SUMMARY
Long non-coding RNAs (lncRNAs) including natural antisense transcripts (NATs) are expressed more extensively than previously anticipated, and have widespread roles in regulating gene expression. Nevertheless, the molecular mechanisms of action of the majority of NATs remain largely unknown. Here we identify a NAT of Low-density lipoprotein receptor-related protein 1 (Lrp1), referred to as Lrp1-AS, that negatively regulates Lrp1 expression. We show that Lrp1-AS directly binds to High mobility group box 2 (Hmgb2) and inhibits the activity of Hmgb2 to enhance Srebp1a-dependent transcription of Lrp1. Short oligonucleotides targeting Lrp1-AS inhibit the interaction of antisense transcript and Hmgb2 protein, and increase Lrp1 expression by enhancing Hmgb2 activity. qRT-PCR analysis of Alzheimer’s disease brain samples and aged-matched controls revealed upregulation of LRP1-AS and downregulation of LRP1. Our data suggest a new regulatory mechanism whereby a NAT interacts with a ubiquitous chromatin-associated protein to modulate its activity in a locus-specific fashion.
INTRODUCTION
Mammalian genomes are more extensively transcribed than expected, giving rise to thousands of long non-coding RNAs (lncRNAs) which are define as RNA transcripts non coding for protein and longer than 200 nt (Bertone et al., 2004; Birney et al., 2007; Carninci et al., 2005; Cheng et al., 2005; Djebali et al., 2012; Kapranov et al., 2007; Yelin et al., 2003). Among lncRNAs, NATs have emerged as a large class of regulatory long ncRNAs (Faghihi and Wahlestedt, 2009; Magistri et al., 2012). NATs are reported for more than 70% of all transcriptional units (Katayama et al., 2005), and 20% of human genes (Cheng et al., 2005; Yelin et al., 2003). We and others have recently shown that functional knockdown of NATs has positive or negative influences on the expression of neighboring protein-coding genes (Carrieri et al., 2012; Faghihi et al., 2008; Katayama et al., 2005; Mahmoudi et al., 2009; Modarresi et al., 2012) thus implying a critical role of NATs in the regulation of gene expression.
LncRNAs are implicated in numerous cellular processes ranging from pluripotency, differentiation, cell-cycle regulation and are often dysregulated in disease states, such as Alzheimer’s disease (AD), coronary artery disease and cancer (Bond et al., 2009; Faghihi et al., 2008; Guttman et al., 2011; Harismendy et al., 2011; Hung et al., 2011; Pastori and Wahlestedt, 2012; Prensner et al., 2011; Velmeshev et al., 2013). Although the mechanisms of lncRNAs as key regulators of gene expression are yet to be fully elucidated, a common emerging theme is that lncRNAs form RNA-protein complexes to exert their regulatory functions. In some cases NATs are reported to modulate DNA accessibility by binding to chromatin-modifying complexes and sequestration of transcription factor, which in turn influence gene expression (Guttman and Rinn, 2012; Pastori et al., 2010; Rinn and Chang, 2012; Wang and Chang, 2011). Therefore, in order to understand the function of lncRNAs it is of crucial importance to identify the interacting proteins.
Low-density lipoprotein receptor-related protein (LRP) 1 is a member of the low-density lipoprotein receptor family, which has a role in a variety of physiological processes including the cellular transport of cholesterol, endocytosis of ligands, and transcytosis across the blood-brain barrier (Lillis et al., 2008). Recently, LRP1 has been implicated in the systemic clearance of AD amyloid-beta (Aβ), and the level of LRP1 expression is critical for AD progression (Deane et al., 2008; Holtzman et al., 2012; Kang et al., 2000; Liu et al., 2007; Shibata et al., 2000). However, little is known about the mechanisms of LRP1 expression regulation. Here, we showed that transcription of LRP1 locus gives rise to both LRP1 mRNA and a spliced NAT of LRP1 gene, which we named it, LRP1-AS. We demonstrated that LRP1-AS negatively regulates LRP1 gene expression through modulating the activity of the non-histone chromatin modifier HMGB2 and we showed that LRP1-AS is elevated in the brains of AD patients where it might repress LRP1 expression.
RESULTS
Identification of Human and Mouse LRP1-AS
To identify putative ncRNAs associated with human and mouse LRP1 gene, we utilized the UCSC Genome Browser to search for Expressed Sequence Tags (ESTs) overlapping human and mouse LRP1 gene and checked for annotated antisense RNAs in Ensembl and AceView databases. We found ESTs from the opposite DNA strand of exon 5 of the human LRP1 gene and exons 5 and 6 of the mouse Lrp1 gene. In human these ESTs correspond to an Ensembl annotated two-exons antisense RNA of 645 bp (RP11-545N8.3) that we named LRP1-AS (Figure 1A). Similarly, in mouse these ESTs correspond to an AceView (Thierry-Mieg and Thierry-Mieg, 2006) annotated two-exons antisense RNA of 1387 bp (sloty) that we named Lrp1-AS (Figure 1B). We found short open reading frames (ORFs) of 141 bp (15 to 155) and 108 bp (226 to 333), in exon 2 of human LRP1-AS. We also found short ORFs of 120 bp (388 to 507) and 117 bp (675 to 791) in exons 2 of mouse Lrp1-AS. The potential polypeptides had no sequence similarity to any other polypeptide sequence in the Genbank protein database, and were not conserved with any predicted polypeptide in the Protein Clusters Database or the Conserved Domain Database. Exon 2 of Lrp1-AS directly overlaps the exon 5 and 6 of Lrp1 by 395 bp, while exon 2 of LRP1-AS directly overlaps the exon 5 of LRP1 by 119bp (Figure S1A). This similar location of LRP1/LRP1-AS and Lrp1/Lrp1-AS has been maintained throughout evolution, indicating that this genomic arrangement might have a biological function. In order to derive general correlation between the reciprocal expression of LRP1 and LRP1-AS we analyzed data from the Developmental Transcriptome project of the BrainSpan atlas (http://brainspan.org/) (Miller et al., 2014). This project consists of RNAseq data profiling up to 16 cortical and subcortical regions across the course of human brain development (13 developmental stages). The analysis of these data revealed a positive correlation between LRP1 and LRP1-AS expression across human brain development (Pearson r = 0.6115; P value < 0.0001) (Figure S1B) and RPKM value for LRP1-AS vary from 1 to 5.9. Depending of the total amount of RNA present in a single cell it has been calculated that one copy of a transcript per cell corresponds to RPKM value between 0.5 and 5 (Mortazavi et al., 2008). Thus, in the human brain, there is approximately one transcript of LRP1-AS per cell. We then used digital PCR (dPCR) to measure the expression of Lrp1 and Lrp1-AS in the mouse brain and in variety of murine cells available in the lab. We noticed a general positive correlation between Lrp1-AS and Lrp1 expression, with Lrp1-AS expressed at higher level in the macrophage cell line RAW264.7 and at a lower level in pancreatic beta cell line MIN6 (Figure S2). Because of the higher expression in RAW264.7, we decided to utilize this cell line as a model to study the mechanism of function of Lrp1-AS.
Figure 1. Genomic organization of the Human LRP1 and Mouse Lrp1 locus.
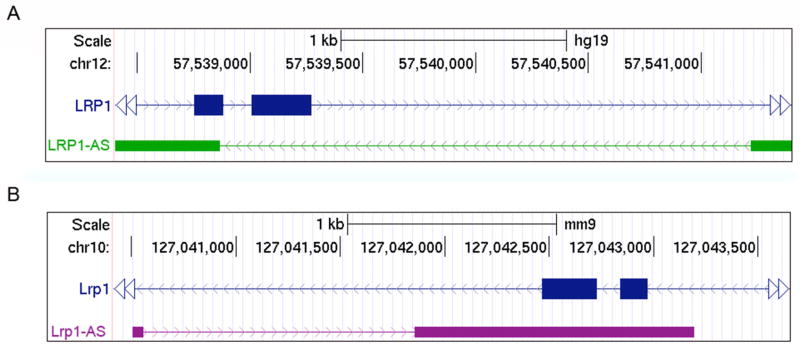
Natural antisense transcripts, human LRP1-AS (A) and mouse Lrp1-AS (B), are in cis transcribed from the opposite strand of human LRP1 gene on chromosome 12, and mouse Lrp1 gene on chromosome 10, respectively. Exons 5 and 6 of LRP1 and Lrp1 are depicted in blue boxes; the exons of LRP1-AS and Lrp1-AS are depicted in green and violet boxes respectively.
Lrp1-AS negatively regulates Lrp1 Expression
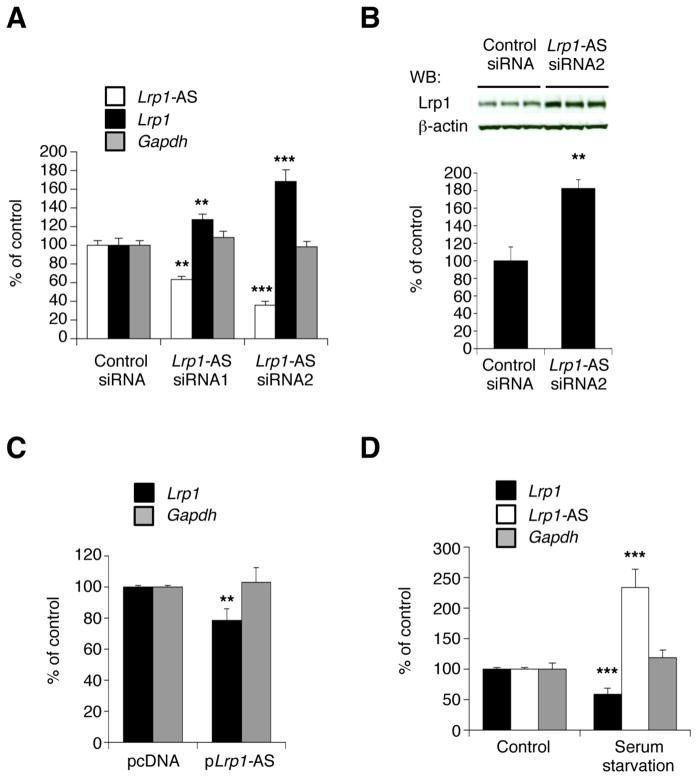
To investigate a possible regulation of Lrp1 levels by Lrp1-AS, we analyzed changes in Lrp1 levels after silencing Lrp1-AS expression. Silencing of Lrp1-AS by two different siRNAs targeting non-overlap regions in exons 2 led to a significant decrease in Lrp1-AS levels in RAW264.7 cells. We observed that the degree of Lrp1-AS downregulation was proportional to Lrp1 mRNA and protein upregulation (Figure 2A and 2B). To confirm that knockdown of Lrp1-AS does not induce non-specific upregulation of other genes, we analyzed changes in Gapdh levels and found no change. We further examined Lrp1 levels after overexpression of Lrp1-AS, and observed a significant decrease in Lrp1 expression (Figure 2C). These data indicated that Lrp1-AS negatively regulates Lrp1 expression, directly or indirectly, at both RNA and protein levels.
Figure 2. Lrp1-AS Negatively Regulates Lrp1 levels.
(A) Lrp1, Lrp1-AS and Gapdh RNA levels after Lrp1-AS silencing by control or two different siRNAs against Lrp1-AS in RAW264.7 cells. RNA levels were measured by quantitative RT-PCR of 24 hr post transfection. Control siRNA has a sequence with no homology to any gene.
(B) Western blot (WB) analysis of Lrp1 and β-actin proteins in RAW264.7 cells treated with siRNA2 against Lrp1-AS (upper). Lrp1 protein levels were densitometrically quantified and normalized by β-actin (lower).
(C) Lrp1 and Gapdh RNA levels after overexpression of Lrp1-AS in RAW264.7 cells. Empty vector (pcDNA) serves as a negative control.
(D) Lrp1-AS, Lrp1 and Gapdh RNA levels of RAW264.7 cells after 12 hr serum starvation.
Mean ± s.d. (n = 3 replicates) are shown in all bar graphs in (A) to (D).**P < 0.01, ***P < 0.001 determined by ANOVA.
Cellular stress such as nutrient deprivation is known to influence the expression of Lrp1 (Annabi et al., 2010). In our cells after serum starvation we observed upregulation of Lrp-AS and downregulation of Lrp1 (Figure 2D). Together, these data showed that changes in Lrp1-AS levels influence Lrp1 expression, and this regulation may be triggered by nutrient deprivation.
Identification of Hmgb2 Binding to Lrp1-AS
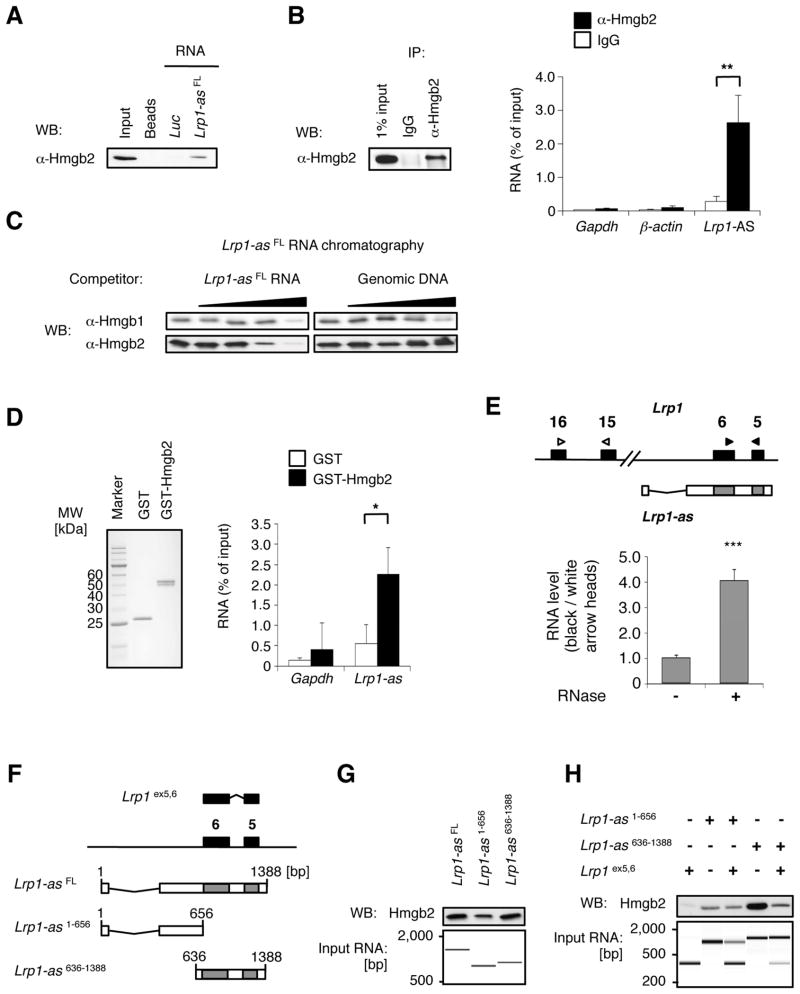
To identify specific proteins associated with Lrp1-AS RNA, we performed RNA chromatography on purified, in-vitro-transcribed, full-length Lrp1-AS RNA, and 1.7-kb Luc mRNA as a negative control, to probe nuclear extracts of RAW264.7 cells. Isolated proteins were run on a gel and visualized with silver staining; one differentially visible protein band was subjected to mass spectrometry, resulting in the identification of Hmgb2 (Figure S3). Consistently, the specificity of Hmgb2 binding to Lrp1-AS was confirmed by Western blotting with specific antibody (Figure 3A). Hmgb1/2 are the most abundant Hmg proteins regulating numerous cellular activities including transcription, and Hmgb2 is highly expressed in lymphoid organs and testes (Ronfani et al., 2001). In the reciprocal experiment, we performed RNA immunoprecipitation (RIP) of endogenous Hmgb2 from RAW264.7 cells extracts. The anti-Hmgb2 IP retrieved associated Lrp1-AS RNA as detected by qRT-PCR, but not nonspecific Gapdh or β-actin (Figure 3B).
Figure 3. Lrp1-AS binds to Hmgb2.
(A) RNase-assisted RNA chromatography on full-length Lrp1-AS FL in RAW264.7 nuclear extracts, visualized by Western blotting (WB) with specific antibody against Hmgb2.
(B) Immunoprecipitation with control IgG or specific antibody against Hmgb2 from RAW264.7 lysates, visualized by WB (left), and co-precipitated RNAs from (B) were detected by qRT-PCR using primer pairs for Lrp1-AS, Gapdh or β-actin (right).
(C) In vitro pulldown of 6XHis-tagged Hmgb2 with Lrp1-ASFL and free nucleic acids as indicated.
(D) In vitro pulldown of endogenous Lrp1-AS with GST or GST-fused Hmgb2 from RAW264.7 nuclear extracts. RNAs were detected by qRT-PCR using primer pairs for Lrp1-AS or Gapdh (right). Equal input proteins were visualized on electrophoresis by CBB stain (left). Mean ± s.d. (n = 3 replicates) are shown in all bar graphs. *P < 0.05, **P < 0.01 determined by ANOVA.
(E) RNase protection assay. The RNA ratios were calculated from qRT-PCR using primer pairs for overlap region (black arrowhead) or non overlap region (white arrowhead) from RAW264.7 nuclear extracts treated with (+) or without (−) RNase A. Mean ± s.d. (n = 3 replicates) are shown. ***P < 0.001 determined by ANOVA.
(F) Schematic illustration of deletion constructs of Lrp1 and Lrp1-AS, used for (C) and (D): Lrp1 construct contains exons 5 and 6 (395 bp) of Lrp1. Lrp1-AS636-1388 construct contains the overlap region between Lrp1 and Lrp1-AS, whereas Lrp1-AS1-656 only contains the 5′ fragment of Lrp1-AS.
(G) RNA chromatography on full length or fragments of Lrp1-AS, as indicated above, in RAW264.7 nuclear extracts, followed by Western blotting (WB, upper). Equal input RNAs were visualized on electrophoresis by Agilent Bioanalyzer (lower).
(H) RNA chromatography on Lrp1-AS fragments, Lrp1 fragment, or Lrp1-AS:Lrp1 fragments hybrid, as indicated above, in RAW264.7 nuclear extracts, followed by Western blotting (WB, upper). Equal input RNAs were visualized on electrophoresis by Agilent Bioanalyzer (lower).
Direct and specific binding of Lrp1-AS RNA to Hmgb2 protein
Recently Hmgb1/2 were shown to bind to a various immunogenic nucleic acids including RNA (Yanai et al., 2009). To examine the direct interaction of Hmgb1/2 with nucleic acids, we purified recombinant Hmgb1/2 proteins from E. coli and subjected to in vitro binding assays with Lrp1-AS RNA. Hmgb1 and Hmgb2 were precipitated by immobilized-full-length-Lrp1-AS RNA, which was inhibited by free Lrp1-AS RNA in a dose-dependent manner (Figure 3C, left). Hmgb1-Lrp1-AS RNA binding was inhibited by genomic DNA, but Hmgb2-Lrp1-AS RNA binding was not affected (Figure 3C, right), suggesting the strong and specific binding of Hmgb2 to Lrp1-AS RNA. Moreover, we pulled down Glutathione S-transferase (GST) fusion of Hmgb2 and found that Lrp1-AS RNA is associated with Hmgb2 protein (Figure 3D). Collectively, these experiments showed that Lrp1-AS RNA directly and specifically binds to Hmgb2.
We and others have previously shown that some lncRNAs including NATs can interact with sense transcripts and make RNA duplex through their overlap regions (Faghihi et al., 2008; Kretz et al., 2013). To test if this is applied to Lrp1-AS, we performed in vivo RNase protection assay by qRT-PCR on Lrp1 RNA. Single-stranded RNA was digested with RNase A, and Lrp1 RNA fragment overlapped with Lrp1-AS was protected (Figure 3E), indicating that Lrp1 and Lrp1-AS are capable of forming an RNA duplex at an overlapping region. To elucidate if the overlap region of Lrp1-AS is functional, we performed RNA chromatography on two deletion mutants of Lrp1-AS, and we observed that the fragment (636-1388) containing the overlap region with Lrp1, strongly bound to Hmgb2 (Figure 3F and G). This result prompted us to investigate whether duplex of Lrp1-AS and Lrp1 may modulate specific binding of Lrp1-AS to Hmgb2. Following RNA chromatography on Lrp1 fragment and in-vitro-hybridized Lrp1-AS-Lrp1 RNA fragments duplex, showed that Lrp1 fragment does not bind to Hmgb2 (Figure 3H, lane1), and that Lrp1 inhibits Lrp1-AS fragment (636-1388) from binding to Hmgb2 (Figure 3H, lane 4 and 5). These data showed that Hmgb2 interacts with the overlap region of Lrp1-AS, but not Lrp1, and this interaction is inhibited by Lrp1.
Lrp1-AS suppresses Hmgb2-enhanced Activity of Srebp1a on Lrp1 transcription
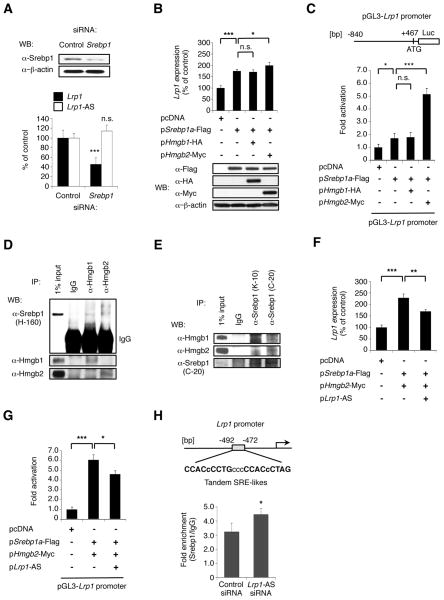
Sterol regulatory element-binding proteins (SREBPs) are a family of three transcription factors that regulate expression of genes involved in lipid homeostasis and glucose metabolism (Jeon and Osborne, 2012; Raghow et al., 2008), and Srebp1/2 are known to interact with Hmgb1/2, and their activity has been reported to be enhanced by Hmgb1 (Najima et al., 2005). Since Srebp2 was not expressed in RAW264.7 cells (data not shown), we focused on Srebp1a/c, which is also known to play a role in transcriptional regulation of Lrp1 (Bown et al., 2011). To investigate a possible regulation of Lrp1 by Hmgb1/2 via Srebp1a/c, we first analyzed changes in Lrp1 levels after altering Srebp1 levels. Silencing of Srebp1 in RAW264.7 cells led to a significant decrease in Lrp1 levels, but not Lrp1-AS (Figure 4A). Overexpression of Srebp1a-flag increased Lrp1 levels (Figure 4B, lane 2), whereas Srebp1c-flag did not change Lrp1 levels (Figure S4A). Together, these data showed that Lrp1 is positively regulated by Srebp1a.
Figure 4. Lrp1-AS reverses the Hmgb2-mediated increase in Srebp1-dependent Lrp1 expression.
(A) Lrp1 and Lrp1-AS levels in RAW264.7 cells transfected with control or Srebp1 siRNA. The control siRNA has a sequence with no homology to any gene. Western bloting (WB) confirms Srebp1 silencing (upper).
(B) Lrp1 levels after overexpression of mature Srebp1a-Flag and/or Hmgb1-HA/2-Myc in RAW264.7 cells (upper). Expression of exogenous proteins was monitored by WB with the indicated antibodies (lower). An antibody to β-actin was used as loading control in (A) and (B).
(C) Luciferase reporter assay after co-transfection of the Lrp1 promoter-coupled luciferase construct (upper, pGK3-Lrp1 pro.) together with overexpression of Srebp1-Flag and Hmgb1-HA/2-Myc in RAW264.7 cells. Firefly luciferase activity was normalized to Renilla luciferase activity.
(D, E) Reciprocal immunoprecipitation (IP) of endogenous Srebp1 or Hmgb1/2 from RAW264.7 cells followed by WB with the indicated antibodies. Rabbit IgG was used as negative control.
(F) Lrp1 levels after overexpression of mature Srebp1a-Flag and Hmgb2-Myc with or without Lrp1-AS RNA in RAW264.7 cells.
(G) Luciferase activity after co-transfection of the Lrp1 promoter-coupled luciferase construct together with overexpression of mature Srebp1-Flag and Hmgb2-Myc with or without Lrp1-AS RNA in RAW264.7 cells. Firefly luciferase activity was normalized to Renilla luciferase activity.
(H) Quantitative chromatin immunoprecipitation (qChIP) analysis with Srebp1 antibody or control IgG from RAW264.7 cells transfected with control or Lrp1-AS siRNA (lower). Diagram of the Lrp1 promoter, showing the two tandem SREBP Responsive Elements-like motifs (SRE-likes in upper) (Bist et al., 1997). Mean ± s.d. (n = 3 replicates) are shown in all bar graphs.*P < 0.05, **P < 0.01, ***P < 0.001 determined by ANOVA.
Next, to assess the effect of Hmgb1/2 on Srebp1 transcriptional activity, we examined Lrp1 levels on overexpression of Hmgb1/2. Hmgb2, but not Hmgb1, increased Srebp1a-dependent Lrp1 upregulation (Figure 4B, lane 3 and 4). Srebp1a and Hmgb2 did not change Lrp1-AS levels (Figure S4B). Moreover, Hmgb2, but not Hmgb1, amplified Srebp1a-flag-induced Lrp1 promoter activity (Figure 4C). To investigate the mechanisms for the functional cooperation between Srebp1a and Hmgb2, we examined their possible interactions. Reciprocal co-immunoprecipitation with specific antibodies and Western blot analyses demonstrated interaction of endogenous mature Srebp1 and Hmgb2 (Figure 4D and E). RNA chromatography on Lrp1-AS RNA failed to detect associated Srebp1, and immunoprecipitation of mature Srebp1 did not retrieve associated Lrp1-AS RNA (Figure S4A and S4B). These showed that Hmgb2 directly binds to Srebp1a in vivo, and enhances its activity. Together, Lrp1 upregulation by Srebp1a occurred at the level of transcription, and Hmgb2 directly enhances this regulation. We next examined whether Lrp1-AS could affect Hmgb2- and Srebp1a-dependent Lrp1 transcriptional activation. Overexpression of Lrp1-AS repressed the potentiating effect of Hmgb2 on Lrp1 upregulation by Srebp1a, but not Srebp1c (Figure 4F). Lrp1-AS decreased Lrp1 promoter activity induced by Hmgb2 and Srebp1a (Figure 4G). Chromatin immunoprecipitation showed that Lrp1-AS depletion increased Srebp1 occupancy at two tandem SRE-like regions within Lrp1 promoter (Figure 4H). To further investigate this regulatory mechanism we measured Lrp1 expression in different tissues of Hmgb2 KO mice (Figure S5A). As expected we observed a significant decrease in Lrp1 expression in the lungs of KO mice compared to wild type. Unexpectedly we observed an increase of Lrp1 expression in the spleen of KO animals and no significant changes in the brain, thymus or testes. Srebp1 expression was significantly increased in the spleen of KO mice (Figure S5B), thus possibly explaining the increase of Lrp1 expression in this organ. Collectively, these data showed that Lrp1-AS inhibits the ability of Hmgb2 to enhance Srebp1a-dependent transcription of Lrp1.
AntagoNAT Reveals Functional Domain of Lrp1-AS Interacting with Hmgb2
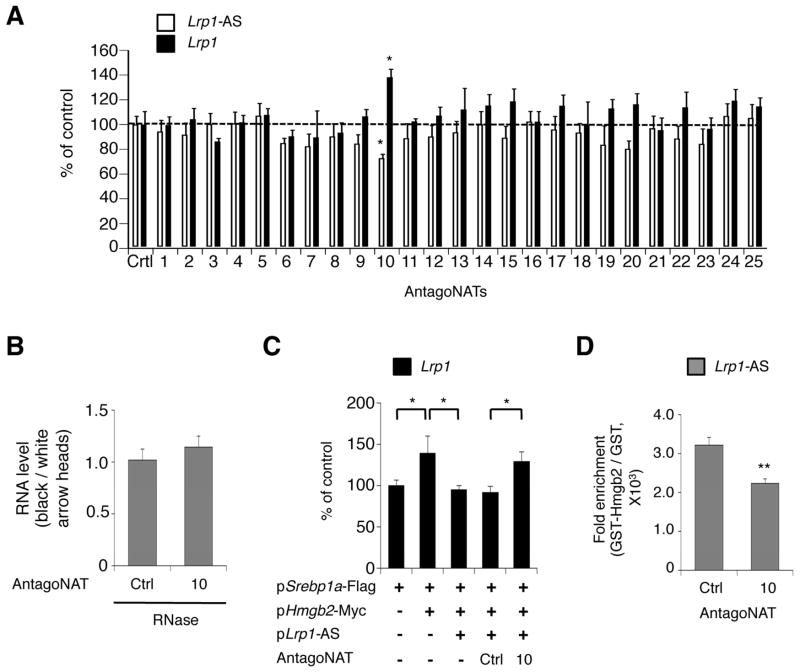
We have previously shown that inhibition of NATs by oligonucleotide (termed as antagoNATs) induces upregulation of their sense transcripts, and open a new possibility that synthetically engineered DNAs could interact with both nucleic acids and protein functional domains to carry out engineered regulatory roles (Modarresi et al., 2012). We also observed that antagoNATs, which target the overlapping region between sense and antisense transcripts, produced the largest response in causing an increase in sense transcript expression(Modarresi et al., 2012).
To test if antagoNATs could interfere the interactions of Lrp1-AS and, either Hmgb2 or Lrp1, we designed a series of 25 antagoNATs that target Lrp1-AS, covering (‘walking’) the entire shared sequence of Lrp1-AS and Lrp-1 (Figure S6A). We observed that antagoNAT10 significantly increased Lrp1 levels and simultaneously decreased Lrp1-AS levels (Figure 5A). We also used 8 additional oligonucleotides, partially sharing sequence with antagoNAT10, to more exactly determine the potential domain of Lrp1-AS regulating Lrp1 levels, and we observed similar effect with antagoNAT10e that was set shifting to 3′ by two bases from antagoNAT10 (Figure S6B and S6C). We further found that locked nucleic acid (LNA)-modified antagoNAT10 was more efficacious in increasing Lrp1 levels (Figure S6D). We then performed in vivo RNase protection assay and found that antagoNAT10 failed to change the levels of preserved RNA (Figure 5B), indicating that the interaction between Lrp1-AS and Lrp1 was not disrupted by antagoNAT10. This led us to test whether antagoNAT10 could modulate the binding of Lrp1-AS to Hmgb2. Co-transfection of AntagoNAT10 reversed the effect of Lrp1-AS on Hmgb2 and Srebp1-induced Lrp1 levels (Figure 5C, lane 4 and 5). The addition of AntagoNAT10, but not AntagoNAT control, efficiently inhibited Lrp1-AS retrieval by GST-Hmgb2 (Figure 5D). Together, these results suggests that antagoNAT against a part of the overlap region of Lrp1-AS, inhibits the interaction between Lrp1-AS and Hmgb2, which is critical for regulating Lrp1 levels (Figure 5E).
Figure 5. Blocking of Lrp1-AS-Hmgb2 Interaction by antagoNATs induces Lrp1 expression.
(A) Lrp1 and Lrp1-AS levels of RAW264.7 cells transfected with control (Ctrl) or specific antagoNATs against Lrp1-AS, tiling the entire overlap region between Lrp1 and Lrp1-AS (as in Figure S6). Control antagoNAT has a sequence with no homology to any gene.
(B) RNase protection assay. The RNA ratios were calculated from qRT-PCR using primer pairs for overlap region (black arrowhead) or non overlap region (white arrowhead) as in Figure 3E from RAW264.7 nuclear extracts transfected with control (Ctrl) or antagoNAT10.
(C) Lrp1 levels after overexpression of mature Srebp1a-Flag, Hmgb2-Myc and Lrp1-AS RNA with control (Ctrl) or AntagoNAT10 in RAW264.7 cells.
(D) The in vitro translated Lrp1-AS was preincubated with control (Ctrl) or antagoNAT10, and pulled down with recombinant GST or GST-fused Hmgb2 as in Figure 3F. Associated Lrp1-AS RNAs were detected by qRT-PCR. Mean ± s.d. (n = 3 replicates) are shown in all bar graphs.*P < 0.05, **P < 0.01 determined by ANOVA.
LRP1-AS dysregulation in Alzheimer’s disease brain
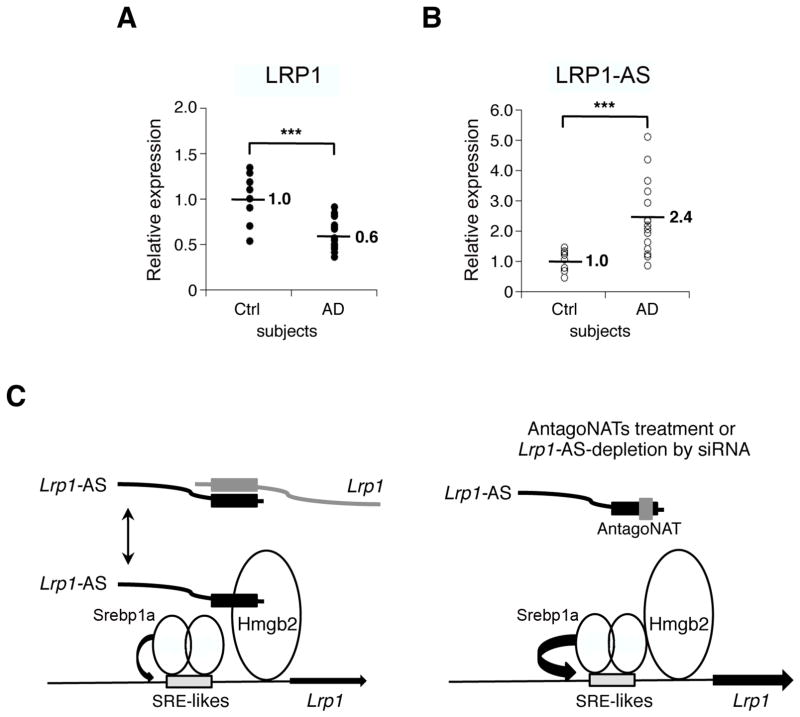
Previous reports have shown that LRP1 levels are decreased in AD subjects, and the level of NAT is induced by diverse cell stressors, and elevated in AD subjects (Bishop et al., 2010; Faghihi et al., 2008; Saxena and Caroni, 2011). This prompted us to test whether LRP1-AS and LRP1 mRNA are dysregulated in the brain of AD patients. To test this hypothesis we performed qRT-PCR using RNA extracted from the superior frontal gyrus of AD patients and age-matched controls to measure the expression of LRP1-AS and LRP1 mRNA. As previously reported we observed decreased expression of LRP1 mRNA levels in AD subjects, while LRP1-AS levels were surprisingly increased (Figure 6A and 6B).
Figure 6. LRP1 and LRP1-AS dysregulation in Alzheimer disease.
Quantitative RT-PCR analysis of LRP1 (A) and LRP1-AS (B) expression in superior frontal gyrus of 15 patients with Alzheimer’s disease (AD) and 8 control subjects (Ctrl). The housekeeping gene β-ACTIN was used as endogenous control; LRP1 and LRP1-AS expression was normalized to Ctrl. ***P<0.001 determined by ANOVA.
(C) A Model Proposing Functional and Physical Interactions Between Lrp1-AS, Srebp1a and Hmgb2 on the Lrp1 Promoter. Hmgb2 can bind with Srebp1a, and induces Lrp1 promoter activity containing SRE-like motifs (depicted in gray boxes). When Lrp1-AS is depleted or treated with antagoNATs, cooperative activation of Lrp1 expression by Hmgb2 and Srebp1a is enhanced.
DISCUSSION
The multiple functions of lncRNAs are just starting to emerge, and the mechanisms through which they mediate their functions are subject to intense investigation. Here, we have identified a NAT, Lrp1-AS that has a critical role in the transcriptional regulation of Lrp1 gene. The Lrp1-AS directly binds to Hmgb2, and inhibits Hmgb2-mediated Srebp1a transcriptional activity on Lrp1. Lrp1-AS function is in turn regulated by Lrp1 mRNA that can base pair with Lrp1-AS forming an RNA duplex which prevents the interaction between Lrp1-AS and Hmgb2. Furthermore antagoNAT against specific domain of Lrp1-AS inhibits the interaction between Lrp1-AS and Hmgb2, suggesting a model whereby a specific regulatory sequence of lncRNA is critical for its function.
In the nucleus, HMGBs are the most abundant regulatory proteins, which dynamically interact with chromatin and influence numerous activities including transcription, replication, repair and genomic stability (Bianchi and Agresti, 2005). HMGBs affect the chromatin fiber as architectural components by competing with histone H1 for chromatin binding sites and weakening its ability to restrict the access of transcription machinery to the chromatin (Bianchi and Agresti, 2005). This HMGBs–H1 interactions might facilitate nucleosome remodeling and regulate accessibility of transcription factors to the nucleosomal DNA in response to external stimuli. The NAT studied here interacts with HMGB2 and may serve as cell-type-specific natural RNA ligand to inhibit HMGB2 to exert their enhancer activities as a specific fine-tuner. These suggest that a set of NATs interacts with ubiquitous regulatory proteins to form specific RNA-protein complexes that coordinate cell-type-specific gene expression patterns.
We and others provide evidence that HMGBs function as an RNA-binding proteins in addition to its long-recognized role as a DNA-binding protein (Yanai et al., 2009). HMGBs bind to chromatin without any known apparent preference for the underlying DNA sequence; their functional specificity could depend on direct interactions with sequence-specific transcriptional factors and bending of the DNA target sequences. However, many proteins that bind nucleic acids could be expected to display at least modest non-specific affinity for other nucleic acids, and even sequence-specific DNA-binding transcription factors appear to bind RNA with at least some sequence specificity. These examples include STAT1 transcription factor inhibited by a ncRNA in MHC expression, and TLS/FUS factor bound to RNA, which is involved in oncogenic chromosomal translocations (Lerga et al., 2001; Perrotti et al., 1998; Peyman, 1999). LncRNAs are cis- or trans-regulators, which can regulate target genes expression by acting as signals, guides or scaffolds to the chromatin through interaction with chromatin proteins to change the epigenetic status of genes (Guttman and Rinn, 2012; Magistri et al., 2012; Rinn and Chang, 2012; Wang and Chang, 2011). In our studies Lrp1-AS interacts with HMGB2 and may serve as cell-type- and locu-specific natural RNA ligand to fine tune HMGB2 activity. The specific sequence of the NAT resembles that of the protein’s alternative target, and therefore, the NAT may compete for the protein binding to the target. This ‘molecular decoy’ model was initially demonstrated in a study showing regulation of E. coli CsrA activity by a regulatory RNA called CsrB (Romeo, 1998). The regulatory RNAs can create new signaling pathways to regulate other transcriptional targets than those original targets. The degree of pairing of RNA–proteins may provide variations on global gene expression in cells, but exactly what regulatory mechanisms allow a NAT to bind to proteins should be elucidated. Complementary experiments may be needed to provide insight into the aspects of NAT sequence required for recognition by HMGB2, and this may make it clear whether there is specific sequence requirement in other NATs recognized by HMGB2. Finally, our study showed that LRP1 and LRP1-AS are discordantly dysregulated in the brain of AD patients compared to controls, where LRP1 is expressed at lower level and LRP1-AS at higher level. Further research will be needed to investigate the functional implication of LRP1-AS in the pathological processes underlying AD but accumulating findings suggest that NATs and other lncRNAs could potentially be pursued as diagnostic markers or therapeutic targets for different human diseases.
EXPERIMENTAL PROCEDURES
Cell culture, RNA interference, Plasmids and AntagoNATs
RAW264.7 cell line (TIB-71, ATCC) was maintained in DMEM supplemented with 10% fetal bovine serum (Atlas Biologicals) and 1% Pen/Strep. Lrp1-AS siRNAs and control siRNA (AM4611) were synthesized by Ambion. Srebp-1 siRNA (sc-36558) was purchased from Santa Cruz Biotechnology. Mouse cDNAs were amplified by PCR (KOD Hot Start DNA Polymerase, Novagen). Full length (1-1388 bp), 5′ (1-656 bp), and 3′ (636-1388 bp) Lrp1-AS and Luciferase gene (from pGL3 Basic Vector, Promega), 3XHA-tagged Hmgb1 and 3XMyc-tagged Hmgb2 were cloned into pcDNA3.1 vector (Invitrogen). pcDNA3.1-Flag-tagged Srebp1a/c were kindly provided by Dr. Timothy Osborne (Sanford-Burnham Medical Research Institute). Cells were transfected with 30nM of siRNA, 100nM of antagoNATs or 1ug of plasmid using Amaxa 4D-Nucleofector (Lonza) according to manufacturer’s instructions, and harvested after 24 or 48 hr post-transfection. All oligonucleotide sequences are listed in Table S1.
RNA Purification and qRT-PCR Analysis
Total RNA from cells was isolated with RNeasy Plus Micro Kit (Qiagen) as per manufacturer’s instructions, and analyzed with 2100 Bioanalyzer (Agilent). Reverse Transcription was performed using SuperScript III (Invitrogen) and random priming. Real time PCR was performed with TaqMan Gene Expression Assays and HT7900 sequence detection system (Applied Biosystems), as previously described (Faghihi et al., 2008; Modarresi et al., 2012). Eukaryotic 18S or mouse β-actin was measured for an internal control and used for normalization. Details of the human brain samples were previously described (Faghihi et al., 2008). For RNA protection assays, cells were lysed in standard radioimmunoprecipitation assay (RIPA) Buffer, incubated with or without RNase A/T1 (Ambion) for 1hr at 37°C and then treated with Proteinase K (Invitrogen) for 30min at 37°C, followed by RNA extraction, as previously described (Faghihi et al., 2008).
GST Pull-Down Assays
GST-tagged Hmgb1/2 were expressed in BL21 Escherichia coli (Novagen) and purified according to standard protocols. Each GST-fusion protein was bound to glutathione beads (GE Healthcare) for 1-hr at room temperature, and then incubated with cell lysates for 2hr at 4°C. After five washes with Phosphate-Buffered Saline (PBS), bound RNAs were extracted and analyzed by qRT-PCR. For in vitro binding assay, full-length Lrp1-AS was transcribed in vitro using MEGAscript T7 Kit (Ambion), treated with DNase I, purified with NucAway spin columns (Ambion), denatured and refolded in RNA structure buffer (Ambion). The folded RNA was incubated with antagoNATs and GST-fusion protein for 1-hr, and the complex was captured with glutathione beads for 1-hr. After five times washes with PBS, bound RNA was extracted and analyzed by qRT-PCR.
RNase-assisted RNA Chromatography
Full-length or fragment in-vitro-transcribed 1.38-kb Lrp1-AS RNA, or 1.8-kb fragment of Luciferase RNA were conjugated to adipic acid dehydrazide agarose beads (Sigma-Aldrich) as described (Michlewski and Caceres, 2011; Wang et al., 2011). Briefly, the complexed beads were incubated with cell lysates for 2 hr at 37°C. After five washes, bound proteins were eluted with RNase A/T1 and V1 (Ambion), and visualized by Silver Staining Kit (GE Healthcare) or detected by Western blotting. Selected band was subjected to mass spectrometry at The Scripps Research Institute. For RNA Pull-Down Assays, 100pmol of in-vitro-transcribed Lrp1-AS RNAs were conjugated to beads, and incubated with 10pmol of recombinant 6XHis-tagged Hmgb1/2 (Prospec) in the presence of increasing amounts of free Lrp1-AS RNA (10, 100, 300, 1000 pmol) or genomic DNA (50, 500, 1500, 5000 ng) for 2 hr at 37°C. After five washes, bound proteins were extracted and subjected to Western blotting.
Western Blotting and Immunoprecipitation (IP)
To prepare protein lysates, cells were harvested, washed, and lysed in standard RIPA Buffer. Total protein concentration was measured by Pierce 660nm protein assay, and Western blotting was performed according to standard protocols. The quantification of signals was performed using Image J software. For IP, cells were cross-linked with 1% formaldehyde for 10 min, followed by the addition of glycine to a final concentration of 0.125 M. After two washes with PBS, cells were lysed with Buffer A (10mM Hepes pH 7.4, 10mM KCl, 1.5mM MgCl2, 0.5mM DTT supplemented with Roche Complete Protease Inhibitor Cocktail), lysed in 0.25% NP40, fractionated by low speed centrifugation. The nuclear pellet was resuspended with Buffer C (20 mM Hepes pH 7.4, 420mM KCl, 4mM MgCl2, 0.5 mM DTT, 10% Glycerol, Roche protease Inhibitor), and sonicated for 7 min with Bioruptor UCD-200 (Diagenode). Combined nuclear and cytoplasmic fractions were mixed with antibody, and incubated for 2hr at 4°C. The complex was captured with Protein A/G beads for 1hr at 4°C. After four times washes with NP-40 Buffer (50 mM Hepes pH 7.4, 100mM KCl, 5mM MgCl2, 0.5 mM DTT, 0.1% NP-40, 10% Glycerol), bound proteins were eluted in 2x Laemmli Sample Buffer, and subjected to Western blotting. For RNA-IP (RIP), bound RNA were extracted with Proteinase K (Ambion) in RIPA Buffer for 1 hr at 45 °C, and analyzed by qRT–PCR. Specific antibodies in the present study include, Lrp1 (EPR3724, Novus Biologicals), Srebp-1 (H-160 for IP and ChIP, K-10 and C-20 for WB, Santa Cruz Biotechnology), Hmgb2 (H9789, Sigma-Aldrich), Flag (M2, Sigma-Aldrich), HA.11 (16B12, Covance), c-myc (9E10, Covance), β-actin (AC-15, Sigma-Aldrich).
Luciferase Reporter Assays
pGL3-Lrp1 promotor (Liu et al., 2007) was kindly provided by Dr. Guojun Bu (Mayo Clinic). Cell extracts after 24 hr transfection were assayed for firefly and renilla (phRL-TK, Promega) luciferase activities in 96-well white plates (Nunc) using Dual-luciferase Reporter Assay System (Promega) and EnVision 2104 Multilabel Reader (PerkinElmer)
Chromatin Immunoprecipitation (ChIP)
Cells were crosslinked with 1% formaldehyde for 10 min, followed by the addition of glycine to a final concentration of 0.125 M. After two washes with PBS, cells was lysed with Lysis Buffer 1 (50mM Hepes pH 7.4, 140mM NaCl, 1mM MgCl2, 0.5% NP-40, 0.25% Triton X-100, 10% Glycerol, Roche protease Inhibitor) for 10 min, followed by Lysis Buffer 2 (10mM Tris pH8.0, 200mM NaCl, 1mM EDTA, 0.5mM EGTA, Roche protease Inhibitor) for 10 min. Next, the nuclear pellet was sonicated for 40min with Bioruptor UCD-200 in Lysis Buffer 3 (20mM Tris pH8.0, 300mM NaCl, 1mM EDTA, 0.5mM EGTA, 0.1% Sodium deoxycholate, 0.5% N-Laurorylsarcosine, Roche protease Inhibitor), and lysed in 1%Triton X-100. The nuclear fraction was mixed with antibody, and incubated overnight at 4°C. The complex was captured with sheep anti-Rabbit IgG-coupled magnetic beads (Dynabeads, Invitrogen) for 1hr at 4°C. After one wash with Low Salt Buffer (20mM Tris pH8.0, 150mM NaCl, 2mM EDTA, 0.1% SDS, 1% Triton X-100), two washes with High Salt Buffer (20mM Tris pH8.0, 400mM NaCl, 2mM EDTA, 0.1% SDS, 1% Triton X-100), five washed with RIPA Buffer and one time with TE Buffer (50mM Tris pH8.0, 10mM EDTA) containing 50mM NaCl, bound DNA was reverse-crosslinked in TE buffer with 1% SDS at 65°C overnight, and treated with RNase A/T1 at 55°C for 30min, and following Proteinase K at 55°C for 1hr. Bound DNA was purified and analyzed by qPCR with Power SYBR Green Master Mix (Applied Biosystems).
Raindance Raindrop digital PCR assay
Digital PCR assays were performed as per manufacturer’s protocols. In brief, 50uL reactions were used with Applied Biosystems 2X Taqman gene expression master mix, 20X primer probe, 25X drop stabilizer, and 100ng of cDNA. Oil droplets were generated on the Raindrop Source chip instrument, then amplified on a BioRad C1000 thermocycler using the following conditions: 50°C for 2min, 95°C for 10min and 40 cycles of 95°C for 15sec, 60°C for 1min, hold at 10°C. Fluorescent droplets were detected on the Raindance Raindrop Sense chip instrument, then analyzed using the Raindance Raindrop Analyst software V2 to count VIC and FAM drops and to prepare output graphs.
Supplementary Material
Acknowledgments
This work was supported by US National Institute of Health awards DA035592, MH084880 and NS071674. Y.Y. was supported by Japan Society for the Promotion of Science (21-8333). Some parts of this study were performed by the authors at The Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Annabi B, Doumit J, Plouffe K, Laflamme C, Lord-Dufour S, Beliveau R. Members of the low-density lipoprotein receptor-related proteins provide a differential molecular signature between parental and CD133+ DAOY medulloblastoma cells. Mol Carcinog. 2010;49:710–717. doi: 10.1002/mc.20645. [DOI] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–535. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown MJ, Jones GT, Harrison SC, Wright BJ, Bumpstead S, Baas AF, Gretarsdottir S, Badger SA, Bradley DT, Burnand K, et al. Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am J Hum Genet. 2011;89:619–627. doi: 10.1016/j.ajhg.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Deane R, Sagare A, Hamm K, Parisi M, Lane S, Finn MB, Holtzman DM, Zlokovic BV. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118:4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Herz J, Bu G. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006312. doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DE, Pietrzik CU, Baum L, Chevallier N, Merriam DE, Kounnas MZ, Wagner SL, Troncoso JC, Kawas CH, Katzman R, et al. Modulation of amyloid beta-protein clearance and Alzheimer’s disease susceptibility by the LDL receptor-related protein pathway. J Clin Invest. 2000;106:1159–1166. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerga A, Hallier M, Delva L, Orvain C, Gallais I, Marie J, Moreau-Gachelin F. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem. 2001;276:6807–6816. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- Lillis AP, Van Duyn LB, Murphy-Ullrich JE, Strickland DK. LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev. 2008;88:887–918. doi: 10.1152/physrev.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, Herz J, Muglia L, Bu G. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56:66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistri M, Faghihi MA, St Laurent G, 3rd, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Najima Y, Yahagi N, Takeuchi Y, Matsuzaka T, Sekiya M, Nakagawa Y, Amemiya-Kudo M, Okazaki H, Okazaki S, Tamura Y, et al. High mobility group protein-B1 interacts with sterol regulatory element-binding proteins to enhance their DNA binding. J Biol Chem. 2005;280:27523–27532. doi: 10.1074/jbc.m414549200. [DOI] [PubMed] [Google Scholar]
- Pastori C, Magistri M, Napoli S, Carbone GM, Catapano CV. Small RNA-directed transcriptional control: new insights into mechanisms and therapeutic applications. Cell Cycle. 2010;9:2353–2362. doi: 10.4161/cc.9.12.12049. [DOI] [PubMed] [Google Scholar]
- Pastori C, Wahlestedt C. Involvement of long noncoding RNAs in diseases affecting the central nervous system. RNA Biol. 2012;9:860–870. doi: 10.4161/rna.20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, Grassilli E, Lozzo RV, Cooper DR, Calabretta B. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. Embo J. 1998;17:4442–4455. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyman JA. Repression of major histocompatibility complex genes by a human trophoblast ribonucleic acid. Biol Reprod. 1999;60:23–31. doi: 10.1095/biolreprod60.1.23. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghow R, Yellaturu C, Deng X, Park EA, Elam MB. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol Metab. 2008;19:65–73. doi: 10.1016/j.tem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Ronfani L, Ferraguti M, Croci L, Ovitt CE, Scholer HR, Consalez GG, Bianchi ME. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development. 2001;128:1265–1273. doi: 10.1242/dev.128.8.1265. [DOI] [PubMed] [Google Scholar]
- Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71:35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- Shibata M, Yamada S, Kumar SR, Calero M, Bading J, Frangione B, Holtzman DM, Miller CA, Strickland DK, Ghiso J, et al. Clearance of Alzheimer’s amyloid-ss(1-40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. J Clin Invest. 2000;106:1489–1499. doi: 10.1172/JCI10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12, 11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmeshev D, Magistri M, Faghihi MA. Expression of non-protein-coding antisense RNAs in genomic regions related to autism spectrum disorders. Mol Autism. 2013;4:32. doi: 10.1186/2040-2392-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.