Abstract
Purpose
To improve triggering efficiency of the prospective respiratory amplitude triggered 4DMRI method and to develop a 4DMRI imaging protocol that can offer T2 weighting for better tumor visualization, good spatial coverage, spatial resolution and respiratory motion sampling within a reasonable amount of time for radiation therapy applications.
Methods and Materials
The respiratory state splitting (RSS) method and the multi-shot acquisition (MSA) method were analytically compared and validated in a simulation study using the respiratory signals from 10 healthy human subjects. The RSS method was identified to be more effective in improving triggering efficiency. It was implemented in the prospective respiratory amplitude triggered 4DMRI. 4DMRI image datasets were acquired from 5 healthy human subjects. Liver motion was estimated using the acquired 4DMRI image datasets.
Results
The simulation study showed that the RSS method was more effective in improving triggering efficiency compared to the MSA method. The average reductions in 4DMRI acquisition time were 36% and 10% for the RSS and MSA methods, respectively. The human subject study showed that T2-weighted 4DMRI with 10 respiratory states, 60 slices at a spatial resolution of 1.5×1.5×3.0 mm3 could be acquired in 9-18 minutes, depending on individual's breath pattern. Based on the acquired 4DMRI image datasets, the ranges of peak-to-peak liver displacements among 5 human subjects were 9.0-12.9 mm, 2.5-3.9 mm and 0.5-2.3 mm in superior-inferior, anterior-posterior and left-right directions, respectively.
Conclusions
We demonstrated that with the RSS method, it was feasible to acquire high quality T2-weighted 4DMRI within a reasonable amount of time for radiation therapy applications.
Keywords: T2 weighted 4DMRI, Amplitude triggered, High spatial and temporal resolution
Introduction
Respiration-induced organ and tumor motion imposes significant challenges in the radiation therapy treatment of abdominal and lung cancers. Studies have shown that peak-to-peak tumor or organ motion can be as large as 2-3 cm (1-5). To ensure sufficient dose coverage to the target, a large treatment margin has to be used (6-9), which unavoidably increases radiation dose to radiosensitive critical structures surrounding the target, causing increased side effects. Reducing the treatment margin without sacrificing dose coverage requires effective motion management, for which accurate knowledge of respiration induced motion is essential.
Four-dimensional computed tomography (4DCT) (10-13) is commonly used to provide respiration-induced motion information. However, for abdominal cancer where the tumor is embedded in the soft tissue, the low tumor tissue contrast of CT images makes 4DCT less useful and therefore limits its applications. To improve soft tissue and tumor tissue contrast, 4D Magnetic Resonance Imaging (4DMRI) has been developed using either retrospective sorting (14-19) or prospective triggering (20, 21) techniques. 4DMRI (T2-weighted) is able to provide accurate tumor motion information, from which internal target volume can be derived for the subsequent treatment planning in a way similar to what 4DCT does but more powerful for abdominal cancers. In addition, tumor motion information is useful in 4D radiation therapy (22), e.g. gated dose delivery.
Compared to retrospective 4DMRI, prospective 4DMRI acquires images during specific respiratory states that meet predefined criteria, resulting in improved acquisition efficacy, reduced acquisition time and increased robustness to breathing irregularity. More importantly, with am amplitude based prospective triggering system, images of a single slice at different respiratory states no longer have to be acquired consecutively within one respiratory cycle. They can be separated into multiple respiratory cycles to allow complete relaxation of magnetization. This overcomes the upper limit placed on repetition time (TR) and therefore can accommodate more MRI sequences including those with better tumor tissue contrast such as T2-weighted MRI (20), allowing motion tracking of the tumor itself. In a feasibility study, Hu et al (20) reported that under prospective guidance based on respiratory amplitude, T2 weighted 4DMRI with 32 slices, 4 states per respiratory cycle and 1.5×1.5×5.0 mm3 spatial resolution could be obtained within a few minutes. However, more respiratory states and slices are typically required for radiation therapy applications.
As numbers of slices and respiratory states increase, 4DMRI image acquisition time increases substantially. While increasing the number of slices prolongs the 4DMRI acquisition time linearly, increasing the number of respiratory states has more dramatic, non-linear effect on the 4DMRI acquisition time, primarily due to frequent trigger missing. Here, trigger missing indicates a situation in which the respiratory amplitude that triggers image acquisition of a particular slice appears before the image acquisition completion of the previous slice and therefore gets ignored by the MRI scanner. As the number of respiratory states increases, the number of slices that needs to be acquired in a respiratory cycle increases (cf. Figure 1a & b). Accordingly, the amount of time allocated for the image acquisition of each slice decreases. When the allocated time becomes less than the required image acquisition time of a slice, overlaps in image acquisition can happen, thereby causing trigger missing events, especially at mid inhalation and mid exhalation as shown in Figure 1 a & b. Frequent trigger missing can reduce triggering efficiency tremendously, thereby increasing the 4DMRI acquisition time because the MRI scanner has to pause and wait for the same respiratory amplitude to appear again, which is at least one respiratory cycle later.
Figure 1.
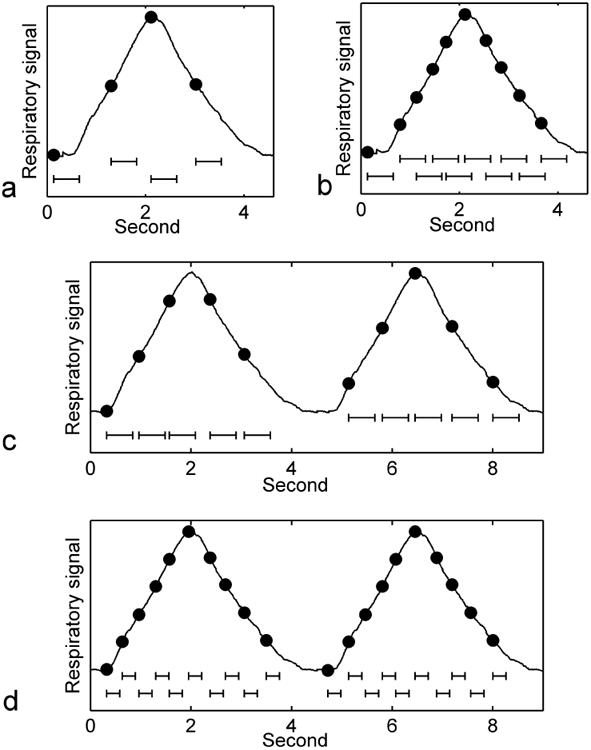
(a) 4 respiratory states in a respiratory cycle, (b) 10 respiratory states in a respiratory cycle, (c) respiratory state splitting method and (d) multi-shot acquisition method to address the trigger missing issue. The dots indicate triggering respiratory amplitudes for individual respiratory states. The line bars under the curve indicate the required image acquisition times.
The purpose of this study is to 1) investigate methods to improve triggering efficiency of the prospective respiratory amplitude triggered 4DMRI by mitigating the trigger missing issue associated with increased number of respiratory states, and 2) develop an imaging protocol to acquire T2 weighted 4DMRI within a reasonable amount of acquisition time to offer good spatial coverage and resolution, and sufficient respiratory motion sampling. Both simulations and experiments on healthy human subjects were performed in this study.
Methods and Materials
Human subjects
10 healthy adult human subjects, including both female and male, were enrolled in this study to validate the methodology. The respiratory signals from 5 human subjects were acquired for the simulation purpose only. Both the respiratory signal and 4DMRI image dataset were acquired for the rest 5 human subjects. This study was approved by the Institutional IRB office.
Potential solutions
To mitigate the trigger missing issue due to the increased number of respiratory states, we could either increase the amount of time allocated for the image acquisition of a single slice (Figure 1c), or decrease the image acquisition time of each respiratory state (Figure 1d). Splitting all respiratory states into multiple groups in an interleaved manner and acquiring each group of respiratory states independently increases the amount of time available for image acquisition of a slice by a factor of the number of groups. This method is referred to as the respiratory state splitting (RSS) method. Although the nominal 4DMRI acquisition time also increases by a factor of the number of groups, the RSS method can effectively reduce the chance of trigger missing, thereby reducing the total 4DMRI acquisition time. Dividing the required k-space data for a single slice into multiple segments and acquiring one segment of k-space data at a time following each respiratory trigger can decrease the image acquisition time at each respiratory state by a factor of the number of segments and therefore mitigate the trigger missing issue. This method is referred to as the multi-shot acquisition (MSA) method.
Analytical comparison
An analytical comparison was performed to compare the efficiencies of the RSS and MSA methods in reducing 4DMRI image acquisition time. Assume that the number of slices is NSlice, the number of respiratory states is NRS, the number of groups in the RSS method is NGroup, the number of shots in the MSA method is NShot, the average respiratory period is TResp, the single shot acquisition time for a 2D slice is TSSAcq and the time between two consecutive shot acquisitions (or triggers) is tAcqInt,_the total 4DMRI acquisition time for the RSS method (TRSS) and the MSA method (TMSA) can be estimated using Equations (1) and (2), respectively.
| (1) |
| (2) |
Here P(·) indicates the chance of trigger missing. If the formula inside the parentheses is true, then P(·) = 1, indicating a trigger missing event. Otherwise, P(·) = 0, indicating no trigger missing. In the equations (1) and (2), the first term represents the 4DMRI acquisition time without considering the trigger missing issue and the second term indicates the added acquisition time caused by the missing triggers.
For a direct comparison, we assume that NGroup = NShot. Therefore, the first term in the two equations becomes identical. The second term differs in the way to calculate the chance of trigger missing, which is P(tAcqInt < TSSAcq) for the RSS method and for the MSA method. Mathematically, the following inequality is always true.
| (3) |
Proof: From the trigger missing point of view and assume NGroup = NShot, tAcqInt in the RSS method shall equal in the MSA method. Since the goal is to estimate the probability of trigger missing, for the moment we can neglect the requirement for MRI contrast formation, e.g. multiple shots for the same slice cannot be acquired consecutively. When tAcqInt > TSSAcq and , for l = 1,…, NShot, then P(·) = 0 for both methods.
Therefore, the above inequality (3) holds true. Due to the fact that , when tAcqInt < TSSAcq, then holds true for at least one shot out of NShot in the MSA method. In such a situation, P(tAcqInt < TSSAcq) = 1 for the RSS method and for the MSA method. Therefore, the above inequality (3) holds true as well.
Based on Equation (1) and (2), as well as Inequality (3), we can reach the conclusion that TRSS is always equal or less than TMSA. The analytical comparison is in favor of the RSS method.
Simulation study
A simulation study was performed using MATLAB (The MathWorks Inc., Natick, MA) to evaluate the performance of the RSS and MSA methods in reducing 4DMRI acquisition time. Respiratory signals were acquired from 10 healthy human subjects (5 prospective and 5 retrospective) using a standard air cushion bellow wrapped around subjects' abdomen. Three triggering strategies (no intervention, RSS and MSA) were applied to the respiratory signals to simulate 4DMRI acquisition times. The 4DMRI acquisition time using the original prospective 4DMRI with no intervention applied to address the trigger missing issue was used as the reference to calculate the percentage reduction in the acquisition time for 4DMRI using the RSS and MSA methods. The parameters used in the simulation study were NSlice = 60, NRS = 10, NGroup = Nshot = 2 and TSSAcq = 380 ms. Respiratory trigger acceptance range was set to be 5% of the respiratory range placed at each triggering respiratory amplitude (or respiratory state). The 5% window was determined empirically based on tradeoff between the expected liver displacement, scan efficiency, and triggering accuracy. When respiratory amplitude falls within the acceptance range, a trigger is sent to start image acquisition. In the simulation study, image acquisition time (either TSSAcq or TSSAcq/NShot) was placed after each respiratory trigger to simulate the image acquisition and no real MRI data acquisition was actually involved. Results of the simulation study are listed in the RESULTS section, which support the superiority of the RSS method.
Human subject study – 4DMRI acquisition
Since both the analytical comparison and the simulation study showed that the RSS method was more effective in reducing 4DMRI acquisition time, we chose this technique for the human subject study and implemented it into a respiratory amplitude triggered T2 weighted TSE (Turbo Spin Echo) 4DMRI sequence (20) using the Philips PARADISE pulse sequence programming environment (Philips Healthcare, Andover, MA, version R2.5.3).
Five healthy human subjects were scanned on a Philips Achieva 1.5T MRI scanner (Philips Healthcare, Andover, MA). The integrated body coil was used as the transmitter and the 4-channel receive-only phase array coil was used as the receiver. An air-filled cushion with a pressure sensor was placed on the abdomen of the human subject and secured by a belt wrapped around the human subject to provide the respiratory signal that triggered the 4DMRI image acquisition. Since respiratory-induced organ motion is mainly in the superior-inferior direction, 4DMRI images were acquired in the sagittal orientation around the abdominal area. Sixty 3 mm slices with no gap were acquired to provide 180 mm coverage in the lateral direction. The in-plane field of view (FOV) was 375 × 260 mm2 and the in-plane spatial resolution was 1.5 × 1.5 mm2. Single-shot spin echo was used to acquire each 2D image. Parallel imaging with an accelerator factor of 2 and partial-k acquisition with 70% k-space coverage were utilized to reduce the acquisition time for each 2D image (TSSAcq) to 380 ms. Within the 380 ms, the first 250 ms was devoted to image acquisition which determined image quality and the next 130 ms was merely waiting time to satisfy the specific absorption rate (SAR) requirement. Ten respiratory states were acquired. They were evenly separated into two groups in an interleaved manner (Figure 1c). The nominal scan time without respiratory triggering was 3 minutes 42 seconds. The acquired 2D images were arranged into 3D volumes based on respiratory states. 3D volumes at 10 respiratory states were assembled together to form a complete 4DMRI image dataset, which was subsequently used to estimate liver motion.
Liver motion estimation
Liver motion was characterized using deformation vector field (DVF) analysis in a manner similar to that reported in previous studies (18, 23). Note that other techniques to measure liver motion have been reported such as template tracking (24) and centroid displacement (25). Because images were acquired on healthy volunteers and no tumors were involved, DVF analysis was appropriate to provide motion information of the entire liver. A non-rigid image registration was applied between the 3D volumetric images at a given respiratory state and those at the end-of-exhalation using an open source MIRT (medical imaging registration tool) software (26). This software utilizes B-spline based non-rigid registration (27). The deformation between the two volumes was represented by a 3D vector matrix, indicating the motion of each voxel relative to its 3D location at the end of exhalation. Because this was a single modality image registration, we employed SSD (Sum of squared differences) as the similarity measurement. The maximum liver motions were calculated using the DVF.
Results
Simulation study
The simulated acquisition times for 4DMRI methods incorporating no intervention, RSS and MSA were listed in Table 1. The average reductions in acquisition time were 36% and 10% for the RSS and the MSA methods, respectively. Figure 2 shows the simulated 4DMRI acquisition from a representative human subject for each of the three methods. The RSS method was more time efficient compared to the other two methods.
Table 1.
Simulated acquisition time and percentage reduction in acquisition time for 4DMRI incorporating no intervention (None), respiratory state splitting (RSS) and multi-shot acquisition (MSA).
| Subject | 4DMRI_None (Min) | 4DMRI_RSS (Min) | 4DMRI_MSA (Min) | Acquisition time reduction (RSS) | Acquisition time reduction (MSA) |
|---|---|---|---|---|---|
| 1 | 20.6 | 13.1 | 18.9 | 36% | 8% |
| 2 | 25.3 | 16.4 | 23.7 | 35% | 6% |
| 3 | 21.8 | 13.5 | 21.6 | 38% | 1% |
| 4 | 20.0 | 12.1 | 17.9 | 39% | 9% |
| 5 | 19.8 | 11.5 | 12.9 | 42% | 35% |
| 6 | 15.3 | 9.5 | 12.0 | 38% | 22% |
| 7 | 22.7 | 15.6 | 18.9 | 31% | 17% |
| 8 | 26.1 | 18.3 | 28.6 | 30% | -9% |
| 9 | 19.8 | 13.1 | 19.1 | 34% | 4% |
| 10 | 14.7 | 8.9 | 13.1 | 39% | 11% |
| Ave | 20.6 | 13.2 | 18.7 | 36% | 10% |
| p-value | --- | 4×10-9 | 0.03 | --- | --- |
Figure 2.
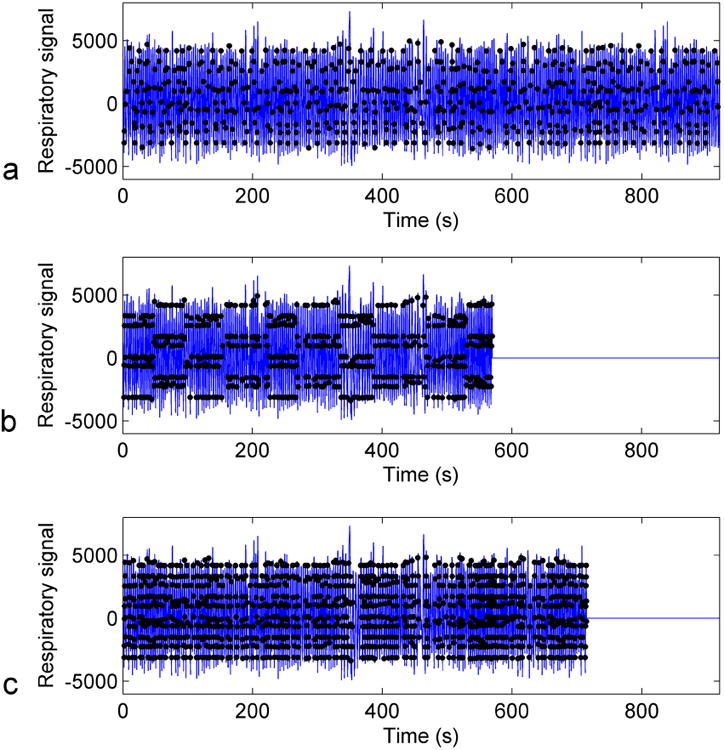
Simulated 4DMRI acquisition using the prospective 4DMRI acquisition method incorporating (a) no intervention, (b) respiratory state splitting method and (c) multi-shot acquisition method.
Human subject study
Using the RSS method, the 4DMRI acquisition times were 9.9, 16.1, 17.8, 13.1 and 8.9 minutes for the 5 healthy human subjects (subject #6 to subject #10 in Table 1), which were within half a minute of the simulated acquisition times. Figure 3 shows images of a representative sagittal slice at the 10 respiratory states (0%, 20%↑, 40%↑, 60%↑, 80%↑, 100%, 80%↓, 60%↓, 40%↓ and 20%↓ where ↑ indicates inhalation and ↓ indicates exhalation) for the 5 human subjects. With the aid of a white line placed on the liver dome at the end of exhalation, the respiration-induced liver motion can be easily visualized. Note that breathing motion may manifest a hysteresis behavior in which lung tissue, as well as the diaphragm, moves in different paths during inhalation and exhalation (28, 29). It was hypothesized that this was caused by pressure imbalances within the lung tissue during breathing (30). The hysteresis motion behavior was observed in subject a (Figure 3a) in which diaphragm motion is relatively linear during inhalation but quite non-linear during exhalation. Figure 4 shows the reconstructed images of a representative coronal plane at the 10 respiratory states for the 5 human subjects. The boundaries of the liver and kidney are smooth, indicating that images in each 3D volume were indeed acquired at the same respiratory state.
Figure 3.
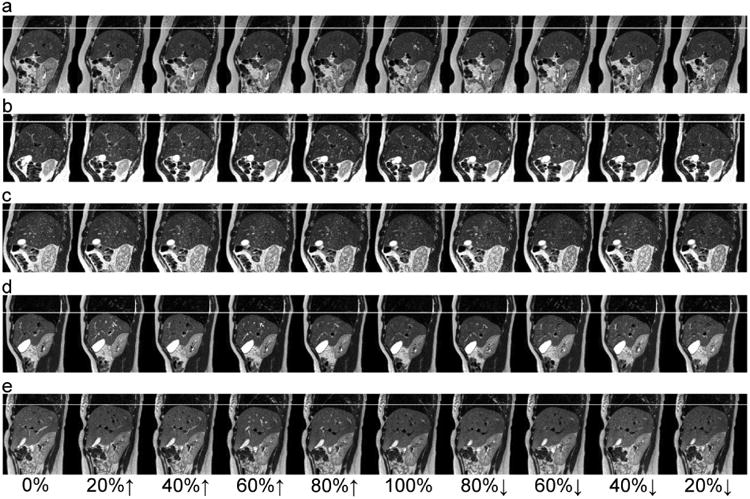
Images of a representative sagittal slice at the 10 respiratory states (4DMRI with respiratory state splitting) for the 5 human subjects. The white line indicates the liver dome at the end of exhalation.
Figure 4.
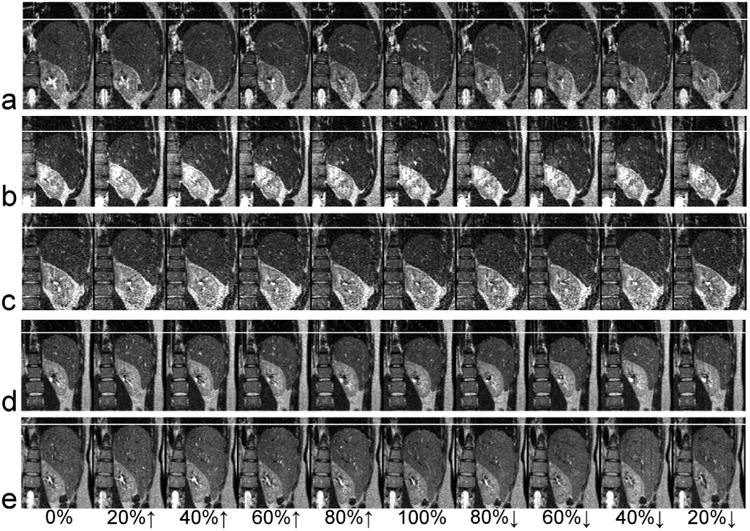
Reconstructed images of a representative coronal plane at the 10 respiratory states (4DMRI with respiratory state splitting) for the 5 human subjects. The white line indicates the liver dome at the end of exhalation.
Liver motion estimation
Figure 5 shows the deformation vectors in the liver on top of the end-of-exhalation images in three orthogonal views. The deformation vectors were obtained from the deformable registration between the end-of-exhalation and end-of-inhalation volumetric images. The arrows started from the locations at the end-of-exhalation and pointed towards the locations at the end-of-inhalation. The liver motion was large in the superior-inferior (SI) direction, medium in the anterior-posterior (AP) direction and small in the left-right (LR) direction. The peak-to-peak liver displacements in SI, AP and LR directions were 11.2 mm, 3.2 mm, 0.8 mm for the subject #1, 10.7 mm, 2.8 mm, and 0.9 mm for the subject #2, 12.1 mm, 3.4 mm, and 0.5 mm for the subject #3, 9.0 mm, 3.9 mm, and 1.5 mm for the subject #4, 12.9 mm, 2.5 mm, and 2.3 mm for the subject #5, respectively.
Figure 5.
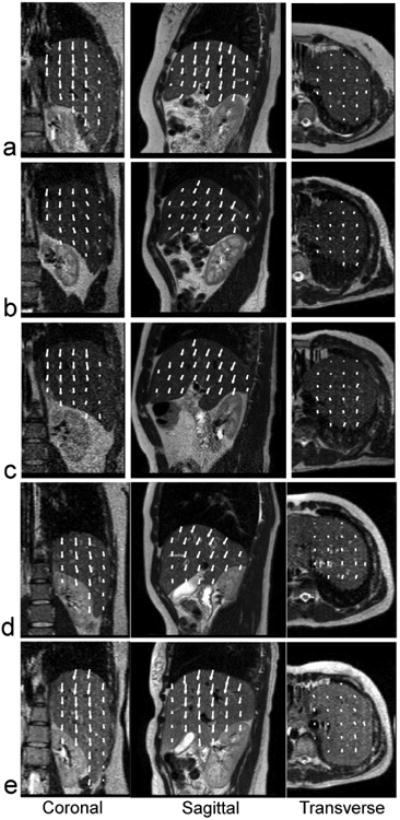
Deformation vectors in liver on top of the end-of-exhalation images in three orthogonal views for the 5 human subjects. The arrows start from the locations at the end-of-exhalation and point towards the locations at the end-of-inhalation.
Discussion
In this work, we have investigated methods (RSS and MSA) to mitigate the trigger missing issue in 4DMRI image acquisition that is often associated with the increased number of respiratory states. The RSS method was identified to be more effective and therefore implemented in the prospective respiratory amplitude triggered 4DMRI pulse sequence and validated using multiple healthy human subjects. It has been shown that with the RSS method, we were able to provide a practical 4DMRI protocol for radiation therapy applications, which offers T2 weighting for improved tumor tissue contrast, 10 respiratory states for sufficient respiratory motion sampling, 60 slices with 3 mm slice thickness for good spatial coverage, 1.5 mm in-plane spatial resolution for tumor delineation and a reasonable acquisition time between 9-18 minutes depending on the individual breathing pattern. We demonstrated that it was possible to estimate the liver motion using the acquired 4DMRI image datasets.
The RSS and MSA methods can reduce the chance of trigger missing and consequently the 4DMRI acquisition time. On the other hand, breaking the acquisition into multiple groups or shots will increase the total 4DMRI acquisition time by a factor of NGroup, or NShot. The net effect of the two competing factors determines the ultimate 4DMRI acquisition time. It is possible that the added scan time caused by using multiple groups (RSS) or shots (MSA) may exceed the reduced scan time resulting from less chance of trigger missing. Under such circumstance, the overall 4DMRI acquisition time may increase. This is observed for subject #8 using the MSA method (Table 1). For the RSS method, the net effect was always a reduction in the 4DMRI acquisition time compared to the original prospective 4DMRI method incorporating no intervention.
In this study, DVF analysis was used to characterize liver motion. The accuracy of the DVF inside the liver, however, is difficult to evaluate due to the lack of contrast and feature, which is a limitation of the current study. In future patient studies, the good tumor tissue contrast of the T2-weighted images will provide a good visualization of the tumor (the most important ROI). The accuracy of the DVF can then be evaluated by comparing tumor motion obtained using the DVF and physician's manual contouring.
For human subjects, breathing patterns show irregularity in both amplitude and period (31, 32). For all respiratory signals used in this study, we didn't notice baseline drift over the long period of time (over 10 minutes). The respiratory baseline may experience drift in a relatively short time range but returns to previous level eventually. Therefore, it may be treated as one type of irregularity in amplitude. The prospective respiratory-amplitude-based 4DMRI acquires images only when pre-determined conditions are met. Therefore, it is less susceptible to breathing irregularity compared to retrospective 4DMRI, at a cost of lengthened acquisition time. The similar rationale has been applied to prospective acquisition for 4DCT (33). One limitation of prospective triggering approach is that if patient's breathing pattern/baseline changes substantially between the preparation and acquisition stages, the acquisition time will be prolonged. If long pauses are observed by the operator, the scan can be restarted to generate a new calibration based on the external surrogate.
Summary.
We investigated methods to improve triggering efficiency of the prospective respiratory amplitude triggered 4DMRI method, implemented the most effective one and validated it using a human study. The new 4DMRI method is able to provide T2 weighting with good spatial coverage, spatial resolution and respiratory motion sampling within a reasonable amount of time for radiation therapy applications. Liver motion was estimated using the acquired T2-weighted 4DMRI image datasets.
Acknowledgments
This work is supported in part by National Institutes of Health grant 1R21CA167092
Footnotes
Conflict of interest: Dr. Caruthers is an employee of Phillips Healthcare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gierga DP, Chen GT, Kung JH, et al. Quantification of respiration-induced abdominal tumor motion and its impact on IMRT dose distributions. Int J Radiat Oncol Biol Phys. 2004;58:1584–1595. doi: 10.1016/j.ijrobp.2003.09.077. [DOI] [PubMed] [Google Scholar]
- 2.Langen KM, Jones DT. Organ motion and its management. Int J Radiat Oncol Biol Phys. 2001;50:265–278. doi: 10.1016/s0360-3016(01)01453-5. [DOI] [PubMed] [Google Scholar]
- 3.Suramo I, Paivansalo M, Myllyla V. Cranio-caudal movements of the liver, pancreas and kidneys in respiration. Acta Radiol Diagn (Stockh) 1984;25:129–131. doi: 10.1177/028418518402500208. [DOI] [PubMed] [Google Scholar]
- 4.Bryan PJ, Custar S, Haaga JR, et al. Respiratory movement of the pancreas: an ultrasonic study. J Ultrasound Med. 1984;3:317–320. doi: 10.7863/jum.1984.3.7.317. [DOI] [PubMed] [Google Scholar]
- 5.Brock KK, Hollister SJ, Dawson LA, et al. Technical note: creating a four-dimensional model of the liver using finite element analysis. Med Phys. 2002;29:1403–1405. doi: 10.1118/1.1485055. [DOI] [PubMed] [Google Scholar]
- 6.Shirato H, Shimizu S, Kunieda T, et al. Physical aspects of a real-time tumor-tracking system for gated radiotherapy. Int J Radiat Oncol Biol Phys. 2000;48:1187–1195. doi: 10.1016/s0360-3016(00)00748-3. [DOI] [PubMed] [Google Scholar]
- 7.Hanley J, Debois MM, Mah D, et al. Deep inspiration breath-hold technique for lung tumors: the potential value of target immobilization and reduced lung density in dose escalation. Int J Radiat Oncol Biol Phys. 1999;45:603–611. doi: 10.1016/s0360-3016(99)00154-6. [DOI] [PubMed] [Google Scholar]
- 8.Rosenzweig KE, Hanley J, Mah D, et al. The deep inspiration breath-hold technique in the treatment of inoperable non-small-cell lung cancer. Int J Radiat Oncol Biol Phys. 2000;48:81–87. doi: 10.1016/s0360-3016(00)00583-6. [DOI] [PubMed] [Google Scholar]
- 9.Kubo HD, Hill BC. Respiration gated radiotherapy treatment: a technical study. Phys Med Biol. 1996;41:83–91. doi: 10.1088/0031-9155/41/1/007. [DOI] [PubMed] [Google Scholar]
- 10.Beddar AS, Kainz K, Briere TM, et al. Correlation between internal fiducial tumor motion and external marker motion for liver tumors imaged with 4D-CT. Int J Radiat Oncol Biol Phys. 2007;67:630–638. doi: 10.1016/j.ijrobp.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Pan T, Lee TY, Rietzel E, et al. 4D-CT imaging of a volume influenced by respiratory motion on multi-slice CT. Med Phys. 2004;31:333–340. doi: 10.1118/1.1639993. [DOI] [PubMed] [Google Scholar]
- 12.Vedam SS, Keall PJ, Kini VR, et al. Acquiring a four-dimensional computed tomography dataset using an external respiratory signal. Phys Med Biol. 2003;48:45–62. doi: 10.1088/0031-9155/48/1/304. [DOI] [PubMed] [Google Scholar]
- 13.Keall PJ, Starkschall G, Shukla H, et al. Acquiring 4D thoracic CT scans using a multislice helical method. Phys Med Biol. 2004;49:2053–2067. doi: 10.1088/0031-9155/49/10/015. [DOI] [PubMed] [Google Scholar]
- 14.Cai J, Chang Z, Wang Z, et al. Four-dimensional magnetic resonance imaging (4D-MRI) using image-based respiratory surrogate: a feasibility study. Med Phys. 2011;38:6384–6394. doi: 10.1118/1.3658737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tryggestad E, Flammang A, Han-Oh S, et al. Respiration-based sorting of dynamic MRI to derive representative 4D-MRI for radiotherapy planning. Med Phys. 2013;40:051909. doi: 10.1118/1.4800808. [DOI] [PubMed] [Google Scholar]
- 16.Tryggestad E, Flammang A, Hales R, et al. 4D tumor centroid tracking using orthogonal 2D dynamic MRI: implications for radiotherapy planning. Med Phys. 2013;40:091712. doi: 10.1118/1.4818656. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Cai J, Wang H, et al. Four-dimensional magnetic resonance imaging using axial body area as respiratory surrogate: initial patient results. Int J Radiat Oncol Biol Phys. 2014;88:907–912. doi: 10.1016/j.ijrobp.2013.11.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Siebenthal M, Szekely G, Gamper U, et al. 4D MR imaging of respiratory organ motion and its variability. Phys Med Biol. 2007;52:1547–1564. doi: 10.1088/0031-9155/52/6/001. [DOI] [PubMed] [Google Scholar]
- 19.von Siebenthal M, Cattin P, Gamper U, et al. 4D MR imaging using internal respiratory gating. Med Image Comput Comput Assist Interv. 2005;8:336–343. doi: 10.1007/11566489_42. [DOI] [PubMed] [Google Scholar]
- 20.Hu Y, Caruthers SD, Low DA, et al. Respiratory amplitude guided 4-dimensional magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2013;86:198–204. doi: 10.1016/j.ijrobp.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokuda J, Morikawa S, Haque HA, et al. Adaptive 4D MR imaging using navigator-based respiratory signal for MRI-guided therapy. Magn Reson Med. 2008;59:1051–1061. doi: 10.1002/mrm.21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G, Citrin D, Camphausen K, et al. Advances in 4D medical imaging and 4D radiation therapy. Technol Cancer Res Treat. 2008;7:67–81. doi: 10.1177/153303460800700109. [DOI] [PubMed] [Google Scholar]
- 23.Williams CL, Mishra P, Seco J, et al. A mass-conserving 4D XCAT phantom for dose calculation and accumulation. Med Phys. 2013;40:071728. doi: 10.1118/1.4811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi X, Diwanji T, Mooney KE, et al. Evaluation of template matching for tumor motion management with cine-MR images in lung cancer patients. Med Phys. 2014;41:052304. doi: 10.1118/1.4870978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gauthier JF, Varfalvy N, Tremblay D, et al. Characterization of lung tumors motion baseline using cone-beam computed tomography. Med Phys. 2012;39:7062–7070. doi: 10.1118/1.4762563. [DOI] [PubMed] [Google Scholar]
- 26.Myronenko A, Song X. Intensity-based image registration by minimizing residual complexity. IEEE Trans Med Imaging. 2010;29:1882–1891. doi: 10.1109/TMI.2010.2053043. [DOI] [PubMed] [Google Scholar]
- 27.Rueckert D, Sonoda LI, Hayes C, et al. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 28.Seppenwoolde Y, Shirato H, Kitamura K, et al. Precise and real-time measurement of 3D tumor motion in lung due to breathing and heartbeat, measured during radiotherapy. Int J Radiat Oncol Biol Phys. 2002;53:822–834. doi: 10.1016/s0360-3016(02)02803-1. [DOI] [PubMed] [Google Scholar]
- 29.Boldea V, Sharp GC, Jiang SB, et al. 4D-CT lung motion estimation with deformable registration: quantification of motion nonlinearity and hysteresis. Med Phys. 2008;35:1008–1018. doi: 10.1118/1.2839103. [DOI] [PubMed] [Google Scholar]
- 30.Low DA, Parikh PJ, Lu W, et al. Novel breathing motion model for radiotherapy. Int J Radiat Oncol Biol Phys. 2005;63:921–929. doi: 10.1016/j.ijrobp.2005.03.070. [DOI] [PubMed] [Google Scholar]
- 31.Glide-Hurst CK, Schwenker Smith M, Ajlouni M, et al. Evaluation of two synchronized external surrogates for 4D CT sorting. J Appl Clin Med Phys. 2013;14:4301. doi: 10.1120/jacmp.v14i6.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruan D, Fessler JA, Balter JM, et al. Real-time profiling of respiratory motion: baseline drift, frequency variation and fundamental pattern change. Phys Med Biol. 2009;54:4777–4792. doi: 10.1088/0031-9155/54/15/009. [DOI] [PubMed] [Google Scholar]
- 33.Langner UW, Keall PJ. Prospective displacement and velocity-based cine 4D CT. Med Phys. 2008;35:4501–4512. doi: 10.1118/1.2977539. [DOI] [PMC free article] [PubMed] [Google Scholar]


