Key Points
We evaluated a cohort of ARL samples for known and novel tumor viruses, revealing the oncogenic herpesvirus EBV as the sole detected infectious agent.
Heterogeneous viral gene expression suggests that variable host immunosurveillance of EBV latency may influence lymphomagenesis.
Abstract
Immunodeficiency dramatically increases susceptibility to cancer as a result of reduced immune surveillance and enhanced opportunities for virus-mediated oncogenesis. Although AIDS-related lymphomas (ARLs) are frequently associated with known oncogenic viruses, many cases contain no known transforming virus. To discover novel transforming viruses, we profiled a set of ARL samples using whole transcriptome sequencing. We determined that Epstein-Barr virus (EBV) was the only virus detected in the tumor samples of this cohort, suggesting that if unidentified pathogens exist in this disease, they are present in <10% of cases or undetectable by our methods. To evaluate the role of EBV in ARL pathogenesis, we analyzed viral gene expression and found highly heterogeneous patterns of viral transcription across samples. We also found significant heterogeneity of viral antigen expression across a large cohort, with many patient samples presenting with restricted type I viral latency, indicating that EBV latency proteins are under increased immunosurveillance in the post-combined antiretroviral therapies era. Furthermore, EBV infection of lymphoma cells in HIV-positive individuals was associated with a distinct host gene expression program. These findings provide insight into the joint host-virus regulatory network of primary ARL tumor samples and expand our understanding of virus-associated oncogenesis. Our findings may also have therapeutic implications, as treatment may be personalized to target specific viral and virus-associated host processes that are only present in a subset of patients.
Introduction
Evidence that viral infection could lead to the development of cancer came early in the 20th century, and many associations between viruses and hematologic malignancies have been identified in a variety of animal models. It has been estimated that close to 12% of all cancers in humans worldwide are caused by viral infection.1 However, only a few viruses have been shown to genuinely cause cancer in humans, and only 3 have been shown to directly cause lymphomas: Epstein-Barr virus (EBV; formally designated human herpesvirus 4), Kaposi sarcoma-associated herpesvirus (KSHV; human herpesvirus 8), and human T-cell lymphotropic virus 1.
Immunodeficient individuals have greatly increased cancer incidence, the pathogenesis of which is thought to be related to disrupted immune surveillance, chronic antigenic stimulation, genetic alterations, cytokine dysregulation, and viral infection.2-4 The viral contribution to these malignancies has been extensively studied, and 2 tumor viruses have been discovered after a targeted search based on the epidemiologic association of specific cancer types and AIDS: KSHV and Merkel cell polyomavirus.5,6 These previous findings have raised the likelihood of novel tumor viruses being discovered in the context of immunosuppression.
Improved combined antiretroviral therapies (CART) as HIV treatment options have resulted in decreased immunosuppression and concomitantly reduced incidence of the highly immunogenic virus-associated malignancies, such as Kaposi sarcoma.7-10 As a result, the proportion of non-Hodgkin lymphomas has increased as a fraction of the total number of AIDS-related cancers.11 Non-Hodgkin lymphomas constitute >50% of all AIDS-defining cancers in developed countries and are the most common cause of cancer-related death in HIV-infected individuals.12 AIDS-related lymphomas (ARLs) are phenotypically and histologically similar to lymphomas of the immunocompetent; however, ARL cases are more frequently associated with known virus mediators of oncogenesis.13-15 For example, although EBV is present in ∼30% of all ARLs, it is more common among diffuse large B-cell lymphomas (DLBCLs) with immunoblastic morphology (>80%) than those with centroblastic morphology (20-40%), and it is present in the vast majority of DLBCLs with presentation in the central nervous system.16 In addition, ∼5% of ARLs are associated with KSHV infection. However, the remaining ∼60% of ARLs have no known viral contribution to lymphomagenesis. Given that non-Hodgkin lymphoma is increased ∼15-fold in the context of AIDS,17 this implies that >55% of ARLs have an etiology directly related to the immunosuppressed host and cannot be ascribed to any known virus.
The permissive environment of the moderately immunosuppressed host and lack of known oncoviruses in a majority of cases motivated us to reexamine the viral and cellular transcriptomes of ARLs, in search of novel transforming viruses.
Methods
Patient samples
Seventy-two formalin-fixed paraffin-embedded (FFPE) ARL samples were collected from New York Presbyterian Hospital–Weill Cornell, the University of Siena, and the AIDS Malignancy Consortium. Cases were included if >80% tumor cells were present and the diagnosis of B-cell lymphoma was confirmed. All samples were obtained with the approval of the institutional review boards at both institutions. Research was conducted in accordance with the Declaration of Helsinki. Sixteen cases with frozen tissue were collected from New York Presbyterian Hospital–Weill Cornell and confirmed to contain >80% tumor cells. The 16 frozen and 9 of the FFPE samples were subjected to RNA extraction and transcriptome sequencing. Independent validation cases shown in Table 2 and Figure 2 were collected from the Hematopathology Laboratory of Weill Cornell Medical College Pathology/The New York Presbyterian Hospital (40 cases), the AIDS Malignancy Consortium (12 cases), The University of Siena (20 cases), and the AIDS Cancer Specimen Resource (140 cases). Tissue diagnosis was made with the use of criteria from the World Health Organization.18 In cases of DLBCL, germinal center B-cell (GCB) vs non-GCB subtype was determined by the Hans algorithm.19,20 Our study of 25 ARL samples is well powered to contain ≥1 sample with a virus if that virus occurs in >10% of total ARL cases (statistical power >0.9).
Table 2.
Pathological subclassification and EBV assessment by EBER ISH and immunohistochemistry for LMP1 and EBNA2 in AIDS-related lymphoma
| AIDS lymphoma subtype | No. | EBV-positive (%) EBER-ISH | Immunophenotype | ||
|---|---|---|---|---|---|
| Latency I [LMP1−EBNA− (% of EBV+)] | Latency II [LMP1+EBNA2− (% of EBV+)] | Latency III [LMP1+EBNA2+ (% of EBV+)] | |||
| DLBCL non-GC | 48 | 27 (56%) | 10 (37%) | 8 (30%) | 8 (30%) |
| DLBCL GCB | 98 | 25 (25%) | 19 (76%) | 3 (12%) | 3 (12%) |
| DLBCL null | 13 | 4 (31%) | 1 (25%) | 3 (75%) | |
| BL | 19 | 10 (53%) | 9 (90%) | 1 (10%) | |
| PBL | 9 | 8 (89%) | 8 (100%) | ||
| PEL (solid variant) | 14 | 12 (86%) | 11 (92%) | 1 (8%) | |
| BCL-U | 4 | 3 (75%) | 3 (100%) | ||
| Polymorphic LPD | 7 | 5 (71%) | 1 (20%) | 1 (20%) | 3 (60%) |
| Total | 212 | 94 (44%) | 61 (65%) | 15 (16%) | 17 (18%) |
These 212 cases were classified as latency I in EBER+ cases when no LMP1 or EBNA2 was expressed, as latency II when these were positive for LMP1 but negative for EBNA2, and as latency III where there was expression of both EBNA2 and LMP1. BCL-U, B-cell lymphoma, unclassifiable, with features of DLBCL and BL; DLBCL null, negative for CD10, BCL6 and MUM1; LPD, lymphoproliferative disorder; PEL, primary effusion lymphoma.
Figure 2.
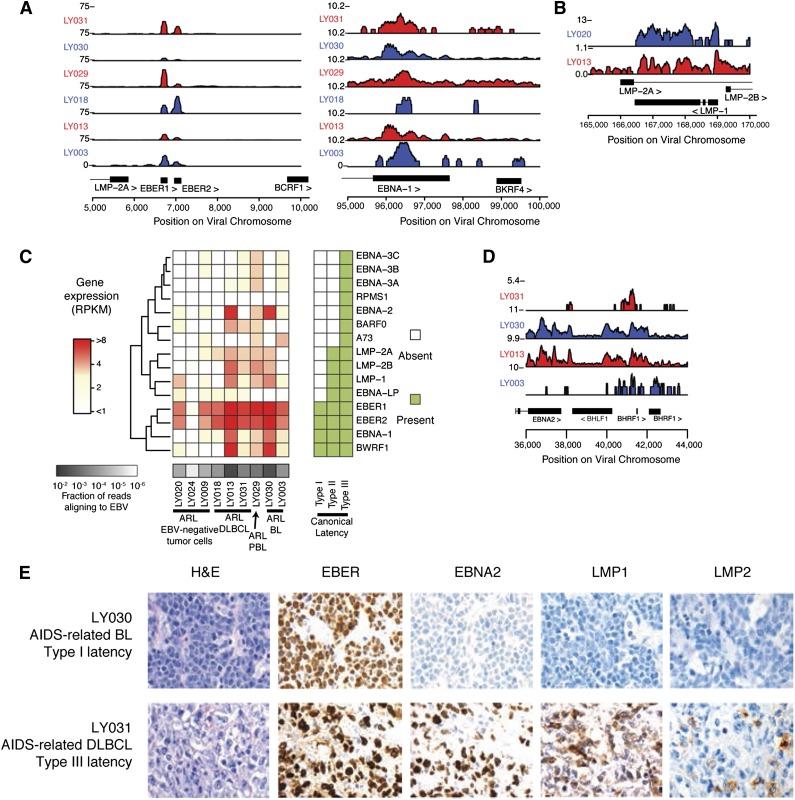
Transcriptional regulation of EBV gene expression demonstrates diversity of latency types. (A) Gene expression is shown as read counts per million reads aligned (y-axis) at canonical type I latency genes EBER1, EBER2, and EBNA1. Rows show individual samples and are colored alternating red/blue to emphasize sample-track pairing. (B) Immunogenic viral genes associated with type II and III latency are also expressed, such as LMP1. (C) EBV gene expression is highly diverse across ARL samples. The heatmap shows gene expression profiles of latency-associated genes (y-axis) across samples (x-axis). Gene expression is in units of RPKM normalized by fraction of reads aligning to EBV (“Methods”). (D) Genes associated with lytic reactivation, such as transcripts at the BamHI H locus, were expressed in primary ARL tumor samples. (E) Immunohistochemical staining for viral proteins EBNA2 and LMP1 was used to classify viral gene expression latency in ARL cases. A BL sample with latency I (upper) and a DLBCL sample with latency III (lower) are representative examples. A complete analysis can be found in Table 2.
Histology
EBV Probe ISH Kit (Leica Microsystems, Wetzlar, Germany; Vision BioSystems Novocastra, Newcastle-upon-Tyne, UK) was used for in situ hybridization (ISH) for EBV RNA (EBER). Cases were considered positive when >20% neoplastic cells were immune reactive, except for BCL-2, where cases were considered positive when >50% of tumor cells had moderate to strong positivity. Immunohistochemical (IHC) studies were performed using monoclonal antibodies to CD-10, BCL-2, BCL-6, MUM-1 (3H2E8; Santa Cruz Biotech, Santa Cruz, CA), and Ki-67 (MIB-1; DakoCytomation, Carpenteria, CA). Nuclear Ki-67 expression was semiquantitative and assessed as the percentage of positive tumor cells. For EBV latency assessment, IHC was done with antibodies to LMP1 (clones CS1-4, Abcam; and clone OT21C, kind gift from Jaap Middeldorp from the VU University Medical Center), EBNA2 (clone PE2; Dako), and LMP2A (15F9; Abcam).
Sequencing and analysis
Sequenced cDNA libraries were constructed from 25 cases using random hexamer primers, ligated to Illumina adaptors, and subjected to high-throughput Illumina sequencing to generate 17 to 150 million 76- to 100-bp paired-end sequence reads per sample. Reads were aligned to the EBV (human herpesvirus 4; GenBank accession no. NC_007605) and human (UCSC hg19) genomes using Bowtie2 with the default “sensitive” parameters for alignment.21 This approach allows multiple read alignment mismatches, which increases coverage of diverse EBV genotypes. We also aligned using Burrows-Wheeler Aligner22 (both default and increased sensitivity parameters) and obtained similar results (supplemental Table 1, available on the Blood Web site). Reads that mapped to the viral genome with low alignment score or to the human genome were considered of ambiguous origin and discarded, as previously described.23
To detect other known and novel viruses, the PathSeq pipeline was used, as previously described.24 Briefly, reads were aligned against all known viral genomes, as downloaded from the National Center for Biotechnology Information, using Bowtie2. To mitigate monoclonal reads inflating counts, we compress all reads that align to a single position in the genome to a single read. This provides more reliable evidence of presence/absence of a given virus or gene transcript but may decrease the dynamic range of estimated gene expression for highly expressed transcripts. In search of novel pathogens, high-quality unalignable reads were formed into contigs using Trinity.25
Viral RNA gene expression values were estimated using cufflinks as reads per kilobase per million reads aligned (RPKM) across all known transcriptional units, as previously described.23 Cufflinks estimates overlapping transcript abundance by the read coverage of unique portions of each transcript. Track plots show read overlap counts normalized to total reads aligning to the viral genome. Normalized heatmaps show RPKM divided by −log(f), where f is the fraction of reads uniquely aligning to the viral genome.
Human RNA gene expression was estimated using Cufflinks.23 Human genes without significant expression (ie, those genes with RPKM <1) were removed from subsequent analyses, resulting in ∼8000 genes for analyses. Principal component analysis clustering identified histological subtype (DLBCL and Burkitt lymphoma [BL]) and sample preparation method (fresh frozen or archival FFPE) as covariates (supplemental Figure 5A-B). We thus used cuffdiff to analyze differential gene expression by subtype and by sample preparation method. Consistency of gene expression signatures was determined through overlap of significant differentially expressed genes (Figure 4C), scatter plot correlation (supplemental Figure 5C), and most differentially expressed rank overlap (supplemental Figure 5D). Overlap statistical enrichment was estimated using the hypergeometric distribution and implemented as a Fisher exact test. Human genome-wide gene expression was summarized by principal component analysis on genes with variance >0.01.
Figure 4.
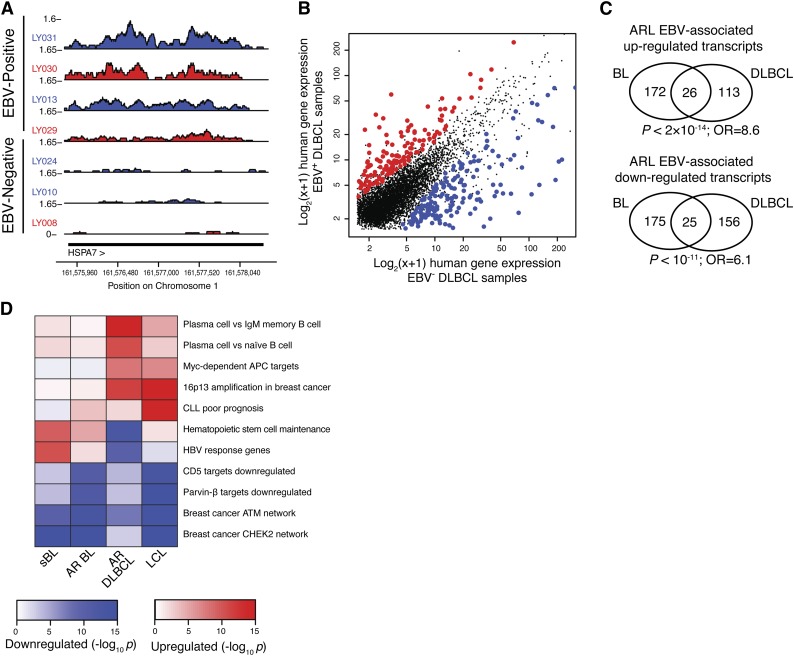
EBV-positive ARL cases have a distinct host gene expression signature. (A) HSPA7 is an example of a gene that is more highly expressed in EBV+ samples (above) compared with EBV− samples (below). (B) Genes consistently up- and downregulated in the presence of EBV are shown in DLBCL cases. Axes show log2(x + 1), where x is the gene expression level in units of RPKM as quantified by cufflinks. (C) EBV-associated host gene expression changes are significantly similar in AIDS-related BL and DLBCL. Significantly differentially expressed upregulated (upper) and downregulated (lower) genes in EBV-positive samples are part of the EBV signature in both BL and DLBCL (hypergeometric test). (D) EBV-associated host gene expression changes cluster into pathways, including plasma cell differentiation, maintenance of hematopoietic stem cells, and genes dysregulated in primary effusion lymphoma. Heatmap shows pathways (y-axis) enriched for host expression signatures associated with EBV in a variety of cellular and host settings (x-axis), including HIV-negative sBL, AR BL, AR DLBCL, and LCLs derived from whole peripheral blood infected with high titers of EBV. Enrichment significance is determined by 1-sided Kolmogorov-Smirnov tests.
Results
AIDS-related lymphoma cohort description, histology, and sample selection
To understand the viral landscape of ARL and determine whether previously unidentified oncogenic pathogens are present in this disease, we established a tissue sample cohort (n = 88 distinct patients) that could be assayed for viral RNA. We selected 25 cases with tissue blocks available for accurate histological subclassification by World Health Organization criteria, which included 19 DLBCLs, 3 BLs, 2 follicular lymphomas, and 1 plasmablastic lymphoma (PBL) (Table 1). Six EBV-positive cases, as determined by EBER ISH, were sequenced and analyzed blindly as a way to assess our ability to detect this virus. Due to known double viral infection in ARL (eg, EBV and KSHV in primary effusion lymphomas), we also analyzed the EBV+ cases for novel pathogens. We excluded KSHV-positive cases from our cohort.
Table 1.
Virus- and host-aligning RNA-seq reads in AIDS-related lymphoma patient cohort
| ID | Diagnosis | Storage | EBV status | Total reads | Unique human reads | Unique EBV reads |
|---|---|---|---|---|---|---|
| LY002 | DLBCL | Frozen | EBV− | 40 642 580 | 4 179 804 | 4 |
| LY003 | BL | Frozen | EBV+ | 58 461 328 | 6 082 519 | 1 094 |
| LY004 | DLBCL | Frozen | EBV− | 45 676 046 | 14 737 943 | 0 |
| LY005 | Follicular lymphoma | Frozen | EBV− | 66 222 036 | 19 694 654 | 0 |
| LY006 | Follicular lymphoma | Frozen | EBV− | 51 384 920 | 6 113 426 | 0 |
| LY007 | DLBCL | Frozen | EBV− | 45 299 480 | 5 195 300 | 0 |
| LY008 | DLBCL | FFPE | EBV− | 70 393 682 | 21 924 559 | 8 |
| LY009 | DLBCL | FFPE | EBV− | 74 302 054 | 24 897 813 | 971 |
| LY010 | DLBCL | FFPE | EBV− | 80 747 336 | 22 747 406 | 0 |
| LY011 | DLBCL | FFPE | EBV− | 79 507 540 | 28 720 368 | 0 |
| LY013 | DLBCL | FFPE | EBV+ | 65 902 794 | 12 861 869 | 114 938 |
| LY014 | DLBCL | Frozen | EBV− | 107 114 152 | 6 542 704 | 0 |
| LY015 | DLBCL | Frozen | EBV− | 165 832 378 | 9 712 379 | 0 |
| LY016 | DLBCL | Frozen | EBV− | 139 967 864 | 7 754 150 | 2 |
| LY017 | DLBCL | Frozen | EBV− | 66 708 934 | 8 132 534 | 0 |
| LY018 | DLBCL | Frozen | EBV+ | 62 481 036 | 8 862 120 | 1 286 |
| LY019 | DLBCL | Frozen | EBV− | 64 447 628 | 4 411 390 | 20 |
| LY020 | DLBCL | Frozen | EBV− | 62 411 645 | 9 436 092 | 512 |
| LY021 | DLBCL | Frozen | EBV− | 48 797 764 | 9 209 151 | 50 |
| LY022 | DLBCL | Frozen | EBV− | 81 856 928 | 9 976 478 | 78 |
| LY023 | DLBCL | Frozen | EBV− | 61 246 428 | 12 269 534 | 81 |
| LY024 | BL | FFPE | EBV− | 52 597 350 | 19 920 974 | 36 |
| LY029 | PBL | FFPE | EBV+ | 52 770 210 | 30 181 777 | 23 584 |
| LY030 | BL | FFPE | EBV+ | 16 461 068 | 10 901 688 | 66 897 |
| LY031 | DLBCL | FFPE | EBV+ | 63 740 374 | 16 484 786 | 7 318 |
Columns shown are internal ID, histological subclass diagnosis, sample storage, EBV EBER ISH status, total RNA-seq reads, and number of stringently unique reads that align to human and EBV.
PathSeq identifies EBV as the only virus present in ARL samples
To identify the viral populations in ARL samples, we performed massively parallel whole transcriptome sequencing of total RNA, enabling detection of non–poly-adenylated and other RNA species in addition to coding transcripts. These data were then analyzed using the PathSeq pipeline, which subtracts all reads that could potentially align to the human genome, aligns remaining reads to all known viruses, and assembles unaligned reads into contigs24 (Figure 1A).
Figure 1.
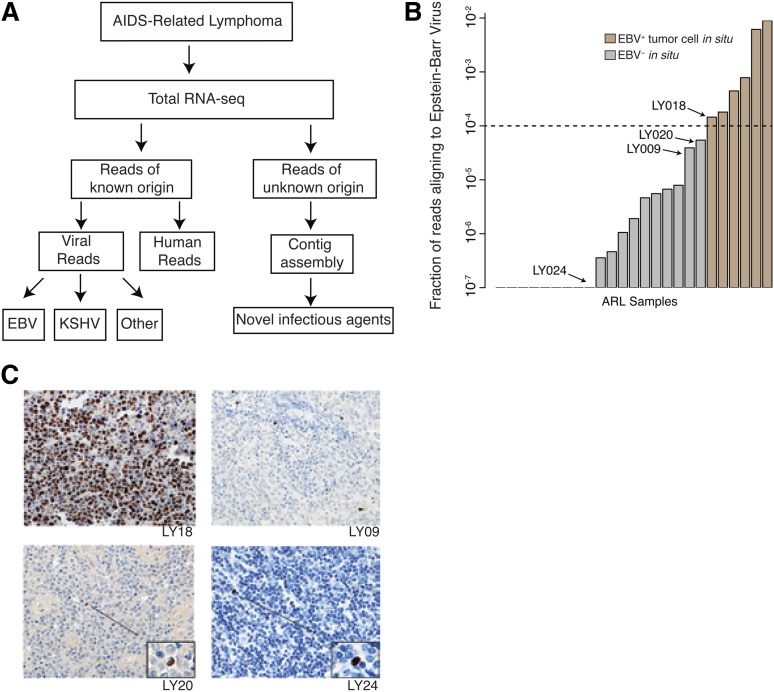
Transcriptome sequencing coupled with the PathSeq computational analysis pipeline is highly sensitive and specific for detecting presence of novel and known viruses in primary AIDS-related lymphoma samples. (A) The PathSeq pipeline profiles total RNA by random hexamer priming and high-throughput sequencing. Reads that do not map to the human genome are aligned against known viral and bacterial genomes. The remaining unmappable reads are assembled into contigs to detect transcripts originating from novel viruses. (B) PathSeq is sensitive and specific for detecting the EBV in tumor cells. Viral read count fraction (y-axis) is shown for all ARL samples (x-axis). Using a threshold of 0.01%, we were able to uniquely identify all EBV-positive cases, which were confirmed by in situ hybridization for the EBV encoded RNA 1 (EBER1) transcript. A complete listing of read counts, viral ISH detection, and case identifiers can be found in Table 1. (C) Samples with <0.01% of reads aligning to EBV were profiled by ISH for EBER1 to determine if any of these cases were mislabeled as EBV negative by PathSeq. While scattered EBV+ cells infiltrated the tumor, tumor cells were in fact negative for EBV. This is exemplified by cases LY09, LY20, and LY24 with morphologically nontransformed tumor infiltrating lymphocytes highlighted. LY18 is shown to demonstrate EBER1 staining of EBV+ samples with EBV RNA in tumor cells.
To assess specificity and sensitivity of our assay, we performed a double-blind control where we analyzed RNA-seq data of samples that were positive and negative for EBV. We determined a simple read cutoff statistic, which was able to identify EBV in all samples that tested positive by EBER RNA ISH while maintaining perfect specificity (Figure 1B). The samples that tested negative for EBER but had reads that aligned to EBER and other viral genes (supplemental Figure 1A) were found to contain tumor infiltrating EBV-infected lymphocytes (Figure 1C). This confirmed that PathSeq coupled with total RNA sequencing is a sensitive and specific assay for presence of viral RNA.
We next determined whether other known viruses were present in our cohort. We identified a subset of samples that contained reads aligning to genic regions of known human adenovirus C and polyomaviruses (supplemental Table 2). Based on previous experience and reports, we tested the sequence libraries for contamination.26,27 Subsequent polymerase chain reaction (PCR) analysis of cDNA sequencing libraries and DNA re-extracted from the original tumor samples revealed that neither adenovirus nor polyomavirus was present in our tumor samples. Namely, the viral-aligning reads were assembled into >100-nt contigs that could be efficiently amplified by PCR. Gel electrophoresis revealed that the sequencing libraries were positive for viral sequence, but the original and re-extracted cDNAs were negative (supplemental Figure 1B-E). Conversely, PCR detected multiple EBV transcripts in the re-extracted cDNA (data not shown). Thus, the sequences represent low-level contamination in the libraries and the samples were in fact devoid of known adeno- and polyomaviruses.
To detect novel viruses that may be contributing to AIDS-related lymphomas, we assembled all unalignable reads into contigs. Although many contigs were formed, all contigs aligned to known human, viral, or bacterial genomes (supplemental Table 3). To increase statistical power and sensitivity for detection, reads across all tumor samples were aggregated; however, this again resulted in contigs with negligible potential for encoding novel viral agents.
EBV gene expression in primary ARL samples is heterogeneous
To characterize viral persistence and potential maintenance of lymphomagenesis, we characterized the viral RNA and protein antigen landscape through transcript expression quantification, ISH, and IHC. EBV-mapping reads aligned to known transcribed regions of the viral genome, with the most prominently expressed transcripts being the RNA Pol III transcribed EBER1 and EBER2 transcripts along with type I latent associated EBNA1 transcripts (Figure 2A). Additional type II and III viral latency gene transcripts were expressed, including immunogenic LMP1 and LMP2 (Figure 2B-C) and an unexpected proportion of samples contained lytic transcripts, most notably being transcribed at the BHLF1 locus (Figure 2D; supplemental Figure 2A). Consistent with previous results,28 our findings suggested a diversity of latency-associated gene expression programs with a subset of cells exhibiting expression of genes corresponding to the lytic program.
It was unclear if the viral protein antigen landscape would reflect the heterogeneity of RNA gene expression, and how intratumoral heterogeneity would contribute to this phenomenon. To characterize viral antigens and intratumoral heterogeneity, we stained for the immunogenic LMP1, LMP2A, and EBNA2 proteins using IHC on ARL tissue microarrays. Many EBV+ cases were positive for ≥1 of these proteins (Table 2; Figure 2E; supplemental Figure 2B); however, there was significant diversity in the intensity of protein staining and the proportion of tumor cells that stained positive (supplemental Figure 2C). Thus, at both the RNA and protein level, EBV latent gene expression is heterogeneous in EBV-positive ARLs and does not necessarily conform to classical latency.
Given the heterogeneity of the viral transcriptional and protein landscape, we sought to confirm our findings across a larger cohort of ARL cases. We characterized viral latency type by LMP1 and EBNA2 immunohistochemistry in a validation cohort of 212 ARL cases (Table 2), 94 (44%) of which were positive for EBV as determined by EBER ISH. Although we expected a large proportion of DLBCLs to have a viral type II or III expression pattern, as suggested by previous studies,29,30 most cases were restricted to viral latency I programs, especially the majority of tumors with GC B-cell origin. Consistent with previous studies, most cases of BL had a latency I pattern.
It was unclear if the EBV transcriptional regulation patterns were unique to the immunosuppressed host environment of ARL. Thus, we performed a meta-analysis of the publicly available viral transcriptome data in HIV-negative sporadic BLs (sBLs),31 which are thought to express type I latency to limit immunogenicity. We found that EBV+ sBL samples presented with robust expression of EBNA1 and other viral genes compatible with type I latency, such as RPMS1 (Figure 3A; supplemental Figure 3A). Consistent with previous reports,28,32,33 we also found significant RNA expression of LMP2 in all EBV-positive sBL samples, and a significant subset also expressed LMP1, BHLF1, BHRF1, BMLF1, and BMRF2 (Figure 3B; supplemental Figure 3B).
Figure 3.
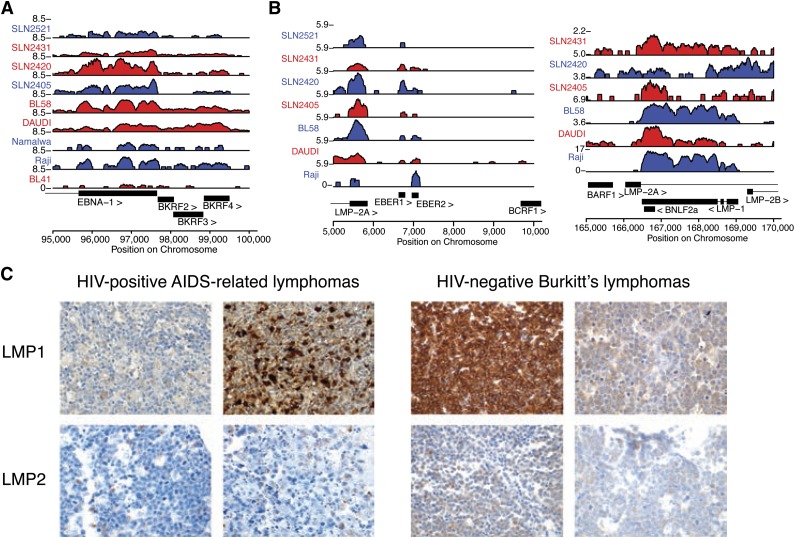
Viral antigen expression variation is comparable in immunodeficient and immunocompetent hosts. (A) Analysis of polyadenylated RNA-seq data confirms that viral expression is detectable in HIV-negative sBL in immunocompetent hosts. Expression of latency I associated EBNA1 is robustly detected in primary samples (prefixed by SLN) and cell lines. (B) LMP1 and LMP2 RNA were detected in most primary HIV-negative sBL tumor samples. SLN2521 is excluded in the right panel due to low read counts. (C) Viral RNA is translated into immunogenic protein expression in a subset of cases (additional analysis in Table 3). IHC stains in 4 representative cases demonstrate that ARL (left) and sBL (right) cases contain highly immunogenic viral proteins. All samples are positive for the respective stains.
We confirmed the active translation of the viral RNA diversity in a subset of tumors of immunocompetent individuals. IHC staining demonstrated presence of LMP1 after stringent positive and negative controls (Figure 3C). We next compared AIDS-related BL (AR BL) to HIV-negative sBL by IHC staining for LMP2A as a test for latency II or III viral gene expression. We assembled a cohort of 84 EBV+ ARL and 59 EBV+ sBL cases and found that 3 of 59 EBV+ sBL samples and 9 of 84 ARL cases showed robust LMP2A staining (Table 3). Taken together, these findings demonstrate a broad set of transcriptional latency programs in vivo in both the immunocompetent and the immunocompromised host.
Table 3.
LMP2A assessment by IHC in AIDS-related and HIV-negative BL
| HIV status | EBV positive by EBER-ISH | LMP2A IHC positive (% of EBV-positive) |
|---|---|---|
| HIV positive | 84 | 9 (11%) |
| HIV negative | 59 | 3 (5%) |
EBV-positive ARL has a distinct host gene expression program
Because viral gene products influence host gene expression patterns that can drive oncogenesis and inhibit apoptosis, we characterized the host expression program associated with the presence of EBV. Total RNA-seq reads were mapped to the human genome, and expression values were estimated using cufflinks as RPKM.34 We confirmed that expression of host genes previously identified as enhanced by EBV were upregulated in EBV-positive cases. These genes included the EBNA3A response gene HSP70B (HSPA7)35 and CD30 (TNFRSF8), which acts as a biomarker for EBV titers in immunosuppressed lymphomas and infectious mononucleosis36 (Figure 4A; supplemental Figure 4A-C). Genome-wide, hundreds of host genes were quantitatively associated with EBV in ≥1 lymphoma histological subtype (DLBCL or BL) (Figure 4B; supplemental Figure 5A-B; supplemental Table 4; “Methods”). These findings were robust to leave-one-out validation, and host genes were associated with EBV in independent sets of samples (supplemental Figure 5C). Intriguingly, differentially regulated genes were significantly similar across the histological subtypes, supporting a consistent role for EBV across multiple ARL malignancies (Figures 4C; supplemental Figure 5D).
We next wanted to understand how the immunodeficient environment influenced the EBV-host regulatory network. We compared the EBV-associated host gene expression changes in ARL vs lymphomas of immunocompetent individuals by analyzing EBV-positive HIV-negative sBL cases.31 We first confirmed the existence of a robust EBV-signature in sBL samples (supplemental Figure 6A). Host genes differentially expressed in the presence of EBV in sBL had highly statistically significant overlap with the EBV-associated gene expression program in BL of HIV+ subjects but not DLBCL (supplemental Figure 6B-C). To determine whether the EBV-associated host gene expression program was enriched for lymphomagenesis pathways, we performed a gene set enrichment analysis on the molecular signatures database MSigDB3.0.37 EBV-positive samples had a statistically significant differential regulation of genes with altered expression in the context of plasma cell differentiation, maintenance of hematopoietic stem cells, and primary effusion lymphomas (Figure 4D; supplemental Figure 7). There also existed a significant correlation between EBV and host gene expression in lymphoblastoid cell lines (LCLs).23 When taken together, these 4 datasets capture the influence of immunosurveillance and malignant cell type morphology on the EBV-associated host gene expression signature. Accordingly, we found that sBL and AR BL cluster, as do the LCL and AIDS-related DLBCL (AR DLBCL) pathway signatures. Viral response pathways (eg, Hepatitis B virus response genes) were more prevalent in sBL than other samples, whereas EBV-associated enhancement of the plasma cell differentiation program was present only in DLBCL and LCLs.
Discussion
Using the sensitive technology of total RNA-seq and the PathSeq analysis pipeline, we evaluated the transcribed viral contribution to AIDS-related lymphoma. Although no novel viruses were detected, we discovered that EBV exploits the immuno-permissive environment of ARL to express viral antigens that are associated with an altered host gene expression program. Meta-analysis of sBLs in immunocompetent individuals revealed that EBV transcriptional regulation is broadly more promiscuous than previously appreciated; however, the immunocompetent immune system exerts a significant selection on viral antigen expression.
Although extensive literature has speculated on the contribution of viruses to increased cancer risk in the immunosuppressed,2,4,38 our observation that no additional pathogens were found in a cohort of 25 ARLs suggests that if any new pathogens exist in this disease, they are relatively rare or tightly latent, without significant RNA expression (“Methods”). Because we examined steady-state RNA sequences in established tumors, we cannot rule out the possibility that EBV or other viruses are transient enablers of cellular transformation, from which frank malignancy can expand and be subsequently maintained independent of initial viral contribution (ie, through so-called “hit-and-run” mechanisms).39 The potential for this mechanism is illustrated by EBV rescue of crippled GC centrocytes, allowing cells with transforming translocations to escape apoptosis,40-42 which can be followed by loss of viral episome after in vitro culture of EBV+ tumors.43-45 AIDS-associated malignancies may also be affected, in some cases, by increased activation of lymphocytes, resulting in greater frequency of DNA-damaged uninfected GC B cells.46,47 This indirect mechanism could be considered analogous to cancers associated with hepatitis C or Helicobacter pylori infections, which may trigger transformation through chronic inflammation, but pathogen sequences or proteins are not required for maintenance of tumorigenesis. Beyond hit-and-run pathogens, lack of immunosurveillance required for removal of sporadic pretumor dysplastic tissue may contribute to cancer susceptibility; however, attributing the increase of AIDS-defining cancers to decreased tumor surveillance begs the question of why there exists such an extensive risk for specific malignancies, whereas non–AIDS-defining cancers are increased only moderately in the HIV-positive or post-transplant immunosuppressed individual.17
Our findings point toward a dramatic increase of immunoselective pressure on viral antigen in the post-CART era, as evidenced by our cohort being predominantly type I latent EBV+ DLBCLs, whereas this histological subclass before CART was associated with a type III latency immunophenotype.30 Furthermore, distinct viral oncogenic mechanisms may be used in specific ARL histological subclasses. For instance, most EBV+ non-GC DLBCLs (63%) maintained type II or III latency, in contrast to only 24% of GCB DLBCLs, suggesting that more active EBV gene expression programs may be required to maintain the proliferative program of this tumor subclass.
Last, our results reveal a diversity of viral and host gene expression programs that may be therapeutic targets in only a subset of cases. Better integration of molecular diagnostics that include viral gene expression, as well as IHC, has the potential to identify patients that may benefit from in-development antilatent EBV protein therapies that interrupt the host-virus joint regulatory network.
Acknowledgments
This project was funded by the Starr Cancer Consortium and National Institutes of Health (NIH) National Cancer Institute grant 1RC2CA148317 (M.M. and E.C.). A.I.O. was supported by the Rebecca Ridley Kry Fellowship of the Damon Runyon Cancer Research Foundation. The AIDS Malignancy Clinical Trials Consortium (NIH National Cancer Institute grant U01CA121947) and the AIDS Cancer Specimen Resource (NIH National Cancer Institute grant 1UM1CA181255) contributed cases of ARL.
Footnotes
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.A., A.I.O., M.M., and E.C. designed research; A.A., A.I.O., and C.S.P. performed statistical and computational analyses; G.B. and E.B. analyzed primary samples and sequencing libraries for contamination and EBV; J.J. provided computational assistance; F.D. prepared samples for RNA-seq; L.L., W.T., G.D.F., and A.C. contributed ARL samples and confirmed the pathological classification; and A.A. and E.C. wrote the manuscript with input from A.I.O. and M.M.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for A.A. is Gilead Sciences, Foster City, CA; for A.I.O. is Department of Epidemiology, University of Alabama at Birmingham, Birmingham, AL. The current affiliation for J.J. is Division of Oncology, Sanofi Aventis Group, Cambridge, MA. The current affiliation for G.D.F. is School of Biological and Chemical Sciences, Queen Mary University of London, London, UK. The current affiliation for A.C. is Department of Pathology and Laboratory Medicine, Weill Cornell Medical College, New York, NY.
Correspondence: Ethel Cesarman, Department of Pathology and Laboratory Medicine, Weill Cornell Medical College, 1300 York Ave, New York, NY 10065; e-mail: ecesarm@med.cornell.edu.
References
- 1.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 2.Silverberg MJ, Chao C, Leyden WA, et al. HIV infection and the risk of cancers with and without a known infectious cause. AIDS. 2009;23(17):2337–2345. doi: 10.1097/QAD.0b013e3283319184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gates AE, Kaplan LD. AIDS malignancies in the era of highly active antiretroviral therapy. Oncology (Williston Park). 2002;16(4):441-451. [PubMed]
- 4.Grulich AE, van Leeuwen MT, Falster MO, Vajdic CM. Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis. Lancet. 2007;370(9581):59–67. doi: 10.1016/S0140-6736(07)61050-2. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 6.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franceschi S, Lise M, Clifford GM, et al. Swiss HIV Cohort Study. Changing patterns of cancer incidence in the early- and late-HAART periods: the Swiss HIV Cohort Study. Br J Cancer. 2010;103(3):416–422. doi: 10.1038/sj.bjc.6605756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biggar RJ, Chaturvedi AK, Goedert JJ, Engels EA, Study HACM HIV/AIDS Cancer Match Study. AIDS-related cancer and severity of immunosuppression in persons with AIDS. J Natl Cancer Inst. 2007;99(12):962–972. doi: 10.1093/jnci/djm010. [DOI] [PubMed] [Google Scholar]
- 9.d’Arminio Monforte A, Sabin CA, Phillips A, et al. Antiretroviral Therapy Cohort Collaboration. The changing incidence of AIDS events in patients receiving highly active antiretroviral therapy. Arch Intern Med. 2005;165(4):416–423. doi: 10.1001/archinte.165.4.416. [DOI] [PubMed] [Google Scholar]
- 10.Ledergerber B, Telenti A, Egger M. Risk of HIV related Kaposi’s sarcoma and non-Hodgkin’s lymphoma with potent antiretroviral therapy: prospective cohort study. Swiss HIV Cohort Study. BMJ. 1999;319(7201):23–24. doi: 10.1136/bmj.319.7201.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Collaboration on HIV and Cancer. Highly active antiretroviral therapy and incidence of cancer in human immunodeficiency virus-infected adults. J Natl Cancer Inst. 2000;92(22):1823–1830. doi: 10.1093/jnci/92.22.1823. [DOI] [PubMed] [Google Scholar]
- 12.Simard EP, Engels EA. Cancer as a cause of death among people with AIDS in the United States. Clin Infect Dis. 2010;51(8):957–962. doi: 10.1086/656416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaidano G, Carbone A, Dalla-Favera R. Genetic basis of acquired immunodeficiency syndrome-related lymphomagenesis. J Natl Cancer Inst Monogr. 1998;(23):95–100. doi: 10.1093/oxfordjournals.jncimonographs.a024181. [DOI] [PubMed] [Google Scholar]
- 14.Said J, Cesarman E, Knowles D. Lymphadenopathy and the lymphoid neoplasms associated with HIV infection and other causes of immunosuppression. In: Orazzi AWL, Foucar K, Knowles DM, eds. Knowles' Neoplastic Hematopathology. Baltimore: Wolters Kuwer/Williams and Wilkins; 2014:584–658. [Google Scholar]
- 15.Carbone A. Emerging pathways in the development of AIDS-related lymphomas. Lancet Oncol. 2003;4(1):22–29. doi: 10.1016/s1470-2045(03)00957-4. [DOI] [PubMed] [Google Scholar]
- 16.Cesarman E. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett. 2011;305(2):163–174. doi: 10.1016/j.canlet.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel P, Hanson DL, Sullivan PS, et al. Adult and Adolescent Spectrum of Disease Project and HIV Outpatient Study Investigators. Incidence of types of cancer among HIV-infected persons compared with the general population in the United States, 1992-2003. Ann Intern Med. 2008;148(10):728–736. doi: 10.7326/0003-4819-148-10-200805200-00005. [DOI] [PubMed] [Google Scholar]
- 18.Swerdlow S, Campo E, Harris N, et al. Lyon, France: IARC Press; 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 19.Hans CP, Weisenburger DD, Vose JM, et al. A significant diffuse component predicts for inferior survival in grade 3 follicular lymphoma, but cytologic subtypes do not predict survival. Blood. 2003;101(6):2363–2367. doi: 10.1182/blood-2002-07-2298. [DOI] [PubMed] [Google Scholar]
- 20.Chadburn A, Chiu A, Lee JY, et al. Immunophenotypic analysis of AIDS-related diffuse large B-cell lymphoma and clinical implications in patients from AIDS Malignancies Consortium clinical trials 010 and 034. J Clin Oncol. 2009;27(30):5039–5048. doi: 10.1200/JCO.2008.20.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arvey A, Tempera I, Tsai K, et al. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12(2):233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostic AD, Ojesina AI, Pedamallu CS, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol. 2011;29(5):393–396. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strong MJ, Xu G, Morici L, et al. Microbial contamination in next generation sequencing: implications for sequence-based analysis of clinical samples. PLoS Pathog. 2014;10(11):e1004437. doi: 10.1371/journal.ppat.1004437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tierney RJ, Shannon-Lowe CD, Fitzsimmons L, Bell AI, Rowe M. Unexpected patterns of Epstein-Barr virus transcription revealed by a high throughput PCR array for absolute quantification of viral mRNA. Virology. 2015;474:117–130. doi: 10.1016/j.virol.2014.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young L, Alfieri C, Hennessy K, et al. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med. 1989;321(16):1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 30.Kersten MJ, Van Gorp J, Pals ST, Boon F, Van Oers MH. Expression of Epstein-Barr virus latent genes and adhesion molecules in AIDS-related non-Hodgkin’s lymphomas: correlation with histology and CD4-cell number. Leuk Lymphoma. 1998;30(5-6):515–524. doi: 10.3109/10428199809057564. [DOI] [PubMed] [Google Scholar]
- 31.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue SA, Labrecque LG, Lu QL, et al. Promiscuous expression of Epstein-Barr virus genes in Burkitt’s lymphoma from the central African country Malawi. Int J Cancer. 2002;99(5):635–643. doi: 10.1002/ijc.10372. [DOI] [PubMed] [Google Scholar]
- 33.Bell AI, Groves K, Kelly GL, et al. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt’s lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J Gen Virol. 2006;87(Pt 10):2885–2890. doi: 10.1099/vir.0.81906-0. [DOI] [PubMed] [Google Scholar]
- 34.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young P, Anderton E, Paschos K, White R, Allday MJ. Epstein-Barr virus nuclear antigen (EBNA) 3A induces the expression of and interacts with a subset of chaperones and co-chaperones. J Gen Virol. 2008;89(Pt 4):866–877. doi: 10.1099/vir.0.83414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haque T, Chaggar T, Schafers J, Atkinson C, McAulay KA, Crawford DH. Soluble CD30: a serum marker for Epstein-Barr virus-associated lymphoproliferative diseases. J Med Virol. 2011;83(2):311–316. doi: 10.1002/jmv.21953. [DOI] [PubMed] [Google Scholar]
- 37.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10(12):878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambinder RF. Gammaherpesviruses and “Hit-and-Run” oncogenesis. Am J Pathol. 2000;156(1):1–3. doi: 10.1016/S0002-9440(10)64697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bechtel D, Kurth J, Unkel C, Küppers R. Transformation of BCR-deficient germinal-center B cells by EBV supports a major role of the virus in the pathogenesis of Hodgkin and posttransplantation lymphomas. Blood. 2005;106(13):4345–4350. doi: 10.1182/blood-2005-06-2342. [DOI] [PubMed] [Google Scholar]
- 41.Mancao C, Altmann M, Jungnickel B, Hammerschmidt W. Rescue of “crippled” germinal center B cells from apoptosis by Epstein-Barr virus. Blood. 2005;106(13):4339–4344. doi: 10.1182/blood-2005-06-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roughan JE, Thorley-Lawson DA. The intersection of Epstein-Barr virus with the germinal center. J Virol. 2009;83(8):3968–3976. doi: 10.1128/JVI.02609-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dittmer DP, Hilscher CJ, Gulley ML, Yang EV, Chen M, Glaser R. Multiple pathways for Epstein-Barr virus episome loss from nasopharyngeal carcinoma. Int J Cancer. 2008;123(9):2105–2112. doi: 10.1002/ijc.23685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Razzouk BI, Srinivas S, Sample CE, Singh V, Sixbey JW. Epstein-Barr Virus DNA recombination and loss in sporadic Burkitt’s lymphoma. J Infect Dis. 1996;173(3):529–535. doi: 10.1093/infdis/173.3.529. [DOI] [PubMed] [Google Scholar]
- 45.Srinivas SK, Sample JT, Sixbey JW. Spontaneous loss of viral episomes accompanying Epstein-Barr virus reactivation in a Burkitt’s lymphoma cell line. J Infect Dis. 1998;177(6):1705–1709. doi: 10.1086/517427. [DOI] [PubMed] [Google Scholar]
- 46.Paiardini M, Müller-Trutwin M. HIV-associated chronic immune activation. Immunol Rev. 2013;254(1):78–101. doi: 10.1111/imr.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong JJ, Amancha PK, Rogers K, Ansari AA, Villinger F. Spatial alterations between CD4(+) T follicular helper, B, and CD8(+) T cells during simian immunodeficiency virus infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. J Immunol. 2012;188(7):3247–3256. doi: 10.4049/jimmunol.1103138. [DOI] [PMC free article] [PubMed] [Google Scholar]