Abstract
We compared two walk-away molecular diagnostic assays, the GeneXpert MRSA Gen 3 assay and the BD-Max MRSA XT assay. A total of 119 prospective swabs and 36 culture-positive samples were tested. Xpert MRSA Gen 3 had sensitivity of 95.7% and specificity of 100% versus 87.5% and 97.1% for BD-Max. The difference in agreement with the enriched culture results was significantly in favor of the Xpert assay (P < 0.02, McNemar nonparametric text).
TEXT
The transmission of methicillin-resistant Staphylococcus aureus (MRSA) from patient to patient in health care settings is a major concern due to a poorer prognosis associated with severe MRSA infections, such as bacteremia (1, 2). The rise in the incidence of methicillin-resistant Staphylococcus aureus infections was initially restricted to a few health care-associated clones and confined to a limited set of patients in high-risk groups. The molecular detection of MRSA directly from clinical samples is complicated by the frequent association of methicillin-susceptible S. aureus with methicillin-resistant coagulase-negative staphylococci. Distinguishing between this association and true MRSA requires the specific detection of the junction of the staphylococcal cassette chromosome in the orfX locus as well as positive detection of the mec gene encoding methicillin resistance (3). With the onset of community-acquired MRSA (4), outpatients and emergency room patients became at risk for MRSA carriage, further increasing the need for determination of carriage of MRSA. More recently, livestock-associated MRSA carrying variant mecC genes further complicated the epidemiology of MRSA by increasing its molecular diversity (5). Microbiology laboratories usually define MRSA phenotypically, but the underlying genetic structures responsible for this phenotype are constantly changing (6–8), and the molecular assays designed several years back are not necessarily relevant to the epidemiology observed today (9–11). A recent comparison of culture and a commercial molecular assay performed in a high-prevalence environment showed the molecular assay to be slightly less sensitive than enriched culture using ChromID MRSA agar but also showed that the results were not significantly different, although the molecular assay yielded results in hours instead of days (12).
Two fully automated walk-away platforms offer molecular detection of MRSA in nasal swabs: the BD-Max (Becton Dickinson, Le Pont de Claix, France) and the GeneXpert (Cepheid, Maurens Scopont, France) platforms. Both tests rely on the same principle (3) and detect the presence of the mecA gene and the mecC gene (mecA/C) as well as the presence of the junction between the staphylococcal cassette chromosome mec element (SCCmec) and the chromosome. However, the sequences of the primers and probes in the two assays are likely distinct, and the extraction processes rely on different principles. The performances of the assays of the previous generations have recently been found to be similar (13), but the release of updated versions warrants a renewed comparison. Moreover, the MRSA clones isolated in Europe and the United States are epidemiologically different and assay performances could be different (14, 15).
We evaluated the performances of early-release versions of Xpert MRSA Gen3 and BD-Max MRSA XT in Hospital Raymond Poincaré, Garches, France, a teaching hospital in the Paris public hospital system (Assistance Publique-Hôpitaux de Paris) affiliated with the Université de Versailles St. Quentin. At the time of the study, Xpert MRSA Gen3 was designated “research use only” (RUO) whereas BD-Max MRSA XT was CE-In Vitro Diagnostics (CE-IVD) marked. Patients in high-prevalence wards (spinal cord rehabilitation, intensive care unit, and bone and joint infection service) are routinely monitored for MRSA carriage, with the anterior nares swabbed on admission and weekly screening thereafter.
A prospective study that included 119 screened patients was performed in the course of 45 days using Copan Transystem Liquid Stuart single swabs to sample both anterior nares. The testing of leftover samples taken in the course of normal care is institutional review board (IRB) exempt according to French law. In order to ensure that all techniques were performed with identical starting materials, each swab was discharged in 500 μl of isotonic saline solution in a CryoFreeze vial (Nalgene). A 100-μl volume of the solution was used for selective culture on ChromID MRSA agar (bioMérieux, Marcy l'Etoile, France) and for overnight enrichment in tryptic soy broth (Bio-Rad, Marnes-La-Coquette, France) at 36°C. A 100-μl volume of the broth was then plated on MRSA selective agar, with review of the plates performed after 24 h and 48 h. A 100-μl volume (each) was used for molecular detection on the BD-Max and Xpert platforms according to each manufacturer's instructions. This process was adapted with minor modifications from the method used by Dalpke et al. (16) and validated using a quality control panel of organisms, including mecC isolates (Methicillin Resistant S. aureus 2013 EQA Programme; Qnostics [Glasgow, United Kingdom]); both assays successfully categorized all 12 samples. This divergence from the procedure recommended by the manufacturers could prove to lead to biased results, although we decided to favor identical starting materials for the two methods rather than the use of different swabs with potentially different starting materials.
Twelve of the 119 swabs prospectively tested were MRSA positive by culture (10%), 11 samples providing an actionable result within 24 h (sensitivity of 91.67%) and 1 requiring broth enrichment. This sample was detected by both the Xpert and BD-Max assays, suggesting that a low inoculum load is not a difficulty for molecular assays. The positive predictive values (PPV) for the Xpert and BD-Max assays were, respectively, 100% and 72.7%, and the negative predictive values (NPV) were 99.0% and 96.1%. One direct-culture-positive swab was detected by neither Xpert nor BD-Max. An isolated MRSA colony was resuspended in saline solution, and 107 CFU was added to the elution reagent and an identical amount to the sample buffer. Both assays were positive for the amplification of the internal control and the mecA/C gene targets but negative for SCC and mec right extremity junction (MREJ), hinting at incomplete primer and probe repertoires for this target. Three samples negative by culture and Xpert MRSA Gen 3 yielded positive MRSA detection by BD-Max MRSA XT. Results of further examination of the real-time fluorescence curves were suggestive of false positives of the detection algorithm as they were undistinguishable from the curves of the negative samples that were run simultaneously. This “false-positive” call is easily detected by review of the graphs as long as a positive sample is included in the run (Fig. 1), but detection of such a call can be challenging in the absence of positive samples because of the lack of fluorescence units and the autoscaling of the graphs.
FIG 1.
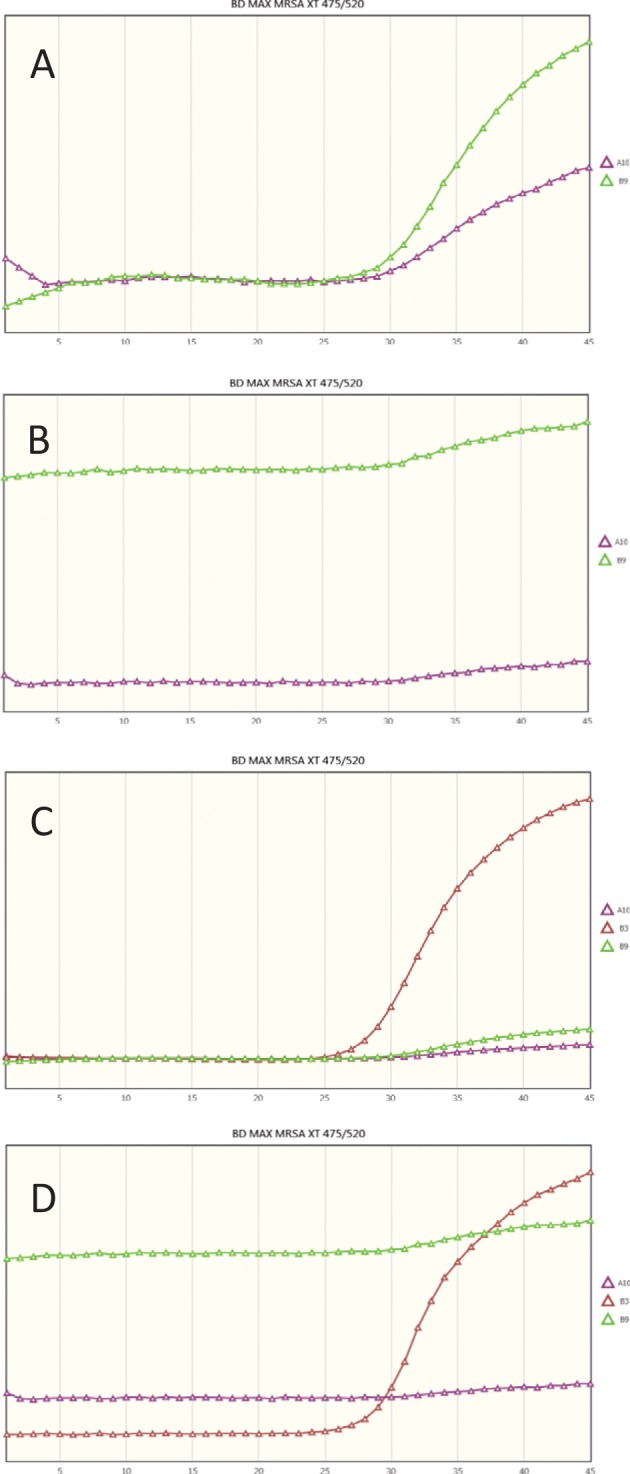
Representative MREJ fluorescence curves generated by the BD-Max MRSA XT in the same amplification and detection run. (A) “Backgrounded” PCR without true positive. (B) “Color-compensated” PCR without true positive. (C) “Backgrounded” PCR with true positive. (D) “Color-compensated” PCR with true positive. A10, MRSA true-negative sample; B3, MRSA true-positive sample; B9, MRSA false-positive sample. The software calls a positive MRSA result in the absence of amplification in the MREJ channel as determined by naked-eye interpretation of the curves. (A) The absence of fluorescence units on the graph and the autoscaling make the call difficult in the backgrounded PCR view in the absence of a positive result in the run. (C and D) In the presence of a true positive (sample B3), the lack of amplification of MREJ in the false positive (sample B9) is readily apparent.
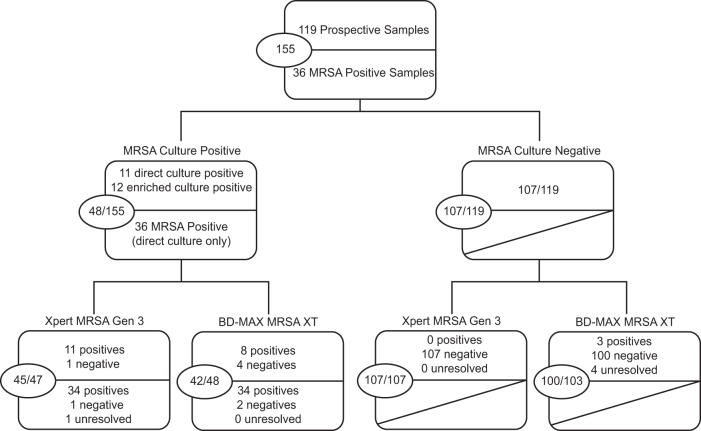
To increase the number of positive samples tested on the molecular platforms, the swabs taken thereafter were similarly discharged in saline solution and the suspension was stored at 4°C for 18 to 24 h until the culture results were known. For 36 direct-culture-positive specimens, the molecular assays were then performed as previously described. The results are displayed in the flow chart (Fig. 2), and the performance of the assays is quantified in Table 1. Overall, the agreement of the Xpert MRSA Gen 3 results with culture results proved to be significantly superior to that of the BD-Max results as determined by the McNemar test (P = 0.023), with better sensitivity and specificity and a lower (0.6% versus 2.6%) and yet not statistically significant number of unresolved samples. Considering the whole study set, the PPV of Xpert MRSA remains at 100% whereas the PPV of BD-Max MRSA XT improves to reach 93.3%. However, the assays were not used strictly according to the manufacturer's recommendation, and only 20% of the total inoculum was tested using each technique. Three of the four samples detected by Xpert MRSA Gen3 had SCC threshold cycle (CT) values greater than 33, suggesting that the sample could possibly have been detected by both platforms using the entire swab. The fourth sample had a CT value of 19.1, suggestive of an incomplete primer-probe repertoire in BD-Max MRSA XT. It is also noteworthy that only half of the nucleic acids extracted from the sample for BD-Max are used for the PCR, the other half being available for storage or further processing.
FIG 2.
Flow chart of the clinical samples included in the study according to ChromID culture status. The top half of each box includes results from the prospective arm of the study. The lower half includes results from the culture-positive samples, only for the MRSA culture-positive specimens. Overall numbers are shown in the circle left at the left side of the box. Unresolved samples (1 for Xpert, 4 for BD-Max) are excluded from the analysis.
TABLE 1.
Results obtained in comparison to enriched culturea
| Assay | No. of samples |
% Sen | % Spe | % PPV | % NPV | % Agreement | |||
|---|---|---|---|---|---|---|---|---|---|
| TP | FP | FN | TN | ||||||
| GeneXpert | 45 | 0 | 2 | 107 | 95.7 | 100 | 100 | 99.0 | 98.7 |
| BD-Max | 42 | 3 | 6 | 100 | 87.5 | 97.1 | 93.3 | 96.1 | 94.0 |
TP, true positive; FP, false positive; FN, false negative; TN, true negative; Sen, sensitivity; Spe, specificity; PPV, positive predictive value; NPV, negative predictive value; Agreement, agreement between each method and the reference method (culture with enrichment). Unresolved samples were excluded from the summarized analysis. Classifying unresolved samples as failed (i.e., negative results as true-positive results and positive results as true-negative results), the sensitivity of Xpert would drop to 93.8% and the specificity of BD-Max would drop to 93.5%. The difference between Xpert agreement with culture and BD-Max agreement with culture is significant (McNemar nonparametric test; P = 0).
These results show that the performance of molecular assays for detection of methicillin resistance in S. aureus needs to be reevaluated periodically to ensure that they continue to match the current and local epidemiology of the organisms. Both of the tests reviewed here have been updated in terms of mecC detection as evidenced by the good performance seen with the mecC quality control isolate. A few isolates that still thwart these assays will be further analyzed in terms of SCC type and genetic makeup of the SCC.
Previous studies have addressed the laboratory efficiency of both platforms using earlier assay versions and have found discrepancies in the turnaround times depending on the number of assays processed and the maximum capacity of the instrument (13). Overall, the hands-on time requirements per sample with the two methods were found to be comparable in our experience. The turnaround time for the Xpert MRSA cartridge is 58 min once it has been loaded, and the throughput is dependent on the number of units installed. The instrument allows each of the slots to be used independently and provides a random access architecture allowing “on demand” testing by personnel with low to medium technical qualifications. The total turnaround time of the BD-Max assay is longer (around 120 min) and varies depending on the number of samples processed simultaneously, since a four-channel robotic arm performs all the pipetting. The instrument is not fully random access and is best suited to work with series of up to 24 samples. The BD-Max offers the interesting possibility of porting laboratory-developed assays to an automated walk-away molecular assay platform.
Overall, the RUO Xpert MRSA Gen 3 assay showed significantly better analytical performance than the BD MAX MRSA XT assay in testing a cohort of hospitalized patients in the setting of a French tertiary care center.
ACKNOWLEDGMENTS
We thank Becton Dickinson for the loan of the BD-Max for evaluation purposes and Cepheid Europe for the loan of the GeneXpert instrument.
This study was funded in part by Cepheid Europe. M.R. was a consultant for Cepheid and Becton Dickinson.
REFERENCES
- 1.Schweizer ML, Furuno JP, Harris AD, Johnson JK, Shardell MD, McGregor JC, Thom KA, Cosgrove SE, Sakoulas G, Perencevich EN. 2011. Comparative effectiveness of nafcillin or cefazolin versus vancomycin in methicillin-susceptible Staphylococcus aureus bacteremia. BMC Infect Dis 11:279. doi: 10.1186/1471-2334-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stryjewski ME, Szczech LA, Benjamin DK Jr, Inrig JK, Kanafani ZA, Engemann JJ, Chu VH, Joyce MJ, Reller LB, Corey GR, Fowler VG Jr. 2007. Use of vancomycin or first-generation cephalosporins for the treatment of hemodialysis-dependent patients with methicillin-susceptible Staphylococcus aureus bacteremia. Clin Infect Dis 44:190–196. doi: 10.1086/510386. [DOI] [PubMed] [Google Scholar]
- 3.Huletsky A, Giroux R, Rossbach V, Gagnon M, Vaillancourt M, Bernier M, Gagnon F, Truchon K, Bastien M, Picard FJ, van Belkum A, Ouellette M, Roy PH, Bergeron MG. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J Clin Microbiol 42:1875–1884. doi: 10.1128/JCM.42.5.1875-1884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kajita E, Okano JT, Bodine EN, Layne SP, Blower S. 2007. Modelling an outbreak of an emerging pathogen. Nat Rev Microbiol 5:700–709. doi: 10.1038/nrmicro1660. [DOI] [PubMed] [Google Scholar]
- 5.Trouillet-Assant S, Rasigade JP, Lustig S, Lhoste Y, Valour F, Guerin C, Aubrun F, Tigaud S, Laurent F. 2013. Ward-specific rates of nasal cocolonization with methicillin-susceptible and -resistant Staphylococcus spp. and potential impact on molecular methicillin-resistant Staphylococcus aureus screening tests. J Clin Microbiol 51:2418–2420. doi: 10.1128/JCM.00491-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dauwalder O, Lina G, Durand G, Bes M, Meugnier H, Jarlier V, Coignard B, Vandenesch F, Etienne J, Laurent F. 2008. Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J Clin Microbiol 46:3454–3458. doi: 10.1128/JCM.01050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heusser R, Ender M, Berger-Bachi B, McCallum N. 2007. Mosaic staphylococcal cassette chromosome mec containing two recombinase loci and a new mec complex, B2. Antimicrob Agents Chemother 51:390–393. doi: 10.1128/AAC.00921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas L, van Hal S, O'Sullivan M, Kyme P, Iredell J. 2008. Failure of the BD GeneOhm StaphS/R assay for identification of Australian methicillin-resistant Staphylococcus aureus strains: duplex assays as the “gold standard” in settings of unknown SCCmec epidemiology. J Clin Microbiol 46:4116–4117. doi: 10.1128/JCM.01146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalpke AH, Hofko M, Zimmermann S. 2012. Comparison of the BD Max methicillin-resistant Staphylococcus aureus (MRSA) assay and the BD GeneOhm MRSA achromopeptidase assay with direct- and enriched-culture techniques using clinical specimens for detection of MRSA. J Clin Microbiol 50:3365–3367. doi: 10.1128/JCM.01496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hombach M, Pfyffer GE, Roos M, Lucke K. 2010. Detection of methicillin-resistant Staphylococcus aureus (MRSA) in specimens from various body sites: performance characteristics of the BD GeneOhm MRSA assay, the Xpert MRSA assay, and broth-enriched culture in an area with a low prevalence of MRSA infections. J Clin Microbiol 48:3882–3887. doi: 10.1128/JCM.00670-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindqvist M, Isaksson B, Grub C, Jonassen TO, Hallgren A. 2012. Detection and characterisation of SCCmec remnants in multiresistant methicillin-susceptible Staphylococcus aureus causing a clonal outbreak in a Swedish county. Eur J Clin Microbiol Infect Dis 31:141–147. doi: 10.1007/s10096-011-1286-y. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Park YJ, Park KG, Jekarl DW, Chae H, Yoo JK, Seo SW, Choi JE, Lim JH, Heo SM, Seo JH. 2013. Comparative evaluation of three chromogenic media combined with broth enrichment and the real-time PCR-based Xpert MRSA assay for screening of methicillin-resistant Staphylococcus aureus in nasal swabs. Ann Lab Med 33:255–260. doi: 10.3343/alm.2013.33.4.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widen R, Healer V, Silbert S. 2014. Laboratory evaluation of the BD MAX MRSA assay. J Clin Microbiol 52:2686–2688. doi: 10.1128/JCM.00237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baud O, Giron S, Aumeran C, Mouly D, Bardon G, Besson M, Delmas J, Coignard B, Tristan A, Vandenesch F, Illes G, Lesens O. 2014. First outbreak of community-acquired MRSA USA300 in France: failure to suppress prolonged MRSA carriage despite decontamination procedures. Eur J Clin Microbiol Infect Dis 33:1757–1762. doi: 10.1007/s10096-014-2127-6. [DOI] [PubMed] [Google Scholar]
- 15.Stegger M, Wirth T, Andersen PS, Skov RL, De Grassi A, Simoes PM, Tristan A, Petersen A, Aziz M, Kiil K, Cirkovic I, Udo EE, del Campo R, Vuopio-Varkila J, Ahmad N, Tokajian S, Peters G, Schaumburg F, Olsson-Liljequist B, Givskov M, Driebe EE, Vigh HE, Shittu A, Ramdani-Bougessa N, Rasigade JP, Price LB, Vandenesch F, Larsen AR, Laurent F. 2014. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. mBio 5:e01044–14. doi: 10.1128/mBio.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalpke AH, Hofko M, Stock C, Zimmermann S. 2014. Evaluation of the BD Max MRSA XT assay for use with different swab types. J Clin Microbiol 52:4343–4346. doi: 10.1128/JCM.02306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]