Abstract
A 22-year-old male developed a recurrent sacral abscess associated with embedded shrapnel following a blast injury. Cultures grew extended-spectrum β-lactamase (ESBL)-producing, carbapenem-susceptible Escherichia coli. Ertapenem was administered, but the infection recurred after each course of antibiotics. Initial surgical interventions were unsuccessful, and subsequent cultures yielded E. coli and Morganella morganii, both nonsusceptible to carbapenems. The isolates were Carba NP test negative, gave ambiguous results with the modified Hodge test, and amplified the blaOXA48-like gene by real-time PCR. All E. coli isolates were sequence type 131 (ST131), carried nine resistance genes (including blaCTX-M-27) on an IncF plasmid, and were identical by genome sequencing, except for 150 kb of plasmid DNA in carbapenem-nonsusceptible isolates only. Sixty kilobases of this was shared by M. morganii and represented an IncN plasmid harboring blaOXA-181. In M. morganii, the gene was flanked by IS3000 and ISKpn19, but in all but one of the E. coli isolates containing blaOXA-181, a second copy of ISKpn19 had inserted adjacent to IS3000. To the best of our knowledge, this is the first report of blaOXA-181 in the virulent ST131 clonal group and carried by the promiscuous IncN family of plasmids. The tendency of M. morganii to have high MICs of imipenem, a blaOXA-181 substrate profile that includes penicillins but not extended-spectrum cephalosporins, and weak carbapenemase activity almost resulted in the presence of blaOXA-181 being overlooked. We highlight the importance of surveillance for carbapenem resistance in all species, even those with intrinsic resistances, and the value of advanced molecular techniques in detecting subtle genetic changes.
INTRODUCTION
Blast injuries account for the overwhelming majority of combat injuries sustained by coalition forces in Iraq and Afghanistan (1). A specific type, dismounted complex blast injury (DCBI), has become significantly more prevalent and can cause extensive damage to the perineum/pelvic areas (2). Contamination of these wounds with environmental debris and contents from the gastrointestinal tract can impair wound healing and serve as sources of infection (2, 3). When these fragments lie close to vital organs or systems, it is often preferable to leave them in place rather than risk surgery (4). Although specific guidelines for infection prevention and prophylactic antibiotics exist for DCBI (2, 5), the increasing incidence of antibiotic-resistant strains has resulted in an ever dwindling arsenal of effective drugs (6).
Escherichia coli is a leading cause of extraintestinal surgical site infections (7, 8). In particular, strains belonging to E. coli sequence type 131 (ST131) (serotype O25:H4) appear to be more virulent, have disseminated worldwide (9, 10), and have a propensity to harbor multiple antibiotic resistance genes (11). A recent study from the United States showed that 67 to 69% of all extended-spectrum β-lactamase (ESBL)-producing E. coli strains tested belonged to this ST (12). The expansion of ESBL-producing strains has resulted in a greater reliance on “last resort” antibiotics, particularly the carbapenems (13), although their efficacies are now compromised by the global spread of carbapenem-hydrolyzing enzymes (14).
While still uncommon, gene-mediated carbapenem resistance in E. coli is increasing worldwide (15). In the United States, it is primarily mediated by the class A Klebsiella pneumoniae carbapenemase (KPC), but strains carrying the metallo-β-lactamase NDM have been reported (16). OXA48-like enzymes (OXA48, -162, -163, -181, -204, and -232) are an emerging concern as they are difficult to detect by conventional methods due to a substrate profile that includes penicillins but not extended-spectrum cephalosporins (with the exception of OXA-163 [17]). Furthermore, OXA-48-like enzymes exhibit low carbapenemase activity compared to class A and B enzymes (18), making their detection even more difficult. One variant, OXA-181, has recently been identified in a K. pneumoniae clinical isolate in the United States (19) and has previously been detected in clinical isolates of Citrobacter freundii, Enterobacter cloacae, E. coli, Klebsiella pneumoniae, and Providencia rettgeri with links to the Indian subcontinent (for a recent review see, reference 18). To date, blaOXA-181 has only been found on plasmids, although information on many of these plasmids is scant. In a C. freundii isolate cultured from a urine sample from a French woman with recent surgery in India, blaOXA-181 was carried on pT-OXA-181, an 83.5-kb IncT plasmid (20). In contrast, blaOXA-181 was harbored on pKP3-A, a 7-kb ColE2-type plasmid in K. pneumoniae KP3, cultured from the wound of a patient in the Sultanate of Oman who had received recent hospital treatment in Tanzania and India (21). The genetic environments surrounding blaOXA-181 were significantly different in both plasmids, but ISEcp1 appears to have played a role in mobilizing blaOXA-181 in both (20, 21).
Here, we report on the in vivo transfer of an IncN plasmid containing blaOXA-181 from Morganella morganii to an ESBL-producing ST131 strain of E. coli, resulting in carbapenem resistance. Unlike other descriptions of the genetic environment surrounding blaOXA-181, mobilization appeared to be facilitated by IS3000. The isolates gave ambiguous results with the modified Hodge Test (mHT) (22) and were negative using the Carba NP test (23).
CASE REPORT
A 22-year-old service member sustained multiple fragment wounds of the pelvis and lower extremities, resulting in sacral and coccygeal fractures, colorectal perforation, and cerebral spinal fluid (CSF) leakage, following a DCBI in Afghanistan (day 1). He was evacuated through Germany to Maryland, where initial groin surveillance swabs (day 6) grew an extended-spectrum β-lactamase (ESBL)-producing E. coli isolate (designated MRSN 17749) (Table 1) susceptible to amikacin, ertapenem, gentamicin, imipenem, piperacillin-tazobactam, and tobramycin. On day 10, aerobic and anaerobic blood culture bottles grew E. coli (MRSN 17758) with the same susceptibilities. Repeat blood cultures were negative on day 13, following initiation of treatment with meropenem. Initial intraoperative tissue wound cultures were negative.
TABLE 1.
Phenotypic characteristics of the clinical isolates in this study
| Straina | Day postinjuryb | Sourcec | MIC (μg/ml) ofd |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SAM | ETP | IPM | MEM | CAZ | FEP | CRO | TZP | |||
| E. coli MRSN 17749 | 6 | Groin swab | 8/4 | <0.5 | <0.25 | <0.25 | 8 | 16 | >32 | ≤2/4 |
| E. coli MRSN 17749T | NAe | NA | >16/8 | 2 | 4 | 2 | 8 | 16 | >32 | >64/4 |
| E. coli MRSN 22624 | 293 | Sacral abscess | >16/8 | 4 | 4 | 2 | 8 | 16 | >32 | >64/4 |
| M. morganii MRSN 22709 | 293 | Sacral abscess | >16/8 | 4 | 8 | 2 | ≤2 | ≤1 | ≤2 | >64/4 |
| E. coli MRSN 22713 | 295 | Epidural abscess | >16/8 | 4 | 4 | 4 | 8 | >16 | >32 | >64/4 |
| K. pneumoniae 11978 | NA | >16/8 | >4 | 16 | 16 | >16 | >16 | >32 | >64/4 | |
The MRSN strain number is the deidentified designation used to track isolates throughout the Multidrug-Resistant Organism Repository and Surveillance Network (MRSN). Klebsiella pneumoniae 11978 is the blaOXA-48 type strain. MRSN 17758, 18675, and 24213 had the same susceptibility profile as MRSN 17749. MRSN 22710, 22711, and 22712 had the same susceptibility profile as MRSN 22624.
Number of days after the initial injury.
Clinical site where the isolate was recovered.
Antibiotic susceptibilities were determined using the BD Phoenix (BD Diagnostics Systems, MD) with NMIC/ID 133 panels (Gram negative), except for the MICs of imipenem and meropenem, which were determined in quintuplicate by broth microdilution (24). CAZ, ceftazidime; CRO, ceftriaxone; ETP, ertapenem; FEP, cefepime; IPM, imipenem; MEM, meropenem; SAM, ampicillin-sulbactam; TZP, piperacillin-tazobactam.
NA, not applicable.
On day 62, the patient developed fever and back pain, and computerized tomography (CT) revealed a presacral abscess. Tissue culture from this area (day 65) grew ESBL-producing E. coli (MRSN 18675) with the aforementioned susceptibilities. Four days later (day 69), the patient was discharged and was treated for a total of 6 weeks with ertapenem (1 g/24 h) for presacral abscess with possible contiguous osteomyelitis. A repeat CT scan 4 weeks into therapy showed a significant decrease in abscess size. One week after completion of ertapenem therapy, the patient presented with recurrent fever and chills, and imaging showed reaccumulation of the presacral abscess. The patient was restarted on ertapenem for presumed recurrence of the same organism and completed an additional 9 weeks of therapy with resultant normalization of inflammatory markers and resolution of symptoms. However, 1 week after antibiotic therapy was discontinued, fever and abdominal pain recurred. CT imaging again showed reaccumulation of the abscess with clear proximity to a sacral fragment. Drainage and fragment removal were planned but aborted after fragment migration presented significant concern for nerve damage/paralysis. An alternate anterior drainage approach was instead pursued via an exploratory laparotomy (already planned colostomy reversal) on day 180. Unfortunately, the sacral fragment could not be safely accessed during the procedure. The repeated abscess drainage grew an ESBL E. coli isolate with the same sensitivities as that described above (MRSN 24213).
Ultimately, the patient underwent posterior sacral hemilaminectomy with abscess drainage and fragment removal (day 293). Cultures from the abscess drainage revealed growth of M. morganii (MRSN 22709), E. coli now resistant to ertapenem and imipenem (MIC of 4) (MRSN 22710, 22711, and 22712), and Bacteroides fragilis. Antibiotic coverage was changed from initial empirical vancomycin and meropenem at the time of clinical decompensation to colistin, tigecycline, and metronidazole, when culture results became available. Within 12 h after the procedure, the patient developed lower-extremity parasthesias, and an emergent magnetic resonance imaging (MRI) revealed anterior-tracking abscess fluid with development of an epidural abscess. This abscess was drained and grew only carbapenem-resistant E. coli (MRSN 22713).
The patient completed 2 weeks of therapy prior to stopping tigecycline for gastrointestinal intolerance and then colistin, 2.5 weeks later, for renal toxicity. Metronidazole was continued for 6 weeks (to day 338). There have been no fevers, elevation of inflammatory markers, or evidence of abscess recurrence as of this writing (day 620).
MATERIALS AND METHODS
Bacterial strains and detection of carbapenemases.
Strain identification and antibiotic susceptibilities were determined using the BD Phoenix (BD Diagnostics Systems, MD) with panels NMIC/ID 133 (Gram negative) and PMIC/ID 107 (Gram positive) in a College of American Pathologists (CAP)-accredited laboratory. MICs of imipenem and meropenem were confirmed using broth microdilution as described previously (24).
Phenotypic detection of carbapenemase production was performed using the mHT (22) and the Carba NP test with various inoculum sizes (23, 25) from overnight cultures on blood agar. Molecular detection of blaOXA48-like genes was performed using real-time PCR and primer pair OXA48-60F (GTAGCAAAGGAATGGCAA) and OXA48-185R (GCCCGTTTAAGATTATTGG), which amplify a 125-bp product from all 6 variants of blaOXA48 described to date (OXA-48, -162, -163, -181, -204, and -232). Real-time PCR was performed in 20-μl volumes with 200 nM primer on a Bio-Rad CFX96 real-time PCR instrument (Bio-Rad Laboratories, Hercules, CA) using SsoAdvanced universal SYBR green supermix (Bio-Rad Laboratories, CA). The cycling parameters were 95°C for 5 min followed by 40 cycles of 95°C for 10 s and 56°C for 10 s. Appropriate positive (K. pneumoniae 11978 [26]) and negative (K. pneumoniae ATCC 1706) controls were incorporated into every test.
S1 nuclease plasmid profiling.
S1 nuclease plasmid profiling was performed using pulsed-field gel electrophoresis (PFGE). Briefly, overnight cultures were diluted in cell resuspension buffer (100 mM Tris-HCl [pH 8], 10 mM EDTA [pH 8]) to an optical density at 600 nm (OD600) of between 0.68 and 0.72, and 200 μl of the resulting suspension was mixed with 200 μl of molten 1.8% agarose and 1 mg/ml of proteinase K. The agarose-cell suspension mixture was allowed to set in disposable plug molds (Bio-Rad Laboratories, Hercules, CA) and incubated overnight at 55°C in lysis buffer (50 mM Tris-HCl [pH 7.5 to 8.0], 50 mM EDTA [pH 8], 10 mg/ml sodium laurylsarcosine) and proteinase K (0.1 mg/ml). Plugs were washed twice in sterile water and three times in Tris-EDTA (TE) buffer at 55°C and then incubated with 50 U of S1 nuclease in 200-μl volumes at 37°C for 2 h. EDTA was added to a final volume of 35 mM and heated to 70°C for 10 min to inactivate the S1 nuclease. One percent PFGE agarose gels were run for 15 h at a gradient of 6 V/cm, an included angle of 120°, and initial and final switch times of 10 s and 40 s, respectively.
Plasmid mating.
Overnight cultures of M. morganii MRSN 22709 (donor) and E. coli MRSN 17749 (recipient) were diluted to an OD600 of 0.6. One milliliter of a 1:1 and 1:10 ratio of donor to recipient was added to 4 ml of LB broth and incubated with shaking for 5 h. Serial dilutions were plated onto Mueller-Hinton agar supplemented with cefepime (64 μg/ml) and imipenem (2 μg/ml) and incubated for 48 h at 37°C. Aliquots were also plated onto Mueller-Hinton agar alone, Mueller-Hinton agar supplemented with cefepime (64 μg/ml), and Mueller-Hinton agar supplemented with imipenem (2 μg/ml). Potential transformants were selected and confirmed as E. coli using the BD Phoenix (BD Diagnostics Systems). Plasmid transfer efficiency was calculated as the ratio of confirmed E. coli transformants to the total number of E. coli cells isolated on Mueller-Hinton agar supplemented with cefepime. One transformant, designated MRSN 17749T, was selected for whole-genome sequencing.
Plasmid stability.
MRSN 22624 and 22713, carrying 4 and 14 copies of pMR3-OXA181, respectively, were subcultured daily for 2 weeks on Mueller-Hinton agar with and without meropenem at a final concentration of 1 μg/ml. Each day, DNA was rapidly extracted (27) from 3 colonies and tested by real-time PCR for the single-copy chromosomal genes yccT and uidA (28) and for three genes carried by pMR3-OXA181: traG (traGRT-F [GTCGGGGAAAACGGTACTC] and traGRT-R [GTGATCGACCGTGACTTC]), IS3000 (IS3000RT-F [CTGACCAAAAGGGATCTGGA] and IS3000RT-R GACTTTCTTTCTCGTCGCAG), and blaOXA-181 (see above for primer sequences). Real-time PCR was performed as described above. Experiments were performed from three biological replicates.
Whole-genome sequencing.
Whole-genome sequencing was performed using an Illumina MiSeq desktop sequencer as previously described (27). Newbler version 2.7 (454 Life Sciences, Branford, CT) was used to assemble MiSeq sequencing reads into de novo contigs and scaffolds. Comparative genomic analyses were performed using Geneious (Biomatters, Auckland, New Zealand) (29). Antimicrobial resistance genes were annotated using ResFinder 2.0 (30), transposons and insertion sequences were identified using ISfinder (31), and potential promoter binding domains were identified using BPROM (32). Plasmid maps were generated using Easyfig 2.1 (33) and Lasergene 11 (DNASTAR, Madison, WI).
Nucleotide sequence accession numbers.
The whole-genome shotgun (WGS) sequences of MRSN 17749, 22464, and 22709 have been deposited in the DDBJ/EMBL/GenBank WGS database under accession no. JRKU00000000, JRKV00000000, and JRKW00000000, respectively. The complete sequence of plasmid pMR3-OXA181 has been deposited in the DDBJ/EMBL/GenBank nucleotide database under accession no. KM660724.
RESULTS
Phenotypic characteristics of clinical isolates.
Nine E. coli isolates and one M. morganii isolate were obtained from the patient over the course of treatment (Table 1). The first E. coli isolate, MRSN 17749, was cultured from a groin surveillance swab 6 days after the initial injury. The isolate had MICs of ertapenem, imipenem, and meropenem of <0.5 μg/ml and was also susceptible to ampicillin-sulbactam (SAM) and piperacillin-tazobactam (TZP). Successive E. coli isolates cultured from different anatomical sites over the following 7 months displayed the same antibiotic susceptibility profile (Table 1). On day 293, multiple E. coli isolates and M. morganii MRSN 22709 were obtained from the sacral region following surgery (Table 1), with MICs of ertapenem, imipenem, and meropenem of ≥2 μg/ml—nonsusceptible by CLSI standards (24). Furthermore, all isolates were no longer susceptible to SAM and TZP (Table 1). The final E. coli isolate, MRSN 22713, was cultured from an epidural abscess 295 days after the initial injury.
Detection of carbapenemase activity.
All carbapenem-nonsusceptible isolates were negative for carbapenemase activity using the Carba NP test, irrespective of inoculum size (23, 25), with the exception of E. coli MRSN 22713. This isolate displayed a very weak color change, from red to red-orange after 2 h, but only when 3 loopfuls were inoculated. The modified Hodge test (mHT) results with both ertapenem and meropenem were difficult to interpret for many of the isolates (see Fig. S1 in the supplemental material). With ertapenem, all E. coli isolates carrying blaOXA-181 were positive, although the characteristic cloverleaf pattern was reduced in all isolates, with the exception of E. coli MRSN 22713, compared to the reference strain, K. pneumoniae ATCC BAA-1705 (see Fig. S1). Conversely, carbapenem-nonsusceptible M. morganii MRSN 22709 was negative. In contrast, the mHT results with meropenem were the opposite: positive with M. morganii MRSN 22709 but negative for all E. coli isolates, with the exception of MRSN 22713 (see Fig. S1). Molecular testing detected a blaOXA-48-like gene in the carbapenem-nonsusceptible E. coli and M. morganii isolates but not in carbapenem-sensitive E. coli.
WGS and plasmid analysis.
Whole-genome sequencing was performed on every E. coli isolate collected from the patient, the single M. morganii isolate, the E. coli transformant MRSN 17749T, and MRSN 22711 and MRSN 22713 after 12 days of serial passage on Mueller-Hinton agar with and without meropenem. The average genome coverage was 85× (±15). All E. coli isolates belonged to ST131, shared >99.9% identity, and had approximately 132 kb of plasmid DNA that was not found in the M. morganii isolate. An additional 150 kb of plasmid DNA was found only in carbapenem-nonsusceptible E. coli isolates, and 60 kb of this was shared with M. morganii. S1 nuclease digestion confirmed the presence of the 132-kb plasmid in all E. coli isolates but also detected two additional plasmids, approximately 90 kb and 60 kb in carbapenem-nonsusceptible E. coli only (Fig. 1). The 60-kb plasmid was also present in M. morganii and carried blaOXA-181 by WGS. Plasmid assembly was greatly facilitated by having sequencing reads from all isolates, including those carrying one, two, or all three of the plasmids identified. Furthermore, pMR3-OXA181 (see below), carrying blaOXA-181, was the only plasmid in M. morgannii and shared no sequence with the other plasmids found in the E. coli isolates. This fortunate set of circumstances allowed pMR3-OXA181 to be assembled into a single closed contig with no ambiguities.
FIG 1.
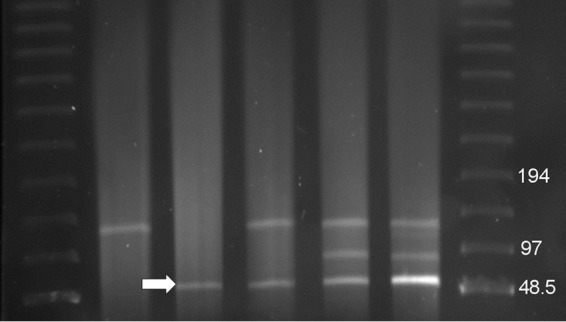
Pulsed-field gel electrophoresis (PFGE) of S1 nuclease-digested DNA. Lanes 1 and 7, PFGE size standard lambda (λ) ladder (Bio-Rad Laboratories): lane 2, E. coli MRSN 17749, carbapenem susceptible; lane 3, M. morganii MRSN 22709, carbapenem nonsusceptible; lane 4, E. coli MRSN 17749T transformant, carbapenem nonsusceptible; lane 5, E. coli MRSN 22624, carbapenem nonsusceptible; lane 6, MRSN 22713, carbapenem nonsusceptible. pMR3-OXA181 is indicated by the white arrow. The MRSN 17769 plasmid profile is representative of MRSN 17758, 18675, and 24213. The MRSN 22624 plasmid profile is representative of 22710, 22711, and 22712, each of which carries 4 copies of pMR3-OXA181. MRSN 22713 carries 14 copies of pMR3-OXA181.
The approximately 132-kb plasmid, designated pMR0713, belonged to incompatibility group F (IncF) and shared closest homology (99.9% identity over 89% of the plasmid) to plasmid pKF3-140 (GenBank accession no. FJ876827.1), isolated from a K. pneumoniae culture collected from the sputum of a patient in Wenzhou, China, in 2006 (34). The plasmid harbored nine antibiotic resistance genes, including genes encoding resistance to aminoglycosides (aadA5, strA, and strB), macrolides (mphA), sulfonamides (sul1 and sul2), tetracyclines (tetA), trimethoprim (dfrA17), and the extended-spectrum β-lactamase (blaCTX-M-27).
The approximately 90-kb plasmid, designated pMR0514, belonged to IncI1 and shared the closest homology (98.6% identity over 90% of the plasmid) to pESBL-12 (GenBank accession no. CP008735), an IncFII plasmid isolated from E. coli. The plasmid had 105 putative or confirmed open reading frames (ORFs), including a conjugal transfer gene cluster, a pilus assembly cluster, and genes involved in plasmid maintenance and stability. The plasmid carried no antibiotic resistance genes.
The final 57,797-bp plasmid, designated pMR3-OXA181, was present in carbapenem-nonsusceptible E. coli and in M. morganii MRSN 22709 only. The plasmid copy number varied from just a single copy in M. morganii MRSN 22709 to approximately 14 copies in MRSN 22713 based on average sequence read coverage (see Fig. S2 in the supplemental material). pMR3-OXA181 belonged to incompatibility group N (IncN) and shared the closest homology to pNDM-BTR (GenBank accession no. KF534788), a plasmid carrying blaNDM-1 from an E. coli isolate cultured in China (Fig. 2A). pMR3-OXA181 and pNDM-BTR share >99.9% identity over 92% of the plasmid, and the inversion of a 7-kb region flanked by IS26 accounts for the majority of the differences in their shared backbone (Fig. 2A).
FIG 2.
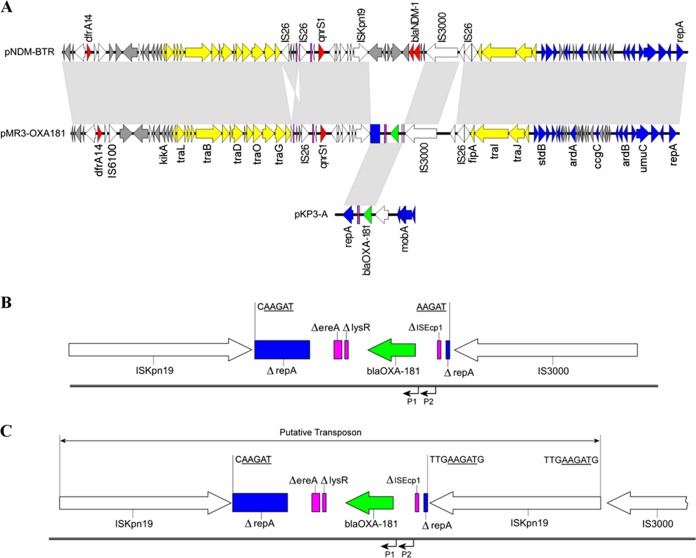
Analysis of pMR3-OXA181 and the blaOXA-181 environment. (A) Alignment of pNDM-BTR, pMR3-OXA181, and pKP3-A. Block arrows indicate confirmed or putative open reading frames (ORFs) and their orientations. Arrow size is proportional to predicted ORF length. Resistance genes are indicated by red arrows, except for blaOXA-181 (green arrow). Conjugal transfer genes are indicated by yellow arrows. DNA mobilization genes are indicted by white arrows. Plasmid mobility, replication, and maintenance genes are indicated by blue arrows. Pseudogenes are indicated by pink rectangles. Hypothetical and unknown genes are indicted by gray arrows. The light gray regions between the three plasmids indicate nucleotide identity of >98% by BLASTN. For clarification, only a subset of gene names are included. Gene nomenclature was assigned based on the closest BLAST match from NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi; last accessed March 2015). (B and C) Genetic environment surrounding blaOXA-181 in M. morganii MRSN 22709 and E. coli MRSN 22711 (B) and E. coli MRSN 22624, 22710, 22712, and 22713 (C). Open reading frames are shown as block arrows and truncated genes by rectangles. Arrows indicate the direction of transcription. blaOXA-181 is represented by a green block arrow. Pink rectangles represent pseudogenes. The disrupted repA gene is indicated by blue rectangles. White block arrows are used to represent ISKpn19 and IS3000. (Note that ISKpn19 is composed of three ORFS [35].) The 5-bp (B) and 9-bp (C) target site duplications, indicative of IS3000 and ISKpn19 transposition, respectively, are indicated. The 5-bp target site duplications have been underlined for clarification. The positions of the two putative promoters upstream of the blaOXA-181 are indicated by black arrows. The putative composite transposon formed by the replicative transposition of ISKpn19 is indicated by a double arrow (C).
pMR3-OXA181 has 90 confirmed or putative open reading frames, including genes encoding a conjugal transfer system and three genes encoding resistance to trimethoprim (dfrA14), quinolones (qnrS1), and β-lactams, including the carbapenems (blaOXA-181). blaOXA-181 is situated on a 3,075-bp region that shares >99% identity with a region in plasmid pKP3-A (21) but has lost approximately 4 kb corresponding to the oriV gene, the mobilization genes mobA, mobB, mocC, and mobD, and all but 265 bp of ISEcp1 (Fig. 2A). Of note, the terminal inverted repeat (IR) and the putative −35 (TTGAAA) and −10 (TACAAT) promoter sequence of ISEcp1 identified in pKP3-A (21) have been retained in pMR3-OXA181. A second putative −35 (TTAGCA) and −10 (TATATT) promoter sequence is also located 20 bp upstream of the blaOXA-181 start codon (Fig. 2B), and the contribution of both these promoters to blaOXA-181 expression remains to be determined. The ΔlysR and Δere fragments identified in pKP3-A have been retained in pMR3-OXA181, but ISKpn19 and IS3000 have inserted into repA, resulting in two repA fragments that are situated at the extremities of the unit. A 5-bp target duplication (AAGAT) beside the 50-bp inverted repeats of ISKpn19 and IS3000 (31) suggests that IS3000 was involved in the initial mobilization of this region (Fig. 2B) (35).
Unexpectedly, analysis of the plasmid in M. morganii MRSN 22709 and the five E. coli strains (Table 1) revealed that in four of the E. coli isolates (MRSN 22624, 22710, 22712, and 22713) a second copy of ISKpn19 had inserted into the region directly downstream of IS3000 (Fig. 2C). Consequently, pMR3-OXA181 was 60,657 bp in these isolates. A 9-bp target duplication at the site of insertion suggests ISKpn19 was mobilized by replicative transposition (31).
pMR3-OXA181 transferred from M. morganii MRSN 22709 to the initial carbapenem-susceptible E. coli isolate MRSN 17749 with an efficiency of 4 × 10−5, resulting in MRSN 17749T. This resulted in increased resistance to carbapenems and β-lactamase inhibitors (Table 1). Plasmid analysis by S1 digestion and WGS of one transformant, MRSN 17749T, confirmed that this corresponded to the transfer of pMR3-OXA181 (Fig. 1).
Stability of pMR3-OXA181.
Normalization of the three plasmid-carried genes traG, IS3000, and blaOXA-181 to the single-copy chromosomal genes yccT and uidA allowed daily monitoring of plasmid copy number by real-time PCR in the presence and absence of meropenem in two strains. MRSN 22711 was isolated prior to the patient being administered meropenem and contained 3 to 4 copies of pMR3-OXA181, based on average read coverage with WGS (see Fig. S1 in the supplemental material). MRSN 22713 was the final E. coli isolate recovered from the patient and was cultured 48 h after the patient was administered meropenem. Unlike all other isolates, MRSN 22713 contained 13 to 14 copies of pMR3-OXA181, was weakly Carba NP positive, and had MICs of meropenem equal to 4 μg/ml (Table 1).
In the absence of meropenem, the plasmid copy number in MRSN 22711 remained stable at 4 copies for 12 days. In MRSN 22713, the plasmid copy number dropped from 13 to 4 after 48 h and remained at 3 (±1) copies for the remaining 10 days. Of interest, in one of the three biological replicates of MRSN 22713, the plasmid copy number dropped to just 1 after 48 h and was no longer detectable after 72 h. Daily passage in the presence of 1 μg/ml meropenem resulted in substantial variation in plasmid copy number. In MRSN 22711, the plasmid number varied from 4 to 8 over the course of the 12 days, even within single colonies isolated from the same plate. A similar result was observed with MRSN 22713, with plasmid copy number dropping from 13 to 8 after 48 h and remaining at 6 (±4) copies for the remaining 10 days. WGS of MRSN 22711 and 22713 after the 12 days supported these findings.
DISCUSSION
Active surveillance for carbapenemases identified the first case of blaOXA48-like genes within the U.S. Military Healthcare System (36). The gene encodes OXA-181, an OXA-48-like protein that differs from OXA-48 by 4 amino acids but shares a similar substrate profile (37). blaOXA-181 was carried on pMR3-OXA181, a 58-kb IncN plasmid (Fig. 2) that likely transferred in vivo from M. morgannii to an ESBL-producing E. coli isolate belonging to the virulent, global ST ST131 (serotype O25:H4) (9).
The mHT was ambiguous and inconsistent (see Fig. S1 in the supplemental material), and all isolates were negative for carbapenemase production using the most recent Carba NP methodology (38), in agreement with previous studies with some OXA181-producing strains (25, 39). The exception was MRSN 22713, which gave a weak red to red-orange color change when excess inoculum was used, as suggested by Tijet and coworkers (25). This was most likely the result of increased OXA-181 production due to the 14 copies of pMR3-OXA181 carried in this isolate (described below).
pMR3-OXA181, carrying blaOXA-181, was carried by carbapenem-nonsusceptible E. coli isolates and M. morganii MRSN 22709 only. The copy number varied from just a single copy in M. morganii MRSN 22709 to 14 in MRSN 22713 (see Fig. S2 in the supplemental material), and this likely modulated OXA-181 production, in accordance with the positive Carba NP test and mHT results with MRSN 22713. It is noteworthy that MRSN 22713 was the last E. coli isolate cultured from the patient (Table 1) and was the only isolate recovered after the patient was administered meropenem. The presence of a more potent carbapenem could help explain why MRSN 22713 had 14 copies of pMR3-OXA181, and this is supported by in vitro data showing significant variation in plasmid copy number when strains were cultured for 12 days in the presence of 1 μg/ml meropenem.
To the best of our knowledge, this is the first description of a blaOXA-48-like gene on an IncN plasmid. This is concerning because IncN plasmids are common among the Enterobacteriaceae and have played an important role in the spread of carbapenemase genes, including blaKPC, blaVIM, and blaNDM (40). pMR3-OXA181 was initially identified in an M. morganii isolate and E. coli from the same wound sample. Previous cultures from the same site grew only E. coli without pMR3-OXA181, suggesting transfer of the plasmid from M. morganii to E. coli in vivo. Epidemiological data suggest that M. morganii was most likely introduced to the wound site from the patient's gastrointestinal tract following an unsuccessful attempt to remove the contaminated sacral fragment via exploratory laparotomy 4 months prior. The patient received prolonged, ineffective ertapenem therapy between the laparotomy and the final surgery, providing the necessary selective pressure to promote in vivo plasmid transfer due to antibiotic use. This is supported by in vitro data showing transfer of pMR3-OXA181 from M. morganii to the initial carbapenem-susceptible E. coli isolate after just 5 h of coincubation.
In this study, a 3.1-kb fragment carrying blaOXA-181 was incorporated into pMR3-OXA181. This region shares >99% homology to the corresponding region in pKP3-A, but an intermediate step has resulted in the removal of the remaining sequence of pKP3-A, including all but 265 bp of ISEscp1. It is noteworthy that the left IR of ISEcp1, the target repeat (ATATA), and the putative promoter identified by Potron and colleagues have also been maintained in pMR3-OXA181, providing additional evidence that pKP3-A, or a similar structure, was the original location of blaOXA-181 (21). The loss of ISEcp1 may have stabilized blaOXA-181 by disrupting ISEcp1 transposase activity, as previously speculated for blaOXA-232, a single-amino-acid variant of OXA-181 (41). Instead of ISEscp1, mobilization of blaOXA-181 into pMR3-OXA181 appeared to be mediated by IS3000, as suggested by the presence of 5-bp target repeats that are consistent with IS3000 insertions (Fig. 2B) (35). The orientation of the two fragments of repA at the termini of the insert suggests that IS3000 initially captured a progenitor plasmid—likely pKP3-A or a derivative—that subsequently lost the missing sequence. Unexpectedly, in four of the five E. coli isolates, a second copy of ISKpn19 was inserted directly downstream of IS3000 (Fig. 2C). A 9-bp target repeat flanking the second copy of ISKpn19 suggests that this occurred via replicative transposition and may represent the formation of a novel composite transposon (Fig. 2C). The insertion of ISKpn19 directly downstream of IS3000 suggests that both IS elements recognize a consensus sequence, possibly within repA, that can be used to facilitate transposition.
Ultimately, surgical removal of the fragment, in combination with tigecycline and colistin, was required to resolve the infection. The presence of blaOXA-181 in M. morganii, a species that tends to have high MICs of imipenem (42), confounds the already difficult task of identifying this carbapenemase (18). Only resistance to ertapenem and the β-lactam/β-lactamase inhibitors alerted personnel to the potential presence of a carbapenemase in this isolate, even though the ambiguous mHT and negative Carba NP test suggested otherwise. As the current reservoir for blaOXA-181 appears to be the Indian subcontinent, it is plausible that the M. morganii isolate was passively acquired during the deployment to Afghanistan and then selected for due to prolonged ertapenem therapy. These data underscore the critical importance of active surveillance for carbapenem resistance in all species, including those with intrinsic resistances, to ensure potential reservoirs for antibiotic resistance plasmids are detected rapidly. Furthermore, advanced molecular techniques should ideally be performed whenever ambiguities are recorded with phenotypic tests.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the U.S. Army Medical Command, the Global Emerging Infections Surveillance and Response System, and the Defense Medical Research and Development Program.
Klebsiella pneumoniae 11978 was kindly provided by Patrice Nordmann.
This material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or reflecting the views of the Department of the Army or the Department of Defense.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04442-14.
REFERENCES
- 1.Schoenfeld AJ, Dunn JC, Bader JO, Belmont PJ Jr. 2013. The nature and extent of war injuries sustained by combat specialty personnel killed and wounded in Afghanistan and Iraq, 2003-2011. J Trauma Acute Care Surg 75:287–291. doi: 10.1097/TA.0b013e31829a0970. [DOI] [PubMed] [Google Scholar]
- 2.Dismounted Complex Blast Injury Task Force. 2011. Dismounted complex blast injury. http://armymedicine.mil/Documents/DCBI-Task-Force-Report-Redacted-Final.pdf Last accessed December 2014.
- 3.Petersen K, Riddle MS, Danko JR, Blazes DL, Hayden R, Tasker SA, Dunne JR. 2007. Trauma-related infections in battlefield casualties from Iraq. Ann Surg 245:803–811. doi: 10.1097/01.sla.0000251707.32332.c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joint Trauma System. 2012. Joint theater trauma system clinical practice guideline: management of high bilateral amputations. http://usaisr.amedd.army.mil/cpgs/High_Bilateral_Amputations_7_Mar_12.pdf Last accessed December 2014.
- 5.Forgione MA, Moores LE, Wortmann GW, Prevention of Combat-Related Infections Guidelines Panel. 2011. Prevention of infections associated with combat-related central nervous system injuries. J Trauma 71:S258–S263. doi: 10.1097/TA.0b013e318227ad86. [DOI] [PubMed] [Google Scholar]
- 6.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 7.Owens CD, Stoessel K. 2008. Surgical site infections: epidemiology, microbiology and prevention. J Hosp Infect 70(Suppl 2):S3–S10. doi: 10.1016/S0195-6701(08)60017-1. [DOI] [PubMed] [Google Scholar]
- 8.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee R, Johnson JR. 2014. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oteo J, Perez-Vazquez M, Campos J. 2010. Extended-spectrum β-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr Opin Infect Dis 23:320–326. doi: 10.1097/QCO.0b013e3283398dc1. [DOI] [PubMed] [Google Scholar]
- 12.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 13.Vardakas KZ, Tansarli GS, Rafailidis PI, Falagas ME. 2012. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum beta-lactamases: a systematic review and meta-analysis. J Antimicrob Chemother 67:2793–2803. doi: 10.1093/jac/dks301. [DOI] [PubMed] [Google Scholar]
- 14.Patel G, Bonomo RA. 2011. Status report on carbapenemases: challenges and prospects. Expert Rev Anti Infect Ther 9:555–570. doi: 10.1586/eri.11.28. [DOI] [PubMed] [Google Scholar]
- 15.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasheed JK, Kitchel B, Zhu W, Anderson KF, Clark NC, Ferraro MJ, Savard P, Humphries RM, Kallen AJ, Limbago BM. 2013. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 19:870–878. doi: 10.3201/eid1906.121515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Castanheira M, Carrer A, Rodriguez CP, Jones RN, Smayevsky J, Nordmann P. 2011. OXA-163, an OXA-48-related class D beta-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother 55:2546–2551. doi: 10.1128/AAC.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 19.Mathers AJ, Hazen KC, Carroll J, Yeh AJ, Cox HL, Bonomo RA, Sifri CD. 2013. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the New World. J Clin Microbiol 51:680–683. doi: 10.1128/JCM.02580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villa L, Carattoli A, Nordmann P, Carta C, Poirel L. 2013. Complete sequence of the IncT-type plasmid pT-OXA-181 carrying the blaOXA-181 carbapenemase gene from Citrobacter freundii. Antimicrob Agents Chemother 57:1965–1967. doi: 10.1128/AAC.01297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4896–4899. doi: 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect 7:88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 23.Dortet L, Brechard L, Poirel L, Nordmann P. 2014. Impact of the isolation medium for detection of carbapenemase-producing Enterobacteriaceae using an updated version of the Carba NP test. J Med Microbiol 63:772–776. doi: 10.1099/jmm.0.071340-0. [DOI] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing. M100-S22 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. 2013. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4578–4580. doi: 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGann P, Courvalin P, Snesrud E, Clifford RJ, Yoon EJ, Onmus-Leone F, Ong AC, Kwak YI, Grillot-Courvalin C, Lesho E, Waterman PE. 2014. Amplification of aminoglycoside resistance gene aphA1 in Acinetobacter baumannii results in tobramycin therapy failure. mBio 5(2):e00915. doi: 10.1128/mBio.00915-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clifford RJ, Milillo M, Prestwood J, Quintero R, Zurawski DV, Kwak YI, Waterman PE, Lesho EP, McGann P. 2012. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One 7:e48558. doi: 10.1371/journal.pone.0048558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salamov VSA. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61-78. In Li RW. (ed), Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 33.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao F, Bai J, Wu J, Liu J, Zhou M, Xia S, Wang S, Yao X, Yi H, Lin M, Gao S, Zhou T, Xu Z, Niu Y, Bao Q. 2010. Sequencing and genetic variation of multidrug resistance plasmids in Klebsiella pneumoniae. PLoS One 5:e10141. doi: 10.1371/journal.pone.0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabate M, Navarro F, Miro E, Campoy S, Mirelis B, Barbe J, Prats G. 2002. Novel complex sul1-type integron in Escherichia coli carrying bla(CTX-M-9). Antimicrob Agents Chemother 46:2656–2661. doi: 10.1128/AAC.46.8.2656-2661.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterman P, Kwak Y, Clifford R, Julius M, Onmus-Leone F, Tsurgeon C, Riley M, Black C, McGann P, Lesho E. 2012. A multidrug-resistance surveillance network: 1 year on. Lancet Infect Dis 12:587–588. doi: 10.1016/S1473-3099(12)70149-4. [DOI] [PubMed] [Google Scholar]
- 37.Potron A, Poirel L, Nordmann P. 2011. Origin of OXA-181, an emerging carbapenem-hydrolyzing oxacillinase, as a chromosomal gene in Shewanella xiamenensis. Antimicrob Agents Chemother 55:4405–4407. doi: 10.1128/AAC.00681-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dortet L, Poirel L, Nordmann P. 2014. Further proofs of concept for the Carba NP test. Antimicrob Agents Chemother 58:1269. doi: 10.1128/AAC.01825-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob Agents Chemother 55:1274–1278. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, Nordmann P. 2013. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D beta-lactamase from Enterobacteriaceae. Int J Antimicrob Agents 41:325–329. doi: 10.1016/j.ijantimicag.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Stock I, Wiedemann B. 1998. Identification and natural antibiotic susceptibility of Morganella morganii. Diagn Microbiol Infect Dis 30:153–165. doi: 10.1016/S0732-8893(97)00243-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.