Abstract
The antipseudomonal efficiency and mechanism of action of a novel engineered antimicrobial peptide, T9W, were evaluated in this study. T9W displayed high activity, with a lethal concentration (LC) of 1 to 4 μM against Pseudomonas aeruginosa, including against ciprofloxacin-, gentamicin-, and ceftazidime-resistant strains, even in the presence of 50 to 300 mM NaCl, 1 to 5 mM Ca2+, or 0.5 to 2 mM Mg2+. The time-kill curve (TKC) analysis demonstrated concentration-dependent activity, with T9W achieving complete killing in less than 30 min at 1× LC and in less than 5 min at 4× LC. Combination TKC analyses additionally demonstrated a synergistic effect with ciprofloxacin and gentamicin. The selectivity of T9W was further supported by its ability to specifically eliminate P. aeruginosa in a coculture with macrophages without toxicity to the mammalian cells. The results from fluorescent measurement indicated that T9W bound to lipopolysaccharide (LPS) and induced P. aeruginosa membrane depolarization, and microscopic observations and flow cytometry further indicated that T9W targeted the P. aeruginosa cell membrane and disrupted cytoplasmic membrane integrity, thereby causing cellular content release leading to cell death. This study revealed the potential usefulness of T9W as a novel antimicrobial agent against P. aeruginosa.
INTRODUCTION
The discovery and development of antibiotics rank among the greatest achievements of medicine (1). Although the introduction and use of these agents played an important role in treating bacterial infections, the emergence and increase in the number of antibiotic-resistant clinical isolates, together with the low rate of development and introduction of new antimicrobials, have been a constant threat to human health. These resistant bacteria are frequently resistant to at least one of the conventional antibiotics, leading to significant increases in morbidity and mortality (2). It is necessary to develop antimicrobials with a completely new mechanism of action to combat these resistant bacteria (3).
Among the resistant bacteria, the Gram-negative bacterium Pseudomonas aeruginosa is an opportunistic pathogen that infects a wide range of animal, plant, and human hosts (4). Compared with other pathogens, P. aeruginosa is very difficult to eradicate because it displays high resistance to a wide variety of antibiotics, including aminoglycosides, fluoroquinolones, and β-lactams, largely because of the low permeability of its outer membrane, which limits the rate of penetration of antibiotic molecules into the cells (5). With increased resistance rates, there is a challenging urgency to find new drugs against P. aeruginosa.
Antimicrobial peptides (AMPs) are small, cationic peptides that play an important role in the innate immune system (6). In most multicellular organisms, AMPs constitute a major component of the ancient, nonspecific innate defense system, forming the first line of defense against invading microbes (7, 8). Compared with conventional antibiotics, AMPs possess multiple modes of action, rapid kill kinetics, broad-spectrum antimicrobial activity, and little host toxicity. Generally, AMPs do not have specific microbial cell targets; instead, it is widely accepted that the cytoplasmic membrane is the main target of AMPs. It is precisely because of this mechanism of action that bacteria have a low potential to develop resistance because the entire membrane structure or membrane lipid composition of a microbe would need to be changed to evade AMPs (9). Consequently, AMPs attract wide and specific attention for the development of a therapeutic candidate for protection against drug-resistant organisms.
Similar to the activity of conventional antibiotics, the broad-spectrum antimicrobial activity of AMPs can have a negative impact on the microbiota, immunity, and health because these agents inhibit or kill pathogens and nonpathogens indiscriminately, thus unfavorably disturbing or disrupting the homeostasis between a healthy microbiota and the immune system (10). The development of single-pathogen therapies to treat highly resistant or totally resistant bacterial pathogens is an unmet need that merits attention (11).
Recently, we determined that a linear, 16-residue α-helical antimicrobial peptide, T9W, has strong and specific activity against P. aeruginosa and low cytotoxicity against human red blood cells (hRBCs) as well as weak or no activity against other Gram-negative bacteria and Gram-positive bacteria, such as Escherichia coli, Salmonella enterica serovar Typhimurium, Staphylococcus aureus, and Streptococcus faecium (12). The value of T9W as an antipseudomonal drug has not been thoroughly evaluated. In comparison, LL37 is the only known human cathelicidin and is effective against a broad spectrum of bacteria, including P. aeruginosa. In addition, LL37 has also been described for its role in neutralization of endotoxins, immunomodulating properties, and inhibition of biofilm formation (13–15). In vitro assays using lipid bilayers indicate that LL37 permeabilizes model membranes by a carpeting or toroidal pore mechanism (16).
We showed here that T9W maintained its P. aeruginosa-targeted activity in the presence of physiological concentrations of sodium chloride and divalent cations and that the susceptibility of P. aeruginosa strains to T9W does not correlate with preexisting resistance to antibiotics. In bacterial cells, we showed that T9W binds to lipopolysaccharide (LPS), dissipates membrane potential, and disrupts membrane integrity. Additionally, we demonstrated that T9W showed synergism with ciprofloxacin and gentamicin when it is used against antibiotic-resistant strains as well as cell selectivity against P. aeruginosa rather than against host cells. Taken together, our data on T9W indicate that there is great potential for developing a new antipseudomonal drug from the synthesis and optimization of a class of antimicrobial agents with a membrane-disruptive mechanism and activity against antibiotic-resistant strains.
MATERIALS AND METHODS
Strains and culture conditions.
P. aeruginosa ATCC 27853 samples were kept in our laboratory, and ATCC 10419, ATCC 21625, and ATCC 21630 were kindly provided by Jianhua Wang (Chinese Academy of Agriculture Sciences, Beijing, China); the clinical antibiotic-susceptible strain LC was obtained from the College of Veterinary Medicine, Northeast Agricultural University (Harbin, China). P. aeruginosa variants resistant to ciprofloxacin (strain LCCI), gentamicin (LCGE), and ceftazidime (LCCE) were developed by a stepwise culture of strain LC with selected antibiotics, and the stability of the acquired resistance was confirmed by at least 30 generations of exponential growth. Clinically isolated P. aeruginosa strains 11441, 11451, 21328, 25349, and 26305 were provided by the 2nd Affiliated Hospital of Harbin Medical University. The strains were stored at −80°C in 15% glycerol. The strains were cultured at 37°C on Mueller-Hinton (MH) agar and grown to the exponential phase in MH broth with rotary shaking at 220 rpm prior to use.
Antimicrobial activity assays.
The MIC was determined using a standard microtiter dilution method with minor modifications, as described previously (17). Briefly, 50 μl of bacterial cells (5 × 105 CFU/ml) in Mueller-Hilton broth (MHB) were incubated in 96-well microtiter plates with 50 μl of serial 2-fold dilutions of the peptides, dissolved in 0.01% (vol/vol) acetic acid and 0.2% (wt/vol) bovine serum albumin (Sigma) to give final concentrations ranging from 256 to 0.5 μM (equivalent to 380 to 1 μg/ml of T9W, 1,152 to 4.5 μg/ml of LL37, 352 to 0.5 μg/ml of polymyxin B, 200 to 0.4 μg/ml amikacin, 100 to 0.2 μg/ml of ciprofloxacin, 148 to 0.3 μg/ml of gentamicin, and 140 to 0.3 μg/ml of ceftazidime). The MICs were defined as the lowest concentration with no visible growth of bacteria from the microtiter plates after 16 h of incubation at 37°C. MICs were determined in duplicate, and each test was reproduced at least three times.
To further evaluate the antimicrobial potency of the peptides, the lethal concentration (LC) was also determined, as described previously (18). Briefly, the bacterial cells were washed three times with sterile phosphate-buffered saline ([PBS] 10 mM, pH 7.2) and resuspended in the same buffer. The bacterial suspensions (ca. 105 CFU/ml) in PBS with or without NaCl, MgCl2, or CaCl2 were incubated with 2-fold dilutions of peptides for 2 h at 37°C as described before (19). Following the treatment, the cell samples were incubated on MH agar to determine the viable bacteria. The surviving colonies were counted the following day, and the lowest concentration of peptides at which there was complete killing was taken as the LC. The results given are mean values of three independent determinations.
Kinetics of bacterial killing.
P. aeruginosa cells (ca. 106 CFU/ml) were treated with peptides at 0×, 1×, 2×, or 4× LC in PBS. Aliquots of 50 μl of the peptide-treated suspension were withdrawn at different times, serially diluted, and plated on MH agar to determine the viable bacterial counts. Synergistic time-kill kinetics analysis was performed by testing T9W and ciprofloxacin, gentamicin, or ceftazidime at 0.5× LC each, as described above, and each test was reproduced at least three times.
Cell toxicity assay.
The hemolytic activity of the peptides was measured as described previously (20). Briefly, fresh and healthy human red blood cells (hRBCs) were collected and then centrifuged at 1, 000 × g for 5 min. The hRBCs were resuspended in PBS (pH 7.2) to attain a dilution of 1% (vol/vol). Next, each 100 μl of hRBC solution was incubated with 100 μl of serial dilutions of peptides dissolved in PBS for 1 h at 37°C. After centrifugation at 1,000 × g for 5 min, the supernatant was transferred to a 96-well microtiter plate, and the release of hemoglobin was monitored by measuring the absorbance at 570 nm. The hRBCs in PBS and 0.1% Triton X-100 were employed as negative and positive controls, respectively. Minimal hemolytic concentration (MHC) was defined as the peptide concentration causing 10% hemolysis.
An MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] staining assay was also performed to determine cell viability in the presence or absence of the peptide. Briefly, RAW 264.7 macrophage cells (ca. 104/well) in RPMI 1640 medium were placed into 96-well plates and then incubated under a fully humidified atmosphere of 95% air and 5% CO2 at 37°C overnight. The next day, the peptides were added to the cell cultures at final concentrations of 0.5 to 256 μM. After incubation for 24 h, the cell cultures were incubated with MTT (50 μl, 0.5 mg/ml) for 4 h at 37°C. The cell cultures were centrifuged at 1,000 × g for 5 min, and the supernatants were discarded. Subsequently, 150 μl of dimethyl sulfoxide was added to dissolve the formed formazan crystals, and the optical density at 492 nm (OD492) was measured using a microplate reader (Tecan GENios F129004; Tecan, Austria).
Cell selectivity assay in the presence of bacteria.
To determine the selectivity of T9W between the bacteria and mammalian cells, the activities of T9W were assayed in a coculture of macrophage cells with P. aeruginosa ATCC 27853. The cells (ca. 105 cells/well in RPMI 1640 medium) were placed into 12-well plates and then incubated at 37°C and 5% CO2 until a monolayer was formed. The medium was aspirated, and a 100-μl suspension of P. aeruginosa (ca. 107 cells/ml) was added to each well; then 900 μl of antibiotic-free medium containing peptides (at a final concentration of 1× LC) was added. The coculture was then incubated at 37°C for 30, 60, or 180 min. To determine bacterial survival, the coculture medium was washed three times with sterile PBS and serially diluted and plated on MH agar. After incubation overnight at 37°C, bacterial counts were determined. For measurement of cell viability after peptide treatment, cells were washed with PBS twice and then incubated in RPMI 1640 medium containing 0.5 mg/ml MTT for 4 h at 37°C. After incubation, percent viability was assessed as described above. As controls, cells were treated with RPMI 1640 medium in the absence (0% cytotoxicity) or presence of bacteria or with 100% lysis buffer (PBS–0.1% Triton X-100; 100% cytotoxicity). These experiments were verified by at least three independent trials, and viability data were averaged. The final toxicity values were expressed as the mean percent toxicity for each test condition minus the percent toxicity in the presence of bacteria alone.
LPS binding assay.
The affinities of the binding of the peptide to LPS were examined in a displacement assay using the fluorescent dye BODIPY-TR-cadaverine (21). The fluorescence of BODIPY-TR-cadaverine is quenched upon binding to LPS, and the displacement of BODIPY-TR-cadaverine by the test peptide results in dequenching of BODIPY-TR-cadaverine fluorescence. Stock solutions of LPS from P. aeruginosa ATCC 27316 (5 mg/ml; Sigma-Aldrich, China) and BODIPY-TR-cadaverine (2.5 mg/ml; Sigma-Aldrich, China) were prepared and diluted in Tris buffer (pH 7.4, 50 mM) to yield a final concentration of 25 μg/ml of LPS and 2.5 μg/ml BODIPY-TR-cadaverine. Then, 2 ml of this LPS–BODIPY-TR-cadaverine mixture was added in a quartz cuvette, and the background fluorescence was recorded (excitation wavelength, 580 nm; emission wavelength, 620 nm). The changes in fluorescence were recorded with an F-4500 fluorescence spectrophotometer (Hitachi, Japan). Polymyxin B was used as a positive control because of its binding to and neutralizing of LPS (22). Each test was performed independently at least three times.
Cytoplasmic membrane depolarization assay.
The ability of the peptides to alter the cytoplasmic membrane electrical potential was determined using the membrane potential-sensitive dye diSC3(5) (3,3′-dipropylthiadicarbocyanine iodide; Sigma-Aldrich, China) as previously described (17). Briefly, P. aeruginosa ATCC 27853 cells were collected from the mid-log-phase culture, washed in HEPES buffer (5 mM HEPES, pH 7.2, and 20 mM glucose), and resuspended in the same buffer with the addition of 0.2 mM EDTA to an OD600 of 0.05. The mixture was incubated in the dark for 2 h at room temperature under shaking (150 rpm) with 0.4 μM diSC3(5) to allow maximal uptake of the fluorescent dye. The osmotic gradient was equilibrated to a final concentration of 0.1 M KCl. A 2-ml cell suspension was added in a 1-cm cuvette, and the desired concentration of the peptide was added. Fluorescence was monitored with an F-4500 fluorescence spectrophotometer (Hitachi, Japan) at an excitation wavelength of 622 nm and an emission wavelength of 670 nm.
CLSM.
P. aeruginosa ATCC 27853 cells (ca. 107 CFU/ml) were incubated with fluorescein isothiocyanate (FITC)-labeled peptide at 1× LC for 30 min, and the bacterial cells were washed three times with PBS with centrifugation at 1,000 × g for 10 min. A smear was made, and the images were captured using a Leica TCS SP2 confocal laser-scanning microscope (CLSM) with a 488-nm band-pass filter for the FITC excitation.
TEM.
The P. aeruginosa ATCC 27853 cells were collected, washed with 10 mM PBS, and resuspended to an OD600 of 0.2. The cells were incubated with the peptides at 37°C for 30 or 60 min at 1× LC. The control was run without peptides. After incubation, the cell pellets were harvested, washed with PBS, and subjected to fixation with 2.5% glutaraldehyde overnight and 2% osmium tetroxide for 70 min at 4°C. Following washing with PBS, the bacterial samples were dehydrated in a graded ethanol series and transferred to 1:1 mixtures of absolute acetone and epoxy resin for 30 min and pure resin and incubated overnight at a constant temperature. Finally, the specimens were sectioned with an ultramicrotome, stained by uranyl acetate and lead citrate, and examined using a Hitachi H-7650 transmission electron microscope (TEM; Japan).
Flow cytometry.
P. aeruginosa ATCC 27853 cells (ca. 107 CFU/ml) were mixed with the peptides at 37°C for 30 min. Then a final concentration of 10 μg/ml of propidium iodide (PI) was added to the bacterial suspension and incubated for 30 min. The bacterial cells were harvested and resuspended in PBS. Flow cytometry was performed using a FACScan (Becton-Dickinson, San Jose, CA).
Statistical analysis.
Values of LC were expressed as means ± standard errors. Differences between the results of the control and the treatment were analyzed using the procedure Proc TTest in SAS, version 9.3 (SAS Institute, Inc., Cary, NC, USA).
RESULTS
T9W is active against P. aeruginosa.
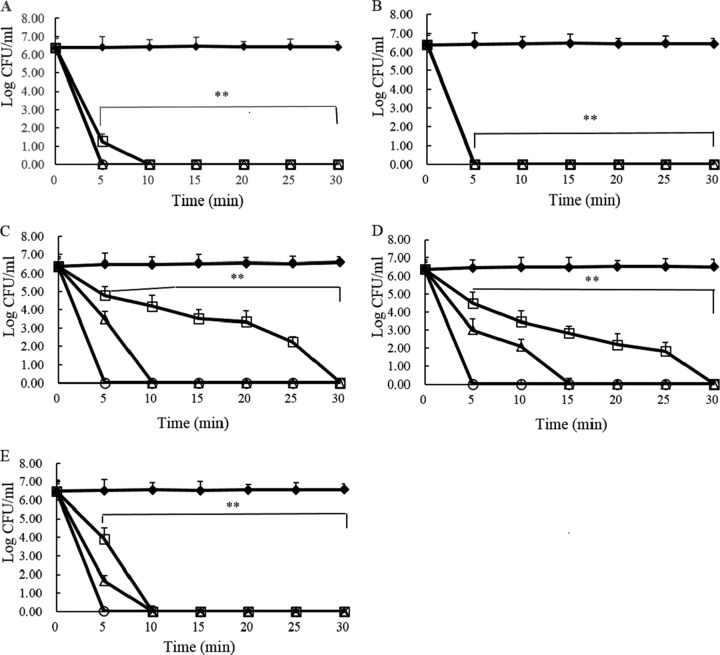
As shown in Table 1, MIC values indicate that T9W activity against P. aeruginosa is comparable to that of other antimicrobials (values ranging from 0.5 to 4 μM or from 0.3 to 4 μg/ml). To further test the ability of T9W to kill P. aeruginosa, bacterial cells were incubated in PBS with or without various concentrations of peptides. We found that T9W significantly killed P. aeruginosa in a concentration-dependent manner, and a concentration of 4 μM, or 9 μg/ml, T9W effectively killed P. aeruginosa at 2 h of incubation (Table 2). This activity of T9W against P. aeruginosa was comparable to that of currently available antibiotics such as polymyxin B, amikacin, gentamicin, and ceftazidime. We examined whether T9W exerts bactericidal activity against antibiotic-resistant strains. The susceptibility assays indicated that the bactericidal concentration of 4 μM, or 9 μg/ml, was adequate for killing ciprofloxacin-, gentamicin-, and ceftazidime-resistant strains, including clinically isolated multidrug-resistant strains (Table 2). To understand the kinetics of the T9W interaction with P. aeruginosa, we compared the time-kill kinetics of one reference strain and three antibiotic-resistant strains at multiples of the LC for a period of 30 min. T9W induced marked bacterial killing similar to that of polymyxin B, beginning at 5 min after exposure, and complete bacterial growth inhibition was observed within 30 min (Fig. 1A and B). These antibiotic-resistant P. aeruginosa strains revealed different killing profiles. At 1× LC, a ceftazidime-resistant strain, LCCE (Fig. 1E), was completely eliminated by T9W. A ciprofloxacin-resistant strain, LCCI (Fig. 1C), and a gentamicin-resistant strain, LCGE (Fig. 1D), were slightly less sensitive to T9W; they were all killed within 30 min. T9W induced dose- and time-dependent growth inhibition, with a bactericidal effect at all the concentrations tested. Together, these data indicate the substantial and fast killing efficiency of T9W against P. aeruginosa.
TABLE 1.
Susceptibility of P. aeruginosa to T9W and comparable antimicrobial agents and their cytotoxicities
| Parameter | T9W | Polymyxin B | Amikacin | Ciprofloxacin | Gentamicin | Ceftazidime |
|---|---|---|---|---|---|---|
| MIC (μg/ml)a | 2–4 | 0.5–1 | 0.4–0.8 | 0.8–1.6 | 0.3–0.6 | 0.3–0.6 |
| MHC (μg/ml)b | >290 | >176 | >100 | >50 | >74 | >70 |
| Cytotoxicity (μg/ml)c | >290 | >176 | >100 | >50 | >74 | >70 |
The MIC was determined as the lowest concentration of the peptide that inhibited bacterial growth of the reference strain P. aeruginosa ATCC 27853.
Minimal hemolytic concentration (MHC) was determined as the lowest concentration of the peptide that caused 10% hemolysis of human red blood cells.
Cytotoxicity was determined as the lowest concentration of the peptide that reduced cell viability of RAW264.7 macrophage cells by 10%.
TABLE 2.
Lethal concentration values of T9W and comparable antimicrobial agents against P. aeruginosa
| Strain | LC (μg/ml)a |
|||||
|---|---|---|---|---|---|---|
| T9W | Polymyxin B | Amikacin | Ciprofloxacin | Gentamicin | Ceftazidime | |
| 27853 | 4 ± 0.9 | 1 ± 0.3 | 1 ± 0.2 | 25 ± 2.6 | 1 ± 0.2 | 1 ± 0.2 |
| 10419 | 9 ± 0.8 | 11 ± 0.7 | 6 ± 0.3 | 25 ± 3.1 | 5 ± 1.0 | 2 ± 1.0 |
| 21625 | 9 ± 1.8 | 6 ± 0.7 | 13 ± 1.8 | 25 ± 2.1 | 19 ± 2.0 | 4 ± 0.4 |
| 21630 | 9 ± 0.8 | 6 ± 0.7 | 1 ± 0.4 | 25 ± 3.4 | 1 ± 0.6 | 1 ± 0.5 |
| LC | 2 ± 0.5 | 6 ± 1.3 | 0.2 ± 0.2 | 3 ± 0.5 | 0.3 ± 0.1 | 2 ± 0.5 |
| LCCI | 4 ± 0.5 | 6 ± 0.5 | 6 ± 2.9 | >100 | 9 ± 1.9 | 9 ± 1.6 |
| LCGE | 9 ± 1.8 | 2 ± 0.5 | >100 | 50 ± 0 | >148 | 9 ± 1.4 |
| LCCE | 9 ± 1.7 | 6 ± 0.5 | 6 ± 0.6 | 25 ± 2.6 | 5 ± 1.0 | >140 |
| 11441 | 2 ± 0.5 | 11 ± 0.5 | 13 ± 2.5 | 50 ± 0 | 37 ± 3.1 | 70 ± 0 |
| 11451 | 4 ± 0.9 | 6 ± 0.9 | 13 ± 2.5 | 25 ± 0.3 | 37 ± 4.1 | 35 ± 4.3 |
| 21328 | 9 ± 1.8 | 11 ± 0.5 | 100 ± 0 | >50 | >74 | >70 |
| 25349 | 4 ± 0.9 | 2 ± 0.5 | 6 ± 0.6 | 25 ± 2.1 | 5 ± 1.6 | 35 ± 2.7 |
| 26305 | 4 ± 0.8 | 2 ± 0.3 | 3 ± 0.6 | 25 ± 2.4 | 2 ± 0.8 | >70 |
Lethal concentration (LC) was defined as the lowest concentration of antimicrobial agent at which there was complete killing. The LCs were derived from representative values (means ± standard errors) of three independent experimental trials.
FIG 1.
Time-kill kinetics of polymyxin B (A) and T9W (B) against P. aeruginosa 27853 and of T9W against P. aeruginosa LCCI (C), P. aeruginosa LCGE (D), and P. aeruginosa LCCE (E) at 0× LC (filled diamonds), 1× LC (open squares), 2× LC (open triangles), and 4× LC (open circles). Dilutions of aliquots taken from 0 to 30 min were plated on MH agar. The graphs were derived from average values of three independent trials. ∗∗, P < 0.001, compared to the value for the control at the same concentration.
T9W is resistant to salts.
To examine whether the antipseudomonal activity of T9W was compromised in the presence of salts, we treated P. aeruginosa with different peptide concentrations under various salt conditions. As indicated in Table 3, T9W maintained its strong activity against P. aeruginosa in the presence of NaCl, CaCl2, or MgCl2 at all the tested concentrations, with LCs ranging from 2 to 4 μM or from 4 to 9 μg/ml. As a control, LL37, a host defense peptide encoded by the only human cathelicidin gene, displayed a significant decrease in activity against P. aeruginosa, with LCs greater than 4 μM in equal or higher physiological concentrations of salts (150 mM for NaCl, 2.5 mM for CaCl2, and 1 mM for MgCl2), as reported previously (19). These data suggest that the activity of T9W might be particularly critical under physiological conditions or in the treatment of infections that disturb the normal salt homeostasis in certain human tissues.
TABLE 3.
Influence of sodium, calcium, and magnesium chloride on antimicrobial activity
| Salt and concn (mM) | LC (μg/ml)a |
|
|---|---|---|
| T9W | LL37 | |
| Control | 4 ± 1.9 | 18 ± 0.2 |
| NaCl | ||
| 50 | 4 ± 0.9 | 18 ± 0.2 |
| 150 | 9 ± 0.9 | 36 ± 0.4 |
| 300 | 9 ± 1.7 | 288 ± 3.3 |
| CaCl2 | ||
| 1 | 4 ± 0.8 | 36 ± 0.4 |
| 2.5 | 9 ± 1.7 | 72 ± 1.1 |
| 5 | 9 ± 0.9 | 576 ± 0 |
| MgCl2 | ||
| 0.5 | 4 ± 0.9 | 18 ± 0.2 |
| 1 | 9 ± 1.7 | 36 ± 0.4 |
| 2 | 9 ± 1.7 | 72 ± 1.1 |
P. aeruginosa ATCC 27853 was incubated with 2-fold serially diluted peptides in the absence or presence of salts at 37°C for 2 h. The lethal concentration (LC) was defined as the lowest concentration of peptide at which there was complete killing. The LCs were derived from representative values (means ± standard errors) of three independent experimental trials.
T9W selectively kills bacteria from mammalian cells.
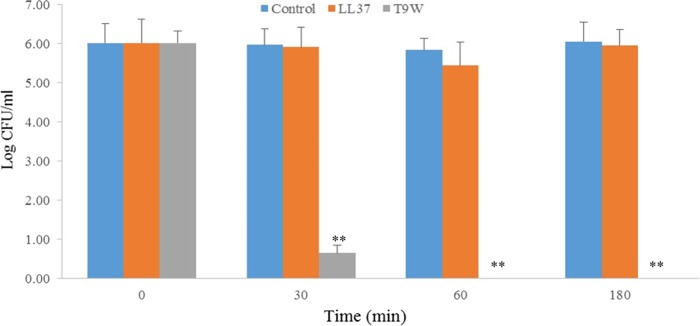
We initially examined the toxicity of T9W against mammalian cells, and we found that T9W was shown to be nontoxic to hRBCs and macrophage cells up to 256 μM or 290 μg/ml (Table 1). To further address the ability of T9W to discriminate bacterial from mammalian cells, a coculture model in which macrophages were infected with P. aeruginosa prior to peptide treatment was utilized as described previously (23). As shown in Fig. 2, T9W decreased the bacterial load by 90% within 30 min and effectively eliminated total extracellular P. aeruginosa in 60 min. By comparison, the activity of the human-derived peptide LL37 against P. aeruginosa in coculture with macrophages was significantly compromised. In addition, these cocultures were also examined for eukaryotic cell viability using an MTT assay. Neither T9W nor LL37 affected the viability of the macrophage cells (0% cytotoxicity [data not shown]) at a concentration of 1× LC. Taken together, these results suggested the high antimicrobial selectivity of T9W as a potentially effective and safe antipseudomonal agent.
FIG 2.
Selective toxicity of T9W in a coculture model. Macrophages (ca. 105 cells/well) were infected with P. aeruginosa 27853 (ca. 107 cells/ml) in RPMI 1640 medium with no bovine fetal serum and antibiotic. The coculture was then treated without or with peptides at 1× LC. The graphs were derived from average values of three independent trials. ∗∗, P < 0.001, compared to the value for the control at the same concentration.
T9W acts synergistically with antibiotics against antibiotic-resistant strains.
A widely accepted approach is to treat serious P. aeruginosa infections with a combination of antimicrobial agents, which enhances antibiotic efficacy and contributes to drug resistance less frequently than monotherapy. We hypothesized that T9W might have synergistic activity with different classes of conventional antibiotics. The in vitro time-kill analysis demonstrated a synergistic effect within 3 h after treatment at 0.5× LC for T9W and ciprofloxacin or within 6 h for T9W and gentamicin (Fig. 3).
FIG 3.
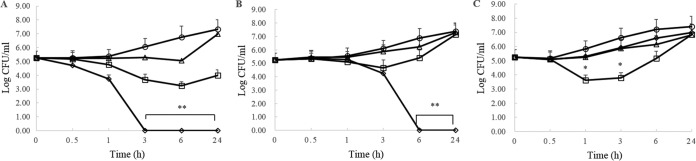
Time-kill kinetics of T9W in combination with ciprofloxacin (A), gentamicin (B), or ceftazidime (C) against P. aeruginosa LCCI (A), P. aeruginosa LCGE (B), or P. aeruginosa LCCE (C). The activity of the combination of 0.5× LC of T9W and 0.5× LC of antibiotic (open diamonds) was compared with the results of 0.5× LC of T9W alone (open squares), 0.5× LC of antibiotic alone (open triangles), or untreated bacteria (open circles). The graphs were derived from average values of three independent trials. ∗, P < 0.01; ∗∗; P < 0.001 (compared to results for the control at the same concentration).
T9W binds to LPS.
LPS, otherwise termed endotoxin, is the major constituent of the outer leaflet of the outer membrane of Gram-negative bacteria. To examine whether T9W has the ability to bind to LPS, a fluorescence-based displacement assay with BODIPY-TR-cadaverine was performed. LL37 showed an effect similar to the one observed with polymyxin B, a decapeptide antibiotic which is known to bind to LPS and is generally used as a reference (22). Significantly, the activity of T9W was dose dependent, and T9W demonstrated the highest potency in displacing BODIPY-TR-cadaverine from its binding to LPS (Fig. 4). These results suggest that T9W effectively binds to the LPS of the P. aeruginosa outer membrane.
FIG 4.
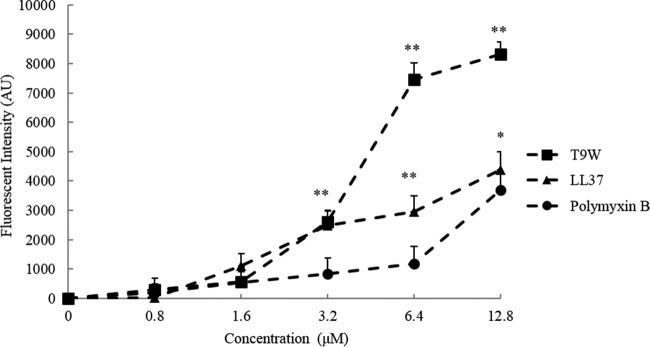
The binding affinity of peptides to the LPS from P. aeruginosa ATCC 27316 (25 μg/ml) was determined by the BODIPY-TR-cadaverine displacement method as described in Materials and Methods. Fluorescence intensity was monitored at an excitation wavelength of 580 nm and an emission wavelength of 620 nm. The graphs were derived from average values of three independent trials. ∗, P < 0.01; ∗∗, P < 0.001 (compared to results with polymyxin B at the same concentration). AU, arbitrary units.
T9W increases cytoplasmic membrane permeability.
To gain insight into the effect of T9W on the P. aeruginosa membrane, we investigated the ability of this peptide to depolarize the bacterial membrane using diSC3(5), a membrane potential-dependent probe. Upon permeabilization of the membrane, the membrane potential is dissipated, and diSC3(5) is released into the medium, causing a consequent increase in fluorescence (17). As a control, LL37 induced an increase in fluorescence intensity in a dose-dependent manner. The addition of T9W triggered a significant increase in fluorescence intensity, indicating rapid membrane depolarization (Fig. 5). These results indicate that T9W effectively induces a loss of membrane potential.
FIG 5.
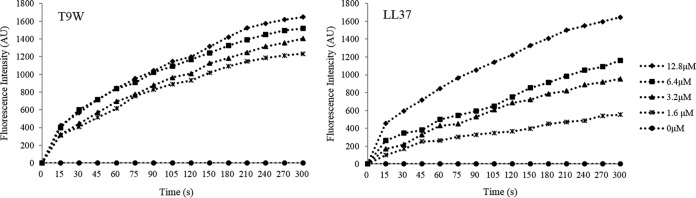
The cytoplasmic membrane potential variation of P. aeruginosa 27853 treated by different concentrations of peptides, as assessed by the release of the membrane potential-sensitive dye diSC3(5). Fluorescence intensity was monitored at an excitation wavelength of 622 nm and an emission wavelength of 670 nm as a function of time. Data plotted are representative average values of three independent experimental trials.
T9W causes membrane damage.
In considering the mechanism of action employed by amphipathic AMPs, we hypothesized that T9W might induce bacterial membrane damage. To monitor the site targeted by T9W in P. aeruginosa, the bacteria were treated with FITC-labeled peptides and observed under a CLSM. As shown in Fig. 6, T9W bound strongly to the surface of the P. aeruginosa cells to a degree comparable to that of LL37, confirming that these peptides interact with the bacterial cell membrane.
FIG 6.
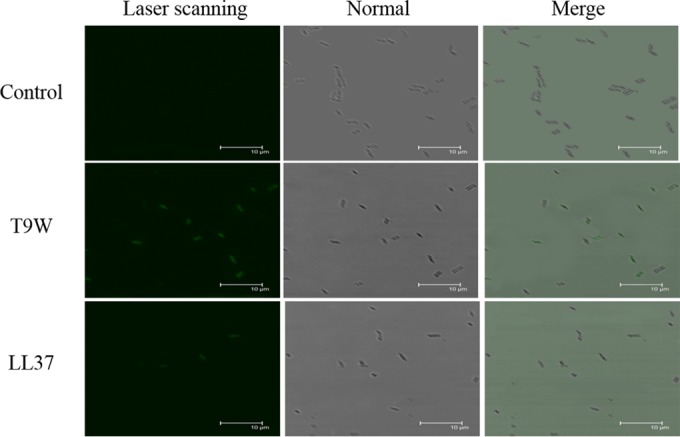
Confocal microscopic images of P. aeruginosa cells. P. aeruginosa 27853 cells were treated with FITC-labeled peptides at 1× LC for 30°C and visualized under laser scanning and transmitted-light scanning (normal) microscopy. Merged profiles are also shown. The control was processed without peptides.
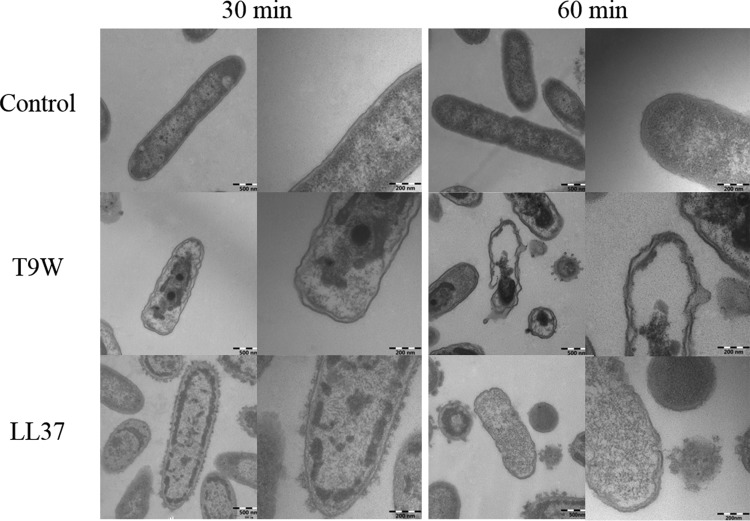
The transmission electron micrographs (TEMs) of P. aeruginosa incubated with the peptides indicated that T9W induced significant rupture of the cytoplasmic membrane after 30 min of exposure, and the complete collapse of the cytoplasmic membrane and dispersion of the intracellular contents induced by T9W were observed after 60 min of exposure (Fig. 7). In contrast, the cell morphology of P. aeruginosa treated by LL37 could be recognized after 30 min, and the dispersion and release of the intracellular contents were observed after 60 min of exposure (Fig. 7). These observations are in agreement with widely accepted models of cationic peptide disruption of the cytoplasmic membrane.
FIG 7.
Transmission electron micrographs of P. aeruginosa cells. P. aeruginosa 27853 cells were treated with peptides at 1× LC and visualized at magnifications of ×50,000 (left panels of each pair) and ×120,000 (right panels of each pair). The control was processed without peptides.
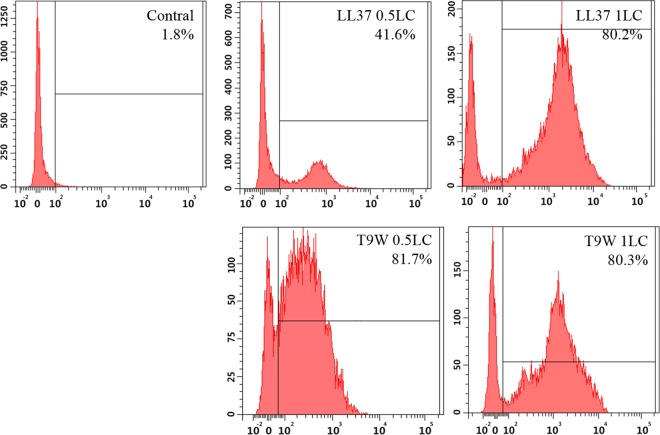
To further investigate cytoplasmic membrane integrity, flow cytometry was used to analyze the cells by staining with PI. PI fluorescently stained the nucleic acids of cells when the cells suffered disruption of the cytoplasmic membrane integrity (24). Compared with the fluorescence released by LL37 treatment, the addition of T9W triggered a significant increase in fluorescence intensity (Fig. 8), indicating strong membrane integrity damage. These results suggest that T9W possesses the ability to damage the P. aeruginosa cell cytoplasmic membrane, resulting in cell death.
FIG 8.
Flow cytometric analysis of membrane damage by PI uptake staining. P. aeruginosa 27853 cells were incubated with peptides for 30 min, and the control was processed without peptides.
DISCUSSION
Compared with other pathogens, P. aeruginosa is very difficult to eradicate because it displays high resistance to a wide variety of currently available antipseudomonal antibiotics, including aminoglycosides, fluoroquinolones, and β-lactams (5). The discovery of new antipseudomonal drugs and/or therapy is an urgent and challenging task. The data in this study demonstrated that T9W had potent activity against P. aeruginosa, even against ciprofloxacin-, gentamicin-, and ceftazidime-resistant strains, that was comparable to the activity of several currently available antipseudomonal drugs. T9W achieved complete killing against ciprofloxacin- and ceftazidime-resistant strains within 5 min and against gentamicin-resistant strains within 30 min, which is a potential functional advantage because rapid and complete peptide clearance in a host environment contributes to shortening the duration of antimicrobial treatment and decreasing the occurrence of antibiotic-resistant genetic mutations or phenotypic variants known as biofilms or persister cells.
Many studies have indicated that Na+ and divalent cations (Ca2+ and Mg2+) might compromise the antimicrobial activity of natural antimicrobial peptides (25–27). In this study, LL37 showed high sensitivity to Na+, Ca2+, and Mg2+ for P. aeruginosa. The high-cationic peptide T9W was shown to be highly potent against P. aeruginosa under similar salt conditions. The underlying salt resistance mechanism of T9W could be attributed to its high net cationic charge and helical stability. The net charge of T9W is +13; thus this high cationicity could overcome the inhibitory effects of monovalent Na+, which could interrupt the electrostatic attraction between the positively charged peptides and the negatively charged membranes (27). In addition, T9W is engineered by substituting threonine for tryptophan in the N terminus of PMAP-36, and this strategic substitution in the hydrophobic face of the amphipathic peptide is responsible for the affinity of T9W for the bacterial membrane, thereby leading to a greater stability of the helical structure; this structural stability resists the countereffect of divalent cations competing for membrane binding with peptides (28). The property of salt resistance displayed by T9W might be particularly critical in the treatment of infections in diseases that might disturb the normal salt homeostasis in certain human tissues.
AMPs have frequently been hypothesized to exhibit cell selectivity. They selectively kill pathogens without being significantly toxic to host cells. Cytotoxicity, a major barrier for the systemic application of AMPs, should be urgently and extensively investigated before expanding the use of AMPs (29). To evaluate the antibacterial selectivity more appropriately, we examined the antimicrobial activity of T9W against P. aeruginosa in a coculture with host cells, and we found that T9W could discriminate against the bacterial cells while sparing the macrophages. These observations provide new information that is critical and useful to the characterization of the therapeutic potential of T9W for systemic applications.
P. aeruginosa, especially multidrug-resistant strains, presents a serious therapeutic challenge for the treatment of infection, and selection of the most appropriate antibiotic is typically complicated; the efficacy of the currently available antipseudomonal drugs is frequently compromised by the ability of P. aeruginosa to develop resistance to multiple classes of antibacterial agents. An urgent but arduous task is to effectively eradicate this opportunistic bacterial pathogen or slow the emergence of resistance to the available drugs (30, 31). To address this need, a combination of antibacterial drugs is the most common strategy used in treating P. aeruginosa infection. Little is known of whether synergistic effects exist between AMPs and antibiotics. Our data indicate that T9W exhibited synergistic activity against resistant P. aeruginosa with ciprofloxacin and gentamicin, suggesting that, in combination, T9W might hold promise for clinical applications.
To gain insight into the molecular mechanism of the action of T9W, we performed a series of molecular and cell assays with a bacterial membrane. We first investigated the ability of T9W to bind to LPS, the main component of the Gram-negative outer membrane, and to permeabilize the cytoplasmic membranes. We showed that T9W bound to the LPS and induced loss of membrane potential. The fluorescence distribution shown by confocal laser scanning microscopy indicated the initial membrane binding. Further research using TEM and flow cytometry demonstrated the morphological alteration of the cytoplasmic membrane and the loss of membrane integrity. We conclude that the target activity allows T9W to bind to the cell membrane, which is followed by disruption of the cytoplasmic membrane, the lethal event leading to bacterial cell death. However, the exact mechanism of action of AMPs can be multiple, and recently the antimicrobial mechanism has been putatively associated with cell wall biogenesis (32), conductive ATP transport (33), and inhibition of DNA and protein synthesis (34, 35). But, here, the fast killing efficiency as well as the fluorescence and the microscopic studies indeed clearly indicated that the major target of the antimicrobial action of T9W is the bacterial membrane. Previous studies have shown that LL37 uses a mechanism by which it causes positive curvature strain and toroidal pore formation, leading to leakage, but does not break the membrane into smaller fragments or micelles to exert its antimicrobial effect (15, 36). T9W can cause membrane permeabilization and intracellular content leakage to a greater extent than LL37, as shown in Fig. 7 and 8. This membrane-disruptive mechanism of action, which is different from that of conventional antibiotics, ensures that T9W is considered a new potential antipseudomonal drug with characteristics that include an ability to kill target cells rapidly and the possibility of a capacity to minimally induce antibiotic-resistant strains.
We demonstrated that an engineered antimicrobial peptide, T9W, exhibited strong bactericidal activity against P. aeruginosa that was comparable to that of the currently available antibiotics and was able to overcome the challenges of physiological serum concentrations of Na+, Mg2+, and Ca2+ more effectively than the human cathelicidin LL37. In addition, T9W showed no adverse effects on mammalian cells and had the ability to effectively eradicate P. aeruginosa from host cells and, synergized with ciprofloxacin and gentamicin, to rapidly kill antibiotic-resistant P. aeruginosa. Last, T9W was shown to bind to the LPS and depolarize the cytoplasmic membrane, damaging membrane integrity and causing intracellular content leakage, leading to cell death. Based on this information, T9W represents a lead molecule for the design and development of peptide-based membrane-disruptive drugs with significant activity and selectivity against P. aeruginosa.
ACKNOWLEDGMENTS
This research was supported by grants from the National Natural Scientific Foundation of China (grant no. 31272453), the National Key Scientific and Technological Project (2013BAD10B03), and the Program for Innovative Research Team of Universities in Heilongjiang Province (2012TD003).
REFERENCES
- 1.Rogers GB, Carroll MP, Bruce KD. 2012. Enhancing the utility of existing antibiotics by targeting bacterial behaviour? Br J Pharmacol 165:845–857. doi: 10.1111/j.1476-5381.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez L, Hancock RE. 2012. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernebro J. 2011. Fighting bacterial infections-future treatment options. Drug Resist Updat 14:125–139. doi: 10.1016/j.drup.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Strateva T, Mitov I. 2011. Contribution of an arsenal of virulence factors to pathogenesis of Pseudomonas aeruginosa infections. Ann Microbiol 61:717–732. doi: 10.1007/s13213-011-0273-y. [DOI] [Google Scholar]
- 5.Breidenstein EB, de la Fuente-Nunez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 7.Radek K, Gallo R. 2007. Antimicrobial peptides: natural effectors of the innate immune system. Semin Immunopathol 29:27–43. doi: 10.1007/s00281-007-0064-5. [DOI] [PubMed] [Google Scholar]
- 8.Maroti G, Kereszt A, Kondorosi E, Mergaert P. 2011. Natural roles of antimicrobial peptides in microbes, plants and animals. Res Microbiol 162:363–374. doi: 10.1016/j.resmic.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen LT, Haney EF, Vogel HJ. 2011. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol 29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Ubeda C, Pamer EG. 2012. Antibiotics, microbiota, and immune defense. Trends Immunol 33:459–466. doi: 10.1016/j.it.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spellberg B, Rex JH. 2013. The value of single-pathogen antibacterial agents. Nat Rev Drug Discov 12:963. doi: 10.1038/nrd3957-c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Shan AS. 2013. Importance of tryptophan along the non-polar helix face of an antimicrobial amphipathic peptide for improving killing efficacy against Pseudomonas aeruginosa. Amino Acids 45:602–603. [Google Scholar]
- 13.Nagant C, Pitts B, Nazmi K, Vandenbranden M, Bolscher JG, Stewart PS, Dehaye JP. 2012. Identification of peptides derived from the human antimicrobial peptide LL-37 active against biofilms formed by Pseudomonas aeruginosa using a library of truncated fragments. Antimicrob Agents Chemother 56:5698–5708. doi: 10.1128/AAC.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noore J, Noore A, Li B. 2013. Cationic antimicrobial peptide LL-37 is effective against both extra- and intracellular Staphylococcus aureus. Antimicrob Agents Chemother 57:1283–1290. doi: 10.1128/AAC.01650-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandamme D, Landuyt B, Luyten W, Schoofs L. 2012. A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell Immunol 280:22–35. doi: 10.1016/j.cellimm.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Sood R, Domanov Y, Pietiainen M, Kontinen VP, Kinnunen PK. 2008. Binding of LL-37 to model biomembranes: insight into target vs host cell recognition. Biochim Biophys Acta 1778:983–996. doi: 10.1016/j.bbamem.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 17.Zhu X, Dong N, Wang Z, Ma Z, Zhang L, Ma Q, Shan A. 2014. Design of imperfectly amphipathic alpha-helical antimicrobial peptides with enhanced cell selectivity. Acta Biomater 10:244–257. doi: 10.1016/j.actbio.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 18.Varkey J, Nagaraj R. 2005. Antibacterial activity of human neutrophil defensin HNP-1 analogs without cysteines. Antimicrob Agents Chemother 49:4561–4566. doi: 10.1128/AAC.49.11.4561-4566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deslouches B, Islam K, Craigo JK, Paranjape SM, Montelaro RC, Mietzner TA. 2005. Activity of the de novo engineered antimicrobial peptide WLBU2 against Pseudomonas aeruginosa in human serum and whole blood: implications for systemic applications. Antimicrob Agents Chemother 49:3208–3216. doi: 10.1128/AAC.49.8.3208-3216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong N, Zhu X, Chou S, Shan A, Li W, Jiang J. 2014. Antimicrobial potency and selectivity of simplified symmetric-end peptides. Biomaterials 35:8028–8039. doi: 10.1016/j.biomaterials.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Ouberai M, El Garch F, Bussiere A, Riou M, Alsteens D, Lins L, Baussanne I, Dufrene YF, Brasseur R, Decout JL, Mingeot-Leclercq MP. 2011. The Pseudomonas aeruginosa membranes: a target for a new amphiphilic aminoglycoside derivative? Biochim Biophys Acta 1808:1716–1727. doi: 10.1016/j.bbamem.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 22.Morrison DC, Jacobs DM. 1976. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry 13:813–818. doi: 10.1016/0019-2791(76)90181-6. [DOI] [PubMed] [Google Scholar]
- 23.Deslouches B, Phadke SM, Lazarevic V, Cascio M, Islam K, Montelaro RC, Mietzner TA. 2005. De novo generation of cationic antimicrobial peptides: influence of length and tryptophan substitution on antimicrobial activity. Antimicrob Agents Chemother 49:316–322. doi: 10.1128/AAC.49.1.316-322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad A, Asthana N, Azmi S, Srivastava RM, Pandey BK, Yadav V, Ghosh JK. 2009. Structure-function study of cathelicidin-derived bovine antimicrobial peptide BMAP-28: design of its cell-selective analogs by amino acid substitutions in the heptad repeat sequences. Biochim Biophys Acta 1788:2411–2420. doi: 10.1016/j.bbamem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 25.Maisetta G, Di Luca M, Esin S, Florio W, Brancatisano FL, Bottai D, Campa M, Batoni G. 2008. Evaluation of the inhibitory effects of human serum components on bactericidal activity of human beta defensin 3. Peptides 29:1–6. doi: 10.1016/j.peptides.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Wu G, Ding J, Li H, Li L, Zhao R, Shen Z, Fan X, Xi T. 2008. Effects of cations and pH on antimicrobial activity of thanatin and s-thanatin against Escherichia coli ATCC 25922 and B. subtilis ATCC 21332. Curr Microbiol 57:552–557. doi: 10.1007/s00284-008-9241-6. [DOI] [PubMed] [Google Scholar]
- 27.Huang J, Hao D, Chen Y, Xu Y, Tan J, Huang Y, Li F, Chen Y. 2011. Inhibitory effects and mechanisms of physiological conditions on the activity of enantiomeric forms of an alpha-helical antibacterial peptide against bacteria. Peptides 32:1488–1495. doi: 10.1016/j.peptides.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Kumar M, Srivastava S. 2011. Effect of calcium and magnesium on the antimicrobial action of enterocin LR/6 produced by Enterococcus faecium LR/6. Int J Antimicrob Agents 37:572–575. doi: 10.1016/j.ijantimicag.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Brogden NK, Brogden KA. 2011. Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int J Antimicrob Agents 38:217–225. doi: 10.1016/j.ijantimicag.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sánchez-Gómez S, Japelj B, Jerala R, Moriyón I, Alonso MF, Leiva J, Blondelle SE, Andrä J, Brandenburg K, Lohner K. 2011. Structural features governing the activity of lactoferricin-derived peptides that act in synergy with antibiotics against Pseudomonas aeruginosa in vitro and in vivo. Antimicrob Agents Chemother 55:218–228. doi: 10.1128/AAC.00904-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foweraker JE, Laughton CR, Brown DF, Bilton D. 2009. Comparison of methods to test antibiotic combinations against heterogeneous populations of multiresistant Pseudomonas aeruginosa from patients with acute infective exacerbations in cystic fibrosis. Antimicrob Agents Chemother 53:4809–4815. doi: 10.1128/AAC.00269-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventos DS, Neve S, Ravn B, Bonvin AM, De Maria L, Andersen AS, Gammelgaard LK, Sahl HG, Kristensen HH. 2010. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 328:1168–1172. doi: 10.1126/science.1185723. [DOI] [PubMed] [Google Scholar]
- 33.Tanida T, Okamoto T, Ueta E, Yamamoto T, Osaki T. 2006. Antimicrobial peptides enhance the candidacidal activity of antifungal drugs by promoting the efflux of ATP from Candida cells. J Antimicrob Chemother 57:94–103. doi: 10.1093/jac/dki402. [DOI] [PubMed] [Google Scholar]
- 34.Hsu CH, Chen C, Jou ML, Lee AY, Lin YC, Yu YP, Huang WT, Wu SH. 2005. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res 33:4053–4064. doi: 10.1093/nar/gki725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krizsan A, Volke D, Weinert S, Strater N, Knappe D, Hoffmann R. 2014. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70 S ribosome. Angew Chem Int Ed Engl 45:12236–12239. doi: 10.1002/anie.201407145. [DOI] [PubMed] [Google Scholar]
- 36.Henzler Wildman KA, Lee DK, Ramamoorthy A. 2003. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry 42:6545–6558. doi: 10.1021/bi0273563. [DOI] [PubMed] [Google Scholar]