Abstract
The OqxAB efflux pump, a plasmid-mediated quinolone resistance (PMQR) determinant, has become increasingly prevalent among members of Enterobacteriaceae over the past decade. To investigate the evolution and dissemination routes of the oqxAB operon, we assessed the prevalence of oqxAB-like elements among various Gram-negative bacterial species and analyzed the genotypic and phenotypic characteristics of organisms harboring such elements. With a comprehensive genotyping approach, a chromosome-based oqxAB operon was detectable in all Klebsiella pneumoniae strains tested, including organisms isolated before the year 1984. Sequence and phylogenetic analyses confirmed that the oqxAB operon in K. pneumoniae isolates was genetically closest to their plasmid-borne counterparts recoverable only from Escherichia coli and Salmonella isolates collected from the year 2003 onward. Chromosomal elements with much lower sequence homology were also found among the Enterobacter spp. but not other Gram-negative species. Contrary to the quinolone resistance phenotypes which were consistently observable among organisms with oqxAB-harboring plasmids, chromosomal oqxAB elements generally did not confer quinolone resistance, except for K. pneumoniae strains, which exhibited a typical oqxAB-mediated phenotype characterized by cross-resistance to olaquindox, chloramphenicol, and the quinolones. Gene expression analysis illustrated that such phenotypes were due to elevated expression of the chromosomal oqxAB operon. Furthermore, transposition of the oqxAB operon from the bacterial chromosome to plasmids was found to result in a >80-fold increase in the level of expression of the OqxAB pump, confirming its status as the first constitutively expressed efflux system located in bacterial mobile elements.
INTRODUCTION
The mobile efflux pump OqxAB, first identified in Escherichia coli in 2003, belongs to the RND family and shares up to 40% homology with other RND-type efflux systems such as AcrAB in Escherichia coli and MexAB in Pseudomonas aeruginosa (1). At the time of its discovery, the gene encoding this pump was located in a conjugative plasmid designated pOLA52 and was found to contribute to phenotypic resistance toward nalidixic acid and chloramphenicol and reduced susceptibility to ciprofloxacin in Escherichia coli (2, 3). Since then, oqxAB has been frequently detected as a plasmid-mediated quinolone resistance (PMQR) determinant among members of Enterobacteriaceae (4–6). Sequencing analysis of pOLA52 initially showed that oqxAB, together with an open reading frame orf68 of unknown function, was flanked by the insertion sequence IS26 (2). A set of corresponding genes that shared 99% nucleotide homology with the oqxAB operon in pOLA52, including an oqxR gene that was genetically identical to the plasmid-borne orf68 element, was subsequently detectable in the genome of Klebsiella pneumoniae, which did not exhibit phenotypic resistance to either nalidixic acid or chloramphenicol (7). More recently, Bialek-Davenet et al. (8) showed that mutations in oqxR induced overexpression of not only oqxAB but also rarA, which encoded the oqxAB transcriptional activator in K. pneumoniae. These findings infer that a mutated oqxR gene is required to elicit overexpression of oqxAB and cross-resistance to quinolone and chloramphenicol in K. pneumoniae. Despite these findings, however, the evolutionary origin of oqxAB-harboring plasmids and the molecular basis of the differential phenotypes observable in organisms harboring the chromosomal and plasmid-borne oqxAB genes remain ill defined. First, although oqxAB was detected frequently in K. pneumoniae, concrete evidence showing that oqxAB is intrinsic to this bacterial species is not available, as failure of oqxAB detection in K. pneumoniae is common (9, 10). In addition, oqxAB homologues in other members of Enterobacteriaceae, including Enterobacter aerogenes and Enterobacter cloacae and some other Klebsiella spp. (7), were also identified, prompting a need to perform cross-species analysis of the pattern of distribution for both chromosomal and plasmid-borne oqxAB-like elements and the respective roles of such elements in conferring phenotypic resistance. Second, whether translocation of the oqxAB genes from chromosome to plasmid results in overexpression of this efflux pump is not clear. Currently, data regarding the expression level of oqxAB in pOLA52 and the regulatory mechanisms concerned are not available.
To address the above issues, we performed a comprehensive assessment of the prevalence of the oqxAB genes in various members of Enterobacteriaceae recovered from different time periods and regions to map the evolution and dissemination routes of this antibiotic resistance determinant. We then performed genetic analysis of oqxAB-like elements recoverable from the test strains to obtain evidence which suggests that the plasmid-borne oqxAB operon originated from the chromosome of K. pneumoniae and evolved to become even more functionally active than their chromosomal counterparts.
MATERIALS AND METHODS
Bacterial isolates.
Eighty-five clinical K. pneumoniae isolates were collected from The Prince of Wales Hospital, Hong Kong, among which 15 were isolated in or before 1984. Another 8 K. pneumoniae isolates were obtained from the Salmonella Genetic Stock Centre (SGSC) at the University of Calgary, Canada (http://people.ucalgary.ca/∼kesander/), including one K. pneumoniae type strain (MGH 78578). Fifty-seven isolates of other bacterial species collected from the SGSC, including Klebsiella oxytoca (n = 8), E. cloacae (n = 27), E. aerogenes (n = 15), Serratia marcescens (n = 3), Serratia odorifera (n = 2), and Serratia liquefaciens (n = 2), and 30 clinical isolates each of Salmonella spp., Pseudomonas aeruginosa, Acinetobacter spp., Vibrio parahaemolyticus, other Vibrio spp., Staphylococcus aureus, and Enterococcus spp. were also included in this study. An oqxAB-positive Salmonella enterica serovar Typhimurium clinical isolate, ST07-37, isolated in 2007 at The Prince of Wales Hospital, Hong Kong, was used in gene expression analysis.
PCR and sequence analysis.
The prevalence of oqxAB among different bacterial species was determined by utilizing multiple PCR primer sets (oqxA, oqxB, hae, oqxAB2, and oqxAB4) targeting different yet overlapping regions of the oqxAB operon in pOLA52 (Fig. 1). The presence of the IS26 element upstream of oqxA was detected by the primer set IS26-oqxA. All PCR amplicons were subjected to nucleotide sequencing for confirmation. Primers used in this study are listed in Table 1. The full length of the oqxAB gene was amplified from the four oldest K. pneumoniae strains (QE137, QE319, QE321, and QE324) isolated in or before the year 1984, followed by nucleotide sequencing and comparison to known oqxAB homologues recovered from K. pneumoniae, K. oxytoca, E. aerogenes, and E. cloacae, as well as plasmid-carried elements recorded in GenBank. Nucleotide and protein BLAST analyses were performed by utilizing the NCBI BLAST services. Sequence alignment and maximum likelihood phylogenetic analysis were conducted by means of MEGA6.06 software (11).
FIG 1.
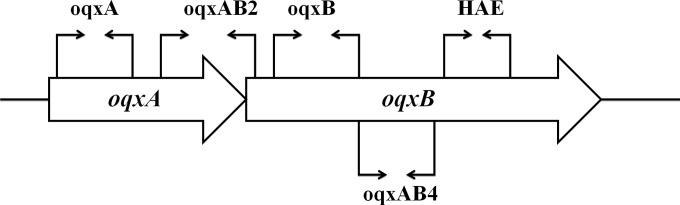
Target regions of the oqxAB operon in pOLA52 subjected to PCR genotyping with 5 primer sets.
TABLE 1.
Primers used in this study
| Primer set | Forward: 5′ to 3′ | Reverse: 5′ to 3′ | Nucleotide position range in oqxAB operona | Reference or study |
|---|---|---|---|---|
| 16S rRNA | CTCCTACGGGAGGCAGCAG | GWATTACCGCGGCKGCTG | 14 | |
| oqxB-RT | TATCTCATTGGCGGCGTGAA | CGCGATTTTGGCGTTGATCT | This study | |
| rarA-RT | GCAGGTGCCACTTCGAATA | GCGCCATCATTCAGGATCT | 15 | |
| oqxR-RT | TAACGAAGCCTGCTCTGCTT | AATGGTTCCGCTAACTCGTG | This study | |
| IS26-oqxA | GCTGTTACGACGGGAGGAG | GGAGACGAGGTTGGTATGGA | 6 | |
| OqxA | CTCGGCGCGATGATGCT | CCACTCTTCACGGGAGACGA | 43−435 | 4 |
| OqxB | TTCTCCCCCGGCGGGAAGTAC | CTCGGCCATTTTGGCGCGTA | 1632−2144 | 4 |
| HAE | GCCTGGTAAGTCGAGATCGG | CTCGAACGGCTATCAGGGAC | 2792−3357 | This study |
| OqxAB2b | ACGGTGTACGTCTACTTTGA | GTCTCGGCAATCACTTTCG | 640−1384 | 16 |
| OqxAB4 | ATCGAGATGGGTTCCGGTAG | TAAACGGACGGAAAATCCAG | 2010−2772 | 16 |
The nucleotide position range was based on that of the oqxAB operon in plasmid pOLA52 (GenBank accession number NC_010378.1).
Also used as a hybridization probe.
Antimicrobial susceptibility testing.
The MICS of five antimicrobials (ciprofloxacin, norfloxacin, nalidixic acid, chloramphenicol, and olaquindox) were determined for all test strains and interpreted according to the CLSI guidelines (12). E. coli ATCC 25922 and ATCC 35218 were used as quality controls.
Southern hybridization.
S1-pulsed-field gel electrophoresis (PFGE) was performed to determine the location of oqxAB in selected strains. Briefly, agarose-embedded DNA was digested with S1 nuclease (New England Biolabs) at 37°C for 1 h. The restriction fragments were separated by electrophoresis in 0.5 M Tris-borate-EDTA buffer at 14°C for 18 h using a Chef Mapper electrophoresis system (Bio-Rad, Hercules, CA) with pulse times of 2.16 to 63.8 s. A phage lambda PFGE ladder (New England Biolabs) was used as a DNA size marker. The gels were stained with GelRed, and DNA bands were visualized with UV transillumination (Bio-Rad). Chromosomal and plasmid DNAs of S. Typhimurium strains were transferred and cross-linked onto nylon membranes and hybridized with DIG-labeled 16S rRNA and oqxAB2 probes using the DIG high prime DNA labeling and detection starter kit I (Roche), following the manufacturer's instructions.
RNA extraction and qRT-PCR.
Total RNA was extracted by the Qiagen Protect bacteria minikit, followed by DNase treatment. The quality and quantity of RNA were determined by using a NanoDrop spectrophotometer. One microgram of RNA was subjected to reverse transcription (RT) using Life Technologies SuperScript III reverse transcriptase; RT-quantitative PCR (qRT-PCR) was performed by using a Bio-Rad iQ5 iCycler and Life Technologies SYBR select master mix. K. pneumoniae strain MGH 78578 was used as a control, and expression levels of the test genes were normalized with that of 16S rRNA.
Nucleotide sequence accession numbers.
Full-length oqxAB sequences from 4 K. pneumoniae isolates (QE137, QE319, QE321, and QE324) were deposited to GenBank (accession numbers KJ875814, KJ875815, KJ875816, and KJ875817, respectively).
Various nucleotide and genome sequences were retrieved from GenBank and used in assessment of the genetic relatedness of oqxAB-like elements: Klebsiella pneumoniae genome sequences MGH 78578 (CP000647.1), XH209 (CP009461.1), PMK1 (CP008929.1), PittNAM01 (CP006798.1), CG43 (CP006648.1), KPNIH31 (CP009876.1), blaNDM-1 (CP009114.1), ATCC BAA-2146 (CP006659.1), 342 (CP000964.1), 1084 (CP003785.1), JM45 (CP006656.1), and KCTC 2242 (CP002910.1); Klebsiella variicola strain At-22 (NC_013850.1); Klebsiella oxytoca strain KCTC 1686 (NC_016612.1); Serratia marcescens genome sequences WW4 (CP003959.1) and SM39 (AP013063.1); Enterobacter aerogenes strain KCTC 2190 (CP002824.1); Enterobacter cloacae genome sequences ATCC 13047 (CP001918.1), EcWSU1 (CP002886.1), and ENHKU01 (CP003737.1); and plasmid sequences pOLA52 (NC_010378.1), pSDB58 (KF840373.1), pHXY (NG_041556.1), and E16 (GQ497565.1).
RESULTS
K. pneumoniae chromosome as origin of oqxAB.
To test the idea that oqxAB originated from K. pneumoniae, where it exists as a chromosomally encoded membrane transporter, five primer sets were used to determine the relative prevalence of oqxAB in different bacterial species. To obtain convincing evidence on the evolutionary origin of oqxAB, we included 15 K. pneumoniae clinical isolates collected in The Prince of Wales Hospital, Hong Kong, in or before the year 1984, which was 10 years earlier than the earliest date when oqxAB was first detected in a plasmid in E. coli (13). This oqxAB genotyping test was regarded as positive if one or more of the five primer sets resulted in successful amplification of oqxAB-like fragments. Based on this criterion, all K. pneumoniae, K. oxytoca, and E. aerogenes isolates, including the 30-year-old strains, were found to be positive, whereas 26 out of 27 E. cloacae strains were also found to contain oqxAB-related genes. The oqxAB-positive rate for Salmonella Typhimurium isolates was 29%. However, no oqxAB-like elements were detectable in Serratia spp., Pseudomonas aeruginosa, Acinetobacter spp., Vibrio parahaemolyticus, other Vibrio spp., Enterococcus spp., and Staphylococcus aureus isolates.
It should be noted that highly variable result patterns for the genotyping tests with five primer sets were observed among the test isolates; hence, only 14 K. pneumoniae strains were positive to all primer sets tested, including 10 collected after 2008. Alignment of primer sequences in known K. pneumoniae genomes suggested that the negative genotyping test results were due to sequence variations rather than to a lack of the priming regions in the respective genomes (results not shown). This idea is supported by our observation that, for each isolate which we defined as oqxAB positive by our oqxAB genotyping approach, at least one primer set targeting the oqxA gene and one targeting the oqxB gene produced a positive result. On the other hand, an association between IS26 and oqxA was not observable in all isolates tested, suggesting that this gene was not introduced into the chromosome of K. pneumoniae by transposition events. S1-PFGE was performed on eight K. pneumoniae isolates for which the positive response rate to different primer sets varied. Only the chromosomal DNA band, but not the plasmid, was observed in these strains. Southern hybridization of the nylon membrane transferred with DNA from the same S1-PFGE gel showed that the oqxAB probe successfully hybridized to the chromosomal DNA band that also was hybridized by the 16S rRNA probe. However, only the 16S rRNA but not the oqxAB probe successfully hybridized to the chromosomal DNA of two control E. coli strains. Taken together, the S1-PFGE and Southern hybridization data further confirm the chromosomal location of oqxAB (results not shown).
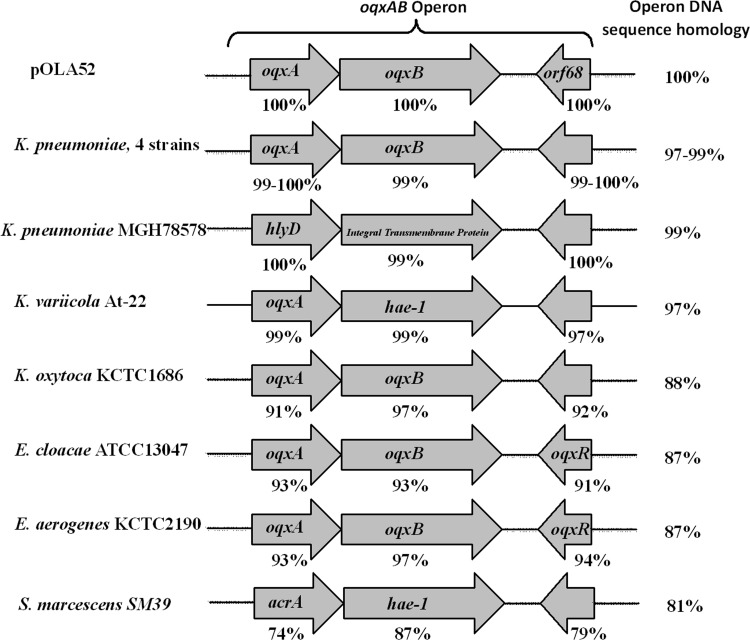
To assess the validity of the genotyping tests, the original oqxAB operon in pOLA52 (GenBank accession number NC_010378.1) was subjected to a BLASTN homology search in the NCBI database. All identical hits were plasmid-borne oqxAB operons in E. coli and Salmonella spp. Chromosomal high homology hits (≥97%) were also identified, but all such elements were membrane transporters (oqxAB homologues) in K. pneumoniae and K. variicola. The rest were intermediate homology hits (81 to 88%) involving K. oxytoca, E. cloacae, E. aerogenes, and Serratia spp. Nucleotide alignment data showed that the oqxAB operon in pOLA52 was 99% and 97% identical to the K. pneumoniae MGH 78578 strain and K. variicola At-22 strain, respectively, but exhibited only 88%, 87%, 87%, and 81% identity to K. oxytoca, E. cloacae, E. aerogenes, and Serratia marcescens, respectively; oqxAB-like elements were not found in organisms not belonging to the family Enterobacteriaceae. On the other hand, deduced amino acid sequences of the OqxA, OqxB, and transcriptional regulator Orf68 proteins encoded by genes located in pOLA52 were 99 to 100% identical to those of K. pneumoniae and K. variicola, but only 91 to 97% identical to those of K. oxytoca, E. cloacae, E. aerogenes, and Serratia marcescens (Fig. 2). Interestingly, the OqxB protein in different bacterial species shared 97 to 100% amino acid homology with pOLA52, indicating that it was more conserved than OqxA. Taken together, the genotyping and sequence alignment data suggest that K. pneumoniae is genetically most related to the plasmid-borne oqxAB genes detectable in E. coli and Salmonella clinical isolates in recent years.
FIG 2.
Amino acid sequence alignment of the oqxAB operon (or its synonyms) from genomes of various control strains and four K. pneumoniae strains recovered in Hong Kong in or before the year 1984, against E. coli plasmid pOLA52. The arrows depict open reading frames (ORF) and their respective orientations. The percentage below each ORF depicts the amino acid sequence homology to a specific gene in the oqxAB operon in pOLA52. The overall nucleotide homology with pOLA52 is shown on the right.
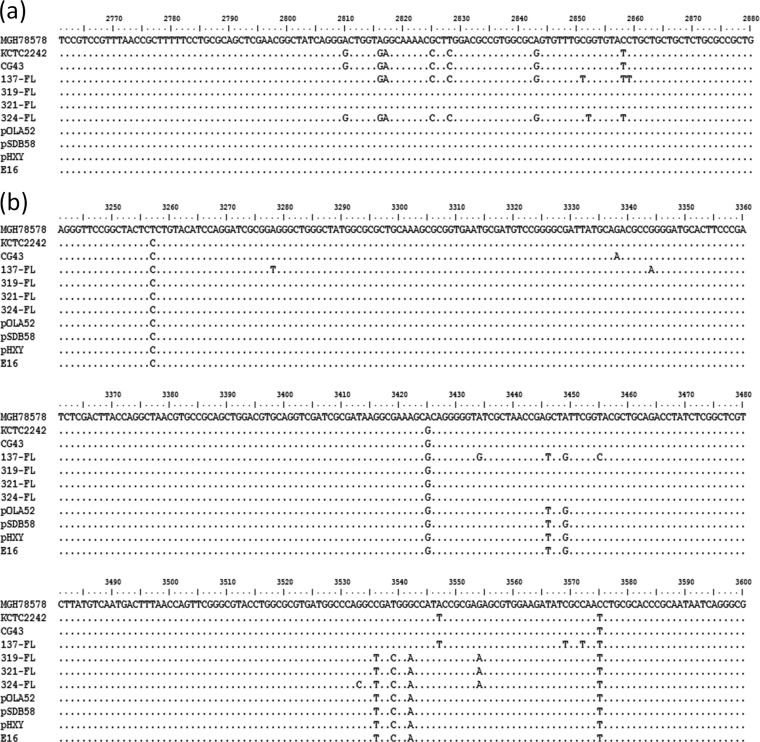
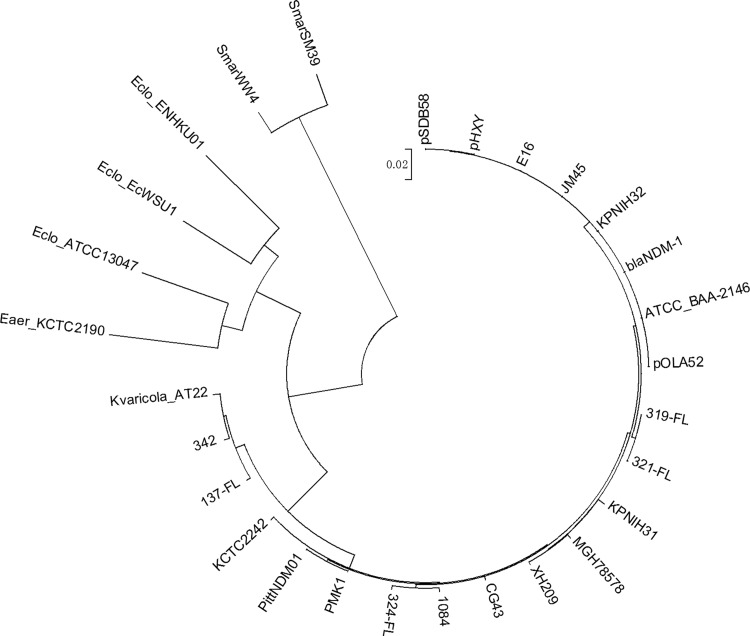
To further investigate the genetic characteristics of the chromosomal oqxAB-like element in K. pneumoniae, the entire oqxAB operon in four of the oldest K. pneumoniae isolates, namely, QE137, QE319, QE321, and QE324, which were isolated in the year 1984 or before, was sequenced and compared to various plasmid-borne and chromosomal oqxAB operons. Consistent with the sequence alignment data, the oqxAB operons in these four isolates were found to share 97 to 99% homology at the nucleotide level and 99 to 100% identity at the amino acid level with pOLA52 (Fig. 2). Importantly, pockets of identical sequence variations or nucleotide polymorphism were observable among the chromosomal and plasmid-borne elements (Fig. 3). Among specific regions of genetic polymorphism, the plasmid-borne element was found to share a higher level of sequence identity with the 30-year-old K. pneumoniae isolates than with the more recent strains, suggesting that the plasmid-borne genes originated from the earlier K. pneumoniae strains. The results of phylogenetic analysis depicting the genetic relationship between various chromosomal and plasmid-borne oqxAB operons lend support to this idea (Fig. 4).
FIG 3.
Nucleotide sequence alignment depicting identical sequence variations in two regions (a and b) of the chromosomal and plasmid-borne oqxAB operon recoverable from K. pneumoniae and Salmonella/E. coli strains, respectively. MGH 78578, KCTC 2242, and CG43: control K. pneumoniae strains. 137-FL, 319-FL, 321-FL, and 324-FL: clinical K. pneumoniae strains isolated in or before the year 1984. pOLA52, pSDB58, pHXY, and E16: plasmids harboring the oqxAB operon, recoverable from Salmonella/E. coli strains. The nucleotide sequence of strain MGH 78578 was used as the reference sequence. Sequence data of the four clinical K. pneumoniae strains were generated in this study. All other data are retrievable from GenBank.
FIG 4.
Phylogenetic tree depicting the genetic relatedness of oqxAB operons retrieved from various sources. OqxAB operons were extracted from the chromosomes of K. pneumoniae (JM45, CG43, MGH 78578, 1084, KPNIH31, KPNIH32, blaNDM-1, ATCC BAA-2146, PMK1, PittNDM01, KCTC 2242, 342, 137-FL, 319-FL, 321-FL, and 324-FL), K. variicola (At-22), Enterobacter cloacae (EcWSU1 and ENHKU01), Enterobacter aerogenes (KCTC 2190 and ATCC 13047), and Serratia marcescens (WW4 and SM39) strains and plasmids recoverable from E. coli and Salmonella (pSDB58, pHXY, E16, and pOLA52) strains.
Relative antimicrobial susceptibility and oqxAB expression profiles of K. pneumoniae and organisms harboring the pOLA52-like plasmid.
With oqxAB being consistently detectable in K. pneumoniae, drug susceptibility phenotypes were checked for the 85 clinical isolates tested in this study, with results being consistent with previous findings that K. pneumoniae clinical isolates were generally susceptible to quinolones (Table 2). Among these 85 K. pneumoniae clinical isolates, only 20 were resistant to chloramphenicol and 28 were resistant to nalidixic acid (MIC of ≥32 μg/ml), 14 of which were also resistant to ciprofloxacin (MIC of ≥4 μg/ml). The majority of the isolates (51 out of 85) had a nalidixic acid MIC of <4 μg/ml, whereas 49 strains had a ciprofloxacin MIC of <0.006 μg/ml. However, high-level resistance to olaquindox, chloramphenicol, and nalidixic acid and reduced susceptibility to ciprofloxacin, a typical resistance phenotype conferred by the oqxAB-harboring plasmid in Salmonella, were observable in only two K. pneumoniae strains isolated during or after the 1990s (strains 94-3 and GN53) (Table 2). We confirmed, by S1-PFGE, that such a phenotype was not caused by extrachromosomal oqxAB elements. On the other hand, bacterial species harboring chromosomal oqxAB homologues such as E. cloacae and E. aerogenes were mostly susceptible to the test agents, suggesting that these homologues did not contribute to drug resistance under the test conditions.
TABLE 2.
Summary of the genotypic and phenotypic characteristics of organisms harboring oqxAB-like elements
| Bacterial species and test population/specific straina | Place/yr(s) of isolation | Location of oqxABb | Nucleotide sequence homology with pOLA52 (%) |
oqxAB genotyping with 5 primer sets (no. [%] or present [+] or absent [−]) |
MIC (mg/liter)c |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| oqxA | oqxB | HAE | oqxAB2 | oqxAB4 | Overall | CIP | NAL | NOR | CHL | OLA | ||||
| K. pneumoniae | ||||||||||||||
| HK (n = 85) | HK/1984–2011 | 67 | 35 | 32 | 83 | 73 | 85 (100) | |||||||
| SGSC (n = 8) | SGSC/1996 | 8 | 4 | 8 | 8 | 8 | 8 (100) | |||||||
| MGH 78578 | Chromosome | 99 | + | − | + | + | + | + | 1 | ≥128 | 4 | ≥128 | 16 | |
| QE319 | HK/1984 | Chromosome | 99 | − | − | + | + | − | + | <0.006 | <4 | <2 | <4 | <16 |
| 94-3d | HK/1994 | Chromosome | NDe | + | − | − | + | − | + | 0.25 | ≥128 | ≤2 | 64 | 512 |
| GN53d | HK/2006 | Chromosome | ND | + | − | + | + | + | + | 4 | ≥128 | ≥64 | ≥128 | 512 |
| 06-2 | HK/2006 | Chromosome | ND | + | − | − | + | + | + | ≤0.006 | ≤4 | ≤2 | ≤4 | ≤16 |
| K. oxytoca (SGSC, n = 8) | SGSC/1996 | 88f | 0 | 0 | 4 | 8 | 8 | 8 (100) | <0.012–0.05 | <4–>128 | <4 | <4 | 8–32 | |
| E. aerogenes (SGSC, n = 15) | SGSC/1996–1999 | 87f | 0 | 10 | 15 | 15 | 4 | 15 (100) | <0.012 | <4–8 | <4 | <4 | 8–64 | |
| E. cloacae (SGSC, n = 27) | SGSC/1996–2000 | 87f | 3 | 15 | 9 | 14 | 15 | 26 (96) | <0.012–0.05 | <4–16 | <4 | <4–16 | 16–128 | |
| S. Typhimurium (n = 17) | HK/2007 | 5 | 5 | 3 | 4 | 4 | 5 (29) | |||||||
| 1792 | HK/2007 | + | + | − | − | + | + | 2 | ≥128 | ND | ≥128 | ≥512 | ||
| 2005 | HK/2007 | Plasmid | 100 | + | + | − | − | + | + | 2 | ≥128 | ND | ≥128 | ≥512 |
| Other speciesg | HK/2006–2010 | Plasmid | 100 | |||||||||||
HK, Hong Kong; SGSC, Salmonella Genetic Stock Centre, Calgary, Canada.
Based on the results of S1-PFGE and Southern hybridization studies.
CIP, ciprofloxacin; NAL, nalidixic acid; NOR, norfloxacin; CHL, chloramphenicol; OLA, olaquindox.
K. pneumoniae strains 94-3 and GN53 exhibited the typical oqxAB-mediated antibiotic resistance profile (resistance to olaquindox, chloramphenicol, and the quinolones) without an oqxAB-harboring plasmid.
ND, not determined.
Based on the nucleotide sequences of the standard strains KCTC 1686, KCTC 2190, and ATCC 13047 for K. oxytoca, E. aerogenes, and E. cloacae, respectively.
All other bacterial species tested, including Serratia spp. (n = 7), Pseudomonas aeruginosa (n = 30), Acinetobacter spp. (n = 30), Vibrio parahaemolyticus (n = 30), other Vibrio spp. (n = 30), Staphylococcus aureus (n = 30), and Enterococcus spp. (n = 30), were oqxAB negative.
To probe the molecular basis of the discrepancy in susceptibility phenotypes observable among various oqxAB-borne organisms, K. pneumoniae strain GN53 and a Salmonella strain harboring the pOLA52 plasmid (ST07-37), which were both olaquindox resistant, were subjected to RT-quantitative PCR analysis, with results showing that the expression levels of both rarA and oqxB in strain GN53 were significantly higher than those of the wild-type strain MGH 78578 (Fig. 5). This finding suggests that the drug resistance phenotypes of this strain were at least partially due to up-regulated expression of oqxAB. Interestingly, expression of oqxR was also moderately elevated, inferring that the effect of its gene product as an oqxAB repressor might have been counteracted by the extraordinarily large amount of the RarA protein produced in this strain. On the other hand, the expression level of oqxB in the Salmonella strain ST07-37 was elevated as much as 85-fold compared to that in the wild-type, drug-sensitive K. pneumoniae strain. Whether such high expression levels of the plasmid-borne oqxAB genes were due to derepression of the oqxAB operon as a result of lack of an OqxR binding site remains to be elucidated.
FIG 5.
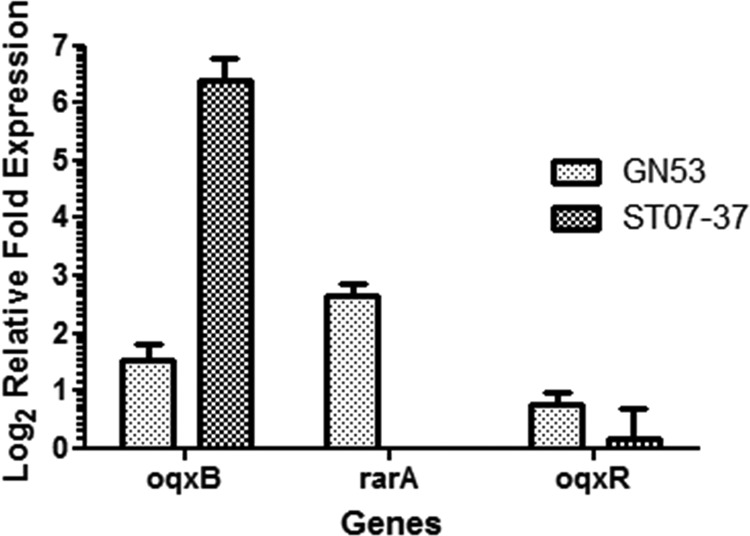
Relative expression levels of the oqxB, rarA, and oqxR genes in the olaquindox-resistant K. pneumoniae strain GN53 and a Salmonella Typhimurium strain ST07-37 harboring a pOLA52-like plasmid. The K. pneumoniae strain MGH 78578 was used as a control.
DISCUSSION
This study highlighted several important issues regarding the evolutionary origin and dissemination features of the PMQR determinant oqxAB. First, although this resistance determinant has become increasingly prevalent among Gram-negative pathogens, our study showed that it is mainly confined to members of Enterobacteriaceae. Second, oqxAB was most prevalent among the Enterobacter spp. and Klebsiella spp. In particular, both the detection rate and level of sequence homology with the oqxAB operon in pOLA52, the original plasmid in which oqxAB was first recovered, approached 100% even in K. pneumoniae strains recovered 10 years earlier than the time of discovery of pOLA52. This finding has important implications for the origin of mobile oqxAB elements. Third, organisms containing chromosome-based and plasmid-borne elements exhibited drastically different levels of gene expression and susceptibility to the quinolones; in particular, the expression level of the plasmid-borne oqxAB operon was >80-fold higher than that of the chromosomal genes.
We postulate that the oqxAB operon, together with a transcriptional regulator (orf68) in the chromosome of K. pneumoniae, was captured by IS26 transposase and transferred to foreign plasmids, which were subsequently disseminated to other bacterial species that do not harbor oqxAB-like elements in the chromosome. It should be noted that, although the oqxAB operon is also prevalent among other species such as Enterobacter spp., a significantly lower degree of sequence homology with the plasmid-borne element was observed, suggesting that these oqxAB homologues were not as readily captured by transposition activities as the K. pneumoniae genes. The underlying principle by which structural differences between various oqxAB homologues affect transposition efficiency remains to be elucidated. In addition, although factors limiting horizontal transfer of existing oqxAB-carrying plasmids to non-Enterobacteriaceae species are not understood, the possibility that the oqxAB operon may be captured and transferred to other types of plasmids that can be taken up by other Gram-negative pathogens should be investigated.
Taken together, our findings indicate that oqxAB or its homologues represent one of the many endogenous efflux systems in K. pneumoniae and Enterobacter spp., a role which is functionally similar to that of acrAB in other members of Enterobacteriaceae. Yet a major difference between oqxAB and other chromosomal efflux genes, as illustrated by findings in this work, is that the oqxAB operon can become plasmid borne via transposition events, during which the oqxAB genes become overexpressed, presumably as a result of loss of the OqxR repressor function. The dissemination patterns of mobile elements harboring overexpressed efflux pumps should be closely monitored.
ACKNOWLEDGMENTS
This work was supported by the Chinese National Key Basic Research and Development 973 Program (2013CB127200) and the Health and Medical Research Fund of the Food and Health Bureau, Hong Kong (12111612 and 13121412 to S.C.).
We thank Julia Ling of the Department of Microbiology, The Chinese University of Hong Kong, for provision of the clinical K. pneumoniae and Salmonella isolates used in this study.
REFERENCES
- 1.Hansen LH, Johannesen E, Burmolle M, Sorensen AH, Sorensen SJ. 2004. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother 48:3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norman A, Hansen LH, She Q, Sorensen SJ. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Hansen LH, Jensen LB, Sorensen HI, Sorensen SJ. 2007. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother 60:145–147. doi: 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- 4.Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. 2009. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother 53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong MH, Chen S. 2013. First detection of oqxAB in Salmonella spp. isolated from food. Antimicrob Agents Chemother 57:658–660. doi: 10.1128/AAC.01144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao J, Chen Z, Chen S, Deng Y, Liu Y, Tian W, Huang X, Wu C, Sun Y, Zeng Z, Liu JH. 2010. Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob Agents Chemother 54:4219–4224. doi: 10.1128/AAC.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan J, Xu X, Guo Q, Zhao X, Ye X, Guo Y, Wang M. 2012. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J Antimicrob Chemother 67:1655–1659. doi: 10.1093/jac/dks086. [DOI] [PubMed] [Google Scholar]
- 8.Bialek-Davenet S, Lavigne JP, Guyot K, Mayer N, Tournebize R, Brisse S, Leflon-Guibout V, Nicolas-Chanoine MH. 2015. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 70:81–88. doi: 10.1093/jac/dku340. [DOI] [PubMed] [Google Scholar]
- 9.Perez F, Rudin SD, Marshall SH, Coakley P, Chen L, Kreiswirth BN, Rather PN, Hujer AM, Toltzis P, van Duin D, Paterson DL, Bonomo RA. 2013. OqxAB, a quinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrob Agents Chemother 57:4602–4603. doi: 10.1128/AAC.00725-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez-Martínez JM, Diaz de Alba P, Briales A, Machuca J, Lossa M, Fernandez-Cuenca F, Rodriguez Bano J, Martinez-Martinez L, Pascual A. 2013. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemother 68:68–73. doi: 10.1093/jac/dks377. [DOI] [PubMed] [Google Scholar]
- 11.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI M100-S23 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Chen X, Zhang W, Pan W, Yin J, Pan Z, Gao S, Jiao X. 2012. Prevalence of qnr, aac(6′)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother 56:3423–3427. doi: 10.1128/AAC.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner S, Pryer KM, Miao VP, Palmer JD. 1999. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 15.Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. 2012. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother 56:4450–4458. doi: 10.1128/AAC.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T, Yokota S, Uchida I, Okubo T, Usui M, Kusumoto M, Akiba M, Fujii N, Tamura Y. 2013. Fluoroquinolone resistance mechanisms in an Escherichia coli isolate, HUE1, without quinolone resistance-determining region mutations. Front Microbiol 4:125. doi: 10.3389/fmicb.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]