Abstract
An outbreak of severe respiratory illness associated with enterovirus D68 (EV-D68) infection was reported in mid-August 2014 in the United States. In this study, we evaluated the diagnostic utility of an EV-D68-specific real-time reverse transcription-PCR (rRT-PCR) that was recently developed by the Centers for Disease Control and Prevention in clinical samples. Nasopharyngeal (NP) swab specimens from patients in a recent outbreak of respiratory illness in the lower Hudson Valley, New York State, were collected and examined for the presence of human rhinovirus or enterovirus using the FilmArray Respiratory Panel (RP) assay. Samples positive by RP were assessed using EV-D68 rRT-PCR, and the data were compared to results from sequencing analysis of partial VP1 and 5′ untranslated region (5′-UTR) sequences of the EV genome. A total of 285 RP-positive NP specimens (260 from the 2014 outbreak and 25 from 2013) were analyzed by rRT-PCR; EV-D68 was detected in 74 of 285 (26.0%) specimens examined. Data for comparisons between rRT-PCR and sequencing analysis were obtained from 194 NP specimens. EV-D68 detection was confirmed by sequencing analysis in 71 of 74 positive and in 1 of 120 randomly selected negative specimens by rRT-PCR. The EV-D68 rRT-PCR showed diagnostic sensitivity and specificity of 98.6% and 97.5%, respectively. Our data suggest that the EV-D68 rRT-PCR is a reliable assay for detection of EV-D68 in clinical samples and has a potential to be used as a tool for rapid diagnosis and outbreak investigation of EV-D68-associated infections in clinical and public health laboratories.
INTRODUCTION
Enteroviruses (EV), belonging to the family Picornaviridae, are small, nonenveloped viruses with a single-stranded, positive-sense RNA genome of approximately 7.5 kb. The genus Enterovirus contains seven species, including enterovirus A to D and rhinovirus A to C, that commonly cause human disease (1, 2). Enterovirus D68 (EV-D68) was first recovered from patients with respiratory illness in California in 1962 (3). Since its initial identification, EV-D68 has been infrequently reported as a cause of human disease, with only 26 cases reported to the Centers for Disease Control and Prevention (CDC) from 1970 through 2005 (4). However, recent studies have suggested an emergence of EV-D68 infection in patients with acute respiratory illness in Asia, Europe, and a few U.S. states since 2009 (5–11). In August 2014, clusters of EV-D68 infection associated with severe respiratory illness were reported in Missouri and Illinois (12). Subsequently, a national outbreak of EV-D68-associated severe respiratory illness was identified. From mid-August 2014 to 15 January 2015, CDC or state public health laboratories confirmed a total of 1,153 people in 49 states and the District of Columbia with respiratory illness caused by EV-D68 (13). EV-D68 was also detected in specimens from 14 patients who died and had samples submitted to CDC for testing and from patients with acute flaccid myelitis in the United States and France (10, 13, 14).
Several FDA-cleared molecular assays are currently available for detection of enterovirus and rhinovirus in clinical samples. These include FilmArray Respiratory Panel multiplex PCR (RP; BioFire, Salt Lake City, UT), the xTAG Respiratory Viral Panel (Lumilex, Austin, TX), the eSensor Respiratory Viral Panel (GenMark, Carlsbad, CA), and the EV assay (Cepheid, Sunnyvale, CA). The FilmArray RP assay uses six sets of broadly reactive primers (namely, EV1, EV2, HRV1, HRV2, HRV3, and HRV4) that amplify RNA from either human rhinovirus (HRV) or enterovirus in a multiplexed reaction, and results are reported as “human rhinovirus/enterovirus.” None of the FDA-cleared molecular assays can reliably differentiate EV-D68 from rhinoviruses or other common serotypes of enterovirus such as coxsackievirus and echovirus, due to the high genetic similarity of their genomes. Confirmation of EV-D68 usually relies on sequencing analysis of the VP1 and/or 5′ untranslated region (5′-UTR) of the EV genome (5, 15). Of these, seminested PCR amplification, followed by analysis of partial VP1 sequences, as described by Nix et al., has an analytical sensitivity of less than 10 EV genome copies (15). Nevertheless, this protocol is time-consuming and costly, which limits its routine use, resulting in a situation where the public health and clinical laboratories of only a few U.S. states have the ability to identify EV-D68 in clinical samples.
In response to the 2014 outbreak of EV-D68-associated severe respiratory illness in the United States, the CDC developed a real-time reverse transcription-PCR (rRT-PCR) assay and made it publically available in October 2014 (16). The aim of this study was to evaluate the diagnostic utility, including the sensitivity, specificity, and positive and negative predictive values, of the EV-D68 rRT-PCR assay by analyzing clinical samples collected from patients during a recent outbreak of EV-D68-associated respiratory illness in the lower Hudson Valley of New York State between August and November 2014.
MATERIALS AND METHODS
Patients.
Beginning with the second week of September 2014, there was a significant increase in the number of pediatric patients with severe respiratory illness who visited the emergency department or were hospitalized at the Maria Fareri Children's Hospital of Westchester Medical Center (WMC). Patients who presented with respiratory symptoms and had a nasopharyngeal (NP) swab specimen that was positive for human rhinovirus or enterovirus (HRV/EV) were included in this study. This included inpatients hospitalized at the Maria Fareri Children's Hospital, patients from a nearby long-term-care facility for children, and outpatients who visited emergency centers or clinics in the lower Hudson Valley. This retrospective study was conducted as part of an infection control and outbreak investigation of EV-D68-associated respiratory illness in the lower Hudson Valley, NY, and a laboratory quality improvement project and did not require Institutional Review Board approval.
FilmArray RP assay.
NP swabs from patients who visited the outpatient clinics or were hospitalized were collected by a standard procedure into tubes with 1 ml each of viral transport medium (Diagnostic Hybrid, San Diego, CA). NP swabs were tested for the presence of HRV/EV and other respiratory pathogens using a FilmArray Respiratory Panel (RP) kit (version 1.7; BioFire Inc., Salt Lake City, UT) in the WMC Clinical Virology Laboratory. NP swab specimens that were positive for HRV/EV by RP were further examined by EV-D68-specific rRT-PCR and DNA sequencing analysis of the VP1 and 5′-UTR of the EV genome.
RNA extraction.
Total RNA was extracted from leftover nasopharyngeal swab specimens (≤140 μl) using a QIAamp Viral RNA minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions, except that no carrier RNA was added to the majority of samples (244 of 285; 85.6%) prior to RNA extraction. Extracted RNA was eluted into 60 μl of buffer and stored at −80°C until use.
EV-D68 real-time RT-PCR assay.
A single-step, EV-D68 2014 outbreak strain-specific real-time reverse transcription-PCR (rRT-PCR) assay was carried out according to the CDC protocol, version 10/14/2014 (16). Briefly, 5 μl of RNA was added to an RT-PCR reaction mixture with a total volume of 25 μl that consisted of 1× reaction buffer and SS III RT/Platinum Taq mix (SuperScript III Platinum one-step quantitative RT-PCR system; Life Technologies, Grand Island, NY), 0.32 μM (each) primer AN887 (5′-CAA ACT CGC ACA GTG ATA AAY CAR CA-3′) and primer AN893 (5′-GTA TTA TTA CTA CTA CCA TTC ACN GCN AC-3′), 0.16 μM probe AN890 (5′-6-carboxyfluorescein [FAM]-GTC CAT TTG AAA AAG TTC TTG TC-black hole quencher 1 [BHQ1]-3′), and 4 mM Mg++ in the final reaction mixture. The reverse transcription was performed at 50°C for 30 min, followed by 2 min at 95°C for polymerase activation and 45 cycles of 95°C for 15 s, 55°C for 1 min, and 72°C for 5 s on an ABI 7500 Fast Dx real-time PCR instrument (Life Technologies). The instrument was used in standard run mode without passive reference dye and analysis with manual threshold setting. RNA diluted 1:1,000 and derived from an NP swab of a patient with EV-D68 previously confirmed by DNA sequencing analysis was used as an external positive control. This positive control and a nontemplate negative control were included with each rRT-PCR run. A result positive for EV-D68 was defined as a sample exhibiting exponential amplification and with a cycle threshold (CT) value of ≤40.0.
Sequencing analysis of the VP1 and 5′-UTR regions.
Seventy-four EV-D68-positive samples and 120 randomly selected EV-D68-negative RNA samples as determined by rRT-PCR were reverse transcribed to cDNA with a QuantiTect reverse transcription kit (Qiagen, Valencia, CA) according to the manufacturer's instructions with oligo(dT) and random primers. For sequencing analysis of the partial VP1 gene, a seminested PCR with expected amplicon sizes of 340 to 393 bp was performed using a HotStarTaq plus master mix kit (Qiagen, Valencia, CA) and primers and PCR conditions as described by Nix et al. (15). For sequencing analysis of the 5′-UTR, primer pair DK001 and DK004 was used for PCR amplification as described previously (5) and yielded an amplicon of approximately 400 bp.
PCR amplicons were sequenced using PCR primers with BigDye Terminator v1.1 sequencing on an ABI 3500xL genetic analyzer (Life Technologies, Grand Island, NY). EV-D68 was confirmed when the VP1 sequence exhibited ≥75% identity to that of strain Fermon (GenBank accession number AY426531). Determination of the rhinovirus species was made by BLASTN analysis with the referenced classification of rhinoviruses (17).
Phylogenetic analysis.
Sequence alignment and construction of a phylogenetic tree based on the sequences of VP1 and 5′-UTR were performed by the use of MEGA 6 (18) with the neighbor-joining method.
Statistical analysis.
One-way analysis of variance (ANOVA) was used with the Kruskal-Wallis test to compare the cycle threshold (CT) values among three groups of patients with different hospitalization statuses using Prism GraphPad software (version 5; GraphPad, La Jolla, CA).
Nucleotide sequence accession numbers.
DNA sequences determined in this study have been deposited into the GenBank database with accession numbers KP742372 to KP742377 for the partial VP1 sequences and KP742378 to KP742384 for the 5′-UTR sequences.
RESULTS
Performance characteristics of EV-D68 rRT-PCR.
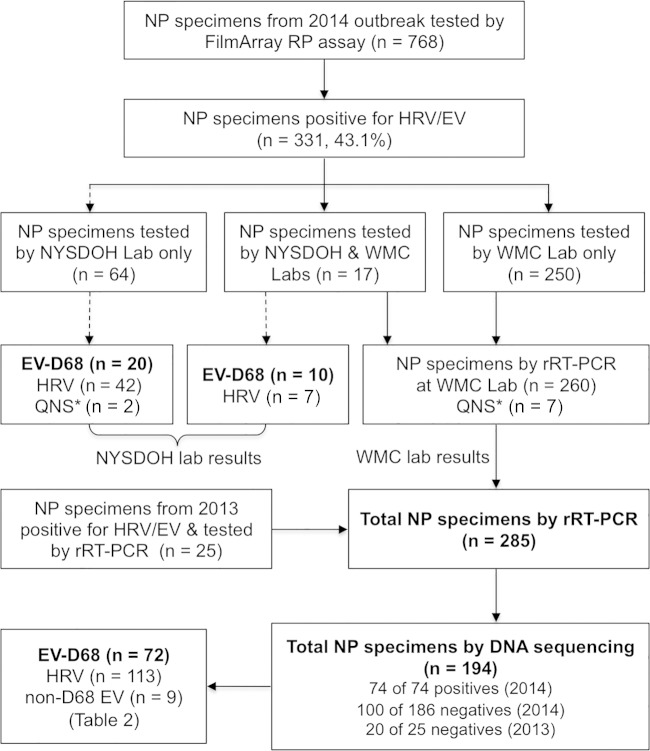
From 7 September 2014 to 31 October 2014, a total of 768 NP specimens were received for testing of respiratory viruses. Of these, 331 (43.1%) of the NP specimens were positive for HRV/EV by the FilmArray RP assay. Eighty-one (n = 81) NP specimens positive for HRV/EV by RP that were mainly collected from 7 to 18 September 2014 were sent to the New York State Department of Health (NYSDOH) Wadsworth Center Laboratory for EV-D68 testing. We were able to retrieve and analyze 260 of 331 (78.6%) HRV/EV-positive NP specimens collected during this outbreak by the use of EV-D68 rRT-PCR. These included 17 samples from which aliquots were sent to NYSDOH Laboratory and 243 of 250 (97.2%) HRV/EV-positive samples collected on or after 18 September 2014 (Fig. 1). In addition, 25 NP samples, which were collected from September to December 2013 and were positive for HRV/EV by the FilmArray RP assay, were also retrieved from a −80°C freezer and analyzed in this study.
FIG 1.
Flow chart of nasopharyngeal (NP) swab specimens analyzed for EV-D68 and rhinoviruses in this study. QNS*, quantity not sufficient (no test results were available for analysis). Specimens examined in this study are shown with arrows with solid lines.
Seventy-four of 260 (28.5%) HRV/EV-positive NP specimens collected during the 2014 outbreak were positive for EV-D68 by rRT-PCR, while 0 of 25 HRV/EV-positive NP specimens collected in 2013 were positive for EV-D68 (P < 0.001). Of 17 NP specimens from the 2014 outbreak that were analyzed by DNA sequencing at the NYSDOH Laboratory and by rRT-PCR and DNA sequencing at the WMC Laboratory, EV-D68 was detected in 10 NP specimens and HRV in the remaining 7 samples by both laboratories, resulting in 100% agreement of the results from the two laboratories.
A total of 194 NP specimens, including all 74 EV-D68-positive specimens and 120 randomly selected EV-D68-negative specimens, were further analyzed by DNA sequencing of partial VP1 and 5′-UTR sequences of the EV genome. Compared to sequence analysis, EV-D68 rRT-PCR showed diagnostic sensitivity and specificity of 98.6% and 97.5%, respectively (Table 1). One NP specimen was positive for EV-D68 by sequencing analysis of the VP1 and 5′-UTR only and was most likely a true false negative by EV-D68 rRT-PCR. Three NP specimens were positive for EV-D68 by rRT-PCR, with CT values of 23.1, 32.2, and 36.1, respectively. For these 3 specimens, a clear mixture of virus sequences was seen for the VP1 amplicons but only rhinoviruses were detected by sequencing analysis of the 5′-UTR. To determine whether EV-D68 was truly present in these samples, we directly sequenced the PCR products of partial VP1 obtained using EV-D68-specific rRT-PCR primers AN887 and AN893. EV-D68-specific sequences were identified for all three samples, which provided evidence of a mixed infection of EV-D68 and HRV in these samples. A mixed infection of EV-D68 and rhinovirus was also confirmed by whole-genome sequencing analysis in one of the NP specimens with a CT value of 23.1 by rRT-PCR (data not shown). The adjusted diagnostic specificity of rRT-PCR was 100%.
TABLE 1.
Comparative results of rRT-PCR versus sequencing analysis of partial VP1 and 5′-UTR of enterovirus genome
| rRT-PCR result | No. of specimens with indicated result of sequencing analysis of VP1 and 5′-UTR |
Total | |
|---|---|---|---|
| EV-D68a | Non-EV-D68b | ||
| Positive | 71 | 3c | 74 |
| Negative | 1 | 119 | 120 |
| Total | 72 | 122 | 194 |
Sensitivity, 98.6%.
Specificity, 97.5%.
For these 3 NP specimens, a mixed infection of EV-D68 and HRV was confirmed by additional DNA sequencing analysis of partial VP1 sequences based on EV-D68-specific PCR primers (see the text for more detail). The adjusted diagnostic specificity of rRT-PCR was 100%.
EV-D68 load and correlation with disease severity.
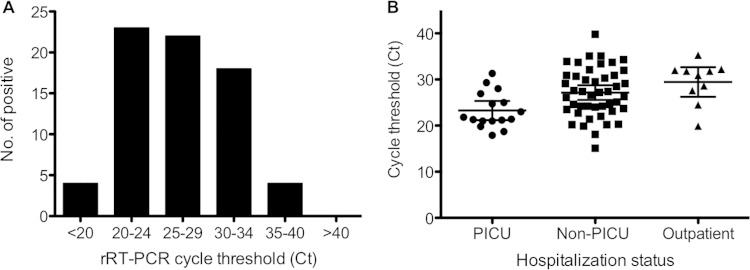
To estimate the EV-D68 RNA load, the rRT-PCR amplification cycle threshold (CT) values for 71 EV-D68-positive NP specimens confirmed by sequencing analysis were retrieved. The CT values of EV-D68-positive specimens ranged from 15.1 to 39.8, with an average CT value of 26.6. The distribution of the CT values among EV-D68-positive specimens is shown in Fig. 2A. It is noteworthy that 67 of 71 (94.4%) EV-D68-positive samples had a CT of ≤35.0, suggesting that the EV-D68 load in these NP specimens was relatively high.
FIG 2.
(A) Distribution of the cycle threshold (CT) of EV-D68-positive specimens confirmed by VP1 sequencing analysis (n = 71). (B) Distribution and mean CT values (± standard deviations) of EV-D68-positive specimens from hospitalized pediatric ICU (PICU) patients, non-PICU patients, or outpatients.
Of 71 patients that were positive by rRT-PCR and confirmed by VP1 sequencing analysis, 48 (67.6%) were male and 23 (32.4%) were female. The ages of the patients ranged from 6 months to 15 years, with an average of 5.5 years. Sixty-one (85.9%) of these patients required hospitalization; 16 (22.5%) were admitted to a pediatric intensive care unit (PICU), and 45 (63.4%) were admitted to non-PICUs. The median CT values for the patients admitted to PICUs and non-PICUs and the outpatients were 21.6, 27.2, and 31.4, respectively (P = 0.0046), suggesting that a higher viral load was present in clinical specimens from patients with more-severe respiratory illnesses (Fig. 2B).
Enterovirus and rhinovirus in NP specimens.
The identity of viruses was determined for 194 NP specimens that were positive for HRV/EV by RP assay based on DNA sequence analysis of the VP1 and 5′-UTR regions (Table 2). No EV-D68 was detected for the 20 specimens collected in 2013 during the same period. Of the 174 HRV/EV-positive NP specimens collected in 2014, enteroviruses and rhinoviruses were detected in 80 (46.0%) and 94 (54.0%), respectively. Overall, EV-D68 was detected in 72 of 174 (41.4%) HRV/EV-positive specimens that were collected during this outbreak and that were examined by DNA sequencing. HRV-C (n = 47, 27.0%) was the predominant rhinovirus, followed by HRV-B (n = 24, 13.8%) and HRV-A (n = 23, 13.2%).
TABLE 2.
Enterovirus subgroups determined by DNA sequencing analysis in nasopharyngeal swab specimens that were positive for human enterovirus/rhinovirus by RP assay
| Organisma | No. (%) of positive specimens (n = 194)b |
||
|---|---|---|---|
| 2013 (n = 20) | 2014 (n = 174) | Total (n = 194) | |
| Enterovirus | |||
| EV-D68 | 0 | 72 (41.4) | 72 (37.1) |
| Coxsackievirus | 0 | 4 (2.3) | 4 (2.1) |
| Echovirus | 1 (5.0) | 3 (1.7) | 4 (2.1) |
| Enterovirus 71 | 0 | 1 (0.6) | 1 (0.5) |
| Rhinovirus | |||
| HRV-A | 5 (25.0) | 23 (13.2) | 28 (14.4) |
| HRV-B | 4 (20.0) | 24 (13.8) | 28 (14.4) |
| HRV-C | 10 (50.0) | 47 (27.0) | 57 (29.4) |
| Total | 20 | 174 (100) | 194 (100) |
HRV-A, human rhinovirus A; HRV-B, human rhinovirus B; HRV-C, human rhinovirus C.
Results included 20 of 25 NP specimens (2013) and 174 of 260 NP specimens (2014) examined by both rRT-PCR and DNA sequencing analysis.
Comparison of VP1 and 5′-UTR sequences for EV-D68 determinations.
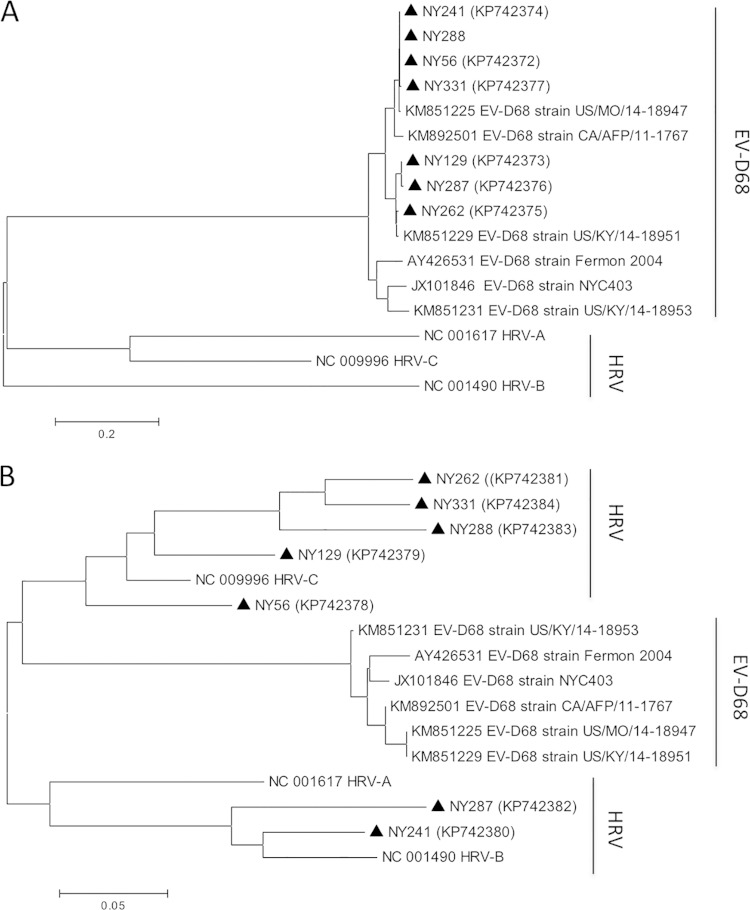
Of 194 NP specimens analyzed by DNA sequencing, 72 were confirmed to be EV-D68 on the basis of partial VP1 sequences. Interestingly, EV-D68 positivity was determined in only 65 (90.2%) of 72 confirmed specimens by sequencing analysis of the 5′-UTR of EV genome; the partial 5′-UTR sequences of the remaining 7 specimens corresponded to HRV by BLASTN analysis. Phylogenetic trees based on the partial VP1 and 5′-UTR sequences of selected EV-D68-positive specimens are shown in Fig. 3. It is unclear whether this discrepant result was due to mixed infections of EV-D68 and rhinoviruses or resulted from recombination in different regions of the EV-D68 genome in these specimens.
FIG 3.
Phylogenetic trees based on partial VP1 (A) and 5′-UTR (B) sequences of selected EV-D68-positive specimens. Specimens from this study are marked with triangles, with accession numbers in parentheses. Specimens NY56 and NY288 had identical partial VP1 sequences. Reference sequences from 6 strains of EV-D68 and 3 strains of HRV and the corresponding accession numbers were included for comparison. The neighbor-joining algorithm was used.
DISCUSSION
With the worldwide emergence of multiple clades of EV-D68 and an outbreak of severe respiratory illness associated with EV-D68 infection in U.S. children, there is an urgent public health need for rapid and accurate detection and identification of EV-68 in clinical samples (6, 8, 9). Unfortunately, most public health and clinical laboratories in North America and Europe lack such capabilities (19). The current reference method to determine the presence of EV-D68 in the United States requires seminested PCR amplification and subsequent analysis of the VP1 sequences, which is not feasible for most clinical laboratories (15). In this study, we validated the diagnostic utility of an EV-D68-specific rRT-PCR assay that was recently developed by the CDC (16) using a large collection of NP swab specimens from patients involved in a recent outbreak of respiratory illness in the lower Hudson Valley in New York State. Among 768 NP specimens examined by the FilmArray RP assay, the test results corresponding to 92 EV-D68-positive patients, including 20 reported by the NYSDOH Wadsworth Center and 72 determined in this study, were confirmed. The probable prevalence of EV-D68 infection during this outbreak was 12.0% (92/768) for our patient population. Compared to VP1 sequence-based reference methods, the EV-D68 rRT-PCR assay showed diagnostic sensitivity of 98.6% and specificity of 97.5%, with estimated positive and negative predictive values of 96.0% and 99.2%, respectively, for this contrived population. To our knowledge, this study is the first report that has independently assessed the performance characteristics of the CDC EV-D68 rRT-PCR protocol in a clinical laboratory setting. Since 67 of 71 (94.4%) confirmed EV-D68 rRT-PCR-positive samples had relatively high viral loads, with CT values of ≤35.0, these samples are most likely to give positive results if they are retested at different clinical and public health laboratories, even after considering potential variations in sample processing and testing protocols. Our data provide strong evidence that the EV-D68 rRT-PCR is a reliable assay for detection of EV-D68 in clinical samples and has the potential to be used routinely in clinical and public health laboratories for rapid diagnosis and outbreak investigation of EV-D68-associated infections.
More than 100 serotypes of rhinoviruses, belonging to three species (HRV-A to HRV-C), with unknown clinical significance have been previously described (17). Our report provides some preliminary data on the molecular epidemiology of rhinovirus-associated infections among patients in our geographic region. HRV-C (29.4%) appears to have been the predominant species in 2013 and 2014, followed by HRV-B (14.4%) and HRV-A (14.4%).
Discrepant topology data in the phylogenetic trees was observed for clinical specimens from 7 patients. While analysis of partial DNA sequencing of the VP1 region confirmed that all 7 patients had EV-D68, rhinovirus-specific DNA sequences were identified on the basis of analysis of the 5′-UTR of the EV genome. These results were most likely due to a mixed infection of EV-D68 and a rhinovirus in the same clinical sample or to recombination of the 5′-UTR sequences between EV-D68 and rhinovirus. Frequent recombination of DNA sequences in the 5′-UTR has been reported among different rhinovirus species and within the same rhinovirus species (17). Since both partial VP1 and 5′-UTR sequences, alone or in combination, have been employed previously for determination of EV-D68 (5, 15), our data imply that analysis of VP1 sequence is more reliable for identification of EV-D68 in clinical samples where a mixed infection may occur.
One limitation of this study was that all clinical samples were collected from patients with respiratory illness in the lower Hudson Valley of New York State in 2013 and during an outbreak in 2014. Nevertheless, comparative analysis of available DNA sequences from patients in St. Louis, Missouri (20, 21), and patients in California (22) suggests that this EV-D68 real-time RT-PCR assay was able to detect other EV-D68 strains associated with a national outbreak of EV-D68 infections in 2014. This is further confirmed by a CDC internal validation study in which 100% sensitivity and 96% specificity were achieved when 134 respiratory specimens from several U.S. states were analyzed (16).
In conclusion, assessment of an EV-D68-specific real-time RT-PCR assay in a diagnostic microbiology laboratory has confirmed that the EV-D68 rRT-PCR assay is a reliable test for rapid detection of EV-D68 infection in NP swab samples from patients with symptomatic respiratory illness. This test, starting from RNA extraction and followed by a single step of combined RT and PCR, takes 3 to 4 h to confirm or rule out an infection with EV-D68. It has the potential to be employed as a tool for routine laboratory diagnosis and outbreak investigation of EV-D68-associated infection in clinical and public health laboratories.
ACKNOWLEDGMENTS
We thank W. A. Nix and M. Steven Oberste from CDC for providing study protocols and NYSDOH Wadsworth Center for EV-D68 testing of our referral samples.
We declare that we have no interests of conflict.
REFERENCES
- 1.Muir P, Kammerer U, Korn K, Mulders MN, Poyry T, Weissbrich B, Kandolf R, Cleator GM, Van Loon AM. 1998. Molecular typing of enteroviruses: current status and future requirements. Clin Microbiol Rev 11:202–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobs SE, Lamson DM, St George K, Walsh TJ. 2013. Human rhinoviruses. Clin Microbiol Rev 26:135–162. doi: 10.1128/CMR.00077-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schieble JH, Fox VL, Lennette EH. 1967. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol 85:297–310. [DOI] [PubMed] [Google Scholar]
- 4.Khetsuriani N, Lamonte-Fowlkes A, Oberst S, Pallansch MA; Centers for Disease Control and Prevention. 2006. Enterovirus surveillance—United States, 1970–2005. MMWR Surveill Summ 55:1–20. [PubMed] [Google Scholar]
- 5.Imamura T, Fuji N, Suzuki A, Tamaki R, Saito M, Aniceto R, Galang H, Sombrero L, Lupisan S, Oshitani H. 2011. Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg Infect Dis 17:1430–1435. doi: 10.3201/eid1708.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokarz R, Firth C, Madhi SA, Howie SR, Wu W, Sall AA, Haq S, Briese T, Lipkin WI. 2012. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol 93:1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meijer A, Van Der Sanden S, Snijders BE, Jaramillo-Gutierrez G, Bont L, Van Der Ent CK, Overduin P, Jenny SL, Jusic E, Van Der Avoort HG, Smith GJ, Donker GA, Koopmans MP. 2012. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology 423:49–57. doi: 10.1016/j.virol.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 8.CDC. 2011. Clusters of acute respiratory illness associated with human enterovirus 68–Asia, Europe, and United States, 2008–2010. MMWR Morb Mortal Wkly Rep 60:1301–1304. [PubMed] [Google Scholar]
- 9.Imamura T, Oshitani H. 3 December 2014, posting date Global reemergence of enterovirus D68 as an important pathogen for acute respiratory infections. Rev Med Virol doi: 10.1002/rmv.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang M, Mirand A, Savy N, Henquell C, Maridet S, Perignon R, Labbe A, Peigue-Lafeuille H. 2014. Acute flaccid paralysis following enterovirus D68 associated pneumonia, France, 2014. Euro Surveill 19:pii=20952 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20952. [DOI] [PubMed] [Google Scholar]
- 11.Imamura T, Suzuki A, Lupisan S, Okamoto M, Aniceto R, Egos RJ, Daya EE, Tamaki R, Saito M, Fuji N, Roy CN, Opinion JM, Santo AV, Macalalad NG, Tandoc A III, Sombrero L, Olveda R, Oshitani H. 2013. Molecular evolution of enterovirus 68 detected in the Philippines. PLoS One 8:e74221. doi: 10.1371/journal.pone.0074221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Midgley CM, Jackson MA, Selvarangan R, Turabelidze G, Obringer E, Johnson D, Giles BL, Patel A, Echols F, Oberste MS, Nix WA, Watson JT, Gerber SI. 2014. Severe respiratory illness associated with enterovirus D68—Missouri and Illinois, 2014. MMWR Morb Mortal Wkly Rep 63:798–799. [PMC free article] [PubMed] [Google Scholar]
- 13.CDC. 18 January 2015, accession date Enterovirus D68 in the United States, 2014. CDC, Atlanta, GA: http://www.cdc.gov/non-polio-enterovirus/outbreaks/ev-d68-outbreaks.html. [Google Scholar]
- 14.Division of Viral Diseases, National Centers for Immunization and Respiratory Diseases, CDC; Division of Vector-Borne Diseases, Division of High-Consequence Pathogens and Pathology, National Center for Emerging and Zoonotic Infectious Diseases, CDC; Children's Hospital Colorado; Council of State and Territorial Epidemiologists. 2015. Notes from the field: acute flaccid myelitis among persons aged ≤21 years—United States, August 1–November 13, 2014. MMWR Morb Mortal Wkly Rep 63:1243–1244. [PMC free article] [PubMed] [Google Scholar]
- 15.Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol 44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CDC. 2014. Enterovirus D68 (EV-D68) 2014 outbreak strain-specific real-time reverse transcription/polymerase chain reaction (rRT-PCR) assay instructions—version 10/14/2014, p 1–13. CDC, Atlanta, GA: http://www.cdc.gov/non-polio-enterovirus/hcp/ev-d68-hcp.html. [Google Scholar]
- 17.Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, Fraser-Liggett CM, Liggett SB. 2009. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science 324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaramillo-Gutierrez G, Benschop KS, Claas EC, De Jong AS, Van Loon AM, Pas SD, Pontesilli O, Rossen JW, Swanink CM, Thijsen S, Van Der Zanden AG, Van Der Avoort HG, Koopmans MP, Meijer A. 2013. September through October 2010 multi-centre study in the Netherlands examining laboratory ability to detect enterovirus 68, an emerging respiratory pathogen. J Virol Methods 190:53–62. doi: 10.1016/j.jviromet.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Wylie KM, Wylie TN, Orvedahl A, Buller RS, Herter BN, Magrini V, Wilson RK, Storch GA. 2015. Genome sequence of enterovirus D68 from St. Louis, Missouri, USA. Emerg Infect Dis 21:184–186. doi: 10.3201/eid2101.141605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown BA, Nix WA, Sheth M, Frace M, Oberste MS. 2014. Seven strains of enterovirus D68 detected in the United States during the 2014 severe respiratory disease outbreak. Genome Announc 2:e01201-14. doi: 10.1128/genomeA.01201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayscue P, Van Haren K, Sheriff H, Waubant E, Waldron P, Yagi S, Yen C, Clayton A, Padilla T, Pan C, Reichel J, Harriman K, Watt J, Sejvar J, Nix WA, Feikin D, Glaser C; Centers for Disease Control and Prevention (CDC) . 2014. Acute flaccid paralysis with anterior myelitis—California, June 2012–June 2014. MMWR Morb Mortal Wkly Rep 63:903–906. [PMC free article] [PubMed] [Google Scholar]