LETTER
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) identification (ID) of Candida yeast isolates is fast and reliable, but the optimal workflow is debated (1, 2). Two different steel target plates are available for the Microflex system (Bruker Daltonics, Bremen, Germany). Ground steel targets (GST) have a highly regular fine surface structure, whereas polished steel targets (PST) are virtually free of any surface structure (3). While fluids sometimes spread to adjacent spots on GST, this is rare on PST. Therefore, PST is favored for routine microbial identifications (4). Here, GST and PST are compared for MALDI-TOF MS-based yeast identification using direct transfer (DT).
Two hundred six Candida isolates previously identified by phenotypic methods (121 C. albicans, 36 C. glabrata; 15 C. parapsilosis, 14 C. tropicalis, 11 C. krusei, 6 C. kefyr, 2 C. guillermondii, and 1 C. norvegiensis isolate) were identified by MALDI-TOF MS in a single laboratory on both GST and PST using the DT method. Briefly, Candida isolates were cultured overnight on blood agar, and each isolate was smeared from the same colony onto two sequential spots on both targets with a wooden toothpick. Subsequently, 1 μl of α-cyano-4-hydroxycinnamic acid (HCCA) matrix (Bruker Daltonics) was overlaid on the spots. Spots were allowed to dry and analyzed by MALDI-TOF MS (MALDI Biotyper [MBT]; Bruker Daltonics) according to the manufacturer's recommendations, using the MBT-BDAL-5627 MSP library from the commercial Bruker Daltonics (BDAL) main spectrum profile (MSP) database.
GST resulted in higher maximum score values than PST (Table 1). Using the manufacturer's recommended cutoff values, higher frequencies of species level identification were found with GST than with PST (P < 0.05, Wilcoxon signed-rank test) (Table 1). To exclude any possible bias linked to this particular laboratory, a set of 15 Candida isolates (4 C. albicans, 4 C. glabrata, and 2 C. dubliniensis isolates and 1 each of C. parapsilosis, C. tropicalis, C. guilliermondii, C. krusei, and C. lusitaniae) were distributed among 5 other laboratories. These Candida isolates were subcultured according to the routine method of each particular laboratory and analyzed on GST and PST using DT and extended direct transfer (eDT), also known as on-plate formic acid extraction (5). The manufacturer's recommended cutoff levels for score values for database matches (using the MBT-BDAL-5627 MSP library) were used to classify identifications as no ID, genus-level ID, or species-level ID. For each isolate, the change of ID level when testing on GST versus PST was assessed using a weighted change in ID level, as this enabled a comparison of the changes in score values in a clinically relevant fashion. Improvement of the ID level from no ID using PST to species-level ID using GST was scored as +3 (very major ID level improvement), from no ID on PST to genus level on GST as +2 (major ID level improvement), and from genus level ID on PST to species level on GST as +1 (ID level improvement). Decreases in ID levels using GST versus PST were scored similarly but with negative signs. No change in ID level was scored as 0. Similar weighted changes in ID levels were calculated for the results obtained with DT on GST using the standard MBT-BDAL-5627 MSP library versus the standard MBT-BDAL-5627 MSP library supplemented with 284 extra yeast MSPs (see below).
TABLE 1.
Distribution of score values obtained for 206 Candida isolates using direct transfer to different types of target plate
| Candida species | No. of isolates with indicated score valuea/total no. tested (%) on: |
|||||
|---|---|---|---|---|---|---|
| Polished steel target |
Ground steel target |
|||||
| <1.700 | 1.700–1.999 | ≥2.000 | <1.700 | 1.700–1.999 | ≥2.000 | |
| C. albicans | 71/121 (59) | 45/121 (37) | 5/121 (4) | 0/121 (0) | 71/121 (59) | 50/121 (41) |
| C. glabrata | 22/36 (61) | 7/36 (19) | 7/36 (19) | 3/36 (8) | 9/36 (25) | 24/36 (67) |
| C. parapsilosis | 9/15 (60) | 5/15 (33) | 1/15 (7) | 2/15 (13) | 9/15 (60) | 4/15 (27) |
| C. tropicalis | 9/14 (64) | 5/14 (35) | 0/14 (0) | 2/14 (14) | 10/14 (71) | 2/14 (14) |
| C. krusei | 3/11 (27) | 5/11 (45) | 3/11 (27) | 1/11 (9) | 2/11 (18) | 8/11 (73) |
| C. kefyr | 1/6 (17) | 3/6 (50) | 2/6 (33) | 2/6 (33) | 1/6 (17) | 3/6 (50) |
| C. guillermondii | 2/2 (100) | 0/2 (0) | 0/2 (0) | 0/2 (0) | 1/2 (50) | 1/2 (50) |
| C. norvegiensis | 1/1 (100) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 1/1 (100) | 0/1 (0) |
Score values of ≥2.000 represent species-level IDs, score values between 1.700 and 1.999 represent genus-level IDs, and score values of <1.700 represent no ID.
Among the 5 laboratories, the species-level identifications improved from a mean of 5.4 per 15 isolates using DT on PST to a mean of 8.2 per 15 isolates using DT on GST (mean difference of 2.8 species-level IDs per 15 isolates, with GST having better results) (Fig. 1A). The improvement of ID levels differed between laboratories, but in all 5 laboratories, the mean weighted ID level change favored GST when DT was used (Fig. 1B). In contrast, using eDT, identifications on PST performed similarly to identifications on GST (Fig. 1A). However, yeast identification using DT on GST performed comparably to eDT on either PST or GST (Fig. 1A).
FIG 1.
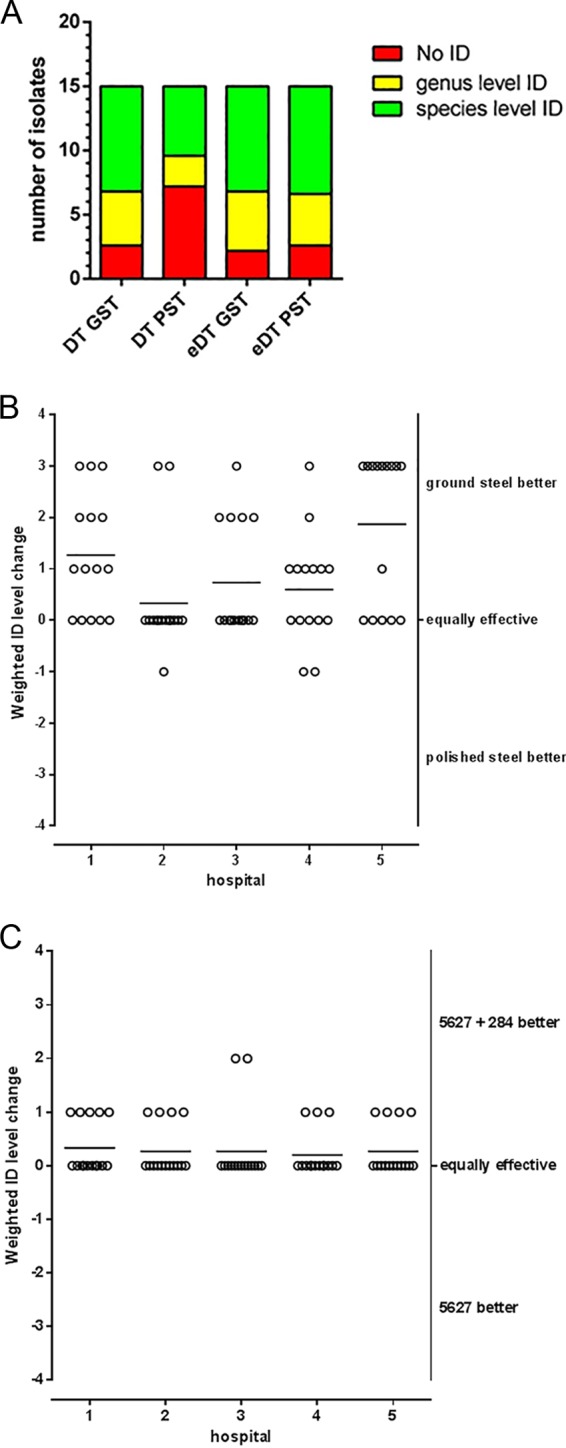
(A) MALDI-TOF MS yeast identification using DT on GST is, on average, as effective as eDT on PST. Mean ID level frequencies for the 15 Candida isolates when tested by 5 independent laboratories are shown. Data are for target plate and method (x axis) versus the numbers of isolates with the indicated results (y axis). Green bars, score value of ≥2.000; yellow bars, score value between 1.700 and 1.999; red bars, score value of <1.7 or no peaks; GST, ground steel target; PST, polished steel target; DT, direct transfer; eDT, extended direct transfer (on-plate formic acid extraction; see reference 5 for details). (B) Yeast identification by MALDI-TOF MS using the commercial Bruker Daltonics database 5627 and DT performed better on GST than on PST in all 5 participating laboratories. Weighted ID level changes (for explanation, see the text) for the five independent laboratories (15 isolates per hospital) are shown. Data are for the hospital (x axis) versus the weighted change in ID level (y axis) per isolate (circles) on GST versus PST. Horizontal lines indicate the mean weighted change in ID level for the 15 isolates per hospital. (C) Database improvement resulted in improved MALDI-TOF MS identifications using DT on GST in all 5 participating laboratories (15 isolates per hospital). Weighted ID level changes (for explanation see the text) for the 5 independent laboratories are shown. Data are for the hospital (x axis) versus the weighted change in ID level (y axis) per isolate (circles) using database 5627 versus database 5627 plus 284 extra yeast MSPs (see the text). Horizontal lines indicate the mean weighted change in ID level for the 15 isolates per hospital.
Several reports have demonstrated that yeast database improvements were associated with better yeast identification results (5–7). However, these investigations were performed with older versions of the main spectrum library that, compared to the MBT-BDAL-5627 MSP library, only contained a few yeast spectra. Here, we investigated whether further database expansion of the MBT-BDAL-5627 MSP library results in better identifications: 226/512 yeast main spectra from the Centraalbureau voor Schimmelcultures (CBS) 512 database are present in the current MBT-BDAL-5627 MSP library, whereas the remaining 284 main spectra from the CBS 512 library are not (5). Using DT on GST, the addition of these 284 spectra further improved the identification results for the 15 clinical yeast isolates (Fig. 1C).
In summary, MALDI-TOF MS identification of clinical yeast isolates using DT on GST performs as well as eDT on PST. Given that the former method is both faster and cheaper, it may be more suitable for routine yeast identification.
REFERENCES
- 1.Clark AE, Kaleta EJ, Arora A, Wolk DM. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorton RL, Seaton S, Ramnarain P, McHugh TD, Kibbler CC. 2014. Evaluation of a short, on-plate formic acid extraction method for matrix-assisted laser desorption ionization–time of flight mass spectrometry-based identification of clinically relevant yeast isolates. J Clin Microbiol 52:1253–1255. doi: 10.1128/JCM.03489-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 2012. Bruker guide to MALDI sample preparation, revision 2. Bruker Daltonik GmbH, Bremen, Germany: https://www.bruker.com/fileadmin/user_upload/8-PDF-Docs/Separations_MassSpectrometry/InstructionForUse/Bruker_Guide_MALDI_Sample_Preparation_Rev2.pdf. [Google Scholar]
- 4.Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol 48:1549–1554. doi: 10.1128/JCM.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlek A, Kolecka A, Khayhan K, Theelen B, Groenewald M, Boel E, Boekhout T. 2014. Interlaboratory comparison of sample preparation methods, database expansions, and cutoff values for identification of yeasts by matrix-assisted laser desorption ionization-time of flight mass spectrometry using a yeast test panel. J Clin Microbiol 52:3023–3029. doi: 10.1128/JCM.00563-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Carolis E, Vella A, Vaccaro L, Torelli R, Posteraro P, Ricciardi W, Sanguinetti M, Posteraro B. 2014. Development and validation of an in-house database for matrix-assisted laser desorption ionization–time of flight mass spectrometry-based yeast identification using a fast protein extraction procedure. J Clin Microbiol 52:1453–1458. doi: 10.1128/JCM.03355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernhard M, Weig M, Zautner AE, Gross U, Bader O. 2014. Yeast on-target lysis (YOTL), a procedure for making auxiliary mass spectrum data sets for clinical routine identification of yeasts. J Clin Microbiol 52:4163–4167. doi: 10.1128/JCM.02128-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


