Abstract
While PCR is the most common method used for detecting Bordetella pertussis in the United States, most laboratories use insertion sequence 481 (IS481), which is not specific for B. pertussis; therefore, the relative contribution of other Bordetella species is not understood. The objectives of this study were to evaluate the proportion of other Bordetella species misidentified as B. pertussis during a period of increased pertussis incidence, determine the level of agreement in Bordetella species detection between U.S. commercial laboratories and the CDC, and assess the relative diagnostic sensitivity of CDC's PCR assay when using a different PCR master mix. Specimens collected between May 2012 and May 2013 were tested at two U.S. commercial laboratories for B. pertussis and B. parapertussis detection. Every fifth specimen positive for IS481 and/or IS1001 with cycle threshold (CT) values of ≤35 was sent to CDC for PCR testing that identifies Bordetella species. Specimens with indeterminate or negative results in the CDC PCR were tested using an alternate PCR master mix. Of 755 specimens, there was agreement in species identification for 83.4% (n = 630). Of the specimens with different identifications (n = 125), 79.2% (n = 99) were identified as indeterminate B. pertussis at CDC. Overall, 0.66% (n = 5) of the specimens were identified as B. holmesii or B. bronchiseptica at CDC. Of 115 specimens with indeterminate or negative results, 46.1% (n = 53) were B. pertussis positive when tested by an alternate master mix, suggesting a possible increase in assay sensitivity. This study demonstrates good agreement between the two U.S. commercial laboratories and CDC and little misidentification of Bordetella species during the 2012 U.S. epidemic.
INTRODUCTION
Pertussis, commonly known as whooping cough, is the most poorly controlled vaccine-preventable bacterial disease in the United States. The number of reported cases has increased in the last 10 years, with more than 48,000 cases reported in 2012 (1, 2). Factors that may contribute to the resurgence of pertussis include waning immunity following vaccination and bacterial adaptation. Improved detection and diagnosis may also have increased due to greater public health awareness and the use of high-sensitivity laboratory diagnostic techniques such as PCR. PCR assays targeting the insertion sequence 481 (IS481), which is present in multiple copies (about 200 to 250 copies) in the Bordetella pertussis genome (3–5), are the most commonly used pertussis diagnostics in the United States. However, the specificity of these assays is compromised because the same target sequence is also present in other Bordetella species such as B. holmesii, which causes pertussis-like illness, and B. bronchiseptica, which occasionally infects humans (4, 6). Consequently, although high IS481 cycle threshold (CT) values may be indicative of low levels of DNA from a patient with pertussis, they may also indicate a misidentified specimen from a patient infected with another Bordetella species or false positivity resulting from DNA cross-contamination (7). Additionally, some laboratories distinguish between B. pertussis and B. parapertussis infection by using a B. parapertussis-specific target (IS1001) in conjunction with IS481 (8–10). However, there are currently no standardized DNA extraction methods or PCR assays for identification of B. pertussis or other Bordetella species (9, 11).
While a few Bordetella species can cause pertussis-like illness, symptomatic patients infected with B. pertussis are reported to experience more-severe symptoms (12). However, patients infected with other Bordetella species who present early in the course of illness may be indistinguishable from those with pertussis (12). Moreover, current vaccines offer little to no protection against infection caused by Bordetella species other than B. pertussis (13, 14). Pertussis is a nationally notifiable disease, but infection by other Bordetella species does not require notification. Thus, misidentification of pertussis, both in the clinic and in the laboratory, may result in unnecessary case investigations by public health personnel. With the increase in reported pertussis disease in the United States, it is important to understand the contribution of infections caused by Bordetella species other than B. pertussis.
Recent studies show that some clinical specimens have been misidentified as positive for B. pertussis and later confirmed as positive for B. holmesii (15–21). B. parapertussis has also been identified as the cause of pertussis-like cough illness during 2008 to 2010 in the United States (22). Because testing for B. holmesii and B. parapertussis is uncommon and the corresponding infection is not notifiable, it is difficult to determine the burden of disease caused by these species. This study aims to identify the proportion of B. holmesii misidentified as B. pertussis and to determine the proportion of suspect pertussis cases caused by B. parapertussis in the United States during a period of increased pertussis incidence. Moreover, we describe the agreement in Bordetella species identification between two commercial laboratories and CDC, evaluate the relationship between the CT values obtained to better inform the qualitative differences among the laboratories, and compare the pertussis diagnostic sensitivity of two PCR master mixes.
MATERIALS AND METHODS
Specimen selection.
Deidentified multiple upper respiratory specimens in a variety of transport media, routinely sent to two commercial laboratories for pertussis testing in the United States were selected for this study based on the following criteria: (i) collected between May 2012 and May 2013 and (ii) tested positive for Bordetella pertussis, B. parapertussis, or B. pertussis/B. parapertussis coinfection. Specimens with CT values of ≤35 for IS481 and/or IS1001 PCR targets were considered positive. Every fifth specimen meeting these criteria was stored frozen, at either −20°C or −80°C, and sent to CDC monthly on dry ice for testing. This project was determined to be public health practice nonresearch. Institutional review for protection of human subjects was not required.
DNA extraction. (i) Commercial laboratories.
Nucleic acids were extracted from 200 μl of upper respiratory specimens on the MagNA Pure LC instrument with the total nucleic acid isolation kit (Roche Diagnostics, Indianapolis, IN). Extracted nucleic acids were eluted with Roche elution buffer in a final volume of 50 μl as described in the kit protocol.
(ii) CDC laboratory.
Nucleic acids were extracted from 200 μl of residual upper respiratory specimens on the MagNA Pure LC or MagNA Pure Compact instruments, with the total nucleic acid isolation kit (Roche Diagnostics). Saline was added to specimens that had insufficient volume for extraction (59/755). Extracted nucleic acids were eluted with Roche elution buffer in a final volume of 100 μl and tested immediately or stored at 4°C overnight before testing.
PCR assays and targets. (i) Laboratory A.
Real-time PCRs were performed on the 3M Integrated Cycler (Focus Diagnostics, Cypress, CA) with the following cycling parameters: one step of 97°C for 2 min, followed by 40 cycles, with 1 cycle consisting of 97°C for 10 s and 60°C for 30 s. Reactions were performed in a final volume of 10 μl, containing 5 μl of extracted DNA and 5 μl PCR master mix (B. pertussis primer pair, B. parapertussis primer pair, and internal-control primer pair). The B. pertussis and B. parapertussis primer pairs were designed to hybridize to regions within insertion sequences IS481 and IS1001, respectively.
(ii) Laboratory B.
PCRs were performed on the RotorGene Q (Qiagen Inc., Alameda, CA) with the following cycling parameters: 50 cycles of 95°C for 5 s, 58°C for 15 s, and 72°C for 20 s. Amplification of IS481 and a B. parapertussis-specific locus of IS1001 was performed using the MultiCode RTx system (Luminex Inc., Madison, WI) for product detection. Reactions were performed in a final volume of 25 μl, containing 10 μl of extracted DNA (including a coextracted internal-control target) and 15 μl of master mix (including primer pairs for all 3 targets). Following amplification, species identification was achieved by melting curve analysis (23).
(iii) CDC laboratory.
Real-time PCRs were performed on the AB7500 (Life Technologies Corp., Carlsbad, CA) with the following cycling parameters: one step of 50°C for 2 min, one step of 95°C for 2 min, followed by 45 cycles, with 1 cycle consisting of 95°C for 15 s and 60°C (or 57°C for ptxS1) for 1 min. Reactions were performed in a final volume of 25 μl, containing 4 μl of extracted DNA and 21 μl of TaqMan Gene Expression PCR master mix (Life Technologies Corp.) containing fluorescently labeled probes targeting IS481, B. parapertussis IS1001 (IS1001), B. holmesii IS1001-like (hIS1001), or ptxS1. The multitarget real-time PCR assay with an algorithm for results interpretation has been previously described (24). A combination of four targets in two separate reactions is employed: a multiplex reaction of three multicopy targets (IS481, IS1001, and hIS1001), and a singleplex reaction with a single-copy target (ptxS1). A test is considered positive for B. pertussis if both IS481 and ptxS1 targets are amplified with CT values of <40 or if IS481 target is amplified alone with CT values of <35 and undetected remaining targets, IS1001, hIS1001, and ptxS1. An indeterminate result for B. pertussis is defined by IS481 amplified with CT values between 35 and 40 (35 ≤ CT < 40) and undetected remaining targets, IS1001, hIS1001, and ptxS1. To explore possible issues with assay diagnostic sensitivity, an alternative master mix which has a polymerase presumably less affected by PCR inhibitors in clinical specimens was also evaluated (25). All specimens that produced an indeterminate or negative PCR result were tested using Quanta PerfeCTa qPCR SuperMix (Quanta BioSciences, Inc., Gaithersburg, MD) as described above.
Statistical analyses.
Statistical analyses were conducted using SAS 9.3 (SAS Institute, Inc., Cary, NC). Categorical variables were compared using chi-square tests for independent proportions. To further investigate the qualitative differences between the assays, CT values from the commercial and CDC laboratories were compared using paired t tests. P values of <0.05 were considered statistically significant. Coefficients of determination (R2) were calculated to measure the strength of the linear relationship between CT values. Concordance correlation coefficients (rc) and their 95% confidence intervals (95% CIs) quantified the agreement between the commercial laboratory and CDC laboratory CT values (26, 27; S. Bai, Concordance correlation coefficient for measuring agreement, The Pennsylvania State University Design and Analysis of Clinical Trials website https://onlinecourses.science.psu.edu/stat509/sites/onlinecourses.science.psu.edu.stat509/files/lesson18/19.2_agreement_concordanc.sas). Deming regression lines were estimated to further describe the relationship between the commercial laboratory and CDC laboratory CT values (28, 29).
RESULTS
Residual positive clinical upper respiratory specimens from 755 patients were sent to CDC from two commercial reference laboratories. All specimens were collected between May 2012 and May 2013 for routine pertussis testing from patients in 38 states and tested positive for B. pertussis, B. parapertussis, or B. pertussis/B. parapertussis coinfection at the commercial laboratories. The majority of clinical specimens (657/755) were sent in universal transport media.
Of the specimens received, 58.7% (443/755) were sent from laboratory A, and 41.3% (312/755) were sent from laboratory B. For analytical purposes, data from both commercial laboratories were combined. Overall, the mean commercial laboratory CT value was 28.24 (range, 12.10 to 35.00). When these specimens were tested using the CDC assay, IS481 was detected in 93.0% (n = 702) of specimens, and the mean IS481 CT value was 29.92 (range, 12.04 to 44.37). B. parapertussis IS1001 was detected by the CDC laboratory in 9.9% (n = 75) of specimens, and the mean IS1001 CT value was 32.93 (range, 17.46 to 43.65).
Comparison between commercial laboratories and CDC PCR testing.
The majority of the clinical specimens were positive for B. pertussis when tested by the commercial laboratories (93.0%) or when tested at CDC by the multitarget assay using the validated PCR master mix (77.2%) (Table 1). The commercial laboratories and CDC identified similar proportions of B. parapertussis and B. pertussis/B. parapertussis coinfections (Table 1). CDC testing identified 13.1% of the specimens as indeterminate B. pertussis, 2.12% as negative for any Bordetella species and 0.40% as positive for B. holmesii. Less than 1% of specimens were identified as positive for B. bronchiseptica or B. pertussis/B. holmesii coinfection (Table 1). Analyses were performed excluding specimens that had insufficient volume for DNA extraction at CDC (59/755), and no substantial differences were found (data not shown).
TABLE 1.
Comparison of PCR results from 755 clinical specimens tested at two U.S. commercial laboratories and at CDC laboratory
| PCR result | Comparison of PCR results [no. of specimens with the indicated PCR result (%)]a |
|||||
|---|---|---|---|---|---|---|
| Laboratory A and CDC laboratory [n = 443 (58.7%)] |
Laboratory B and CDC laboratory [n = 312 (41.3%)] |
Both laboratories and CDC laboratory [n = 755 (100%)] |
||||
| Lab A | CDC | Lab B | CDC | Lab A plus B | CDC | |
| B. pertussis | 414 (93.5) | 326 (73.6) | 288 (92.3) | 257 (82.4) | 702 (93.0) | 583 (77.2) |
| Indeterminate B. pertussis | – | 72 (16.3) | – | 27 (8.65) | – | 99 (13.1) |
| B. parapertussis | 29 (6.6) | 26 (5.87) | 19 (6.1) | 22 (7.05) | 48 (6.36) | 48 (6.36) |
| Negative | – | 15 (3.39) | – | 1 (0.32) | – | 16 (2.12) |
| B. pertussis and B. parapertussis | – | 1 (0.23) | 5 (1.6) | 2 (0.64) | 5 (0.66) | 3 (0.40) |
| B. holmesii | – | 2 (0.45) | – | 1 (0.32) | – | 3 (0.40) |
| B. bronchiseptica | – | 1 (0.23) | – | 1 (0.32) | – | 2 (0.26) |
| B. pertussis and B. holmesii | – | – | – | 1 (0.32) | – | 1 (0.13) |
Percentages may not add to 100% due to rounding. –, no data to report.
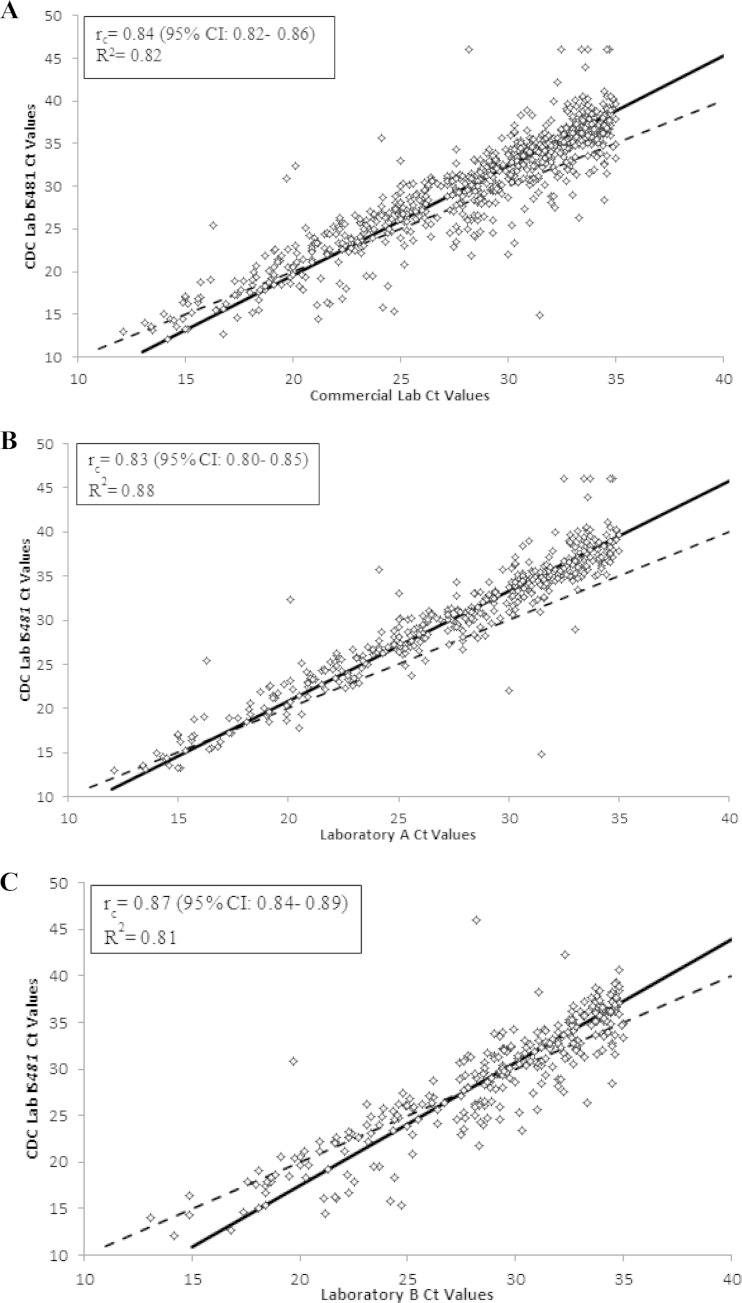
Of the 702 clinical specimens identified as B. pertussis by the commercial laboratories, there was high reproducibility between the commercial laboratory and CDC laboratory IS481 CT values (rc = 0.84; 95% CI, 0.82 to 0.86) (Fig. 1A). Laboratory A and laboratory B had similar levels of reproducibility with the CDC laboratory (rc = 0.83 and rc = 0.87, respectively) (Fig. 1B and C). Additionally, there was a moderate reproducibility between the commercial laboratory IS1001 CT values and CDC laboratory IS1001 CT values among the 48 specimens identified by the commercial laboratories as B. parapertussis (rc = 0.75; 95% CI, 0.61 to 0.85). Reproducibility was higher between laboratory B and CDC than between laboratory A and CDC (rc = 0.90 and rc = 0.65, respectively).
FIG 1.
Scatterplots of the commercial laboratory cycle threshold (CT) values versus CDC IS481 CT values. Specimens with undetected IS481 were assigned a CT value of 46. The dashed line is the line of identity, and the solid black line is the Deming regression line. (A) Commercial laboratories A plus B CT values versus CDC IS481 CT values (n = 702). The Deming regression line was y = 1.29x − 6.26. (B) Laboratory A CT values versus CDC IS481 CT values (n = 414). The Deming regression line was y = 1.25x − 4.19. (C) Laboratory B CT values versus CDC IS481 CT values (n = 288). The Deming regression line was y = 1.32x − 8.86.
Overall, CDC testing resulted in a different result interpretation for 16.6% (125/755) of the specimens (Table 2). The commercial laboratories identified 95.2% (119/125) of these specimens as positive for B. pertussis. However, the CDC laboratory identified 79.2% (99/125) of specimens as indeterminate B. pertussis and 12.8% (16/125) as negative. Laboratory A had a higher percentage of disagreement with the CDC laboratory than did laboratory B (P = 0.0004).
TABLE 2.
Discrepancies in identification of Bordetella pertussis in 125 specimens when comparing PCR results from two U.S. commercial laboratories and CDC laboratory
| PCR result of commercial laboratories A plus B | PCR result of CDC laboratory | No. of specimens (%) with the indicated PCR resultsa |
||
|---|---|---|---|---|
| Lab A (n = 91) | Lab B (n = 34) | Both labs (n = 125) | ||
| B. pertussis (n = 119) | Indeterminate B. pertussis | 72 (79.1) | 27 (79.4) | 99 (79.2) |
| Negative | 14 (15.4) | 1 (2.94) | 15 (12.0) | |
| B. holmesii | 2 (2.20) | 1 (2.94) | 3 (2.40) | |
| B. pertussis/B. holmesii | – | 1 (2.94) | 1 (0.80) | |
| B. bronchiseptica | – | 1 (2.94) | 1 (0.80) | |
| B. parapertussis (n = 3) | B. pertussis/B. parapertussis | 1 (1.10) | – | 1 (0.80) |
| B. bronchiseptica | 1 (1.10) | – | 1 (0.80) | |
| Negative | 1 (1.10) | – | 1 (0.80) | |
| B. pertussis/B. parapertussis (n = 3) | B. parapertussis | – | 3 (8.82) | 3 (2.40) |
Percentages may not add to 100% due to rounding. –, no data to report.
CDC testing using an alternate PCR master mix.
Of the 125 specimens with discordant results, 115 were identified as indeterminate B. pertussis or negative by the CDC laboratory, where the single-copy, less-sensitive ptxS1 target was negative. These 115 specimens were additionally tested using an alternate PCR master mix to explore assay diagnostic sensitivity-related issues by using a master mix less affected by inhibitors. Laboratory A sent 75.7% (87/115) of these specimens. The mean commercial laboratory CT value was 33.31 (range, 30.10 to 35.00), the mean CDC validated master mix IS481 CT value was 37.49 (range, 35.03 to 41.12), and the mean CDC alternate master mix IS481 CT value was 35.16 (range, 31.15 to 44.89). When the specimens were tested by the alternate master mix, the mean difference between the commercial laboratory CT value and CDC IS481 CT value was −1.83 units, and the mean difference between the CDC validated and alternate master mixes IS481 CT values was 2.47 units. Both of these differences were statistically significant (P < 0.0001).
The commercial laboratories had identified 99.1% (114/115) of the CDC indeterminate or negative results as B. pertussis positive. Using the validated CDC master mix, 86.1% (99/115) were identified as indeterminate B. pertussis. The percentage of indeterminate B. pertussis specimens decreased to 42.6% (49/115) when testing was performed by the alternate PCR master mix. Remarkably, 46.1% (53/115) were reclassified as B. pertussis positive. Among those 53 B. pertussis-positive specimens, 6 specimens amplified the single-copy ptxS1 target with CT values of <40. Both PCR master mixes identified similar percentages of negative specimens (13.9% and 11.3%, respectively) (see Fig. S1 in the supplemental material). The result interpretations for 54.8% (63/115) of these specimens were different when tested by the CDC assay using the validated versus alternate master mixes. Similarly, the interpretations for 53.9% (62/115) differed when specimens were tested by the commercial laboratories compared to CDC using the alternate master mix. These differences are described in Table 3.
TABLE 3.
Shift in result interpretation for pertussis PCR in 115 specimens after CDC testing with an alternate PCR master mix
| Shift in result interpretation | No. of specimens (%)a |
|||
|---|---|---|---|---|
| CDC laboratory validated master mix vs. alternate master mix (n = 115) | Commercial laboratory vs. CDC alternate master mix |
|||
| Lab A (n = 87) | Lab B (n = 28) | Both labs (n = 115) | ||
| Total overall | 63 (54.8) | 50 (57.5) | 12 (42.9) | 62 (53.9) |
| Indeterminate B. pertussis to B. pertussis | 52 (82.5) | – | – | – |
| Indeterminate B. pertussis to negative | 4 (6.35) | – | – | – |
| B. pertussis to indeterminate B. pertussis | – | 39 (78.0) | 10 (83.3) | 49 (79.0) |
| B. pertussis to negative | – | 10 (20.0) | 2 (16.7) | 12 (19.4) |
| Negative to indeterminate B. pertussis | 6 (9.52) | – | – | – |
| Negative to B. pertussis | 1 (1.59) | – | – | – |
| B. parapertussis to negative | – | 1 (2.00) | – | 1 (1.61) |
The total overall data are shown in boldface type. Percentages may not add to 100% due to rounding. –, no data to report.
DISCUSSION
Using a prospective sampling scheme, we examined the relative incidence of Bordetella species during the months of May 2012 to May 2013, a period of increased pertussis case reporting in the United States. The clinical specimens in this study were submitted to the commercial laboratories from 38 different states, and thus are representative of the majority of the country. The predominant species identified was B. pertussis. In earlier unpublished work, CDC tested suspect pertussis clinical specimens collected from November 2010 to January 2011 by the same multitarget real-time PCR assay used in the present study. Of the 411 specimens, 24.6% (101/411) were identified as B. parapertussis and 4.14% (17/411) were identified as B. holmesii (data not published). In contrast, the present study identified only 6.36% (48/755) as B. parapertussis and 0.40% (3/755) as B. holmesii. Our results indicate that the pertussis cases reported in 2012 were likely attributed to B. pertussis infection.
We found good agreement in pathogen identification between the commercial laboratories and CDC. Of the 125 specimens with discordant results, 79.2% were categorized as indeterminate B. pertussis and 12.8% as negative by the CDC laboratory (Table 2). The indeterminate B. pertussis identification is given to specimens that generate IS481 CT values between 35 and 40 (35 ≤ CT < 40) but are negative for all other targets employed in the CDC multitarget PCR assay (24). The disagreement in specimen identification is likely the result of several factors, including differences in assay protocols, diagnostic sensitivity, interpretation criteria, DNA degradation following storage or transportation of residual specimens, and differences in elution volumes during DNA extraction.
The CT value evaluation was conducted in this study to better inform the qualitative differences of PCR pertussis testing identified between the laboratories. The rc values reported incorporate both accuracy and precision to produce an estimate of the reproducibility between two sets of values (26, 27). We found excellent reproducibility when comparing the commercial laboratory CT values to the IS481 CT values from the 702 specimens originally identified as B. pertussis. There was moderate reproducibility in the CT values obtained for the IS1001 target for the 48 specimens originally identified as B. parapertussis. The lower agreement can likely be explained by the small B. parapertussis IS1001 CT sample size.
Recent work has reported an increased sensitivity when assays were performed with Quanta master mixes (25). Thus, the possibility of differences in assay sensitivity was further explored by repeating the CDC assay using the alternate Quanta PerfeCTa qPCR SuperMix for 115 specimens identified as indeterminate B. pertussis or negative. We found changes in identifications when using different master mixes at CDC in 63 (54.8%) of the specimens. IS481 CT values were 2.47 units lower when the alternate master mix was used, on average. For 59 (93.7%) of these specimens, the changes were sufficient to shift the interpretations to another category (i.e., B. pertussis indeterminate to B. pertussis positive or negative to B. pertussis indeterminate) (Table 3). These data suggest that discrepancies found in this study may also be attributed to differences in assay diagnostic sensitivity.
The CDC multitarget real-time PCR assay has been validated using the Life Technologies TaqMan Gene Expression master mix (24, 30). Due to the high copy number of IS481 in the B. pertussis genome, employing a more sensitive master mix could present challenges for routine clinical interpretation if diagnostic specificity is adversely impacted. However, for a single-copy gene target such as ptxS1, increasing the assay sensitivity may improve detection without substantially increasing the risk of false-positive results. With improved sensitivity, interpretation algorithms and cutoff values for each target would need to be reevaluated for clinical relevance. More-comprehensive studies should be conducted to determine whether transitioning to an alternate master mix increases the sensitivity of the CDC assay without compromising its specificity.
Our findings were likely influenced by our specimen selection criteria. While specimens with a CT of ≤35 were considered positive for B. pertussis or B. parapertussis at the commercial laboratories, due to reasons previously explained (sample degradation and differences in elution volumes and assay sensitivity), the same CT value is not equivalent across all laboratories. Moreover, having the same CT value does not represent the same bacterial load in the specimen tested. Therefore, employing a ≤35 CT cutoff for specimen selection may explain some of the differences in results observed between the commercial laboratories and CDC.
While this study set out to determine whether other Bordetella species were in fact a causative agent of suspected pertussis cases during a period of increased pertussis incidence, its design did not allow us to address the relative contribution of false-positive diagnostic test results to the total pertussis burden or to evaluate whether commercial laboratories are overdiagnosing pertussis. We found that during our sampling time frame, a small proportion of Bordetella species other than B. pertussis were circulating in the United States, and the significant burden of pertussis disease in the United States was attributable to B. pertussis. The overall agreement in specimen identification and reproducibility of CT values between the laboratories indicates a high standard of PCR testing practices for pertussis in the United States.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gladys Gonzalez-Aviles for conducting the initial retrospective analysis cited in this study and Freda Lyde for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03567-14.
REFERENCES
- 1.Centers for Disease Control and Prevention (CDC). 2012. Pertussis epidemic–Washington, 2012. MMWR Morb Mortal Wkly Rep 61(28):517–522. [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC). 2014. Summary of notifiable diseases: United States, 2012. MMWR Morb Mortal Wkly Rep 61(53):1–121. [PubMed] [Google Scholar]
- 3.Glare EM, Paton JC, Premier RR, Lawrence AJ, Nisbet IT. 1990. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J Clin Microbiol 28:1982–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MTG, Churcher CM, Bentley SD, Mungall KL, Cerdeno-Tarraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 5.Harvill ET, Goodfield LL, Ivanov Y, Meyer JA, Newth C, Cassiday P, Tondella ML, Liao P, Zimmerman J, Meert K, Wessel D, Berger J, Dean JM, Holubkov R, Burr J, Liu T, Brinkac L, Kim M, Losada L. 2013. Genome sequences of 28 Bordetella pertussis U.S. outbreak strains dating from 2010 to 2012. Genome Announc 1(6):e01075-13. doi: 10.1128/genomeA.01075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reischl U, Lehn N, Sanden GN, Loeffelholz MJ. 2001. Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J Clin Microbiol 39:1963–1966. doi: 10.1128/JCM.39.5.1963-1966.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandal S, Tatti KM, Woods-Stout D, Cassiday PK, Faulkner AE, Griffith MM, Jackson ML, Pawloski LC, Wagner B, Barnes M, Cohn AC, Gershman KA, Messonnier NE, Clark TA, Tondella M-LC, Martin SW. 2012. Pertussis pseudo-outbreak linked to specimens contaminated by Bordetella pertussis DNA from clinic surfaces. Pediatrics 129:e424–e430. doi: 10.1542/peds.2011-1710. [DOI] [PubMed] [Google Scholar]
- 8.Arbefeville S, Levi MH, Ferrieri P. 2014. Development of a multiplex real-time PCR assay for the detection of Bordetella pertussis and Bordetella parapertussis in a single tube reaction. J Microbiol Methods 97:15–19. doi: 10.1016/j.mimet.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Gao FX, Mahoney JC, Daly ER, Lamothe W, Tullo D, Bean C. 2014. Evaluation of a multitarget real-time PCR assay for detection of Bordetella species during a pertussis outbreak in New Hampshire in 2011. J Clin Microbiol 52:302–306. doi: 10.1128/JCM.01656-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zouari A, Smaoui H, Brun D, Njamkepo E, Sghaier S, Zouari E, Felix R, Menif K, Ben Jaballah N, Guiso N, Kechrid A. 2012. Prevalence of Bordetella pertussis and Bordetella parapertussis infections in Tunisian hospitalized infants: results of a 4-year prospective study. Diagn Microbiol Infect Dis 72:303–317. doi: 10.1016/j.diagmicrobio.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Tatti KM, Martin SW, Boney KO, Brown K, Clark TA, Tondella ML. 2013. Qualitative assessment of pertussis diagnostics in United States laboratories. Pediatr Infect Dis J 32:942–945. doi: 10.1097/INF.0b013e3182947ef8. [DOI] [PubMed] [Google Scholar]
- 12.Mattoo S, Cherry JD. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18:326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Weyrich LS, Lavine JS, Karanikas AT, Harvill ET. 2012. Lack of cross-protection against Bordetella holmesii after pertussis vaccination. Emerg Infect Dis 18:1771–1779. doi: 10.3201/eid1811.111544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He QS, Viljanen MK, Arvilommi H, Aittanen B, Mertsola J. 1998. Whooping cough caused by Bordetella pertussis and Bordetella parapertussis in an immunized population. JAMA 280:635–637. doi: 10.1001/jama.280.7.635. [DOI] [PubMed] [Google Scholar]
- 15.Njamkepo E, Bonacorsi S, Debruyne M, Gibaud SA, Guillot S, Guiso N. 2011. Significant finding of Bordetella holmesii DNA in nasopharyngeal samples from French patients with suspected pertussis. J Clin Microbiol 49:4347–4348. doi: 10.1128/JCM.01272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pittet LF, Emonet S, Francois P, Bonetti EJ, Schrenzel J, Hug M, Altwegg M, Siegrist CA, Posfay-Barbe KM. 2014. Diagnosis of whooping cough in Switzerland: differentiating Bordetella pertussis from Bordetella holmesii by polymerase chain reaction. PLoS One 9:e88936. doi: 10.1371/journal.pone.0088936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van den Bossche D, De Bel A, De Smet D, Heylen O, Vekens E, Vandoorslaer K, Soetens O, Pierard D. 2013. Prevalence of Bordetella holmesii and Bordetella bronchiseptica in respiratory tract samples from Belgian patients with pertussis-like symptoms by sensitive culture method and mass spectrometry. Acta Clin Belg 68:341–348. doi: 10.2143/ACB.3341. [DOI] [PubMed] [Google Scholar]
- 18.Miranda C, Porte L, Garcia P. 2012. Bordetella holmesii in nasopharyngeal samples from Chilean patients with suspected Bordetella pertussis infection. J Clin Microbiol 50:1505. doi: 10.1128/JCM.06747-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guthrie JL, Robertson AV, Tang P, Jamieson F, Drews SJ. 2010. Novel duplex real-time PCR assay detects Bordetella holmesii in specimens from patients with pertussis-like symptoms in Ontario, Canada. J Clin Microbiol 48:1435–1437. doi: 10.1128/JCM.02417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodgers L, Martin SW, Cohn A, Budd J, Marcon M, Terranella A, Mandal S, Salamon D, Leber A, Tondella M-L, Tatti K, Spicer K, Emanuel A, Koch E, McGlone L, Pawloski L, LeMaile-Williams M, Tucker N, Iyer R, Clark TA, DiOrio M. 2013. Epidemiologic and laboratory features of a large outbreak of pertussis-like illnesses associated with cocirculating Bordetella holmesii and Bordetella pertussis-Ohio, 2010-2011. Clin Infect Dis 56:322–331. doi: 10.1093/cid/cis888. [DOI] [PubMed] [Google Scholar]
- 21.Bottero D, Griffith MM, Lara C, Flores D, Pianciola L, Gaillard ME, Mazzeo M, Zamboni MI, Spoleti MJ, Anchart E, Ruggeri D, Sorhouet C, Fiori S, Galas M, Tondella ML, Hozbor DF. 2013. Bordetella holmesii in children suspected of pertussis in Argentina. Epidemiol Infect 141:714–717. doi: 10.1017/S095026881200132X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherry JD, Seaton BL. 2012. Patterns of Bordetella parapertussis respiratory illnesses: 2008-2010. Clin Infect Dis 54:534–537. doi: 10.1093/cid/cir860. [DOI] [PubMed] [Google Scholar]
- 23.Menard A, Lehours P, Sarlangue J, Bebear C, Megraud F, de Barbeyrac B. 2007. Development of a real-time PCR for the identification of Bordetella pertussis and Bordetella parapertussis. Clin Microbiol Infect 13:419–423. doi: 10.1111/j.1469-0691.2006.01659.x. [DOI] [PubMed] [Google Scholar]
- 24.Tatti KM, Sparks KN, Boney KO, Tondella ML. 2011. Novel multitarget real-time PCR assay for rapid detection of Bordetella species in clinical specimens. J Clin Microbiol 49:4059–4066. doi: 10.1128/JCM.00601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz MH, Waller JL, Napoliello RA, Islam MS, Wolff BJ, Burken DJ, Holden RL, Srinivasan V, Arvay M, McGee L, Oberste MS, Whitney CG, Schrag SJ, Winchell JM, Saha SK. 2013. Optimization of multiple pathogen detection using the TaqMan Array Card: application for a population-based study of neonatal infection. PLoS One 8:e66183. doi: 10.1371/journal.pone.0066183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin LI. 1989. A concordance correlation-coefficient to evaluate reproducibility. Biometrics 45:255–268. doi: 10.2307/2532051. [DOI] [PubMed] [Google Scholar]
- 27.Lin L. 2003. Measuring agreement, p 561–567. In Chow S. (ed), Encyclopedia of biopharmaceutical statistics, 2nd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 28.Deal AM, Pate VW, El Rouby S. 2009. A SAS macro for Deming regression, paper CC-014. SESUG 2009: The Proceedings of the SouthEast SAS Users Group, Birmingham, AL, 2009 http://analytics.ncsu.edu/sesug/2009/CC014.Deal.pdf. [Google Scholar]
- 29.Linnet K. 1993. Evaluation of regression procedures for methods comparison studies. Clin Chem 39:424–432. [PubMed] [Google Scholar]
- 30.Tatti KM, Tondella ML. 2013. Utilization of multiple real-time PCR assays for the diagnosis of Bordetella spp. in clinical specimens. Methods Mol Biol 943:135–147. doi: 10.1007/978-1-60327-353-4_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.