Abstract
Vibrio parahaemolyticus is commonly resistant to ampicillin, yet the mechanisms underlying this phenomenon are not clear. In this study, a novel class A carbenicillin-hydrolyzing β-lactamase (CARB) family of β-lactamases, blaCARB-17, was identified and found to be responsible for the intrinsic penicillin resistance in V. parahaemolyticus. Importantly, blaCARB-17-like genes were present in all 293 V. parahaemolyticus genome sequences available in GenBank and detectable in all 91 V. parahaemolyticus food isolates, further confirming the intrinsic nature of this gene.
TEXT
Vibrio parahaemolyticus is a major causative agent of gastroenteritis in areas with high seafood consumption rates and has recently become pandemic due to the emergence of the serotype O3:K6 (1). In Hong Kong, V. parahaemolyticus is the leading cause of food-borne illnesses due to the high rate of seafood consumption among the population (2). Although most cases of infections are self-limiting, fatality can occur among immunocompromised patients or those with debilitating medical conditions such as liver disease or diabetes (3). Antibiotics such as ciprofloxacin can be used for the treatment of infections caused by V. parahaemolyticus strains, but the choice of antibiotics should be based on the antimicrobial susceptibilities of the organism. V. parahaemolyticus is commonly considered highly susceptible to virtually all antimicrobials except for penicillins. However, mechanisms mediating the development of penicillin resistance in V. parahaemolyticus are not clear. In this study, we identified a novel carbenicillin-hydrolyzing β-lactamase (CARB) from chromosome 2 of V. parahaemolyticus and showed that the product of this gene is responsible for the intrinsic resistance to penicillins in V. parahaemolyticus.
A novel potential β-lactamase gene, blaV110, with a length of 852 bp was identified through bioinformatics analysis of the whole-genome sequence of V. parahaemolyticus V110, which was shown to be resistant to ampicillin (4). The full-length novel β-lactamase gene was amplified by PCR using primer set F-GCTGAGAGCTCATGAAAAAGTTA, R-CGTAGGATCCTTAACTTTCTTTGTAGTGC and then cloned into Escherichia coli BL21 and tested for the MICs of various β-lactams according to CLSI standards (5). E. coli BL21 isolates expressing the novel β-lactamase gene exhibited MICs of 256, 512, 256, and 1,024 μg/ml toward penicillin G, ampicillin, carbenicillin, and piperacillin, respectively. The BlaV110 enzyme appears to be susceptible to β-lactamase inhibitors such as clavulanic acid, sulbactam, and tazobactam (Table 1). To verify whether the penicillin resistance phenotype was attributed to the expression of blaV110, we further purified a truncated form (60 to 852 bp) of this β-lactamase in which the signal peptide was removed and designated mBlaV110. The mBlaV110 protein was purified through several steps, including a Ni-nitrilotriacetic acid (NTA) column, thrombin treatment to remove the His tag, and a size exclusion column as previously described (6) (see Fig. S1 in the supplemental material). The purity of this protein was higher than 99%, and the yield of the purified mBlaV110 was about 2.4 mg/liter. Kinetic constants were determined for mBlaV110 as previously described (7), and very high catalytic activities on ampicillin, penicillin G, carbenicillin, and piperacillin were noted; however, this enzyme exhibited extremely low activities to other β-lactams tested (Tables 2). In general, mBlaV110 exhibited similar Km values with all penicillins, cefepime, and cefpirome tested but variable kcat values to these penicillins, some cephalosporins, aztreonam, and imipenem (Tables 2). The kinetic data were highly consistent with the MICs of E. coli isolates carrying blaV110 genes. Collectively, our data suggested that BlaV110 is an active β-lactamase that mediates the resistance to penicillins in the V. parahaemolyticus V110 strain.
TABLE 1.
MICs of different β-lactams on V. parahaemolyticus V110 parental strain and E. coli carrying pET28-blaV110
| Antibiotica | MIC (μg/ml) against bacterial strain |
||
|---|---|---|---|
| V. parahaemolyticus V110 | E. coli pET28-blaV110 | E. coli pET-28 | |
| Penicillin G | 512 | 256 | <1 |
| Ampicillin | 128 | 512 | 1 |
| AMP/CLA (2:1) | 2 | <1 | <1 |
| AMP/SUL (1:1) | <1 | 4 | <1 |
| Carbenicillin | 256 | 256 | 2 |
| Piperacillin | 256 | 1,024 | <1 |
| PIP/TAZ (10:1) | 0.06 | 1 | 0.5 |
| Cephalothin | 8 | 1 | 0.03 |
| Cefuroxime | 8 | 0.25 | 0.03 |
| Cefotaxime | 0.06 | 0.004 | 0.004 |
| Cefepime | 1 | 0.06 | 0.06 |
| Cefpirome | 0.25 | 0.015 | 0.015 |
| Aztreonam | 4 | 0.008 | 0.004 |
| Imipenem | 0.03 | 0.25 | 0.008 |
AMP, ampicillin; CLA, clavulanic acid; SUL, sulbactam; PIP, piperacillin; TAZ, tazobactam.
TABLE 2.
Kinetic constants of mBlaV110 toward different β-lactams
| Antibiotic | mBlaV110 |
||
|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (s−1μM−1) | |
| Penicillin G | 110.4 ± 19.31 | 2,320 ± 189.2 | 21.01 |
| Ampicillin | 235.8 ± 40.44 | 2,068 ± 170.9 | 8.77 |
| Carbenicillin | 113.9 ± 28.61 | 1,233 ± 177.4 | 10.83 |
| Piperacillin | 32.6 ± 8.87 | 450.7 ± 50.73 | 13.82 |
| Cefuroxime | NHa | <0.01 | |
| Cefotaxime | NH | <0.01 | |
| Cefepime | 104.3 ± 25.67 | 0.74 ± 0.08 | 7.10 × 10−3 |
| Cefpirome | 22.6 ± 4.77 | 0.98 ± 0.07 | 4.34 × 10−2 |
| Aztreonam | NH | <0.01 | |
| Imipenem | NH | <0.01 | |
NH, no detectable hydrolysis was observed with 1 μM purified mBlaV110 and up to 500 μM substrate.
Protein BLAST of BlaV110 showed 99% homology with PSE-4 from V. parahaemolyticus (8, 9) and high homology (54%) with PSE-4 from Pseudomonas spp. PSE-4 is an alternative name for CARB-1, which belongs to the CARB-type family originally identified from Pseudomonas aeruginosa, Acinetobacter, and Vibrio cholerae (10–12). CARBs, also known as carbenicillin-hydrolyzing β-lactamases, have been found to disperse widely among distantly related bacteria, mostly by mobile genetic elements (10, 13, 14). Similarly, BlaV110 also mediated resistance to ampicillin, penicillin G, carbenicillin, and piperacillin. Therefore, BlaV110 was designated a novel member of the CARB family, blaCARB-17 (GenBank accession number KJ934265).
Analysis of its genetic environment showed that the blaCARB-17 gene was located on chromosome 2 of V. parahaemolyticus V110. Several putative genes including those encoding transporters and enzymes are located upstream and downstream of blaCARB-17 (see Fig. S2 in the supplemental material). The blaCARB-17-like genes were also identified in chromosome 2 of 5 V. parahaemolyticus isolates with completed whole-genome sequences in GenBank (see Fig. S2). The genetic environments of the blaCARB-17-like genes in these isolates were very similar but not identical. There were no mobile genetic elements, such as integrase and transposase, within their genetic environments, and the blaCARB-17-like genes were not located in any genomic islands, suggesting that blaCARB-17 genes may be intrinsic to V. parahaemolyticus. Further analyses of the other 292 whole-genome annotation reports of V. parahaemolyticus available in GenBank identified blaCARB-17-like β-lactamases in 280 out of the 292 V. parahaemolyticus strains. blaCARB-17-like β-lactamase genes were also identified in the other 12 V. parahaemolyticus strains, but they either showed one nucleotide deletion within the full-length blaCARB-17-like genes (10 strains) or showed longer nucleotide fragment deletion at the N termini (2 strains). The latter two strains have very low numbers of proteins in their annotation reports, implying that the lack of full-length blaCARB-17-like genes may be due to the sequencing coverage issue. Taken together, complete genome analysis suggested that all V. parahaemolyticus strains intrinsically harbor blaCARB-17 and its variants. To further prove the intrinsic nature of blaCARB-17, 39 and 52 V. parahaemolyticus strains isolated from seafood in Shenzhen and Hong Kong, respectively, were screened for the presence of blaCARB-17-like genes using primers targeting the full length of blaCARB-17. These isolates were confirmed to be V. parahaemolyticus through screening for the presence of tlh and atpA genes as well as API20E assays (bioMérieux). All isolates were resistant to ampicillin with the exception of 10 isolates with ampicillin MICs of 16 μg/ml. All strains showed positive amplifications for blaCARB-17, and these genes were further confirmed to be blaCARB-17-like genes by sequencing. These data further confirmed the intrinsic nature of blaCARB-17-like genes in V. parahaemolyticus.
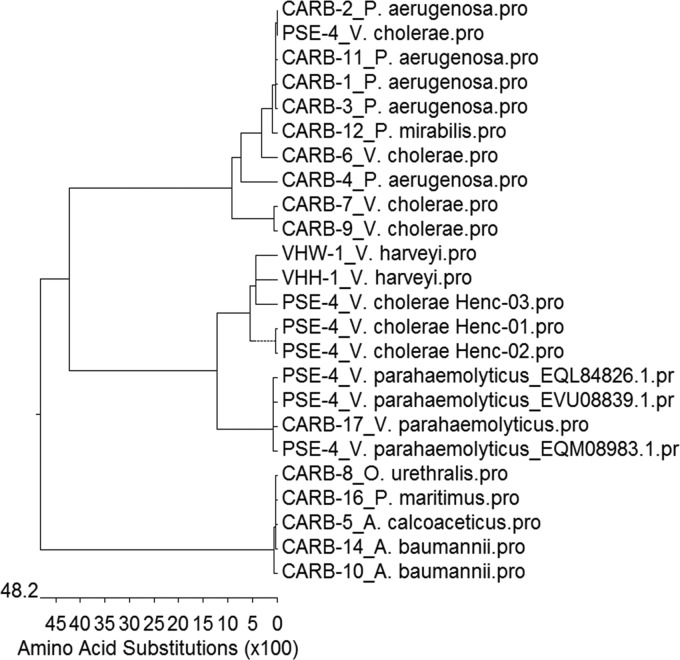
The current nomenclature for the CARB family of β-lactamases is confused with the PSE family in the literature. CARBs are divided into two subgroups, namely the CARB and RTG subgroups (http://www.lahey.org/Studies/). Based on the functional characterization of novel CARB-17 and its variants, the CARB family of β-lactamases can be separated into three distinct squares through further phylogenetic analyses of all CARB β-lactamases. The first square contains CARB-17 and its closely related variants, the second square contains mainly the previously identified narrow-spectrum CARB family from Pseudomonas spp. and V. cholerae, and the third square contains the broad-spectrum RTG subgroup (Fig. 1).
FIG 1.
Phylogenetic tree of CARB family and several related β-lactamases. CARB subgroups, including CARB-1 (PSE-4), CARB-2 (PSE-1), CARB-3, CARB-4, CARB-6, CARB-7, CARB-9, and CARB-11 (PSE-5) to CARB-15, are narrow-spectrum β-lactamases that hydrolyze penicillins (15, 16). RTG subgroups, including CARB-5 (RTG-2), CARB-8 (RTG-3), and CARB-10 (RTG-4), consist of an RTG triad as reported for the GN79 (RTG-1) from Proteus mirabilis (11, 14, 17, 18). Most of the CARB families were not subjected to functional characterization except CARB-10, which has been shown to hydrolyze cefepime and cefpirome and become an extended-spectrum CARB enzyme (14). CARB-17 exhibited 99% homology to PSE-4 from V. parahaemolyticus and approximately 80% to PSE-4 from V. cholerae Henc, as well as VHW-1 and VHH-1 from Vibrio harveyi through BLAST analysis (19). The latter two β-lactamases have been shown to be active on penicillins (19).
In conclusion, this study identified a novel class A β-lactamase, blaCARB-17, which is responsible for the intrinsic resistance to penicillins in V. parahaemolyticus. The data facilitate clear grouping for the whole family of the CARB class of β-lactamases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kathy Po for providing the V. parahaemolyticus isolates for blaCARB-17 gene screening, Yuqian Wu for his help with blaCARB-17 cloning and expression, and Edward Chan for critical reading of the manuscript.
This work was supported by the Chinese National Key Basic Research and Development (973) Program (grant 2013CB127200) and the Health and Medical Research Fund from the Food and Health Bureau, Government of the Hong Kong SAR (grant HMRF:13121422 to S.C.).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00047-15.
REFERENCES
- 1.Shevchuk VB, Gebesh VV, Alekseenko VV, Dobroshtan EV, Padchenko AG. 1986. Clinical aspects of acute intestinal infection caused by Vibrio parahaemolyticus. Vrach Delo 6:114–116. (In Russian.) [PubMed] [Google Scholar]
- 2.Scott L, McGee P, Walsh C, Fanning S, Sweeney T, Blanco J, Karczmarczyk M, Earley B, Leonard N, Sheridan JJ. 2009. Detection of numerous verotoxigenic E. coli serotypes, with multiple antibiotic resistance from cattle faeces and soil. Vet Microbiol 134:288–293. doi: 10.1016/j.vetmic.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Hou CC, Lai CC, Liu WL, Chao CM, Chiu YH, Hsueh PR. 2011. Clinical manifestation and prognostic factors of non-cholerae Vibrio infections. Eur J Clin Microbiol Infect Dis 30:819–824. doi: 10.1007/s10096-011-1162-9. [DOI] [PubMed] [Google Scholar]
- 4.Liu M, Chen S. 2013. Draft genome sequence of Vibrio parahaemolyticus V110, isolated from shrimp in Hong Kong. Genome Announc 1:e00300. doi: 10.1128/genomeA.00300-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing. CLSI M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Chiou J, Leung TY, Chen S. 2014. Molecular mechanisms of substrate recognition and specificity of New Delhi metallo-beta-lactamase. Antimicrob Agents Chemother 58:5372–5378. doi: 10.1128/AAC.01977-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moali C, Anne C, Lamotte-Brasseur J, Groslambert S, Devreese B, Van Beeumen J, Galleni M, Frere JM. 2003. Analysis of the importance of the metallo-beta-lactamase active site loop in substrate binding and catalysis. Chem Biol 10:319–329. doi: 10.1016/S1074-5521(03)00070-X. [DOI] [PubMed] [Google Scholar]
- 8.Nasu H, Iida T, Sugahara T, Yamaichi Y, Park KS, Yokoyama K, Makino K, Shinagawa H, Honda T. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J Clin Microbiol 38:2156–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 10.Partridge SR, Brown HJ, Hall RM. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob Agents Chemother 46:1288–1294. doi: 10.1128/AAC.46.5.1288-1294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choury D, Szajnert MF, Joly-Guillou ML, Azibi K, Delpech M, Paul G. 2000. Nucleotide sequence of the bla(RTG-2) (CARB-5) gene and phylogeny of a new group of carbenicillinases. Antimicrob Agents Chemother 44:1070–1074. doi: 10.1128/AAC.44.4.1070-1074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melano R, Petroni A, Garutti A, Saka HA, Mange L, Pasteran F, Rapoport M, Rossi A, Galas M. 2002. New carbenicillin-hydrolyzing beta-lactamase (CARB-7) from Vibrio cholerae non-O1, non-O139 strains encoded by the VCR region of the V. cholerae genome. Antimicrob Agents Chemother 46:2162–2168. doi: 10.1128/AAC.46.7.2162-2168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philippon AM, Paul GC, Thabaut AP, Jacoby GA. 1986. Properties of a novel carbenicillin-hydrolyzing beta-lactamase (CARB-4) specified by an IncP-2 plasmid from Pseudomonas aeruginosa. Antimicrob Agents Chemother 29:519–520. doi: 10.1128/AAC.29.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potron A, Poirel L, Croize J, Chanteperdrix V, Nordmann P. 2009. Genetic and biochemical characterization of the first extended-spectrum CARB-type beta-lactamase, RTG-4, from Acinetobacter baumannii. Antimicrob Agents Chemother 53:3010–3016. doi: 10.1128/AAC.01164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lachapelle J, Dufresne J, Levesque RC. 1991. Characterization of the blaCARB-3 gene encoding the carbenicillinase-3 beta-lactamase of Pseudomonas aeruginosa. Gene 102:7–12. doi: 10.1016/0378-1119(91)90530-O. [DOI] [PubMed] [Google Scholar]
- 16.Petroni A, Melano RG, Saka HA, Garutti A, Mange L, Pasteran F, Rapoport M, Miranda M, Faccone D, Rossi A, Galas MF. 2004. CARB-9, a carbenicillinase encoded in the VCR region of Vibrio cholerae non-O1, non-O139 belongs to a family of cassette-encoded beta-lactamases. Antimicrob Agents Chemother 48:4042–4046. doi: 10.1128/AAC.48.10.4042-4046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi I, Tsukamoto K, Harada M, Sawai T. 1983. Carbenicillin-hydrolyzing penicillinases of Proteus mirabilis and the PSE-type penicillinases of Pseudomonas aeruginosa. Microbiol Immunol 27:995–1004. doi: 10.1111/j.1348-0421.1983.tb02934.x. [DOI] [PubMed] [Google Scholar]
- 18.Mammeri H, Poirel L, Mangeney N, Nordmann P. 2003. Chromosomal integration of a cephalosporinase gene from Acinetobacter baumannii into Oligella urethralis as a source of acquired resistance to beta-lactams. Antimicrob Agents Chemother 47:1536–1542. doi: 10.1128/AAC.47.5.1536-1542.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teo JW, Suwanto A, Poh CL. 2000. Novel beta-lactamase genes from two environmental isolates of Vibrio harveyi. Antimicrob Agents Chemother 44:1309–1314. doi: 10.1128/AAC.44.5.1309-1314.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.