Abstract
Plasmids play a key role in the horizontal spread of antibiotic resistance determinants among bacterial pathogens. When an antibiotic resistance plasmid arrives in a new bacterial host, it produces a fitness cost, causing a competitive disadvantage for the plasmid-bearing bacterium in the absence of antibiotics. On the other hand, in the presence of antibiotics, the plasmid promotes the survival of the clone. The adaptations experienced by plasmid and bacterium in the presence of antibiotics during the first generations of coexistence will be crucial for the progress of the infection and the maintenance of plasmid-mediated resistance once the treatment is over. Here we developed a model system using the human pathogen Haemophilus influenzae carrying the small plasmid pB1000 conferring resistance to β-lactam antibiotics to investigate host and plasmid adaptations in the course of a simulated ampicillin therapy. Our results proved that plasmid-bearing clones compensated for the fitness disadvantage during the first 100 generations of plasmid-host adaptation. In addition, ampicillin treatment was associated with an increase in pB1000 copy number. The augmentation in both bacterial fitness and plasmid copy number gave rise to H. influenzae populations with higher ampicillin resistance levels. In conclusion, we show here that the modulations in bacterial fitness and plasmid copy number help a plasmid-bearing bacterium to adapt during antibiotic therapy, promoting both the survival of the host and the spread of the plasmid.
INTRODUCTION
Antibiotic resistance in pathogenic bacteria is currently one of the most pressing problems in public health. Resistant bacteria produce infections that are difficult to clear, raising the mortality rates and economic costs associated with them (1, 2). Plasmids can disseminate resistance determinants through horizontal gene transfer, playing a key role in the acquisition of antimicrobial resistance by pathogenic bacteria (3, 4). The carriage of plasmids, however, entails a fitness cost to the bacterial host (5), imposing negative selection for a plasmid-bearing clone in the absence of antibiotics. Previous studies have analyzed how bacteria compensate for the cost produced by plasmids in the long term (6, 7). However, little information is available regarding the adaptations experienced by bacterium and plasmid during short periods of time (8). An antibiotic treatment selects the clones carrying a resistance plasmid and gives them the opportunity to evolve during a short period of time in a competition-free environment, due to the death of antibiotic-susceptible clones. The adaptations in both the antibiotic resistance level and the fitness cost produced by the plasmid will determine the fate of the plasmid-bearing clones during and after the antibiotic treatment.
In this work, we investigated the short-term evolution of plasmid-mediated antibiotic resistance using Haemophilus influenzae, an important human pathogen producing otitis, pneumonia, and meningitis (9). We developed a model system using H. influenzae strain Rd KW20 (Rd) transformed with the small plasmid pB1000 (Rd/pB1000) to analyze the modifications undergone by bacterium and plasmid in the presence of antibiotics. pB1000 is a small ColE1-type plasmid carrying the blaROB-1 β-lactamase gene and conferring resistance to aminopenicillins (10). ColE1-type plasmids belong to the MOBP5 family of mobilizable plasmids (11) and have increasing relevance as antibiotic resistance carriers in the bacterial families Enterobacteriaceae and Pasteurellaceae (10, 12–22). pB1000 has been described in a wide range of members of the Pasteurellaceae family, including animal pathogens such as Pasteurella multocida and Haemophilus parasuis, as well as in human clinical isolates of H. influenzae in Spain, Italy, the United States, and Australia (10, 17–20).
Using experimental evolution, we simulated an antibiotic treatment and investigated the adaptations undergone by plasmid and host in the model system Rd/pB1000. Specifically, we measured (i) the evolution of bacterial fitness, (ii) the effects of the antibiotic on plasmid copy number, and (iii) the fluctuations in the antibiotic resistance level over time. We show here that the modifications in the fitness of the host bacterium and in the plasmid copy number contribute to an augmentation of the antibiotic resistance phenotype of Rd/pB1000.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and antibiotic susceptibility determination.
H. influenzae Rd was transformed by electroporation with pB1000 from P. multocida BB1038 (17) as previously described (18) using a Gene Pulser apparatus (Bio-Rad, USA). Rd and Rd/pB1000 were cultured on chocolate agar PolyViteX plates (bioMérieux, France) and in Haemophilus test medium (HTM) broth (Francisco Soria Melguizo, S.A., Spain) with shaking at 125 rpm at 37°C under microaerophilic conditions (5% CO2). A detailed description of the populations analyzed during the experimental evolution assay is provided in Table 1. We determined the stability of plasmid pB1000 in H. influenzae Rd by propagating five populations of Rd/pB1000 in HTM for 200 generations, plating serial dilutions of the final cultures, and replica plating a total of 100 colonies on chocolate agar plates containing 100 mg/liter of ampicillin. The presence of pB1000 was confirmed by PCR in a subset of 20 of those colonies (18). The MICs of the antibiotics tested were determined in HTM using the broth microdilution method according to the CLSI guidelines (23). We used H. influenzae ATCC 49247 and ATCC 49766 as control strains to determine the resistance levels of parental strains H. influenzae Rd KW20 and H. influenzae Rd KW20/pB1000. In the subsequent MIC determinations, we used H. influenzae Rd KW20 and H. influenzae Rd KW20/pB1000 as control strains. To obtain a good discrimination of the MIC differences among populations, we used 2-fold-increasing concentrations of ampicillin from 0.125 to 256 mg/liter and then we increased the concentration by 256 mg/liter in each following dilution up to 4,096 mg/liter. Antibiotics were provided by Sigma (Sigma-Aldrich, United Kingdom).
TABLE 1.
Populations analyzed in this study
| Populationa | Population evolved from | Daysb | Presence of ampicillin |
|---|---|---|---|
| Rd-10 | Rd | 10 | No |
| Rd/pB1000-10 | Rd/pB1000 | 10 | No |
| Rd/pB1000A-10 | Rd/pB1000 | 10 | Yes |
| Rd-20 | Rd-10 | 10 (20) | No |
| Rd/pB1000-20 | Rd/pB1000-10 | 10 (20) | No |
| Rd/pB1000A-20 | Rd/pB1000A-10 | 10 (20) | No |
| Rd-20/pB1000 | Rd-20 (transformed with pB1000) | NAc | NA |
There were five replicates per population.
Numbers in parentheses represent the accumulated days of evolution from the original parental strain Rd or Rd/pB1000.
NA, not applicable.
Fitness determination.
The fitness cost of plasmid pB1000 was determined by competition experiments between H. influenzae Rd KW20 and H. influenzae Rd KW20/pB1000 in HTM medium in three independent experiments. Strains were grown for 16 h at 37°C and 5% CO2 in HTM, and then 106 CFU of H. influenzae Rd were mixed with 106 CFU of H. influenzae Rd/pB1000 in 2 ml of HTM. The mix was grown at 37°C, 5% CO2, and 125 rpm for 24 h, and 106 CFU were transferred to 2 ml of fresh HTM every 24 h (1/1,000 dilution) for 6 days. Samples were taken at time zero and every 24 h over 6 days. For each sample, aliquots were plated on nonselective chocolate agar, and the proportion of resistant colonies was deduced by replica plating of 50 to 100 colonies on chocolate agar plates containing 100 mg/liter of ampicillin. The competition index (CI) was calculated every day as the ratio between the CFU of the resistant and susceptible strains at t1 divided by the same ratio at t0 (24). The selection coefficient, s, was calculated as the slope of the linear regression model s = ln(CI)/t, where time (t) was measured in bacterial generations, calculated as the log2 of the dilution factor (25). The relative fitness (w) was calculated with the formula w = 1 + s. Competition experiments were done in triplicate, and the selection coefficient per generation was calculated as the average s from the three independent experiments. The selection coefficient estimates the difference between the relative fitnesses of the two competitors over the entire competition experiment. For the determination of the fitness of evolving populations, Rd/pB1000 lines were competed against the parental Rd strain and Rd parental strain and evolved lines were competed against Rd/pB1000 using the same conditions described above. The fitness for each evolving line was calculated relative to the parental Rd strain.
Determination of plasmid copy numbers.
The copy numbers of plasmids were determined essentially as described by San Millan et al. (26). Quantitative PCR (qPCR) was performed using a My iQ single-color real-time PCR detection system (Bio-Rad, USA). The original pB1000 copy number in Rd was determined by qPCR (in triplicate) from three independent DNA extractions. To analyze the average plasmid copy numbers per cell in the different populations, DNA extractions were done for each evolving population and qPCR was performed for each extraction in triplicate. DNA extractions were performed from 2 ml of HTM broth cultures at an optical density at 600 nm (OD600) of approximately 0.9 and using a QIAamp DNA minikit (Qiagen, USA). DNA was quantified using an Eppendorf BioPhotometer (Eppendorf, Germany). Restriction enzyme-digested total DNA is a better template source than nondigested total DNA for plasmid quantification by qPCR (27). Linearizing the plasmids increases accessibility to plasmid DNA template, preventing copy number from being underestimated (27). Therefore, 1 μg of DNA from each sample was digested using 20 units of PstI (TaKaRa, Japan) for 2 h at 37°C. PstI was inactivated at 60°C for 15 min. No PstI targets are present in the amplification products. We developed a specific qPCR for the plasmid-carried blaROB-1 gene (primers CCAATTTCTGTTCATTCGGTAAC [forward] and CATAAGCAAAGCGTTCATCTG [reverse]; amplicon size, 195; efficiency, 98.2%; r2 = 0.999) and the monocopy chromosomal gene rpoB (primers TTGGTTTGTCATAGATATTC [forward] and AATGATGAGCGATTTATTC [reverse]; amplicon size, 199; efficiency, 98.7%; r2 = 0.999) to compare the ratio of plasmid and chromosomal DNAs. The efficiency of the reactions was calculated from the standard curve generated by performing qPCR with five 8-fold dilutions of template DNAs in triplicate (working range of DNA concentration, ∼0.2 ng/μl to 50 fg/μl). We performed the qPCRs using the Bio-Rad iQ SYBR green Supermix (Bio-Rad, USA) at a final DNA concentration of 10 pg/μl. The amplification conditions were as follows: initial denaturation for 10 min at 94°C, followed by 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 58.7°C (blaROB-1) or 52°C (rpoB), and extension for 1 min at 72°C. Interrun calibration samples were used to normalize the results from different plates of each qPCR. To calculate the copy number of plasmid per chromosome we used the formula cn = [(1 + Ec)CTc/(1 + Ep)CTp] × (Sc/Sp), where cn is the plasmid copy number per chromosome, Sc and Sp are the sizes of the chromosomal and plasmid amplicons (in base pairs), respectively, Ec and Ep are the efficiencies of the chromosomal and plasmid qPCRs (relative to 1), respectively, and CTc and CTp are the threshold cycles of the chromosomal and plasmid reactions, respectively.
Next-generation sequencing.
We performed deep sequencing on DNA samples from populations using a SOLiD v4 sequencer (Life Technologies, USA). Fragment libraries were generated following the standard protocol. Briefly, 3 μg total bacterial DNA for each isolate was sonicated to yield fragments of approximately 200 bp. Fragments were end repaired and barcoded using the provided Library Builder kit (product number 4463762; Life Technologies, USA) and the associated Library Builder robot (product number 4463592; Life Technologies, USA). Paired-end (PE) reads of 75 bp plus 35 bp were generated for each library; >10 million reads were able to be mapped to the parental genome reference in all cases, correlating with at least 300× average coverage. The LifeScope genomic analysis software v2.5.1 (Life Technologies, USA) was used for mutation detection and single-nucleotide polymorphism (SNP) calling.
Statistical analysis.
Analyses were performed using R (version 2.14.1). The influence of the plasmid copy number and bacterial fitness on the antibiotic resistance levels of the different lines was analyzed by fitting a linear mixed-effects model using the lmer function of the lme4 package of the R statistical software package (R 2.14.1; http://www.r-project.org/). Fitness and plasmids were treated as continuous explanatory variables and MIC as a response. The time points (days 10 and 20) and the different lines (5 per treatment) were included as random factors to correct for the nonindependence of data from the same line or time point. Significance was assessed with the chi-square test and model fit checked by visual inspection of the residuals. Model parameters were estimated with restricted maximum-likelihood methods (28).
RESULTS AND DISCUSSION
Experimental design.
H. influenzae Rd was transformed with the plasmid pB1000 (17). Plasmid pB1000 presented high stability in H. influenzae Rd; after 200 generations of antibiotic-free culture, all the clones tested maintained the plasmid. To analyze the potential plasmid-bacterium adaptation during an antibiotic treatment we established the following experimental design (see Fig. S1 in the supplemental material). Five replicate populations of Rd/pB1000 (here called populations Rd/pB1000A) were evolved independently in an experiment with two different stages (Table 1; see Fig. S1 in the supplemental material). Stage one emulated a 10-day antibiotic therapy: populations Rd/pB1000A were propagated for 10 days with a daily transfer using a dilution factor of 1,000 (approximately 10 generations per day) with 64 mg/liter of ampicillin. This corresponds to 1/8 of the Rd/pB1000 MIC, and it is similar to the peak concentration achieved in patients under standard ampicillin dosing regimens (29, 30). In stage two, we investigated readaptation to a nonselective scenario, propagating the populations for 10 further days in the absence of antibiotic pressure.
We conducted two different control experiments: (i) to analyze the influence of ampicillin in Rd/pB1000 evolution, we propagated five replicate populations of this strain in the absence of ampicillin for 20 days (populations Rd/pB1000) (Table 1; see Fig. S1 in the supplemental material), and (ii) to determine the influence of pB1000 in the evolution of H. influenzae Rd, we propagated five populations of plasmid-free Rd strain in the absence of antibiotic pressure for 20 days (populations Rd) (Table 1; see Fig. S1 in the supplemental material). We measured three parameters at the beginning of the experiment and at days 10 and 20 for every population: (i) bacterial fitness, (ii) ampicillin resistance level, and (iii) plasmid copy number (average number of plasmids per bacterial chromosome in the population).
Evolution of bacterial fitness.
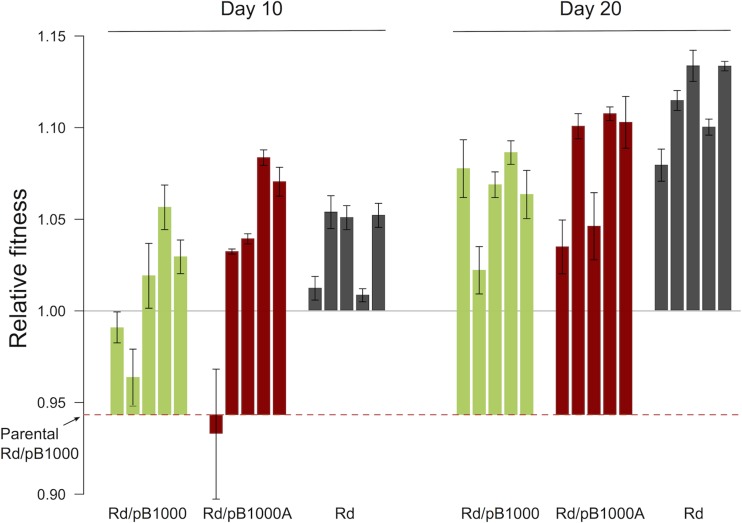
We measured the fitness cost produced by pB1000 in Rd using competition experiments between Rd and Rd/pB1000 without antibiotics. The fitness of Rd/pB1000 was calculated using the selection coefficient relative to the parental H. influenzae Rd (s) (see Materials and Methods). Plasmid pB1000 produced a decrease in fitness of 5.67% (s = −0.056; standard deviation [SD] = 0.0038) under our experimental conditions. Next, we estimated the fitness of each population at days 10 and 20. One important question was to be answered with these experiments: will Rd/pB1000 fitness increase fast enough during the simulated antibiotic therapy to overcome the cost entailed by the plasmid? In Fig. 1 we present the relative fitness values in the different populations throughout the experiment. We observed an increase in relative fitness over time in the three treatments (Rd, Rd/pB1000, and Rd/pB1000A). Interestingly, the initial difference in fitness due to pB1000 disappeared after the first 10 days of the experiment, as indicated by the absence of differences in fitness among the Rd, Rd/pB1000, and Rd/pB1000A populations (by analysis of variance [ANOVA], P = 0.377, F = 0.837, and df = 1, 13). At day 20 there were still no differences in relative fitness between the plasmid-free populations and Rd/pB1000A populations (by the two-sample t test, P = 0.114, t = 1.81, and df = 6.9). The plasmid-bearing populations evolved without ampicillin for 20 days showed lower fitness than the plasmid-free populations evolved under the same conditions (two-sample t test, P = 0.013, t = 3.21, and df = 8).
FIG 1.
Fitness of the evolving populations. Relative fitnesses of the different populations compared to that of the plasmid-free ancestor H. influenzae Rd KW20 (mean ± standard error of the mean [SEM]) are shown. The red dashed line represents the fitness of the parental H. influenzae Rd KW20 carrying plasmid pB1000 (Rd/pB1000). The green bars represent the relative fitnesses of the five Rd/pB1000 populations propagated in the absence of antibiotics. The red bars represent the relative fitnesses of the five Rd/pB1000A populations propagated for 10 days with ampicillin and for 10 further days without ampicillin. The dark gray bars represent the relative fitnesses of the five Rd populations propagated in the absence of antibiotics and plasmid.
H. influenzae Rd was able to overcome the fitness cost imposed by pB1000 in only 10 days, and, in the populations treated with ampicillin, after 20 days there were still no differences in fitness compared to those of the plasmid-free control populations. These results show that Rd/pB1000 compensates for the plasmid cost during the simulated antibiotic treatment, increasing the subsequent stability of the plasmid in the bacterial population after the therapy.
Increase in ampicillin resistance levels over time.
The MICs of ampicillin were determined for the Rd and Rd/pB1000 parental strains and for the different populations at days 10 and 20 (Fig. 2; see Fig. S1 in the supplemental material). The Rd strain showed an MIC of 0.25 mg/liter. In this strain, the MIC remained constant in the populations throughout the experiment. The MIC of ampicillin in the parental Rd/pB1000 was 512 mg/liter (SD = 0 mg/liter). Rd/pB1000A populations propagated with ampicillin showed an MIC of 2,150 mg/liter (SD = 465 mg/liter) after 10 days, which was significantly higher than that of the parental strain (two-tailed paired t test, P = 0.001, t = −7.88, and df = 4). The resistance level in Rd/pB1000A populations was also higher than that in the control populations of Rd/pB1000 cultured for 10 days in the absence of antibiotic (1,075 mg/liter [SD = 458 mg/liter]; two-sample t test, P = 0.007, t = −3.64, and df = 7.9). We therefore conclude that the presence of ampicillin induced an increase in the antibiotic resistance level of Rd/pB1000. The control populations of Rd/pB1000 at day 10 showed no significant increase in the MIC of ampicillin compared to that for the parental strain (two-tailed paired t test, P = 0.089, t = −2.24, and df = 4), although the mean MIC in these populations was unexpectedly higher than that in the parental Rd/pB1000 strain (Fig. 2).
FIG 2.
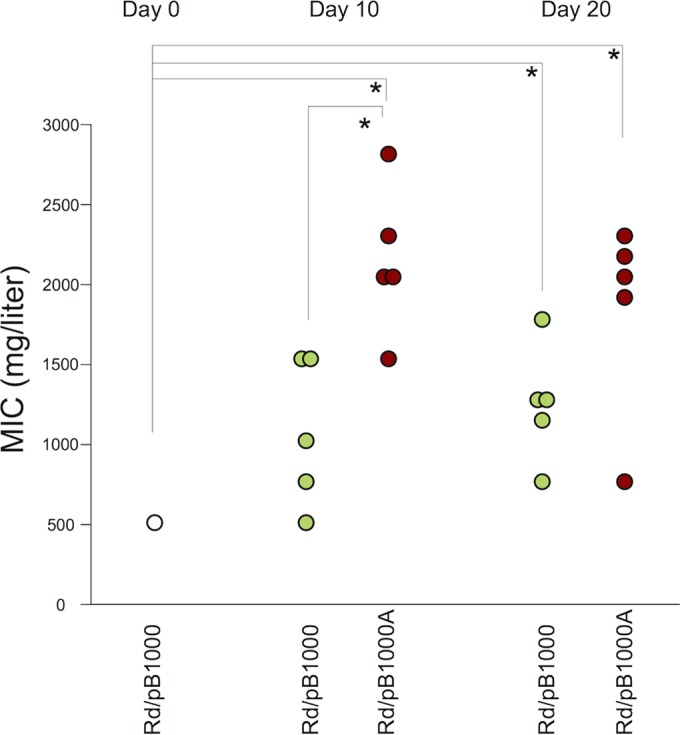
Evolution of antibiotic resistance level. The MICs (mg/liter) of ampicillin in the different populations at days 0, 10, and 20 are shown. The white circle represents the MIC of the parental H. influenzae Rd KW20 carrying plasmid pB1000. The green circles represent the MICs of the five populations of Rd/pB1000 (propagated in the absence of ampicillin), and the red circles indicate the MICs of the five populations of Rd/pB1000A (evolved for 10 days with ampicillin and 10 further days without ampicillin). Asterisks denote significant differences (P < 0.05).
After the antibiotic treatment, Rd/pB1000A populations were further propagated for 10 days in the absence of ampicillin. The MIC after the total 20 days (average = 1,843 mg/liter, SD = 618 mg/liter) remained higher than that for the parental strain (two-tailed paired t test, P = 0.012, t = −4.35, and df = 4) and similar to that at day 10 (two-tailed paired t test, P = 0.801, t = 0.26, and df = 6.5). Interestingly, Rd/pB1000 populations evolved in the absence of ampicillin during 20 days also showed a significant increase in their antibiotic resistance level compared to parental Rd/pB1000 (average = 1,252 mg/liter, SD = 363 mg/liter; two-tailed paired t test, P = 0.01, t = −4.56, and df = 4). These data suggest that plasmid-bearing populations were able to increase antibiotic resistance by two different mechanisms. One mechanism was associated with the presence of ampicillin and appeared at day 10 in Rd/pB1000A, while the other mechanism became evident in the absence of antibiotics and was significant after 20 days (Fig. 2).
Ampicillin leads to an increase in plasmid pB1000 copy number.
We first analyzed the mechanisms behind the increase in the antibiotic resistance of Rd/pB1000 linked to the presence of ampicillin. We completely sequenced the plasmid pB1000, carrying the blaROB-1 β-lactamase, from the five populations of Rd/pB1000A at day 10 (propagated with ampicillin, Rd/pB1000A-10) and also from two of the control populations evolved in the absence of the antibiotic (Rd/pB1000-10). We observed no mutation in any of the seven plasmids compared to the parental pB1000. Previous reports have shown that β-lactamase-mediated ampicillin resistance in H. influenzae can increase by concomitant mutations in ftsI gene, coding for the penicillin binding protein 3, the target of ampicillin (9, 19). We sequenced the ftsI gene from 15 clones (3 clones from each Rd/pB1000A-10 population) (19), and we found no mutations. Therefore, mutations in blaROB-1 or ftsI were not responsible for the increase in ampicillin resistance in the populations evolved under antibiotic pressure.
An alternative explanation for the augmentation in the ampicillin resistance level is an increase in the copy number of plasmid pB1000, which would increase the number of copies of the blaROB-1 gene per cell and therefore the expression of the β-lactamase (31). The augmentation of the antibiotic resistance level due to an elevated plasmid copy number has been reported before in small staphylococcal plasmids (32). Plasmid pB1000 showed an average copy number of 44.5 (SD = 9.9) in the parental H. influenzae Rd/pB1000. In Fig. 3 we present the plasmid copy numbers in the different populations of Rd/pB1000 and Rd/pB1000A. Interestingly, there is a significant increase in pB1000 copy number at day 10 in the populations propagated with ampicillin (average = 104.8, SD = 33.8) compared both to the parental strain (two-sample t test, P = 0.014, t = −3.73, and df = 5) and to the populations evolved in the absence of antibiotics (average = 31.06, SD = 15.7) (two-sample t test, P = 0.005, t = −4.42, and df = 5.6). We found a positive correlation between the pB1000 copy number and the MIC of ampicillin in the Rd/pB1000 and Rd/pB1000A populations at day 10 (Pearson's test, r = 0.675, P = 0.032, t = 2.59, and df = 8). This result supports the notion that the increase in plasmid copy number could contribute to the rise in the ampicillin resistance level. To analyze the genetic basis for the increase of pB1000 copy number, we performed deep sequencing of DNA samples from four of the experimental populations at days 10 and 20 (see File S1 in the supplemental material). None of the mutations detected was conclusively associated with the increase in plasmid copy numbers. In File S1 in the supplemental material, we present the analysis and interpretation of the results.
FIG 3.
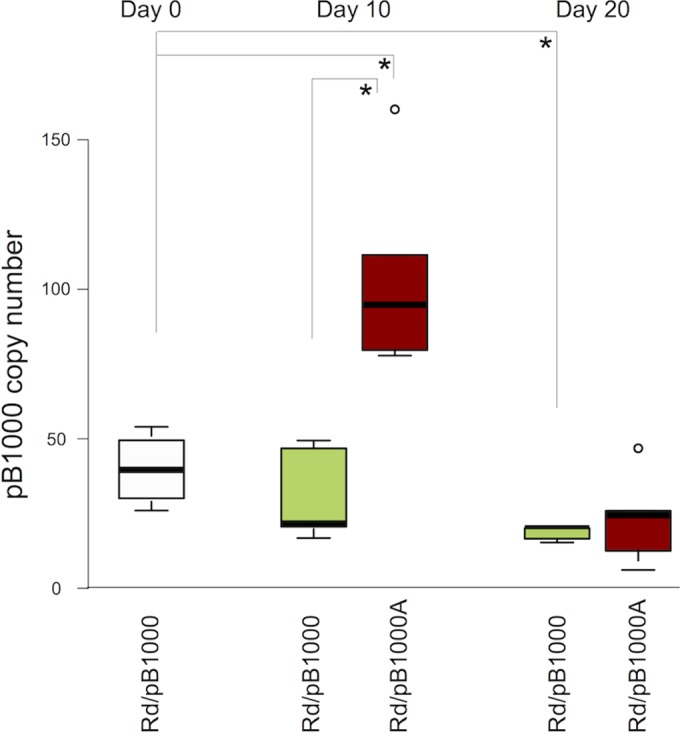
Modification of the copy numbers of plasmid pB1000 over time. A graphic representation of the distribution of plasmid copy numbers at days 0, 10, and 20 is shown. The line inside the box marks the median. The upper and lower hinges correspond to the 25th and 75th percentiles. The upper and lower whiskers extend to the highest and lowest values. The outliers are plotted separately as circles on the chart. The white box represents the pB1000 copy number in the parental Rd/pB1000. The red boxes indicate the pB1000 copy numbers in the five populations of Rd/pB1000A (evolved for 10 days with ampicillin and 10 further days without ampicillin), and the green boxes indicate the pB1000 copy numbers in the five populations of Rd/pB1000 (propagated in the absence of ampicillin). Asterisks denote significant differences (P < 0.05).
Once the presence of ampicillin is removed, the pB1000 copy number decreases at day 20 in Rd/pB1000A (average = 23.80, SD = 14.8) to levels similar to those for the parental strain (two-sample t test, P = 0.057, t = 2.37, and df = 5.8) (Fig. 3). In addition, in the control populations of Rd/pB1000 evolving in the absence of antibiotic, the plasmid copy number appears to decay progressively, showing a significant reduction at day 20 compared to that in the parental strain (average = 18.43, SD = 2.3) (two-sample t test, P = 0.041, t = 4.4807, and df = 2.1). Therefore, plasmid copy number was not responsible for the increase in the antibiotic resistance levels in the plasmid-bearing populations at day 20.
Our experiments showed how the presence of a β-lactam antibiotic produced an increase in the copy number of the small plasmid pB1000 correlating with the rise of the antibiotic resistance levels of the strain. On the other hand, in the absence of ampicillin, the plasmid copy number showed a progressive decrease, which, in principle (33, 34), could contribute to ameliorate the fitness cost associated with pB1000 when no ampicillin resistance is required. These results indicate that variation in plasmid copy number could help bacteria to adapt to different environments.
Enhancement of bacterial fitness increases the ampicillin resistance level in plasmid-bearing clones.
A second mechanism, independent from the presence of ampicillin, is involved in the increase in ampicillin resistance. This mechanism is observed only in strains carrying the antibiotic resistance plasmid pB1000. Previous studies have shown that the improvement in fitness in bacteria carrying antibiotic resistance mechanisms can entail a rise in the antibiotic resistance level of a strain (35, 36). To test for the possibility that the general increase in fitness was responsible for the augmentation of the ampicillin resistance level in the populations carrying pB1000, we used the evolved lines of the plasmid-free Rd strain. The five lines of Rd-20 propagated without the plasmid showed a significant increase in relative fitness over the experiment (average = 11.23%, SD = 2.3%) (Fig. 1). If the general increase in fitness is responsible for part of the rise in the antibiotic resistance level of plasmid-bearing populations, it is reasonable to think that transforming the evolved high-fitness Rd-20 lines with pB1000 would produce strains with higher levels of resistance than the parental Rd/pB1000. Hence, we transformed one clone per Rd-20 population with the original plasmid pB1000 (Rd-20/pB1000). First, we determined the copy number of pB1000 in the transformants (average = 16.47, SD = 6.52), and it was lower than that in the parental Rd/pB1000 (two-sample t test, P = 0.021, t = 4.36, and df = 5.7) but similar to the copy number in the evolved Rd/pB1000-20 (two-sample t test, P = 0.554, t = −0.63, and df = 5.6). Second, we investigated the relative fitness of the Rd-20/pB1000 strains, and it was approximately 11% higher than that of the original Rd/pB1000 strain (Fig. 4). Finally, the MIC of ampicillin of the five Rd-20/pB1000 constructions (average = 900 mg/liter SD = 50.93 mg/liter) was higher than that of the original strain Rd/pB1000 (average = 512 mg/liter SD = 0 mg/liter) (two-sample t test, P < 0.001, t = 38.66, and df = 4) (Fig. 4). Given that the pB1000 copy number is lower in Rd-20/pB1000 than in Rd/pB1000, the fact that the MIC of ampicillin increased significantly provides strong evidence that the general increase in fitness is responsible for an augmentation in the antibiotic resistance level in the lines carrying the plasmid pB1000.
FIG 4.
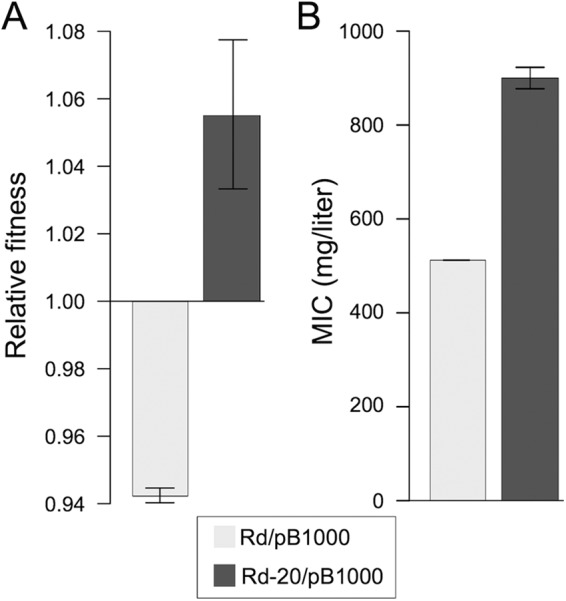
Enhancement in bacterial fitness increases antibiotic resistance levels in plasmid-bearing clones. Rd populations propagated for 20 days were transformed with the original plasmid pB1000 (Rd-20/pB1000). (A) Relative fitnesses (average ± SEM) of the original Rd/pB1000 (light gray bars) and Rd-20/pB1000 clones (one clone per population) (dark gray bars) compared to that of the plasmid-free ancestor H. influenzae Rd KW20. (B) MICs (average ± SEM) of ampicillin for the same clones.
In order to test whether the increase in relative fitness could affect the level of resistance to antibiotics other than ampicillin, we determined the MICs of two antibiotics clinically relevant in H. influenzae infections, chloramphenicol and tetracycline (9), for all the populations at the end of the experiment. The MIC for the parental Rd and Rd/pB1000 strains was 0.5 mg/liter for both antibiotics. There were no changes in the MIC of chloramphenicol or tetracycline in the evolved populations. This result agrees with the initial observation of the fitness-mediated increase in ampicillin being linked to the presence of the ampicillin resistance plasmid pB1000. Therefore, it seems that in this system, the presence of the antibiotic resistance mechanism is required for the increase in fitness to have an effect on the resistance level.
Plasmid copy number and bacterial fitness contribute independently to the increase in ampicillin resistance of Rd/pB1000.
The data presented here suggested that both the increase in plasmid copy number and the increase of bacterial fitness are involved in the augmentation of the antibiotic resistance level of Rd/pB1000 (Fig. 5). To analyze whether both mechanisms contributed to the resistance phenotype in the evolving plasmid-bearing lines, we fitted a linear mixed-effects model where the relative fitness and pB1000 copy number of the Rd/pB1000A and Rd/pB1000 populations (at days 10 and 20) were treated as continuous explanatory variables and the resistance level (MIC of ampicillin) as a response (Fig. 5). We included two random factors to correct for the nonindependence of data from the same line (at different time points) or from the same time point (days 10 and 20) in different lines. Both the plasmid copy number (chi-square test, P = 0.004, χ2 = 8.39, and df = 1) and the bacterial fitness (chi-square test, P = 0.007, χ2 = 7.38, and df = 1) had a significant effect on the antibiotic resistance level of the populations. There was no interaction between the two variables (chi-squared test, P = 0.571, χ2 = 0.32, and df = 1), supporting the hypothesis that plasmid copy number and fitness contribute independently to the increase of the ampicillin resistance level in H. influenzae Rd carrying plasmid pB1000.
FIG 5.
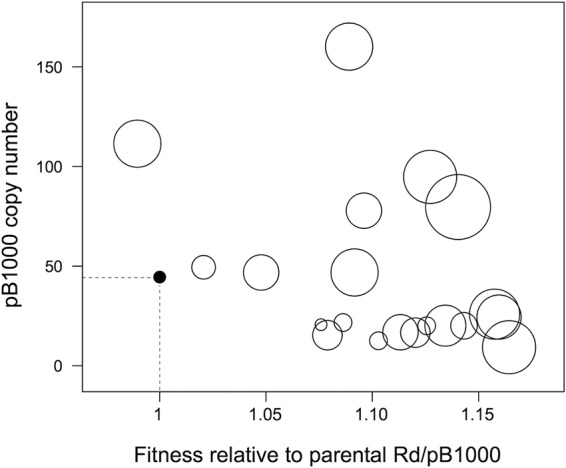
Plasmid copy number and bacterial fitness enhance antibiotic resistance. A representation of the effect of plasmid copy numbers (y axis) and increase in relative fitness (relative to that of the parental H. influenzae Rd KW20 carrying plasmid pB1000) (x axis) on the ampicillin resistance levels of the 20 populations of H. influenzae Rd KW20 carrying plasmid pB1000 (five Rd/pB1000 and five Rd/pB1000A at days 10 and 20) analyzed in this study is shown. The solid circle defined by the dashed lines represents the parental Rd/pB1000. The sizes of the circles are proportional to the MIC of ampicillin of the given population. Both plasmid copy number and bacterial fitness contribute independently to the ampicillin resistance level in the populations.
Conclusion.
This study provides new evidence of the potential effect of antibiotic therapies in the short-term adaptation of resistance plasmids and their bacterial hosts. The cost produced by plasmids is transient and can be rapidly ameliorated, suggesting that plasmids produce a substantial cost only when newly acquired by a bacterial host. This rapid plasmid-host adaptation could help explain how plasmids persist in bacterial populations over long periods, even in the absence of selection for plasmid-encoded traits or horizontal transfer (8). We also showed how plasmid copy number varies over time, facilitating bacterial adaptation to changing environments. Finally, we observed that the increase in both the plasmid copy number and bacterial fitness contribute to enhance the antibiotic resistance level in H. influenzae.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by REDEEX-2 (MICINN, BFU 2011-14145-E) and EU projects EvoTAR 282004-FP7 and EFFORT 613754-FP7.
We thank E. Frago for help with the statistical analyses and N. Montero for excellent technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00235-15.
REFERENCES
- 1.CDC. 2013. Antibiotic resistance threats in the United States. http://www.cdc.gov/drugresistance/threat-report-2013/. [PubMed]
- 2.Gonzalez Zorn B, Escudero JA. 2012. Ecology of antimicrobial resistance: humans, animals, food and environment. Int Microbiol 15:101–109. doi: 10.2436/20.1501.01.163. [DOI] [PubMed] [Google Scholar]
- 3.Gogarten JP, Townsend JP. 2005. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol 3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 4.Nikaido H. 2009. Multidrug resistance in bacteria. Annu Rev Biochem 78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogwill T, MacLean RC. 2015. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl 8:284–295. doi: 10.1111/eva.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison E, Brockhurst MA. 2012. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol 20:262–267. doi: 10.1016/j.tim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Bouma JE, Lenski RE. 1988. Evolution of a bacteria/plasmid association. Nature 335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- 8.San Millan A, Peña-Miller R, Toll-Riera M, Halbert ZV, McLean AR, Cooper BS, MacLean RC. 2014. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat Commun 5:5208. doi: 10.1038/ncomms6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tristram S, Jacobs MR, Appelbaum PC. 2007. Antimicrobial resistance in Haemophilus influenzae. Clin Microbiol Rev 20:368–389. doi: 10.1128/CMR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.San Millan A, Escudero JA, Catalan A, Nieto S, Farelo F, Gibert M, Moreno MA, Dominguez L, Gonzalez-Zorn B. 2007. Beta-lactam resistance in Haemophilus parasuis is mediated by plasmid pB1000 bearing blaROB-1. Antimicrob Agents Chemother 51:2260–2264. doi: 10.1128/AAC.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarado A, Garcillán-Barcia MP, de la Cruz F. 2012. A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One 7:e40438. doi: 10.1371/journal.pone.0040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao V, Lambert T, Courvalin P. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum beta-lactamase CTX-M-17. Antimicrob Agents Chemother 46:1212–1217. doi: 10.1128/AAC.46.5.1212-1217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zioga A, Whichard JM, Kotsakis SD, Tzouvelekis LS, Tzelepi E, Miriagou V. 2009. CMY-31 and CMY-36 cephalosporinases encoded by ColE1-like plasmids. Antimicrob Agents Chemother 53:1256–1259. doi: 10.1128/AAC.01284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pallecchi L, Riccobono E, Sennati S, Mantella A, Bartalesi F, Trigoso C, Gotuzzo E, Bartoloni A, Rossolini GM. 2010. Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob Agents Chemother 54:678–682. doi: 10.1128/AAC.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CY, Lindsey RL, Strobaugh TP, Frye JG, Meinersmann RJ. 2010. Prevalence of ColE1-like plasmids and kanamycin resistance genes in Salmonella enterica serovars. Appl Environ Microbiol 76:6707–6714. doi: 10.1128/AEM.00692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehrenberg C, Hopkins KL, Threlfall EJ, Schwarz S. 2007. Complete nucleotide sequence of a small qnrS1-carrying plasmid from Salmonella enterica subsp. enterica Typhimurium DT193. J Antimicrob Chemother 60:903–905. doi: 10.1093/jac/dkm283. [DOI] [PubMed] [Google Scholar]
- 17.San Millan A, Escudero JA, Gutierrez B, Hidalgo L, Garcia N, Llagostera M, Dominguez L, Gonzalez-Zorn B. 2009. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob Agents Chemother 53:3399–3404. doi: 10.1128/AAC.01522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.San Millan A, Garcia-Cobos S, Escudero JA, Hidalgo L, Gutierrez B, Carrilero L, Campos J, Gonzalez-Zorn B. 2010. Haemophilus influenzae clinical isolates with plasmid pB1000 bearing blaROB-1: fitness cost and interspecies dissemination. Antimicrob Agents Chemother 54:1506–1511. doi: 10.1128/AAC.01489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.San Millan A, Giufré M, Escudero JA, Hidalgo L, Gutierrez B, Cerquetti M, Gonzalez-Zorn B. 2011. Contribution of ROB-1 and PBP3 mutations to the resistance phenotype of a β-lactamase-positive amoxicillin/clavulanic acid-resistant Haemophilus influenzae carrying plasmid pB1000 in Italy. J Antimicrob Chemother 66:96–99. doi: 10.1093/jac/dkq392. [DOI] [PubMed] [Google Scholar]
- 20.Tristram SG, Littlejohn R, Bradbury RS. 2010. blaROB-1 presence on pB1000 in Haemophilus influenzae is widespread, and variable cefaclor resistance is associated with altered penicillin-binding proteins. Antimicrob Agents Chemother 54:4945–4947. doi: 10.1128/AAC.00263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lancashire JF, Terry TD, Blackall PJ, Jennings MP. 2005. Plasmid-encoded TetB tetracycline resistance in Haemophilus parasuis. Antimicrob Agents Chemother 49:1927–1931. doi: 10.1128/AAC.49.5.1927-1931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Toro M, Rodríguez I, Rojo-Bezares B, Helmuth R, Torres C, Guerra B, Sáenz Y. 2013. pMdT1, a small ColE1-like plasmid mobilizing a new variant of the aac(6′)-Ib-cr gene in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 68:1277–80. doi: 10.1093/jac/dkt001. [DOI] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th ed Approved standard M100-S19 CLSI, Wayne, PA. [Google Scholar]
- 24.Björkman J, Andersson DI. 2000. The cost of antibiotic resistance from a bacterial perspective. Drug Resist Updat 3:237–245. doi: 10.1054/drup.2000.0147. [DOI] [PubMed] [Google Scholar]
- 25.Lenski RE. 1991. Quantifying fitness and gene stability in microorganisms, p 173–192. In Ginzburg LR. (ed), Assessing ecological risks of biotecnology. Butterworth-Heinemann, Boston, MA. [DOI] [PubMed] [Google Scholar]
- 26.San Millan A, Heilbron K, MacLean RC. 2014. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J 8:601–612. doi: 10.1038/ismej.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Providenti MA, O'Brien JM, Ewing RJ, Paterson ES, Smith ML. 2006. The copy-number of plasmids and other genetic elements can be determined by SYBR-Green-based quantitative real-time PCR. J Microbiol Methods 65:476–487. doi: 10.1016/j.mimet.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Pinheiro JC, Bates DM. 2000. Mixed-effects models in S and S-PLUS. Springer Science & Business Media, Berlin, Germany. [Google Scholar]
- 29.Meyers BR, Wilkinson P, Mendelson MH, Walsh S, Bournazos C, Hirschman SZ. 1991. Pharmacokinetics of ampicillin-sulbactam in healthy elderly and young volunteers. Antimicrob Agents Chemother 35:2098–2101. doi: 10.1128/AAC.35.10.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacy MK, Lu W, Xu X, Tessier PR, Nicolau DP, Quintiliani R, Nightingale CH. 1999. Pharmacodynamic comparisons of levofloxacin, ciprofloxacin, and ampicillin against Streptococcus pneumoniae in an in vitro model of infection. Antimicrob Agents Chemother 43:672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlin BE, Nordström K. 1977. R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid 1:1–7. doi: 10.1016/0147-619X(77)90003-8. [DOI] [PubMed] [Google Scholar]
- 32.Lüthje P, von Köckritz-Blickwede M, Schwarz S. 2007. Identification and characterization of nine novel types of small staphylococcal plasmids carrying the lincosamide nucleotidyltransferase gene lnu(A). J Antimicrob Chemother 59:600–606. doi: 10.1093/jac/dkm008. [DOI] [PubMed] [Google Scholar]
- 33.Patnaik PR. 2000. An evaluation of models for the effect of plasmid copy number on bacterial growth rate. Biotechnol Lett 22:1719–1725. doi: 10.1023/A:1005696401254. [DOI] [Google Scholar]
- 34.Harrison E, Koufopanou V, Burt A, Maclean RC. 2012. The cost of copy number in a selfish genetic element: the 2-μm plasmid of Saccharomyces cerevisiae. J Evol Biol 25:2348–2356. doi: 10.1111/j.1420-9101.2012.02610.x. [DOI] [PubMed] [Google Scholar]
- 35.Albarracín Orio AG, Piñas GE, Cortes PR, Cian MB, Echenique J. 2011. Compensatory evolution of pbp mutations restores the fitness cost imposed by β-lactam resistance in Streptococcus pneumoniae. PLoS Pathog 7:e1002000. doi: 10.1371/journal.ppat.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcusson LL, Frimodt-Møller N, Hughes D. 2009. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog 5:e1000541. doi: 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.