Abstract
Fifteen percent of all methicillin-resistant Staphylococcus aureus (MRSA) clonal complex 398 (CC398) human carriers detected in The Netherlands had not been in direct contact with pigs or veal calves. To ensure low MRSA prevalence, it is important to investigate the likely origin of this MRSA of unknown origin (MUO). Recently, it was shown that CC398 strains originating from humans and animals differ in the presence of specific mobile genetic elements (MGEs). We hypothesized that determining these specific MGEs in MUO isolates and comparing them with a set of CC398 isolates of various known origin might provide clues to their origin. MUO CC398 isolates were compared to MRSA CC398 isolates obtained from humans with known risk factors, a MRSA CC398 outbreak isolate, livestock associated (LA) MRSA CC398 isolates from pigs, horses, chickens, and veal calves, and five methicillin-susceptible Staphylococcus aureus (MSSA) CC398 isolates of known human origin. All strains were spa typed, and the presence or absence of, scn, chp, φ3 int, φ6 int, φ7 int, rep7, rep27, and cadDX was determined by PCRs. The MRSA CC398 in humans, MUO, or MRSA of known origin (MKO) resembled MRSA CC398 as found in pigs and not MSSA CC398 as found in humans. The distinct human MSSA CC398 spa type, t571, was not present among our MRSA CC398 strains; MRSA CC398 was tetracycline resistant and carried no φ3 bacteriophage with scn and chp. We showed by simple PCR means that human MUO CC398 carriers carried MRSA from livestock origin, suggestive of indirect transmission. Although the exact transmission route remains unknown, direct human-to-human transmission remains a possibility as well.
INTRODUCTION
In The Netherlands, the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) is low (1), and Dutch MRSA strains display broad clonal diversity (2). One exception is the livestock-associated (LA) clone (clonal complex 398 [CC398]), a major clonal reservoir in pigs and veal calves (3) and subsequently in people with occupational exposure to animals. The reported number of MRSA CC398 strains has been around 40% of MRSA strains reported to the Dutch MRSA surveillance since 2008 (2, 4). However, only 78% of reported CC398 strains are found through screening of patients with direct (occupational) contact to pigs or veal calves at hospital admission (a risk factor introduced in 2006) (5).
The remaining MRSA CC398 carriers do not comply with the described risk factors in the Dutch MRSA guideline: industrial contact with live pigs, veal calves, or broiler chickens regardless of whether or not this contact was occupational and/or residence of the individual on such a farm. Currently 15% (352/2,312) of all Dutch and 15% (24/164) of all Danish MRSA CC398 carriers have not been in direct contact with pigs or veal calves (2, 4). In The Netherlands, these MRSA CC398 carriers are considered a MRSA of unknown origin (MUO) subgroup (MUO CC398), with MUO being any MRSA reported to the MRSA surveillance without known risk factors as defined in the Dutch MRSA guideline (4).
The reservoir or transmission route of MUO CC398 still remains unknown: possible transmission routes are direct animal-to-human transmission of animal sources not included as risk factors in the MRSA guideline (due to being unknown or having a limited effect on the population as a whole), indirect animal-to-human transmission through the environment, e.g., by dust or air vehicle (6, 7) or animal products such as meat (8), or human-to-human transmission (9). Hospital outbreaks of CC398 have been described, illustrating the potential of human-to-human transmission by this clonal complex (10). Although the general thought is that long-term colonization of CC398 strains in humans is rare, it was recently shown that CC398 strains from animal origin can survive in a human nose for at least 21 days, suggesting their ability to colonize humans (11). MUO CC398 is therefore an important topic, and the necessity to elucidate the origin of MRSA CC398 strains in humans without direct contact to pigs or veal calves (MUO CC398) is clear.
From the genomic analyses on CC398 isolates of different origins, it can be concluded that the origin of CC398 is most likely human (12, 13). There are indications that methicillin-susceptible Staphylococcus aureus (MSSA) CC398 switched hosts in the past as a result of human-animal interactions (12, 14) and that it adapted to animals by losing several mobile genetic elements (MGEs) while gaining other MGEs, including resistance to tetracyclines and methicillin, before being reintroduced in humans as MRSA (3, 15).
McCarthy et al. showed that CC398 strains from humans in contact with animals differed from strains isolated from humans without contact with animals. The differences were seen in MGE-located genes, e.g., φ3 int, chp, scn, rep27, φ7 int, and cadDX for humans and rep7 and φ6 int for pigs, in addition to genes encoding resistance to tetracycline and trimethoprim (14). We therefore hypothesized that the presence of these MGE-encoding genes with resistance to tetracycline and trimethoprim-sulfamethoxazole might be used as a cheap and fast method to compare MUO CC398 isolates with isolates from humans (MSSA and MKO CC398) and animals (MRSA CC398) to predict the origin of the MUO CC398 in The Netherlands.
We showed that MUO and MKO isolates resembled CC398 isolates of animal origin more closely than those of human origin, indicating that these MUO CC398 isolates most probably originated from livestock. (This work was presented in part as a poster at the International Symposium on Staphylococci and Staphylococcal Infections (ISSSI), Lyon, France, 2012 [16].)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
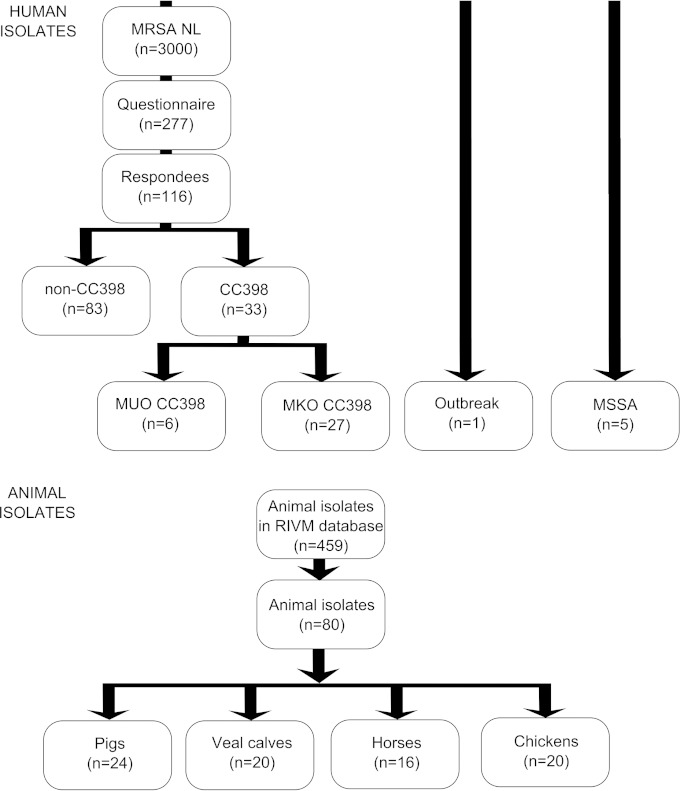
In total, 119 isolates were included in the study (Fig. 1). All isolates were predicted to have a CC398 background, based on multilocus variable-number tandem-repeat analysis (MLVA) typing (http://www.mlva.net/). MLVA is the choice of the National Institute for Public Health and the Environment (RIVM), due to costs and also the agreement between the results for MLVA and multilocus sequence type (MLST). Only sequence types (STs) belonging to the same MLST clonal complex were grouped by MLVA. Furthermore, the spa types associated with MLST clonal complexes show a remarkable agreement with those of the MLVA complexes (17). The MLVA complexes were therefore named in accordance with the MLST complexes. The MLVA complex 398 is thus equal to the MLST clonal complex 398. The isolates were all from The Netherlands and from 2009, except for an outbreak strain from 2007 and five MSSA isolates from human origin, previously described and isolated at the Erasmus MC in the period of 1998-1999 and 2002 (13, 18). All S. aureus CC398 isolates were stored at −80°C and grown on sheep blood agar plates (RBS) (Becton, Dickinson & Co., Belgium) at 37°C overnight.
FIG 1.
Flowcharts. (Top) Human isolates. MUO is MRSA without known risk factors as described by the Dutch national guideline. MKO is MRSA with known risk factors as described by the Dutch national guideline. MKO CC398 represents a pig or veal calf farmer, a person with direct contact to pigs and/or veal calves and/or who lives on a pig or veal calf farm, or a broiler chicken handler. (Bottom) Animal isolates. RIVM, the National Institute for Public Health and the Environment, Bilthoven, The Netherlands.
Bacterial strain selection from animals.
The 80 MRSA strains of animal origin included in this study were previously collected from livestock: pigs, n = 24; veal calves, n = 20; chickens, n = 20; and horses, n = 16. The pig isolates were from apparently healthy animals and originated from eight different farms across The Netherlands that were screened as part of a pilot for a later study by Broens et al. (19). The healthy veal calves were sampled at arrival on three Dutch farms (20). The horse strains were nearly all (94%) samples from diseased horses that visited the Utrecht University equine clinic, the remaining 6% being from healthy horses. The chicken isolates were obtained from a study in six broiler slaughterhouses, where broilers from 40 flocks arriving at the slaughterhouses were sampled in the pharynx after stunning (21). S. aureus isolates were spa typed by the RIVM according to the method of Harmsen et al. (22). From the available livestock MRSA isolates (n = 459), the largest variability in spa types was chosen (n = 80) (Fig. 2); whether an isolate was from screening or a clinical case was not a selection criterion. This resulted in a selection with both screening and clinical isolates.
FIG 2.
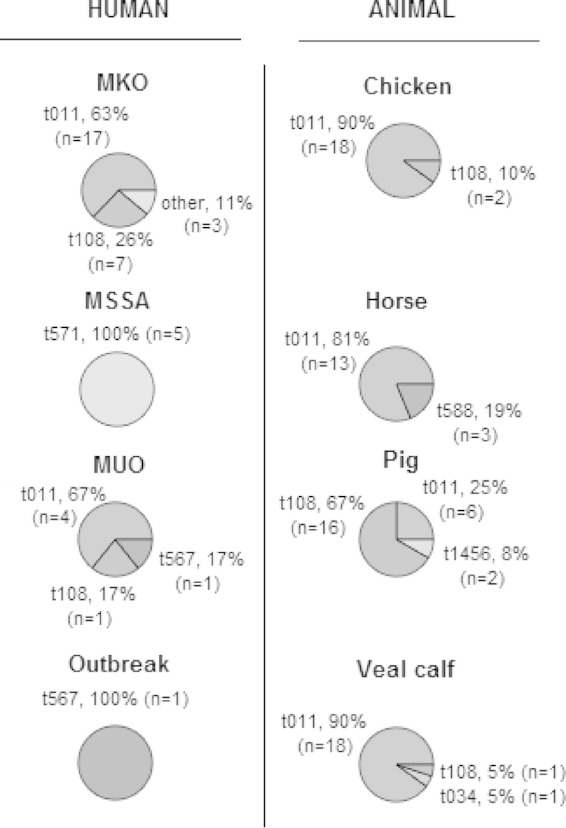
Selected spa types for human and animal groups. MKO, MRSA of known origin (known risk factors described in the Dutch MRSA guideline); MSSA, methicillin-susceptible Staphylococcus aureus; MUO, MRSA of unknown origin (unknown risk factors not described in the Dutch MRSA guideline). The outbreak isolate was described by Wulf et al. (9).
Bacterial strain selection from humans.
The MRSA strains of human origin included an outbreak strain (n = 1), MUO (n = 6), and MKO (n = 27). The outbreak strain reported in 2007 was chosen because it caused nine secondary cases (both patients and health care workers) in a single Dutch hospital after MRSA was cultured from a diabetic foot ulcer of a patient on a surgical ward (10). Both MUO (n = 6) and MKO (n = 27) were from a previous study, in which an extended questionnaire was send to MRSA carriers. Five MSSA isolates were also of human origin. These isolates were previously described and isolated at the Erasmus MC in the period of 1998-1999 and 2002 (13, 18).
Extended questionnaire study.
Around 3,000 MRSA strains have been reported to the Dutch national MRSA surveillance by medical microbiological laboratories from The Netherlands with epidemiological data on applicable risk groups (2, 5). Potential MUO carriers reported to the surveillance were approached by extended questionnaire. The questionnaire was set up to determine the known risk factors for MRSA, as described in the Dutch Working Party on Infection Prevention (WIP) guideline on MRSA (see Table S1 in the supplemental material) (4), and also included further questions on risk factors as described in the literature, as found by a PubMed search up to 1 January 2010, using search keywords “MRSA” and “risk factor” (see Table S2 in the supplemental material).
S. aureus genotyping, detection of expression of β-hemolysin, and DNA isolation.
After an overnight culture on RBS plates, the hemolysin patterns were determined to detect the expression of β-hemolysin. No expression of β-hemolysin indicates the insertion of the φ3 bacteriophage into the bacterial genome, as φ3 inserts on the site that codes for β-hemolysin (23). DNA was isolated using a MagNA Pure system (Roche) according to the protocol supplied by the manufacturer.
Mobile genetic elements.
The presence or absence of MGEs was determined by PCRs specific for cadDX, φ3 int, scn, chp, rep7, rep27, φ6 int, and φ7 int (14, 23, 24). Primers for φ3 int (forward primer, TCCGGCTTCTTTGAAAATGT; reverse primer, CCGGAAAACCTACGAAGTCA; amplicon size, 220 to 323 bp; annealing temperature, 50°C) and cadDX (forward primer, TGATGTGATCTGTGTACATGAGGA; reverse primer, TGATGTGAAGTTGAAGCAACA; amplicon size, 207 bp; annealing temperature, 60°C) were designed with Primer3 software (http://frodo.wi.mit.edu/). All amplified PCR products were visualized by agarose gel (1.2%) electrophoresis (see Tables S3 and S4 in the supplemental material).
Antimicrobial susceptibility.
To determine the antimicrobial susceptibility of S. aureus strains, the standard disc diffusion method was applied using Oxoid antimicrobial susceptibility test discs (Thermo Fisher Scientific, Waltham, MA, USA) on Mueller-Hinton (MH) agar plates. The Clinical and Laboratory Standards Institute (CLSI) breakpoints were used for tetracycline (zone diameter breakpoints: susceptible [S], ≥19 mm; intermediate [I], 15 to 18 mm; resistant [R], ≤14 mm) and trimethoprim-sulfamethoxazole (zone diameter breakpoints: S, ≥16 mm, I, 11 to 15 mm; R, ≤10 mm) (25).
Statistical analysis.
Statistical analysis was performed with SAS Enterprise Guide software (version 4.2; SAS Institute, Inc., NC, USA) using 2 × 2 tables and Fisher's exact test. P values of <0.01 were considered significant to correct for multiple testing. A comparison was made between animal and human hosts and between human epidemiological subgroups. Isolates were clustered transversally, using the Jaccard coefficient, on the presence of MGEs, β-hemolysin expression, and susceptibility to tetracycline and trimethoprim-sulfamethoxazole. The dendrogram was created based on unweighted-pair group method using average linkages (UPGMA) with Jaccard similarity.
RESULTS
A total of 277 suspected MUO carriers from all of The Netherlands were approached by an extended questionnaire; 42% (116) responded and 33 were defined as CC398 carriers. Of these 33 CC398 strains, 6 were MUO (CC398), and these were confirmed to have had no contact with pigs, veal calves, or (broiler) chickens in the year before questioning. The MUO CC398 carriers were found to reside in the Dutch province Noord Brabant, where there is generally more pig farming.
All MUO and MKO CC398 strains were distinctly different from human MSSA CC398 strains based not only on spa type, but also on β-hemolysin expression, tetracycline resistance, and the lack of φ3 int, scn, and chp genes. The human MRSA CC398 strains resembled animal MRSA CC398 strains (Fig. 3). The presence of cadDX and rep27, which are considered human-associated genetic markers as they are highly prevalent in human MSSA and significantly less prevalent in animal MRSA (14), were absent in the MUO strains and few in the MKO strains: cadDX, 0/6 for MUO and 9/27 for MKO; and rep27, 0/6 for MUO and 4/27 for MKO. In horse and pig isolates, cadDX was almost absent, while in veal calf and chicken isolates, cadDX was found frequently: 16/20 (80%) for veal calves and 12/20 (60%) for chickens. rep27 was absent in horse and veal calf isolates and only incidentally found in chickens and pigs: 8% (2/24) for pigs and 20% (4/20) for chickens. All 119 isolates of both MRSA and MSSA were similar in their full susceptibility to trimethoprim-sulfamethoxazole. Also, there was no significant difference between the MUO and MKO strains in rep7, rep27, φ6 int, and cadDX content, resulting in MUO and MKO strains clustering together, despite some minor differences in MGE content.
FIG 3.

Results of β-hemolysin screening, PCR typing, and susceptibility testing. Isolates were clustered transversally, using the Jaccard coefficient, on the presence of MGEs, β-hemolysin expression, and susceptibility to tetracycline and trimethoprim-sulfamethoxazole. The dendrogram was created based on UPGMA with Jaccard similarity. The epidemiological subtypes in humans are MKO (MRSA of known origin: MRSA with known risk factors for acquisition) and MUO (MRSA of unknown origin: MRSA with unknown risk factors for acquisition). MUO 2007 are MUO isolates according to the 2007 guideline definition but no longer included under the 2012 guideline definition. MUO 2012 are MUO isolates according to the current guideline of December 2012. Outbreak represents an isolate involved from a MRSA CC398 outbreak in a Dutch hospital as described by Wulf et al. (9). Mobile genetic elements: chp (gene encoding chemotaxis-inhibiting protein [CHIPS], which is found in the φ3 bacteriophage that contains the immune evasion complex [IEC] that chp is sometimes part of); scn (gene encoding staphylococcal complement inhibitor [SCIN], which is found in the φ3 bacteriophage that contains the IEC that scn is sometimes part of); φ3 int (integrase gene of bacteriophage 3); φ6 int (integrase gene of bacteriophage 6); φ7 int (integrase gene of bacteriophage 7); rep7 (replication protein 7); rep27 (replication protein 27); and cadDX (operon of gene cadX [cad operon regulatory protein], which encodes resistance against the heavy metal cadmium). Antimicrobial susceptibility: tetracycline susceptibility testing; Trim./sulfa., trimethoprim-sulfamethoxazole susceptibility testing.
When the combined data for MUO and MKO isolates were compared to data for the animal isolates, it was clear that the human isolates were less often φ6 int positive than the MRSA isolates from veal calves or horses (P < 0.01), more often rep7 positive than the isolates from horses (P < 0.01), more often rep27 positive than the isolates from pigs (P < 0.01), and less often cadDX positive than the isolates from veal calves and chickens (P < 0.01). No significant differences between the MRSA isolates from the human subgroups (MUO, MKO, and outbreak) were found for rep27. Interestingly, the hospital outbreak strain lacked any previously mentioned human- or pig-associated markers (rep7, rep27, φ3, φ6 int, and cadDX) but displayed tetracycline resistance. MGE variation within a single spa type was observed for human and animal isolates (Fig. 3).
DISCUSSION
The human MRSA CC398 isolates (MUO and MKO) in this study resembled animal MRSA CC398 isolates more than they resembled human MSSA CC398 isolates, because they were β-hemolysin producers and tetracycline resistant, had similar MGE patterns, and had spa types similar to those found in animals. Furthermore, our MUO isolates almost always clustered together with MKO isolates in cluster analysis. The similarity between MUO CC398, MKO CC398, and animal MRSA CC398 strains suggests that these MUO CC398 strains belong to the same MRSA clade originating in animals and that these MUO CC398 strains are not part of the MSSA CC398 clade detected in humans. Stegger et al. found two distinct phylogenetic clades based on single nucleotide polymorphisms (SNPs), revealing a basal human clade and a more derived livestock clade (26). Although no whole-genome sequencing or SNP analysis was done, the outcome of our cheaper and quicker MGE-based method strongly suggests that these MUO CC398 strains belong to the livestock clade with MKO CC398 and MRSA CC398 from animals and not to the MSSA CC398 clade found in humans. The lack of risk factors in our MUO CC398 carriers suggests spread of animal MRSA CC398 strains by means other than those currently described in the MRSA guideline.
The exact mode of transmission remains unknown. An indirect route of transmission is the most likely mode of transmission for these MUO CC398 strains, taking into consideration where these MUO CC398 carriers live, their lack of contact with pigs and veal calves, and also their lack of contact with horses and chickens. Since living in areas with a high density of pigs (6) and private farm visits (27) were risk factors for livestock-associated MRSA carriage, modes of indirect transmission are most likely through contamination of areas in which people live and interact. Considering the survival of S. aureus bacteria in the environment and subsequent spread by air over large distances (7), transmission by air is a possibility (28), as well as transmission by vectors such as rodents (29). Nevertheless, transmission by human-to-human contact cannot be ruled out: of the six MUO CC398 carriers investigated by extended questionnaire in this study, one MUO CC398 carrier had had contact with a MRSA carrier (who or which MRSA type was unknown) outside the family or household, while another had visited a farm without contact to animals. Neither is currently considered an at risk event. In the Dutch guideline, MRSA-positive household members are considered a risk, but contact in the community outside the household or hospital is not.
We know that a pig MRSA CC398 lacking φ3 can survive up until 21 days in a human nose (11), whereas φ3 is currently considered to be the marker for human host adaptation (12). The successful outbreak isolate reported by Wulf et al. (10) lacked φ3 as well. Human host adaptation is explained by more than φ3 alone, or host adaptation might not have to be as extensive to facilitate transmission. In regard to the outbreak isolate, further research is necessary to determine what makes this outbreak isolate so different and successful compared to other human MRSA isolates.
Despite no significant difference between MUO and MKO for MGE-encoding genes, there were slight differences observed for cadDX and rep7 between the MUO and MKO. Furthermore, rep7-positive isolates were as common in MKO as in animal strains, unlike MUO, which showed significantly less rep7 than among pig strains (P < 0.01). rep7 and rep27 genes are typical of small plasmids carrying resistance genes, and rep7 is reported to be associated with the tetracycline resistance gene, tetK (30). We also observed MGE variation within single spa types within humans or animals, as can best be seen in t011 and t108, fitting the known relative stability of MGE (31).
As for the limitations of this study, we do not know whether our isolates were obtained from persistent MRSA carriers or transient carriers (contaminated humans). Follow-up data are important to better understand host adaptation, but since this was a retrospective study, carrier data over time was unfortunately not available. This study's strength is the questionnaire that allowed discrimination between MRSA CC398 carriers with and without known risk factors, regardless of guideline changes. The number of MUO CC398 carriers in this study are few as 28% (33/116) of respondents were CC398 carriers, and only 21% (6/33) fit the MUO definition. However, the number of MUO CC398 isolates per year in The Netherlands is just over 5%, which means on average an additional 150 Dutch people with a MRSA CC398 strain while lacking the risk factors as described in the Dutch MRSA guideline (4).We showed by simple PCR means that the MUO CC398 carriers in this study carry MRSA CC398 strains of livestock origin. This finding is suggestive of an indirect transmission route, possibly the environment (air or water) or through fomites, but we cannot rule out direct human-to-human transmission. Although, the reported numbers of MUO CC398 strains in The Netherlands are currently still small, the problem may increase, giving rise to more CC398 transmission and human host adaptation.
Supplementary Material
ACKNOWLEDGMENTS
We thank all colleagues who provided strains, accompanying data, and advice.
This study was financed by ZonMw (The Netherlands Organization for Health Research and Development).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02702-14.
REFERENCES
- 1.Bode LG, Wertheim HF, Kluytmans JA, Bogaers-Hofman D, Vandenbroucke-Grauls CM, Roosendaal R, Troelstra A, Box AT, Voss A, van Belkum A, Verbrugh HA, Vos MC. 2011. Sustained low prevalence of meticillin-resistant Staphylococcus aureus upon admission to hospital in The Netherlands. J Hosp Infect 79:198–201. doi: 10.1016/j.jhin.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Lekkerkerk WS, van de Sande-Bruinsma N, van der Sande MA, Tjon ATA, Groenheide A, Haenen A, Timen A, van den Broek PJ, van Wamel WJ, de Neeling AJ, Richardus JH, Verbrugh HA, Vos MC. 2012. Emergence of MRSA of unknown origin in the Netherlands. Clin Microbiol Infect 18:656–661. doi: 10.1111/j.1469-0691.2011.03662.x. [DOI] [PubMed] [Google Scholar]
- 3.Cuny C, Friedrich A, Kozytska S, Layer F, Nubel U, Ohlsen K, Strommenger B, Walther B, Wieler L, Witte W. 2010. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int J Med Microbiol 300:109–117. doi: 10.1016/j.ijmm.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Werkgroep Infectie Preventie (WIP). 2012. WIP guideline MRSA. Werkgroep Infectiepreventie. http://www.rivm.nl/Documenten_en_publicaties/Professioneel_Praktisch/Richtlijnen/Infectieziekten/WIP_Richtlijnen/Actuele_WIP_Richtlijnen/Ziekenhuizen/WIP_richtlijn_MRSA_ZKH.
- 5.Haenen A, Pluister CN, van Luit M, Bosch T, Heck MEOC, de Greeff S, Neeling AJ, et al. 2012. Surveillance of MRSA in The Netherlands in 2011, p 198–203. In Infectieziekten Bulletin, vol 23, no 7 Netherlands National Institute for Public Health and the Environment, Bilthoven, The Netherlands. (In Dutch.) http://www.rivm.nl/dsresource?objectid=rivmp:185877&type=org&disposition=inline&ns_nc=1. [Google Scholar]
- 6.Feingold BJ, Silbergeld EK, Curriero FC, van Cleef BA, Heck ME, Kluytmans JA. 2012. Livestock density as risk factor for livestock-associated methicillin-resistant Staphylococcus aureus, the Netherlands. Emerg Infect Dis 18:1841–1849. doi: 10.3201/eid1811.111850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz J, Friese A, Klees S, Tenhagen BA, Fetsch A, Rosler U, Hartung J. 2012. Longitudinal study of the contaminantion of air and of soil surfaces in the vicinity of pig barns by livestock-associated methicillin-resistant Staphylococcus aureus. Appl Environ Microbiol 78:5666–5671. doi: 10.1128/AEM.00550-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buyukcangaz E, Velasco V, Sherwood JS, Stepan RM, Koslofsky RJ, Logue CM. 2013. Molecular typing of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) isolated from animals and retail meat in North Dakota, United States. Foodborne Pathog Dis 10:608–617. doi: 10.1089/fpd.2012.1427. [DOI] [PubMed] [Google Scholar]
- 9.Cuny C, Kock R, Witte W. 2013. Livestock associated MRSA (LA-MRSA) and its relevance for humans in Germany. Int J Med Microbiol 303:331–337. doi: 10.1016/j.ijmm.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Wulf MW, Markestein A, van der Linden FT, Voss A, Klaassen C, Verduin CM. 2008. First outbreak of methicillin-resistant Staphylococcus aureus ST398 in a Dutch hospital, June 2007. Euro Surveill 13(9):pii=8051 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8051. [PubMed] [Google Scholar]
- 11.Slingerland BC, Tavakol M, McCarthy AJ, Lindsay JA, Snijders SV, Wagenaar JA, van Belkum A, Vos MC, Verbrugh HA, van Wamel WJ. 2012. Survival of Staphylococcus aureus ST398 in the human nose after artificial inoculation. PLoS One 7:e48896. doi: 10.1371/journal.pone.0048896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3(1):e00305–11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Belkum A, Melles DC, Peeters JK, van Leeuwen WB, van Duijkeren E, Huijsdens XW, Spalburg E, de Neeling AJ, Verbrugh HA, Dutch Working Party on Surveillance and Research of MRSA-SOM. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg Infect Dis 14:479–483. doi: 10.3201/eid1403.070760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy AJ, van Wamel W, Vandendriessche S, Larsen J, Denis O, Garcia-Graells C, Uhlemann AC, Lowy FD, Skov R, Lindsay JA. 2012. Staphylococcus aureus CC398 clade associated with human-to-human transmission. Appl Environ Microbiol 78:8845–8848. doi: 10.1128/AEM.02398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huijsdens XW, van Dijke BJ, Spalburg E, van Santen-Verheuvel MG, Heck ME, Pluister GN, Voss A, Wannet WJ, de Neeling AJ. 2006. Community-acquired MRSA and pig-farming. Ann Clin Microbiol Antimicrob 5:26. doi: 10.1186/1476-0711-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.2012 Poster no. P 12–157. 15th International Symposium on Staphylococci and Staphylococcal Infections [ISSSI], Lyon, France, 26 to 30 August 2012. [Google Scholar]
- 17.Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, van Santen-Verheuvel MG, van der Heide HG, Grundmann H, Heck ME, de Neeling AJ. 2009. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS One 4:e5082. doi: 10.1371/journal.pone.0005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Wamel WJ, Hansenova Manaskova S, Fluit AC, Verbrugh H, de Neeling AJ, van Duijkeren E, van Belkum A. 2010. Short term micro-evolution and PCR-detection of methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398. Eur J Clin Microbiol Infect Dis 29:119–122. doi: 10.1007/s10096-009-0816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broens EM, Graat EA, Van der Wolf PJ, Van de Giessen AW, De Jong MC. 2011. Prevalence and risk factor analysis of livestock associated MRSA-positive pig herds in The Netherlands. Prev Vet Med 102:41–49. doi: 10.1016/j.prevetmed.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Graveland H, Wagenaar JA, Verstappen KM, Oosting-van Schothorst I, Heederik DJ, Bos ME. 2012. Dynamics of MRSA carriage in veal calves: a longitudinal field study. Prev Vet Med 107:180–186. doi: 10.1016/j.prevetmed.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Mulders MN, Haenen AP, Geenen PL, Vesseur PC, Poldervaart ES, Bosch T, Huijsdens XW, Hengeveld PD, Dam-Deisz WD, Graat EA, Mevius D, Voss A, Van De Giessen AW. 2010. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol Infect 138:743–755. doi: 10.1017/S0950268810000075. [DOI] [PubMed] [Google Scholar]
- 22.Harmsen D, Claus H, Witte W, Rothganger J, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J Bacteriol 188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy AJ, Witney AA, Gould KA, Moodley A, Guardabassi L, Voss A, Denis O, Broens EM, Hinds J, Lindsay JA. 2011. The distribution of mobile genetic elements (MGEs) in MRSA CC398 is associated with both host and country. Genome Biol Evol 3:1164–1174. doi: 10.1093/gbe/evr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing; 22rd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 26.Stegger M, Liu CM, Larsen J, Soldanova K, Aziz M, Contente-Cuomo T, Petersen A, Vandendriessche S, Jimenez JN, Mammina C, van Belkum A, Salmenlinna S, Laurent F, Skov RL, Larsen AR, Andersen PS, Price LB. 2013. Rapid differentiation between livestock-associated and livestock-independent Staphylococcus aureus CC398 clades. PLoS One 8:e79645. doi: 10.1371/journal.pone.0079645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bisdorff B, Scholholter JL, Claussen K, Pulz M, Nowak D, Radon K. 2012. MRSA-ST398 in livestock farmers and neighbouring residents in a rural area in Germany. Epidemiol Infect 140:1800–1808. doi: 10.1017/S0950268811002378. [DOI] [PubMed] [Google Scholar]
- 28.Uhlemann AC, Knox J, Miller M, Hafer C, Vasquez G, Ryan M, Vavagiakis P, Shi Q, Lowy FD. 2011. The environment as an unrecognized reservoir for community-associated methicillin resistant Staphylococcus aureus USA300: a case-control study. PLoS One 6:e22407. doi: 10.1371/journal.pone.0022407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Himsworth CG, Miller RR, Montoya V, Hoang L, Romney MG, Al-Rawahi GN, Kerr T, Jardine CM, Patrick DM, Tang P, Weese JS. 2014. Carriage of methicillin-resistant Staphylococcus aureus by wild urban Norway rats (Rattus norvegicus). PLoS One 9:e87983. doi: 10.1371/journal.pone.0087983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy AJ, Lindsay JA. 2012. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol 12:104. doi: 10.1186/1471-2180-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsay JA, Knight GM, Budd EL, McCarthy AJ. 2012. Shuffling of mobile genetic elements (MGEs) in successful healthcare-associated MRSA (HA-MRSA). Mob Genet Elements 2:239–243. doi: 10.4161/mge.22085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.