Summary
Replication stress induced by nucleotide deficiency plays an important role in cancer initiation. Replication stress in primary cells typically activates the cellular senescence tumor suppression mechanism. Senescence bypass correlates with development of cancer, a disease characterized by metabolic reprogramming. However, the role of metabolic reprogramming in cellular response to replication stress is unknown. Here we report that ATM plays a central role in regulating cellular response to replication stress by shifting cellular metabolism. ATM inactivation bypasses senescence induced by replication stress triggered by nucleotide deficiency. This was due to restoration of dNTP levels through both upregulation of the pentose phosphate pathway via increased G6PD activity and enhanced glucose and glutamine consumption. These phenotypes were mediated by a coordinated suppression of p53 and upregulation of c-MYC downstream of ATM inactivation. Our data indicate that ATM status couples replication stress and metabolic reprogramming during senescence.
Introduction
Replication stress induced by deficiency in cellular dNTP levels is an important early event during cancer initiation (Bester et al., 2011), while its bypass correlates with cancer progression (Bester et al., 2011; Zeman and Cimprich, 2014). Replication stress causes DNA damage accumulation and genomic instability (Bester et al., 2011; Burhans and Weinberger, 2007; Zeman and Cimprich, 2014), which is a hallmark of cancer (Negrini et al., 2010). Notably, activation of oncogenes is known to decrease dNTP levels and consequently triggers replication stress (Aird et al., 2013; Bartkova et al., 2006; Di Micco et al., 2006; Mannava et al., 2013). In normal diploid cells, activation of oncogenes, and the subsequent replication stress, causes a tumor suppressive, stable cell growth arrest termed cellular senescence (Yaswen and Campisi, 2007). Indeed, oncogene-induced suppression of nucleotide metabolism via suppression of ribonucleotide reductase M2 (RRM2) underlies the observed replication stress and the associated DNA damage response (DDR) during senescence (Aird et al., 2013). Therefore, senescence suppresses tumors initiated by replication stress (Bester et al., 2011; Zeman and Cimprich, 2014). dNTP biosynthesis relies on glucose and glutamine consumption, which are at the heart of cancer metabolism (Ward and Thompson, 2012). However, the role of metabolic reprogramming in response to replication stress is unknown. Here we report that ATM status couples replication stress and metabolic reprogramming during senescence.
Results
Knockdown of ATM bypasses replication stress-induced senescence
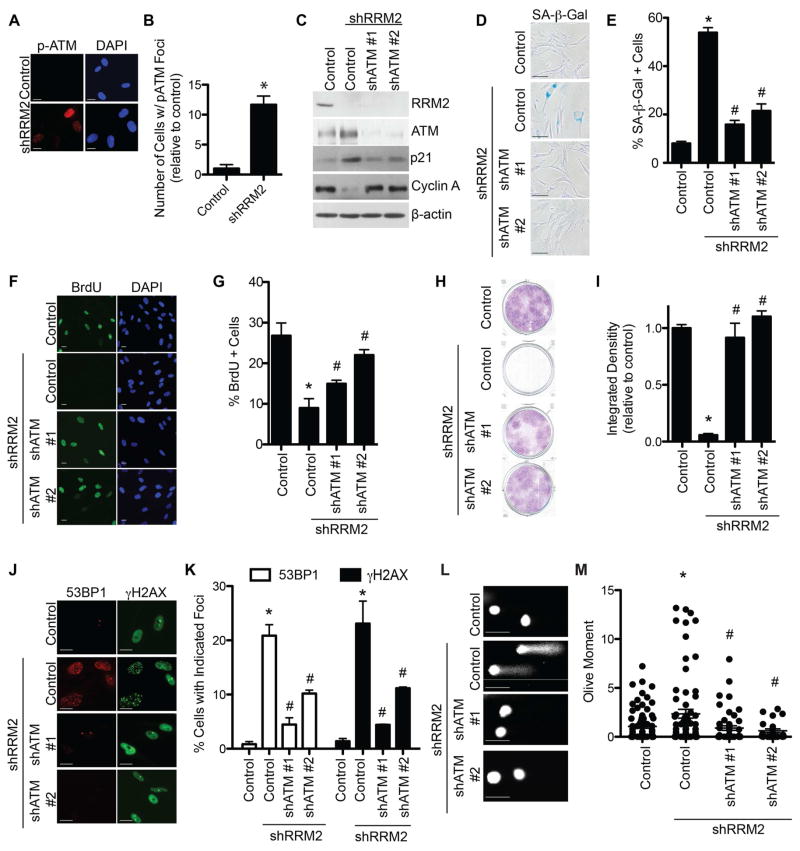
Suppression of RRM2, which depletes the levels of all four dNTPs, underlies replication stress observed during oncogene-induced senescence (Aird et al., 2013). This induces a robust DDR and ultimately a stable senescence-associated cell growth arrest. The replication stress sensors ATR and ATM are activated by oncogenes during senescence (Di Micco et al., 2006). We sought to determine whether ATM and/or ATR are regulated during senescence induced by short hairpin mediated RRM2 knockdown (shRRM2). shRRM2 significantly activated both ATM and ATR, as demonstrated by immunofluorescence using phospho-specific antibodies (Figures 1A–B and S1A). Next, we examined whether these proteins are necessary for the observed senescence. We knocked down ATM or ATR in combination with RRM2 knockdown using two independent short hairpin RNAs for ATM (shATM) or ATR (shATR). shATM in combination with shRRM2 suppressed senescence markers such as p21 expression (Figure 1C) and senescence-associated β-galactosidase (SA-β-Gal) activity (Figure 1D–E). This correlated with an increase in cell proliferation markers such as cyclin A expression (Figure 1C), BrdU incorporation (Figure 1F–G) and apparent cell growth as determined by focus formation assays (Figure 1H–I). Similar results were observed when ATM was inhibited by the specific inhibitor KU55933 (Figure S1B–C). shATM suppressed DDR induced by shRRM2 as determined by a decrease in γH2AX and 53BP1 foci formation (Figure 1J–K). Notably, this is in contrast to its positive role in DNA repair but consistent with the idea that DDR contributes to senescence induced by replication stress. ATM phosphorylates H2AX during foci formation, although other kinases can also phosphorylate H2AX (Yuan et al., 2010). To confirm that the observed decrease in γH2AX foci formation was due to decreased DDR instead of a dependence of its phosphorylation by ATM, we directly measured the extent of DNA damage in these cells by comet assay. shATM significantly decreased the extent of DNA damage induced by shRRM2 (Figure 1L–M). In contrast, neither two independent shATRs nor the ATR inhibitor VE822 was able to suppress senescence and its associated DDR induced by shRRM2 (Figure S1D–P). Interestingly, shRRM2/shATR cells had an even more robust senescent phenotype than shRRM2 alone as indicated by a higher SA-β-gal activity and a lower focus formation ability (e.g., Figures S1F and S1J). Likewise, shRRM2 failed to induce senescence in primary patient fibroblasts with mutated ATM (Figure S1Q–S) but not ATR (Figure S1T–V). Similar to previous reports (Bartkova et al., 2006; Di Micco et al., 2006), RAS-induced senescence was suppressed by shATM (Figure S1W–X). These results demonstrate that loss of ATM, but not ATR, bypasses senescence induced by replication stress, which correlates with the suppression of DDR. In this context, loss of ATM suppresses DDR induced by replication stress, a function different from its canonical, positive role in DNA repair.
Figure 1. Knockdown of ATM bypasses senescence and suppresses DNA damage induced by RRM2 knockdown. See also Figure S1.
(A) Primary human IMR90 cells were infected with a lentivirus encoding shRRM2 or control. Cells were stained for p-ATM (Ser1981) by immunofluorescence. DAPI staining was used to visualize nuclei. (B) Quantification of (A). 200 cells from each of the indicated groups were quantified for p-ATM foci positive cells (n=3). (C) Primary human IMR90 cells were infected with a lentivirus encoding shRRM2 alone or in combination with lentivirus encoding two independent shATMs. Cells were examined for expression of RRM2, ATM, p21 and β-actin by immunoblotting. (D) Same as (C), but stained for SA-β-Gal activity. (E) Quantification of (D). 100 cells from each of the indicated groups were quantified for SA-β-Gal positivity (n=3). (F) Same as (C), but cells were labeled with BrdU for 1 h and BrdU incorporation was visualized by immunofluorescence. DAPI staining was used to visualize nuclei. (G) Quantification of (F). 200 cells from each of the indicated groups were quantified for BrdU positivity (n=3). (H) Same as (C) but an equal number of cells were seeded in 6-well plates, and focus formation was determined by crystal violet staining 14 days later. (I) Quantification of (H). The intensity of foci formed was quantified using NIH ImageJ software (n=3). (J) Same as (C), but cells were examined for 53BP1 and γH2AX foci formation. Scale bars = 5μm. (K) Quantification of (J). 200 cells from each of the indicated groups were quantified for 53BP1 and γH2AX foci positivity (n=3). (L) Same as (C), but comet assay was performed. (M) Quantification of (L). The extent of DNA damage was quantified as Olive Moment using CometScore software (n=100). *p<0.05 shRRM2 vs. control; #p<0.05 shRRM2/shATM vs. shRRM2. Error bars represent SEM. Scale bars = 10μm unless otherwise specified.
Knockdown of ATM rescues dNTP levels and aberrant DNA replication
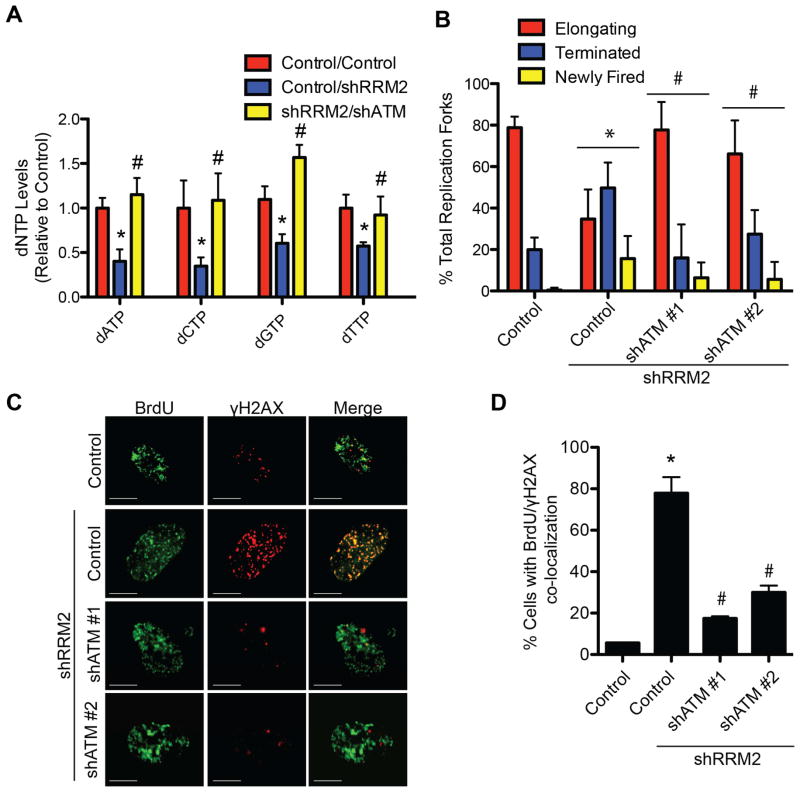
We next sought to determine the effect of knockdown of ATM on cellular dNTP levels. shATM significantly rescued the dNTPs compared with shRRM2 alone (Figure 2A). This correlated with a significant rescue of aberrant replication dynamics induced by shRRM2 as determined by DNA combing analysis (Figure 2B). Collapsed replication forks are characterized by co-localization of pulse labeled BrdU and γH2AX (Groth et al., 2010). shATM significantly reduced the co-localized BrdU and γH2AX induced by shRRM2 (Figure 2C–D). shATR did not affect aberrant replication dynamics induced by shRRM2 (Figure S2), which correlated with the inability of shATR to suppress senescence (Figure S1D–N). These results demonstrate that knockdown of ATM rescues dNTPs, which correlates with the suppression of aberrant replication dynamics and DDR.
Figure 2. Knockdown of ATM rescues replication stress by restoring cellular dNTP levels. See also Figure S2.
(A) Primary IMR90 cells were infected with a shRRM2-expressing lentivirus alone or in combination with a shATM (#1)-expressing lentivirus. dNTP levels were quantified day 1 post-drug selection (n=3). (B) Same as (A). DNA fiber analysis was conducted to observe replication fork dynamics in the indicated cells at day 1 post-drug selection. The percentage of elongating, terminated, or newly fired replication forks was quantified in the indicated cells (n=3). (C) Same as (B), but cells were labeled with BrdU for 15 min and the collapsed replication forks were visualized by co-localized BrdU and γH2AX as determined by immunofluorescence using a confocal microscope. Scale bars = 5μm. (D) Quantification of (C). 200 cells from each of the indicated groups were quantified for BrdU and γH2AX co-localization positive cells (n=3). Note that more than 10/nuclei of co-localized BrdU and γH2AX foci was considered positive. *p<0.05 shRRM2 vs. control; #p<0.05 shRRM2/shATM vs. shRRM2. Error bars represent SEM.
Knockdown of ATM increases substrate availability for dNTP biosynthesis through enhanced glutamine and glucose uptake and metabolism
We next sought to determine the mechanism whereby loss of ATM increases dNTPs. Ribonucleotide reductase (RNR) is involved in de novo dNTP synthesis (Figure S3A) (Nordlund and Reichard, 2006; Reichard, 1988). We first sought to determine whether the increase in dNTPs was due to the salvage pathway, which does not rely on RNR (Blakley and Vitols, 1968; Murray, 1971; Reichard, 1988). We used 3-AP, which inhibits de novo dNTP synthesis by inhibiting both RRM2 and RRM2B, two enzymes necessary for de novo dNTP synthesis (Finch et al., 2000; Finch et al., 1999). Suppression of the de novo pathway reversed the ability of shATM to bypass senescence (Figure S3B–C). We next sought to determine whether the rescue of dNTP levels is due to a compensation of shRRM2 by an increase in RRM2B expression. shATM did not increase RRM2B expression in shRRM2 cells (Figure S3D). These results suggest that de novo dNTP synthesis and RRM2B activity is necessary for the observed senescence bypass.
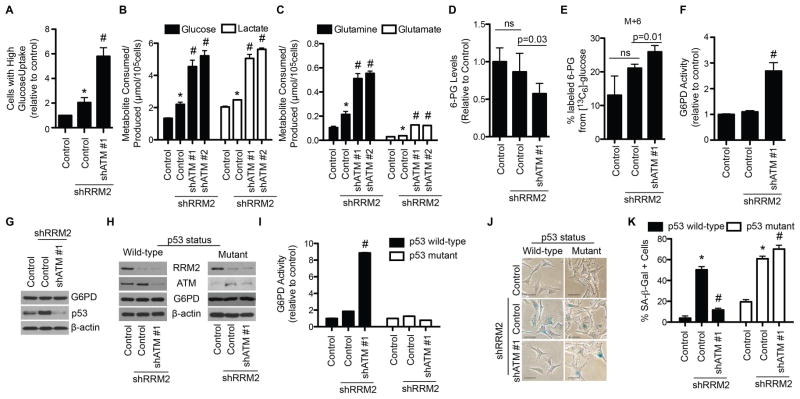
Substrates for dNTP synthesis are derived from consumption of glucose and glutamine (Figure S3A). Since ATM is a tumor suppressor, we hypothesized that knockdown of ATM may increase glucose and glutamine consumption, a hallmark of cancer metabolism, which increases substrate availability for dNTP synthesis. We used 2NBDG, a fluorescent glucose analog, to determine whether shATM increased glucose uptake in shRRM2 cells. shATM in combination with shRRM2 significantly increased glucose uptake (Figure 3A). Metabolite profiling also showed a significant increase in glucose consumption (Figure 3B). Lactate levels were significantly increased in the media, suggesting glucose utilization (Figure 3B). Both glutamine consumption and utilization (glutamate secretion) were also significantly increased (Figure 3C). Similar results were also observed using KU55933, an ATM inhibitor (Figure S3E–F). Likewise, shATM also increased glucose and glutamine consumption and utilization in RAS-expressing cells (Figure S3G–I). These results were observed in multiple cell types (Figure S3J–M). Thus, knockdown of ATM increases substrate availability for dNTP biosynthesis by enhancing glucose and glutamine uptake and metabolism.
Figure 3. Senescence bypass by ATM knockdown correlates with an enhanced glucose and glutamine consumption and a metabolic shift towards the pentose phosphate pathway through an increase in G6PD activity. See also Figure S3.
(A) Primary IMR90 cells were infected with a shRRM2-expressing lentivirus alone or in combination with a shATM-expressing lentivirus, and glucose uptake was determined by incubating cells with a fluorescent glucose analog (2NBDG) followed by flow cytometry (n=3). Cells were gated for high glucose uptake based on fluorescence. (B–C) Same as (A). Media was harvested, and glucose consumption and lactate production (B) or glutamine consumption and glutamate production (C) were quantified (n=3). (D–G) Same as (A). Cells were subjected to the following analysis: liquid chromatography followed by mass spectrometry (LC-MS), and shown are the relative 6-phosphogluconate (6-PG) levels normalized to cell number (n=3) (D); [13C6]-glucose labeling for 30min. Shown is the percent of 13C-labeled 6-PG (n=3) (E); glucose-6-phosphate dehydrogenase (G6PD) activity (n=3) (F); and immunoblotting of G6PD, p53 and β-actin (G). Error bars represent SD. (H) Melanoma cells with known p53 status were infected with a shRRM2-expressing lentivirus alone or in combination with a shATM (#1)-expressing lentivirus, and RRM2, ATM, G6PD, and β-actin protein expression was determined by immunoblotting. (I) Same as (H), but G6PD activity was determined (n=3). (J) Same as (H) but cells were examined for SA-β-Gal activity. Scale bars = 10μm. (K) Quantification of (J). 100 cells from each of the indicated groups were quantified for SA-β-Gal positivity (n=3). *p<0.05 shRRM2 vs. control; #p<0.05 shRRM2/shATM vs. shRRM2. Error bars represent SEM unless otherwise indicated.
Knockdown of ATM increases PPP activity through p53-mediated regulation of G6PD activity
Glucose is metabolized and shunted into different metabolic pathways (Vander Heiden et al., 2009). We used liquid chromatography tandem mass spectrometry (LC-MS/MS) to determine changes in metabolites of multiple metabolic pathways downstream of glucose. There was a significant decrease in the pentose phosphate pathway (PPP) metabolite 6-phosphogluconate (6-PG) in shRRM2/shATM cells (Figure 3D), suggesting that PPP metabolites are being consumed to a greater extent in these cells. To determine whether glucose is being utilized by the PPP, we performed stable isotope tracer analysis using [13C6]-glucose. There was an increase in the fractional proportion of 13C-labeled 6-PG in shRRM2/shATM cells (Figure 3E). These findings suggest that the observed increase in glucose uptake was at least in part utilized by the PPP in these cells for dNTP biosynthesis.
Glucose-6-phosphate dehydrogenase (G6PD) is the rate-limiting enzyme in the PPP, and its activity metabolizes glucose-6-phosphate (G-6-P) into 6-PG for dNTP synthesis (Figure S3A) (Patra and Hay, 2014). We sought to determine whether G6PD is regulated by ATM knockdown in shRRM2 cells. G6PD activity was significantly increased in shATM/shRRM2 cells compared to shRRM2 alone (Figure 3F). Similar results were observed using KU55933, an ATM inhibitor (Figure S3N) or in RAS/shATM cells (Figure S3O). There was no change in G6PD protein expression (Figure 3G). Notably, wild-type p53 has been shown to negatively regulate G6PD activity (Jiang et al., 2011), and knockdown of ATM significantly decreased p53 levels in shRRM2 cells (Figure 3G). Therefore, we sought to determine whether p53 levels contributed to G6PD activity in the context of shATM-mediated senescence bypass. We used melanoma cell lines with known p53 mutational status. Knockdown of ATM (Figure 3H) significantly increased G6PD activity in shRRM2-expressing p53 wild-type, but not mutant, melanoma cells (Figure 3I). Knockdown of p53 (shp53) in combination with shRRM2 in wild-type melanoma cells increased G6PD activity compared to shRRM2 alone (Figure S3P). p53 status, and therefore G6PD activity, correlated with the ability of shATM to bypass senescence in melanoma cells induced by shRRM2 (Figure 3J–K). These results were observed in multiple p53 wild-type and mutant cell lines (Figure S3Q–T), demonstrating this is not a cell line specific effect. Notably, shRRM2 alone did not decrease G6PD activity (Figure 3F), even though we observed an increase in p53 expression (Figure 3G). This suggests that G6PD activity is also regulated by another p53-independent mechanism in shRRM2-expressing cells. Indeed, G6PD activity is known to be positively regulated by ATM-mediated phosphorylation of HSP27 (pHSP27) (Cosentino et al., 2011). Consistently, pHSP27 was increased in shRRM2 cells (Figure S3U), which correlates with activation of ATM by shRRM2 (Figure 1A–B). These data support that G6PD activity is regulated in a context dependent manner through a balance between p53 and HSP27, and when ATM is inhibited, downregulation of p53 correlates with an increase in G6PD activity. We conclude that loss of ATM leads to an increase in G6PD activity through abrogation of p53-mediated suppression.
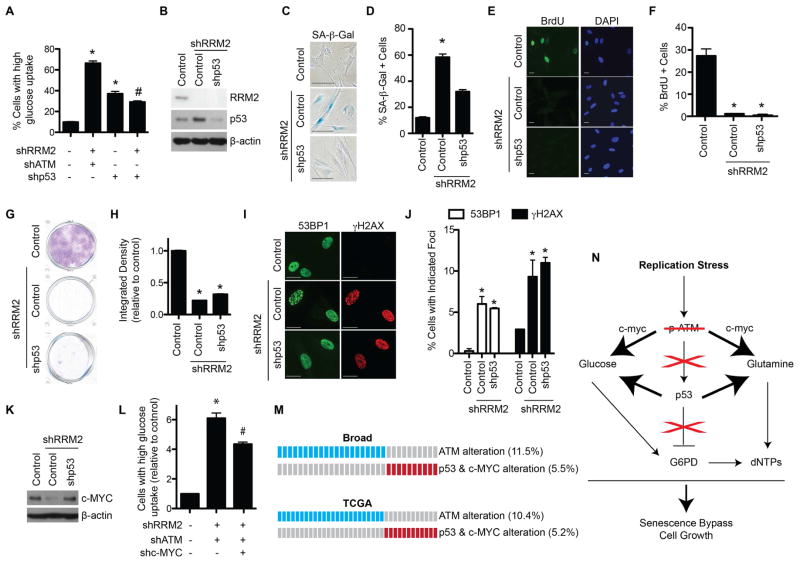
Suppression of p53 is known to affect metabolism (Cairns et al., 2011; Schwartzenberg-Bar-Yoseph et al., 2004). Indeed, shp53 in combination with shRRM2 increased glucose uptake (Figure 4A) and glucose and glutamine consumption compared to shRRM2 alone (Figure S4A–B). Therefore, we sought to determine whether shp53 phenocopies shATM. shp53 (Figure 4B) only partially suppressed senescence phenotypes such as causing a decrease in SA-β-Gal activity (Figure 4C–D). However, shp53 did not fully bypass the senescence-associated cell growth arrest, as demonstrated by BrdU incorporation (Figure 4E–F) and focus formation assays (Figure 4G–H). This correlated with the inability of shp53 to reduce markers of DNA damage (Figure 4I–J). These results suggest that p53 suppression is necessary but not sufficient for bypassing senescence induced by replication stress.
Figure 4. ATM knockdown cooperatively inhibits p53 and upregulates c-MYC in senescence-bypassed cells. See also Figure S4 and Table S1.
(A) Primary IMR90 cells were infected with lentivirus expressing the indicated shRNAs. Glucose uptake was determined by incubating cells with a fluorescent glucose analog (2NBDG) followed by flow cytometry. Cells were gated for high glucose uptake based on fluorescence (n=3). (B) Primary IMR90 cells were infected with a shRRM2-expressing lentivirus alone or in combination with a shp53-expressing lentivirus, and RRM2, p53 and β-actin protein expression was determined by immunoblotting. (C) Same as (B), but SA-β-Gal activity was determined. (D) Quantification of (C). 100 cells from each of the indicated groups were quantified for SA-β-Gal positivity (n=3). (E) Same as (B), but cells were labeled with BrdU for 1 h, and BrdU incorporation was determined by immunofluorescence. DAPI staining was used to visualize nuclei. (F) Quantification of (E). 200 cells from each of the indicated groups were quantified for BrdU positivity (n=3). (G) Same as (B), but an equal number of cells were seeded in 6-well plates and focus formation was determined by crystal violet staining 14 days later. (H) Quantification of (G). The intensity of foci was quantified using NIH ImageJ software (n=3). (I) Same as (B) but 53BP1 and γH2AX foci were observed by immunofluorescence. (J) Quantification of (I). 200 cells from each of the indicated groups were quantified for 53BP1 and γH2AX foci positive cells (n=3). (K) Primary IMR90 cells were infected with an shRRM2-expressing lentivirus alone or in combination with a shATM-expressing lentivirus, and c-MYC and β-actin protein expression was determined by immunoblotting. (L) Same as (K), but cells were also infected with a shc-MYC expressing lentivirus, and glucose uptake was determined by incubating cells with a fluorescent glucose analog (2NBDG) followed by flow cytometry (n=3). Cells were gated for high glucose uptake based on fluorescence. (M) Publically available lung adenocarcinoma databases from cBioPortal were analyzed for ATM, p53, and c-MYC status. Blue boxes indicate patients with ATM mutation or deletion. Red boxes indicate patients with p53 mutation/deletion and c-MYC amplification. (N) Schematic of senescence bypass induced by shATM. Replication stress induced by nucleotide deficiency activates ATM. If ATM is inhibited, p53 is not activated, which abrogates its inhibition of G6PD. Additionally, c-MYC expression is increased, which along with lower p53 expression, leads to increased glucose and glutamine consumption. The convergence of increased substrates and increased G6PD activity leads to increased dNTP levels, which allows for DNA replication and proliferation. *p<0.05 vs. control; #p<0.05 vs. shRRM2/shATM. Error bars represent SEM. Scale bars = 10μm.
Knockdown of ATM cooperatively regulates p53 and c-MYC to increase substrate availability
shp53 in combination with shRRM2 increased glucose uptake but to a lesser extent compared to that of shATM (Figure 4A). This suggests that an additional pathway is implicated in the observed shift in cellular metabolism induced by shATM. c-MYC plays a major role in cellular metabolism (Dang et al., 2009). c-MYC is a known regulator of RAS-induced senescence (Land et al., 1983; Sinn et al., 1987). Thus, we examined c-MYC protein levels. shATM in combination with shRRM2 significantly increased c-MYC protein expression compared to shRRM2 alone (Figure 4K). This was observed in multiple cell lines (Figure S4C). Notably, shp53 did not increase c-MYC expression (Figure S4D).
We next determined the mechanism underlying the observed c-MYC upregulation by shATM. No change in c-MYC mRNA expression was observed in senescence-bypassed cells (Figure. S4E). p27 has been implicated in negatively regulating c-MYC protein stability post-translationally (Maclean et al., 2007). Bypass of senescence by shATM correlated with downregulation of p27 (Figure S4F). This correlated with an increase in c-MYC protein stability (Figure S4G–H). To further demonstrate the role of c-MYC upregulation in the observed senescence bypass, we simultaneously knocked down ATM, RRM2, and c-MYC (Figure S4I). These cells showed decreased glucose uptake and glutamine consumption compared to senescence-bypassed shATM/shRRM2 cells (Figure 4L and S4J). These results support that increased c-MYC expression cooperates with decreased p53 to induce metabolic reprogramming to allow the senescence bypass by shATM.
Our results indicate that ATM converges on the p53 and c-MYC pathways to regulate senescence. We found that ATM mutation/deletion is mutually exclusive from p53 mutation/deletion and c-MYC amplification in multiple tumor types (Figure 4M and Table S1). These data support the notion that ATM functions in the same pathway as p53 and c-MYC in cancers. Overall, our data support a model whereby loss of ATM affects both p53 and c-MYC to bypass the senescence-associated cell growth arrest to drive cell proliferation (Figure 4N).
Discussion
Senescence induced by oncogenes is characterized by increased glucose consumption that shunts glucose towards the TCA cycle and away from aerobic glycolysis and presumably the PPP (Kaplon et al., 2013; Li et al., 2013; Mazurek et al., 2001). Consistently, we observed increased glucose and glutamine consumption in cells undergoing senescence induced by shRRM2 (Figure 3A–C and S3), suggesting that replication stress itself may cause changes in cellular metabolic pathways similar to those induced by oncogenes. Knockdown of ATM in combination with shRRM2 further increased both glucose and glutamine consumption. These data are consistent with the idea that ATM inactivation further drives senescence-associated metabolic reprogramming over a threshold that is necessary to support the proliferation of senescence-bypassed cells in cancer. Our data support the notion that the increased glucose was shunted into the PPP because we observed an increase in 13C-labeling of the PPP metabolite 6-PG after incubation with [13C6]-glucose (Figure 3E). In this context, ATM functions as a tumor suppressor by inhibiting cancer-associated metabolic reprogramming.
In addition to an increase in metabolic substrates for dNTP synthesis, ATM knockdown also increased activity of the PPP pathway rate-limiting enzyme G6PD. Increased G6PD activity correlated with decreased p53 expression (Figure 3F–G). p53 downregulation is known to shift cellular metabolism (Cairns et al., 2011). Knockdown of ATM decreased p53 (Figure 3G), which correlated with increased glucose and glutamine consumption (Figure 3B–C). Knockdown of p53 did not increase glucose uptake to the same extent as ATM knockdown (Figure 4A). Indeed, knockdown of p53 was not sufficient to fully overcome senescence (Figure 4). Consistently, there was also an increase in c-MYC expression induced by ATM knockdown (Figure 4K). c-MYC is among genes that are upregulated in ATM knockout mice (Yan et al., 2006). In the present study, the observed increase in c-MYC expression was post-transcriptional and correlated with a decrease in p27 expression, a known negative regulator of c-MYC protein stability (Figure S4F). Indeed, c-MYC stability was increased in cells with knockdown of both RRM2 and ATM compared to knockdown of RRM2 alone (Figure S4G–H). These data support a model whereby downregulation of p53 and upregulation of c-MYC cooperate to enhance glucose and glutamine consumption, which accounts for the observed increase in substrate availability for the dNTP biosynthesis induced by ATM knockdown (Figure 4N).
Our data show that ATM plays a central role in coupling replication stress and metabolic reprogramming. ATM suppresses cancer-associated metabolic reprogramming to prevent bypass of the senescence tumor suppression mechanism, where its inactivation suppresses DDR induced by replication stress. This is different from its classical role in DNA repair where its activation suppresses cancer by preventing genomic instability. Replication stress induced by nucleotide deficiency plays an important role in the early stages of cancer development; therefore, this mechanistic insight will have broad implications for understanding replication stress and metabolic reprogramming in cancer.
Experimental Procedures
Cells and culture conditions
IMR90 human fibroblasts were cultured according to the ATCC in low oxygen (2%) as previously described (Tu et al., 2011). Experiments were performed on IMR90 between population doubling #25–35. Human melanoma cell lines (kind gift from Dr. Meenhard Herlyn) were cultured as previously described (Satyamoorthy et al., 1997).
Reagents, plasmids, and antibodies
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO). pLKO.1-shRNA plasmids were obtained from Open Biosystems (Waltham, MA). The mature sense sequences are: shRRM2: 5′-CGGAGGAGAGAGTAAGAGAAA-3′; shATM #1: 5′-CGTGTCTTAATGAGACTACAA-3′; shATM #2: 5′-TGATGGTCTTAAGGAACATCT-3′; shp53: 5′-GAGGGATGTTTGGGAGATGTA-3′; and shc-MYC: 5′-CCTGAGACAGATCAGCAACAA-3. The following antibodies were obtained from the indicated suppliers: mouse anti-phospho-ATM (Rockland, Gilbertsville, PA), goat anti-ATM (Bethyl, Montgomery, TX), goat anti-RRM2 (Santa Cruz Biotechnology), mouse anti-Cyclin A (Novocastra), mouse anti-γH2AX (Millipore, Billerica, MA), rabbit anti-53BP1 (Bethyl), mouse anti-BrdU FITC (BD Biosciences, San Jose, CA), mouse anti-p53 (Calbiochem, Billerica, MA), rabbit anti-G6PD (Sigma-Aldrich), rabbit anti-p21 (Abcam, Cambridge, MA), rabbit anti-c-MYC (Cell Signaling, Danvers, MA), and mouse anti-β-actin (Sigma-Aldrich).
Lentivirus infections
Lentivirus was packaged using the Virapower Kit from Invitrogen (Carlsbad, CA) following the manufacturer’s instructions as described previously (Li et al., 2010; Tu et al., 2011). Cells infected with viruses encoding the puromycin-resistance gene were selected in 1 μg/ml puromycin.
Immunofluorescence, BrdU labeling, single cell gel electrophoresis (comet assay), and SA-β-gal staining
Immunofluorescence staining, BrdU labeling, and single cell gel electrophoresis (comet assay) for cultured cells was performed as described previously using antibodies described above (Tu et al., 2011). Confocal microscopy was used for co-localization of BrdU and γH2AX using a Leica TCS SPII Scanning Confocal Microscope (Buffalo Grove, IL). The comet assay was analyzed using the CometScore software (TriTek, Sumerduck, VA). SA-β-Gal staining was performed as previously described (Dimri et al., 1995).
Focus formation assay
For focus formation, an equal number of cells (3000 cells/well) was inoculated in 6-well plates and cultured for additional 2 weeks. Focus formation was visualized by staining the plates with 0.05% crystal violet as previously described (Tu et al., 2011). Integrated density was determined using NIH ImageJ software.
DNA combing analysis
DNA combing was performed as previously described (Aird et al., 2013). DNA replication forks were scored as elongating, terminated, or newly fired as previously described (Aird et al., 2013; Bartkova et al., 2006).
Measurement of dNTP concentrations in cells
Samples were harvested and dNTP levels were measured as previously described (Aird et al., 2013; Wilson et al., 2011).
Flow cytometry for glucose uptake
Cells were incubated with 5μM 2NBDG (Invitrogen) for 2 h. After rinsing with PBS, 2NBDG positive cells were run on a LSRII (14-color; Becton-Dickinson, East Rutherford, NJ), and data were analyzed using FlowJo Software (Ashland, OR).
YSI Metabolite Measurements
Glucose and glutamine consumption and lactate and glutamate production were measured using a YSI 7100 Bioanalyzer (Yellow Springs, OH). Briefly, the same number of cells was seeded in 12-well plates and 24 h later, the media was changed. Twenty-four hours later, the media was harvested and cells were counted to normalize for proliferation.
C13-glucose labeling and Liquid chromatography-mass spectrometry analysis
To extract metabolites, media was aspirated and cells were quenched by the direct addition of 1 mL −80 °C 4:1 methanol:water (v/v). Plates were placed at −80 °C for 20 min then scraped and transferred into tubes. Samples were pulse sonicated on ice for 30 sec at a rate of 1 pulse/sec prior to centrifugation at 16,000g at 4°C for 10 min. The supernatants were transferred to clean glass tubes and evaporated to dryness under nitrogen. Dried residues were resuspended in 100 μL of mobile phase A for LC-MS analysis. For labeling studies, cells were grown in media omitting glucose supplemented with 1 mg/mL [13C6]-glucose.
For liquid chromatography-mass spectrometry, separations were performed using an Agilent 1200 series HPLC pump and autosampler (Agilent Technologies, Santa Clara, CA). Analytes were separated by reverse-phase ion-paring chromatography using a Phenomenex Kinetex Luna C18 column (250 × 2.1 mm I.D., 3 μm). N,N-Diisopropylethylamine (DIPEA) was used as the ion-pairing reagent. Solvent A was 400 mM 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) and 10 mM DIPEA in water and solvent B was 300 mM HFIP and 10 mM DIPEA in methanol. The linear gradient conditions were as follows: 2% B at 0 min, 2% B at 3 min, 10%B at 32 min, 95% B at 38 min, 2%B at 39 min, followed by a 6 min equilibration. Analyses were conducted using an Agilent Technologies 6460 Triple Quadrupole mass spectrometer with a JetStream ESI source, in the negative mode. The samples were maintained at 4°C and injections of 10 μL were made for all runs. The column effluent was diverted to waste for the first 5 min and the last 10 min of the analyses. The Agilent 6460 operating conditions were as follows: gas temperature was set at 275 °C and the gas flow was set to 8 L/min. Sheath gas temperature was 400 °C and the sheath gas flow was set to 10 L/min. The capillary voltage was set to 3500 V. The nozzle voltage was set to 1000 V.
Glucose-6-phosphate dehydrogenase activity assay
Cells were harvested by trypsinization and resuspended in cold PBS. Cells were sonicated and then centrifuged at 16,000rpm for 10 min at 4°C. The supernatant was transferred to new tubes, and the combined activity of G6PD and 6-phosphogluconate dehydrogenase (6PGD) was measured by the rate of conversion of NADP+ to NADPH in the presence of glucose-6-phosphate (G6P). The activity of 6PGD alone was then measured by the conversion of NADP+ to NADPH in the presence of 6-phosphogluconate (6PG). G6PD activity was calculated as the difference of these two activities. Cell lysates were added to the reaction buffer containing 50mM Tris and 1 mM MgCl2, pH 8.1, NADP+ (100 μM) and G6P (200 μM) or 6PG (200 μM). The absorbance at 341nm was read 10 min later. Enzyme activities were normalized to protein concentration.
Statistical analysis
Graphpad Prism Version 5.0 was used to perform statistical analyses. The student’s t test was used to determine p values of raw data. A p value <0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by NIH/NCI grants (R01CA160331 and P50CA174523 to R.Z., T32CA9171-35 to K.M.A.), a DoD award (OC093420 to R.Z.), and NIH/NIEHS grants (P30ES013508 to A.J.W. and I.A.B., T32ES019851 to A.J.W.). Support of Core Facilities used in this study was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute.
Footnotes
Author Contributions
K.M.A. designed experiments. K.M.A., A.J.W., N.W.S., J.V.L., and S.S. conducted experiments. K.M.A., A.J.W., N.W.S., K.E.W., and R.Z. analyzed data. Q.L. performed statistical analysis. I.A.B. and K.E.W. also designed and supervised experiments. R.Z. conceived and supervised the study. K.M.A., K.E.W., and R.Z. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, Wu H, Wei Z, Wagner SN, Herlyn M, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell reports. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakley RL, Vitols E. The control of nucleotide biosynthesis. Annu Rev Biochem. 1968;37:201–224. doi: 10.1146/annurev.bi.37.070168.001221. [DOI] [PubMed] [Google Scholar]
- Burhans WC, Weinberger M. DNA replication stress, genome instability and aging. Nucleic Acids Res. 2007;35:7545–7556. doi: 10.1093/nar/gkm1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. The EMBO journal. 2011;30:546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch RA, Liu M, Grill SP, Rose WC, Loomis R, Vasquez KM, Cheng Y, Sartorelli AC. Triapine (3-aminopyridine-2-carboxaldehyde- thiosemicarbazone): A potent inhibitor of ribonucleotide reductase activity with broad spectrum antitumor activity. Biochem Pharmacol. 2000;59:983–991. doi: 10.1016/s0006-2952(99)00419-0. [DOI] [PubMed] [Google Scholar]
- Finch RA, Liu MC, Cory AH, Cory JG, Sartorelli AC. Triapine (3-aminopyridine-2-carboxaldehyde thiosemicarbazone; 3-AP): an inhibitor of ribonucleotide reductase with antineoplastic activity. Adv Enzyme Regul. 1999;39:3–12. doi: 10.1016/s0065-2571(98)00017-x. [DOI] [PubMed] [Google Scholar]
- Groth P, Auslander S, Majumder MM, Schultz N, Johansson F, Petermann E, Helleday T. Methylated DNA causes a physical block to replication forks independently of damage signalling, O(6)-methylguanine or DNA single-strand breaks and results in DNA damage. Journal of molecular biology. 2010;402:70–82. doi: 10.1016/j.jmb.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nature cell biology. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Li H, Cai Q, Godwin AK, Zhang R. Enhancer of zeste homolog 2 promotes the proliferation and invasion of epithelial ovarian cancer cells. Molecular cancer research: MCR. 2010;8:1610–1618. doi: 10.1158/1541-7786.MCR-10-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Durbin KR, Sweet SM, Tipton JD, Zheng Y, Kelleher NL. Oncogene-induced cellular senescence elicits an anti-Warburg effect. Proteomics. 2013;13:2585–2596. doi: 10.1002/pmic.201200298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean KH, Kastan MB, Cleveland JL. Atm deficiency affects both apoptosis and proliferation to augment Myc-induced lymphomagenesis. Molecular cancer research: MCR. 2007;5:705–711. doi: 10.1158/1541-7786.MCR-07-0058. [DOI] [PubMed] [Google Scholar]
- Mannava S, Moparthy KC, Wheeler LJ, Natarajan V, Zucker SN, Fink EE, Im M, Flanagan S, Burhans WC, Zeitouni NC, et al. Depletion of deoxyribonucleotide pools is an endogenous source of DNA damage in cells undergoing oncogene-induced senescence. The American journal of pathology. 2013;182:142–151. doi: 10.1016/j.ajpath.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Zwerschke W, Jansen-Durr P, Eigenbrodt E. Metabolic cooperation between different oncogenes during cell transformation: interaction between activated ras and HPV-16 E7. Oncogene. 2001;20:6891–6898. doi: 10.1038/sj.onc.1204792. [DOI] [PubMed] [Google Scholar]
- Murray AW. The biological significance of purine salvage. Annu Rev Biochem. 1971;40:811–826. doi: 10.1146/annurev.bi.40.070171.004115. [DOI] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nature reviews Molecular cell biology. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends in biochemical sciences. 2014;39:347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K, DeJesus E, Linnenbach AJ, Kraj B, Kornreich DL, Rendle S, Elder DE, Herlyn M. Melanoma cell lines from different stages of progression and their biological and molecular analyses. Melanoma Res. 1997;7(Suppl 2):S35–42. [PubMed] [Google Scholar]
- Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer research. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- Tu Z, Aird KM, Bitler BG, Nicodemus JP, Beeharry N, Zia B, Yen TJ, Zhang R. Oncogenic Ras regulates BRIP1 expression to induce dissociation of BRCA1 from chromatin, inhibit DNA repair, and promote senescence. Dev Cell. 2011;21:1–15. doi: 10.1016/j.devcel.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science (New York, NY) 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PM, Labonte MJ, Russell J, Louie S, Ghobrial AA, Ladner RD. A novel fluorescence-based assay for the rapid detection and quantification of cellular deoxyribonucleoside triphosphates. Nucleic Acids Res. 2011;39:e112. doi: 10.1093/nar/gkr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Zhu C, Liu N, Jiang Y, Scofield VL, Riggs PK, Qiang W, Lynn WS, Wong PK. ATM controls c-Myc and DNA synthesis during postnatal thymocyte development through regulation of redox state. Free radical biology & medicine. 2006;41:640–648. doi: 10.1016/j.freeradbiomed.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Yaswen P, Campisi J. Oncogene-induced senescence pathways weave an intricate tapestry. Cell. 2007;128:233–234. doi: 10.1016/j.cell.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS letters. 2010;584:3717–3724. doi: 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nature cell biology. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.