinberg I, Gorovitz-Harris B, Lundstrom H, Borovok I, Aharonowitz Y, Sjoberg BM, Cohen G. J Bacteriol. 2007;189:5012–5021. doi: 10.1128/JB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin JE, Imlay JA. Molecular microbiology. 2011;80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torrents E, Eliasson R, Wolpher H, Graslund A, Reichard P. J Biol Chem. 2001;276:33488–33494. doi: 10.1074/jbc.M103743200. [DOI] [PubMed] [Google Scholar]
- 42.Hava DL, Camilli A. Molecular microbiology. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 43.Yarbro JW. Semin Oncol. 1992;19:1–10. [PubMed] [Google Scholar]
- 44.Cheton PL, Archibald FS. Free radical biology & medicine. 1988;5:325–333. doi: 10.1016/0891-5849(88)90104-9. [DOI] [PubMed] [Google Scholar]
- 45.Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. Molecular microbiology. 2007;65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 46.Sun X, Xiao CL, Ge R, Yin X, Li H, Li N, Yang X, Zhu Y, He X, He QY. Proteomics. 2011;11:3288–3298. doi: 10.1002/pmic.201000396. [DOI] [PubMed] [Google Scholar]
- 47.Boal AK, Cotruvo JA, Jr., Stubbe J, Rosenzweig AC. Science. 2010;329:1526–1530. doi: 10.1126/science.1190187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furia TE. CRC Handbook of Food Additives. 2nd edn CRC Press; p. 1972. [Google Scholar]
- 49.Huang M, Parker MJ, Stubbe J. J Biol Chem. 2014 doi: 10.1074/jbc.R114.596684. DOI: 10.1074/jbc.R114.596684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cotruvo JA, Jr., Stubbe J. Metallomics : integrated biometal science. 2012;4:1020–1036. doi: 10.1039/c2mt20142a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torrents E. Front Cell Infect Microbiol. 2014;4:52. doi: 10.3389/fcimb.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roca I, Torrents E, Sahlin M, Gibert I, Sjoberg BM. J Bacteriol. 2008;190:4849–4858. doi: 10.1128/JB.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. PLoS Pathog. 2011;7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.