Abstract
A major challenge in microbial biofilm control is biocide resistance. Phenotypic adaptations and physical protective effects have been historically thought to be the primary mechanisms for glutaraldehyde resistance in bacterial biofilms. Recent studies indicate the presence of genetic mechanisms for glutaraldehyde resistance, but very little is known about the contributory genetic factors. Here, we demonstrate that efflux pumps contribute to glutaraldehyde resistance in Pseudomonas fluorescens and Pseudomonas aeruginosa biofilms. The RNA-seq data show that efflux pumps and phosphonate degradation, lipid biosynthesis, and polyamine biosynthesis metabolic pathways were induced upon glutaraldehyde exposure. Furthermore, chemical inhibition of efflux pumps potentiates glutaraldehyde activity, suggesting that efflux activity contributes to glutaraldehyde resistance. Additionally, induction of known modulators of biofilm formation, including phosphonate degradation, lipid biosynthesis, and polyamine biosynthesis, may contribute to biofilm resistance and resilience. Fundamental understanding of the genetic mechanism of biocide resistance is critical for the optimization of biocide use and development of novel disinfection strategies. Our results reveal genetic components involved in glutaraldehyde resistance and a potential strategy for improved control of biofilms.
INTRODUCTION
Poor control of biofilm growth is a major concern in many industries, including health care, food production, and oil and gas (1–4). For example, medical device-associated biofilms are the source of 60% to 70% of nosocomial infections (5). In the oil and gas industry, biofilms present a serious hazard to infrastructure through corrosion and reduced oil quality (6). Biocides are typically used to control and inactivate the biofilms, but resistance to biocides decreases the efficacy of disinfection. Biocide resistance was historically believed to be rare (7), but numerous reports of microbial biofilms resistant to biocides, including chlorine, quaternary ammonium compounds, and aldehydes (8–12), indicate a more widespread phenomenon. Biofilms are inherently more resistant to biocide treatments (8–12), a feature generally attributed to physical mechanisms, such as limited penetration of biocides through exopolysaccharides, reactivity and absorption of biocides to biofilm matrix, and phenotypic adaptations (13–15). Recent reports suggest that genetic factors may also contribute to biocide resistance; for example, efflux pumps were shown to contribute to biocide resistance, indicating the potential for genetically mediated biocide resistance mechanisms (10, 11). The cross-resistance between biocides and antibiotics provides further evidence that biocide resistance may be mediated by genetic factors (15, 16). The control and design of effective biofilm disinfection strategies require understanding of the mechanisms of action of antimicrobials and biocide resistance of biofilms, specifically, the genetically modulated response of microbial biofilms.
Glutaraldehyde is a biocide commonly used to control microbial growth in hospitals, water treatment, oil and gas, food production, and other industries (1–4). Although widely used, to the best of our knowledge, the genetic response of a bacterial biofilm to glutaraldehyde has not been reported. Mechanistically, glutaraldehyde is thought to act by cross-linking proteins and lipids on the outer surface of the cell (4, 17). Resistance to glutaraldehyde has been reported in several bacterial species (18–23), and evidence indicates that genetic factors may contribute to glutaraldehyde resistance. For example, a glutaraldehyde-resistant Pseudomonas aeruginosa was recently isolated from endoscopes associated with an outbreak and was shown to acquire a resistance pattern that remained stable over several passages, indicating the involvement of genetic factors (18, 19). Outer membrane porins are thought to contribute to glutaraldehyde resistance by reducing glutaraldehyde penetration and decreasing binding of glutaraldehyde (20). Specifically, mutants in the major porin MspA of Mycobacterium smegmatis were found to display increased resistance to glutaraldehyde (20); however, this mechanism appears to be species specific, as outer membrane proteins in Pseudomonas fluorescens do not contribute to glutaraldehyde resistance (22). Furthermore, a study conducted previously in our laboratory demonstrated that the exposure of bacterial cells to a high-salinity environment leads to a transcriptionally regulated enhanced resistance toward glutaraldehyde (24). Collectively, these studies strongly suggest genetic, possibly multifactorial, regulation of glutaraldehyde resistance in bacteria; however, the mechanism is not clear. Given that glutaraldehyde is a widely used biocide, it is necessary to elucidate microbial biofilm responses to glutaraldehyde at the genetic level to inform proper biocide application.
To understand the genetic response and mechanistic basis of glutaraldehyde resistance, we systematically investigated the effect of glutaraldehyde exposure on P. fluorescens biofilms and examined the P. fluorescens biofilm transcriptome while exposed to glutaraldehyde using RNA-seq. Biocidal activity of glutaraldehyde was measured on P. fluorescens biofilms of various ages. Furthermore, phenotypic assays and reverse transcriptase quantitative PCR (qRT-PCR) were used to confirm the observed genetic response. Finally, we demonstrated that the observed genetic effects were applicable to P. aeruginosa PAO1 and a mucoid P. aeruginosa isolate from biofilms of a patient with cystic fibrosis (PAmuc).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. fluorescens (ATCC 13525), P. aeruginosa PAO1, and a P. aeruginosa mucoid clinical isolate from a patients with cystic fibrosis (PAmuc) (a gift from George O'Toole, Geisel School of Medicine, Dartmouth College, Hanover, NH) were maintained as laboratory glycerol stock. All strains were routinely cultured on cetrimide agar and Luria-Bertani (LB) broth.
Biofilm assays.
Overnight cultures of P. fluorescens were diluted 100-fold in LB broth. The inoculant (200 μl) was then placed in each well of 96-well plates and incubated at room temperature for 24 to 120 h. The medium in the plates was replaced every 24 h. To determine the susceptibility of biofilm to glutaraldehyde, the biofilms were washed with phosphate-buffered saline (PBS) after 24, 48, 72, 96, and 120 h and treated with glutaraldehyde solution prepared in PBS for 10 min. The biofilms were washed again with PBS posttreatment. Biofilm viability was quantified by adding 200 μl of 250 μg/ml 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) solution in PBS to each well and incubating for 1 h at 25°C. Produced formazan was dissolved in 200 μl of dimethyl sulfoxide (DMSO), as described previously (12). Absorbance at 570 nm was recorded and presented as the mean absorbance of three independent biological replicates ± the standard deviation (SD). In addition, total biofilm formation at 24 and 72 h was measured by staining with 0.3% crystal violet for 20 min, as described previously (25). Excess stain was removed by three washes with PBS, biofilm-associated stain was dissolved with 200 μl DMSO, and absorbance at 570 nm was recorded. The mean absorbance ± SD for three biological replicates is presented.
Biofilm regrowth assay.
The regrowth potential of P. fluorescens biofilms following glutaraldehyde treatment was measured using 72-h-old biofilms. Biofilms grown for 72 h were chosen based on the results of viability assays after glutaraldehyde exposure. Biofilms were treated with glutaraldehyde and washed with PBS. Fresh LB broth (200 μl) was added to each well in the 96-well plates and incubated at room temperature for an additional 24 h. Biofilm viability was determined by the MTT assay as detailed in “Biofilm assays.”
RNA-seq analysis of P. fluorescens exposed to glutaraldehyde.
Overnight cultures of P. fluorescens were diluted 100-fold in LB broth to prepare the inoculant. Biofilms were grown by placing 2 ml inoculant per well in 6-well plates followed by incubation at room temperature for 72 h. A total of six biofilm experiments were set up on different days, starting with six different overnight cultures. After 72 h, biofilms were washed with PBS and treated with 0 or 62.5 mg/liter of glutaraldehyde for 10 min. The glutaraldehyde solution was completely removed, and 1 ml TRIzol (Life Technologies, Carlsbad, CA) per well was added. The biofilms were completely lysed in TRIzol by pipetting and were collected in fresh tubes. RNA was immediately extracted according to the manufacturer's protocol and treated with Turbo DNase (Life Technologies). For each sequencing run, RNA from three biological replicates was extracted and processed for RNA-seq analysis. Two sequencing runs were performed and analyzed to obtain the final analysis.
For each replicated sequencing run, RNA samples from three biological replicates of glutaraldehyde or PBS were pooled and processed using the ScriptSeq complete kit (bacteria) (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's instructions. Briefly, 1 μg each of DNase-treated RNA samples was treated with RiboZero to remove rRNA, purified by ethanol precipitation, and dissolved in 10 μl RNase-free water. rRNA-depleted samples were mixed with the RNA fragmentation solution and cDNA synthesis primer provided in the ScriptSeq kit, and cDNA was synthesized using StarScript reverse transcriptase. The cDNA was di-tagged and purified using the Agencourt AMPure purification kit (Beckman Coulter, Indianapolis IN). Finally, the di-tagged cDNA samples were PCR amplified using the Failsafe PCR enzyme and barcoded reverse primers provided with the ScriptSeq kit per the manufacturer's protocol. The amplified and dual-tagged cDNA samples were purified using Agencourt AMPure XP beads and quantified using Bioanalyzer and Qubit.
Sequencing.
The di-tagged, barcoded cDNA samples were sequenced on an Illumina MiSeq sequencer (San Diego, CA). Equimolar amounts of cDNA from PBS- and glutaraldehyde-treated samples were pooled and diluted to 2 nM. Each pooled sample was then denatured using fresh 0.2 N NaOH for 5 min at room temperature and further diluted according to the manufacturer's instructions. Each final library was spiked with 5% PhiX genome and sequenced using a MiSeq v2 reagent kit (500 cycles) (Illumina, San Diego, CA).
Bioinformatics.
Differential gene expression analysis was conducted using the RNA-seq algorithm on the CLC Genomics Workbench 6.5.1 (CLC bio, Aarhus, Denmark) (26, 27). Reads with quality scores of <Q30 and length of <70 nucleotides were discarded. The reads were mapped to the Pseudomonas fluorescens strain SBW25 genome (GenBank accession number NC_012660). Reads mapping to rRNA were manually removed from all sequencing runs, and reads per kilobase per million (RPKM) were calculated (28). The data were further normalized by scaling (29). Differentially expressed genes were identified by comparing normalized gene reads between PBS- and glutaraldehyde-treated biofilm samples using Baggerley's test on proportions for replicated experiments (30). The differential expression was considered significant at a corrected false discovery rate (FDR) of P ≤ 0.001. The genes that were consistently induced or repressed in the two sequencing runs were considered differentially expressed and reported. Induced and expressed genes were mapped to KEGG pathways and Swiss-Prot and Protein Information Resource (SP-PIR) using the online tool Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (http://david.abcc.ncifcrf.gov/) to identify differentially expressed pathways (31).
qRT-PCR of selected genes.
Primers were designed using Primer 3 online software (32). The relative transcript level of selected genes was measured by qRT-PCR as previously described (33) (see Table S1 in the supplemental material). Briefly, a set of three independent biological replicates of P. fluorescens biofilms was grown and treated with 0, 31.25, 62.5, and 125 mg/liter glutaraldehyde in 6-well plates. RNA was extracted with TRIzol as described for transcriptome analysis (see above). The cDNA was prepared from Turbo DNase-treated RNA using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA) and purified with Agencourt AMPure XP beads. cDNA (25 ng) from each sample was amplified with 10 pmol target primers using SsoAdvanced universal SYBR green supermix (Bio-Rad Laboratories, Inc.) for 35 amplification cycles on a CFX Connect real-time system (Bio-Rad Laboratories, Inc.). All measurements were done on three biological replicates consisting of two technical replicates each. The quantification cycle (Cq) values for primers were normalized against those of rpoD. Fold change in gene expression was calculated by 2(−ΔΔCT) (34) and expressed as fold change ± SD.
Efflux pump inhibitor treatment.
P. fluorescens biofilms were grown in 96-well plates as described in “Biofilm assays.” Four different efflux pump inhibitors (EPIs) were used in the study, namely, 1-(1-naphthylmethyl)-piperazine (NMP), phenylalanine-arginine β-naphthylamide (PAβN), carbonyl cyanide m-chlorophenylhydrazone (CCCP), and chlorpromazine (Cpz). The 72-h-old biofilms were washed with PBS and treated with glutaraldehyde, EPI, or glutaraldehyde plus EPI for 10 min. The glutaraldehyde concentrations used were 0, 15.125, 32.25, 62.5, 125, and 250 mg/liter. Two different concentrations of NMP (100 and 200 mg/liter), PAβN (25 and 50 mg/liter), CCCP (25 and 50 mg/liter), and Cpz (25 and 50 mg/liter) were used in the assays. Following exposure, biofilm viability was quantified by MTT staining, as described in “Biofilm assays,” and presented as the mean ± SD of three biological replicates.
P. aeruginosa PAO1 and PAmuc biofilms were grown in 96-well plates by placing 200 μl of 30-fold-diluted overnight culture in fresh LB medium. P. aeruginosa PAO1 biofilms were grown at room temperature. PAmuc did not form any appreciable biofilms at room temperature; therefore, biofilms were grown at 37°C. Four EPIs, NMP (200 mg/liter), PAβN (50 mg/liter), CCCP (50 mg/liter), and Cpz (50 mg/liter), were utilized in combination with 0, 15.125, 32.25, 62.5, 125, and 250 mg/liter glutaraldehyde to treat biofilms for 10 min and stained with MTT as described above. The means ± SD of three biological replicates are presented.
Sequence data accession number.
All data associated with the RNA-seq analysis have been deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE64448.
RESULTS
Mature biofilms show increased resistance to glutaraldehyde.
The resistance of P. fluorescens biofilms to glutaraldehyde treatment was determined over a range of eight concentrations spanning glutaraldehyde doses of 0 to 1,000 mg/liter. The resistance of P. fluorescens biofilms to glutaraldehyde treatment increased with maturity (Fig. 1A and see Fig. S1 in the supplemental material). The biofilms were least resistant to glutaraldehyde after 24 h of growth and most resistant after 120 h of growth. An increase in glutaraldehyde concentration from 62.5 mg/liter to 125 mg/liter demonstrated the largest percent change in biofilm viability (33%, 26%, 23%, 27%, and 10% in 24-, 48-, 72-, 96-, and 120-h-old biofilms, respectively). Therefore, for the purpose of this study, 125 mg/liter was considered the MIC and 62.5 mg/liter the subinhibitory concentration. Additionally, cell viability in 72-h-old biofilms following glutaraldehyde exposure was determined by plate count. Results showed that 62.5 and 125 mg/liter glutaraldehyde caused 3 and 5 log10 reductions in cell count, respectively (see Fig. S2 in the supplemental material). The highest tested concentration of glutaraldehyde was 1,000 mg/liter, with the expectation that this dose would be effective in completely inactivating all biofilm cells. However, as presented in Fig. 1A, a dose of 1,000 mg/liter lost effectiveness against more mature biofilms (96 and 120 h). As 72-h-old biofilms were moderately mature and showed some resistance to glutaraldehyde treatment, further studies were conducted using 72-h-old biofilms in conjunction with a subinhibitory dose of glutaraldehyde (62.5 mg/liter). To independently verify the results, P. fluorescens biofilms were grown for 24 and 72 h and stained with 0.3% crystal violet. Crystal violet staining demonstrated that 72-h-old biofilms were more resistant than 24-h-old biofilms (see Fig. S1 in the supplemental material). Furthermore, glutaraldehyde exposure did not appear to significantly reduce total biomass (see Fig. S1), which may be attributed to short exposure time in our experiments. Previously, significant changes in P. fluorescens biofilm biomass were observed following ≥12 h of glutaraldehyde exposure (21).
FIG 1.
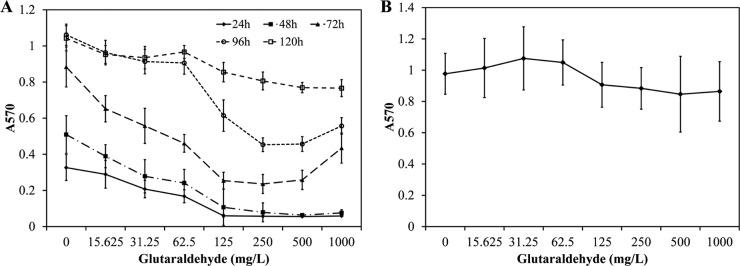
(A) Effect of glutaraldehyde on P. fluorescens biofilm inactivation. Error bars indicate standard deviation of three biological replicates. (B) Regrowth of a 72-h-old biofilm following exposure to glutaraldehyde. The 72-h-old biofilms were treated with various glutaraldehyde concentrations for 10 min at room temperature. Following treatment, biofilms were washed and regrown in fresh medium. The viability was measured after 24 h of regrowth. A570, absorbance at 570 nm.
The ability of 72-h-old P. fluorescens biofilms to recover after glutaraldehyde treatment was measured by exposing them to various concentrations of glutaraldehyde for 10 min. As presented in Fig. 1B, biofilm activity for all treatment levels recovered to control condition levels (P > 0.05) 24 h posttreatment.
P. fluorescens biofilm transcriptomic response to glutaraldehyde.
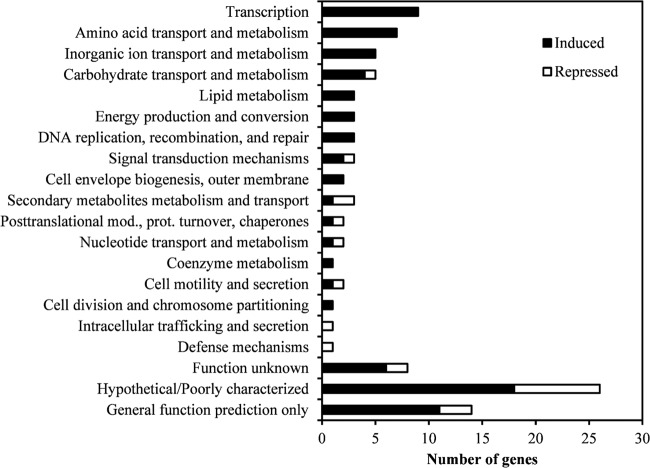
To determine the genetic response of surviving P. fluorescens biofilm cells, we performed an RNA-seq analysis on 72-h-old biofilms after glutaraldehyde or PBS exposure. A summary of sequencing results is presented in Table S2 in the supplemental material. A comparison of glutaraldehyde-exposed biofilms with PBS-exposed biofilms revealed that a total of 101 genes showed a >2-fold change (22 genes repressed and 79 genes induced). The classification of differentially expressed genes by COG (cluster of orthologous groups) revealed that genes in 20 COG categories were differentially expressed (Fig. 2; see also Table S3 in the supplemental material). The most abundant categories were transcription, amino acid transport and metabolism, inorganic ion transport and metabolism, carbohydrate transport and metabolism, lipid metabolism, and energy production and conversion (Fig. 2).
FIG 2.
COG classification of the differentially regulated genes of P. fluorescens 72-h-biofilm cells treated with 62.5 mg/liter glutaraldehyde.
Functional classification of differentially regulated genes revealed that genes involved in, for example, phosphonate degradation, lipid metabolism, efflux pumps, and polyamine biosynthesis were differentially regulated. The phosphonate degradation pathway in our data set was represented by the genes PFLU3910 (2.6-fold), phnD (2.5-fold), phnP (2.5-fold), phnN (−9.2-fold), and PFLU1868 (5.8-fold) (see Table S3 in the supplemental material). PFLU3910 appears to be an ortholog of phnR, a regulator of the phosphonate degradation pathway (35), whereas phnD, phnN, phnP, and PFLU1868 encode a phosphonate ABC transporter substrate-binding protein, a phosphorous compound metabolism-related ATP-binding protein, a carbon-phosphorus lyase complex accessory protein, and an ATPase component of an ABC-type phosphonate transport system, respectively (36).
Lipid metabolism was represented by differential regulation of PFLU0565 (3.5-fold), PFLU3981 (2.2-fold), PFLU0450 (2.03-fold), PFLU5785 (2.04-fold), and PFLU2771 (14.1-fold) (see Table S3 in the supplemental material). PFLU0565 shares 91% protein identity with the Pseudomonas protegens CHA0 FabR protein, a repressor of unsaturated fatty acid synthesis (37), whereas PFLU3981 appears to be a FadR ortholog, an activator of unsaturated fatty acid synthesis (37). Together, FabR and FadR regulate the unsaturated fatty acid synthesis. Induction of lipid biosynthesis genes PFLU0450, PFLU5785, and PFLU2771 was observed (see Table S3).
Two genes involved in multidrug efflux were induced. The PFLU3876 (2.2-fold) and PFLU2929 (14.8-fold) genes were overexpressed upon glutaraldehyde exposure. PFLU2929 and PFLU3876 appear to encode the outer membrane component of the multidrug efflux system and multidrug resistance protein A, respectively. PFLU3876 shows 79% protein similarity to PA5159 of P. aeruginosa PAO1 encoding a multidrug transporter and appears to contain an emrA (PRK15136) domain. PFLU2929 shows 78% protein similarity to oprN (PA2495) of P. aeruginosa PAO1. Since the pump component genes are typically linked and coexpressed to constitute a tripartite pump, we searched the P. fluorescens genome for putative partners of PFLU2929 and PFLU3876 in their neighboring region. PFLU2929 was immediately followed by PFLU2930 and PFLU2931, which are predicted to encode a putative cation efflux protein and HlyD family secretion protein. In addition, PFLU3876 is located between PFLU3875 and PFLU3877, which are predicted to encode multidrug resistance protein B and a hypothetical protein with an outer membrane lipoprotein of the NodT family efflux transporter domain. Furthermore, the genetic arrangement of these genes was similar to MexEF-OprN and PA5158-PA5160. PFLU2930 and PFLU2931 were induced 14.59-fold (P = 0.03) and 90.27-fold (P = 0.06), respectively. These genes were filtered out of our initial analysis due to our stringent cutoff value of an FDR-corrected P value of <0.001. On the other hand, PFLU3875 (−1.38-fold; P = 1.0) and PFLU3877 (1.52-fold; P = 0.00) showed a <2-fold change.
The data further suggest that a polyamine biosynthetic pathway was induced upon glutaraldehyde exposure. Specifically, three genes, PFLU0811 (2.2-fold), PFLU0293 (2.3-fold), and PFLU1724 (2.7-fold), which encode putative ornithine decarboxylase, agmatine deiminase, and a hypothetical protein similar to spermidine synthase, respectively, were induced. In addition, PFLU2340, which encodes a putative polyamine ABC transporter, was induced 8.6-fold upon glutaraldehyde exposure (see Table S3 in the supplemental material).
Glutaraldehyde exposure induces expression of genes involved in multidrug transport.
We examined the relative expression levels of PFLU2929 and PFLU3876 in three independent biological replicates of P. fluorescens biofilms following glutaraldehyde exposure using qPCR. The 72-h-old biofilms were exposed to 0, 31.25, 62.5, and 125 mg/liter glutaraldehyde, and the relative expressions of PFLU2929 and PFLU3876 were measured. The results showed that PFLU2929 and PFLU3876 were significantly induced upon glutaraldehyde exposure (Fig. 3). PFLU2929 and PFLU3876 showed approximately 19-fold and 27-fold changes, respectively, at 62.5 mg/liter glutaraldehyde. The relative expression level of PFLU2929 was similar to the change observed in RNA-seq data (≈15-fold), but the relative expression level recorded for PFLU3876 was considerably higher at 27-fold when measured by qPCR versus by RNA-seq (2.2-fold). Increasing the concentration of glutaraldehyde from 31.25 mg/liter to 62.5 mg/liter resulted in significant (P < 0.05, two-tailed t test) increases in expression levels, but a further increase in glutaraldehyde concentration did not result in significant increases in the expression levels of the two genes. DNA gyrase subunit A coding gene (gyrA) was used as an internal control in our experiments, as relative expression levels of gyrA were found to be unaltered by glutaraldehyde exposure (Fig. 3).
FIG 3.
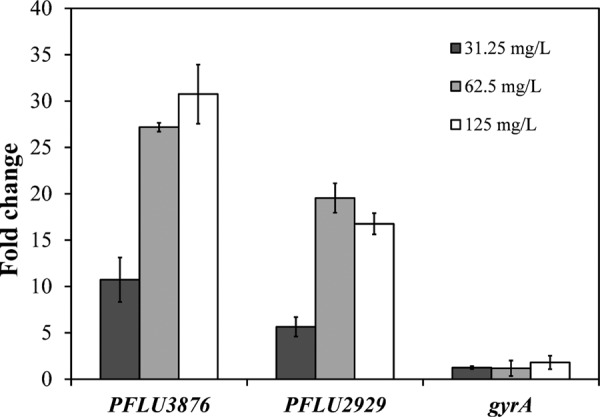
qRT-PCR of putative efflux pumps PFLU2929 and PFLU3876 in 72-h-old biofilm following glutaraldehyde exposure. Fold changes over 0 mg/liter glutaraldehyde exposure. gyrA, internal control.
Chemical inhibition of efflux pumps potentiates glutaraldehyde activity.
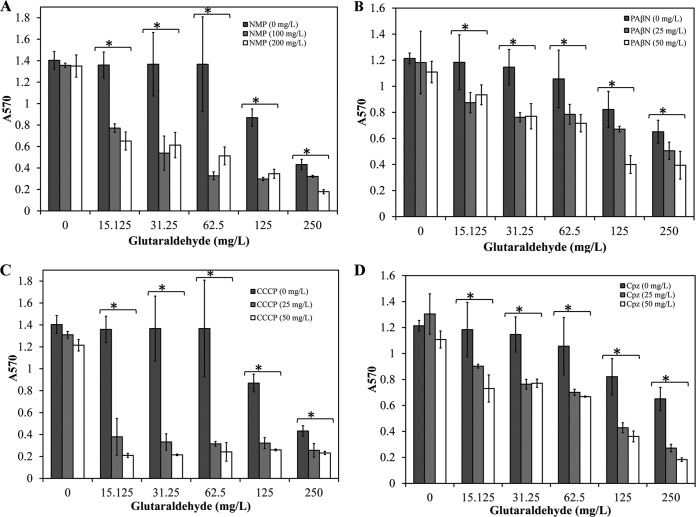
PFLU2929 and PFLU3876 likely encode efflux pump components that may affect glutaraldehyde effusion and impair bactericidal activity. To test this hypothesis, we chemically inhibited efflux pumps using EPIs in 72-h-old P. fluorescens biofilms in combination with glutaraldehyde. Two concentrations of four different EPIs were used in conjunction with six concentrations (0, 15.125, 31.25, 62.5, 125, and 250 mg/liter) of glutaraldehyde. The doses for EPIs were determined based on published literature (38–41). The addition of each EPI resulted in significant improvement in biofilm inactivation by glutaraldehyde (Fig. 4). NMP and CCCP were more effective at potentiating glutaraldehyde activity than were PAβN and Cpz. For instance, a combination of 25 mg/liter CCCP and 15.125 mg/liter glutaraldehyde or 100 mg/liter NMP and 62.5 mg/liter glutaraldehyde resulted in >70% biofilm inactivation. A similar level of biofilm inactivation was achieved by 50 mg/liter PAβN and 250 mg/liter glutaraldehyde or 25 mg/liter Cpz and 250 mg/liter glutaraldehyde. However, none of the tested combinations resulted in complete inactivation of the biofilm. Furthermore, we did not observe a significant difference between the tested concentrations of each EPI, indicating that lower doses of the tested EPIs may be sufficient to potentiate the glutaraldehyde activity.
FIG 4.
Effect of efflux pump inhibitors NMP (A), PAβN (B), CCCP (C), and Cpz (D) on glutaraldehyde activity against 72-h P. fluorescens biofilms. Star, significant difference at P = 0.05 from biofilm treated within each respective concentration of glutaraldehyde.
To further test if efflux pumps contribute to glutaraldehyde resistance in other pseudomonad species, we measured the biofilm viability at 72 h in P. aeruginosa strains PAO1 and PAmuc. P. aeruginosa PAO1 had ∼35% more biofilm at 72 h than did PAmuc (Fig. 5A and B). P. aeruginosa PAO1 and PAmuc biofilms were treated with the four EPIs in combination with six concentrations of glutaraldehyde as described above. In agreement with the observations for P. fluorescens 72-h-old biofilms, we observed decreased viability in P. aeruginosa PAO1 and PAmuc 72-h-old biofilms treated with combinations of EPIs and glutaraldehyde compared to those treated with glutaraldehyde alone (Fig. 5A and B).
FIG 5.
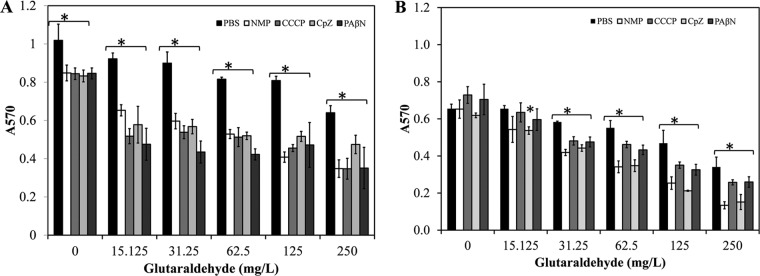
Effect of efflux pump inhibitors on P. aeruginosa strain PAO1 (A) and clinical isolate PAmuc (B) 72-h biofilms. Efflux pump inhibitors NMP, CCCP, Cpz, and PAβN were added at 200, 50, 50, and 50 mg/liter, respectively. Star, significant difference at P = 0.05 from biofilm treated only with respective concentration of glutaraldehyde.
DISCUSSION
The goal of the present study was to understand the genetic responses of P. fluorescens and P. aeruginosa biofilms to glutaraldehyde exposure. Biofilms are known to demonstrate increased antimicrobial resistance compared to planktonic cells (21, 42). In the present study, we examined the response of P. fluorescens biofilms to glutaraldehyde, a widely used disinfectant in many industries. As expected, microbial resistance to glutaraldehyde treatment increased with the age of the biofilm (Fig. 1A). A concentration of glutaraldehyde typically used for surface disinfection is ∼100 mg/liter (43). Our data suggest that this dose may not be adequate for complete inactivation of P. fluorescens biofilms. Given that most biofilms encountered in the environment are more mature than our tested biofilm, it is likely that they will demonstrate a higher resistance to glutaraldehyde and possibly to other surface disinfectants. Although these experiments were conducted in 96-well plates, the results obtained may be valid for mature biofilms present on industrial and medical devices, as 96-well-grown biofilms reportedly develop some characteristic features of mature biofilms (44, 45). These results emphasize that antimicrobial resistance of mature biofilms should be studied more intensively. Biocide resistance is a multifactorial process, and mature biofilms contain higher levels of exopolysaccharides and proteins that may limit penetration of glutaraldehyde (46). Further work is needed to clearly define the role of two efflux pumps encoded by PFLU2929-PFLU2931 and PFLU3875-PFLU3877 in glutaraldehyde resistance and to elucidate the role of metabolic pathways in Pseudomonas biofilm formation.
To gain mechanistic insight into glutaraldehyde resistance, we examined the transcriptomes of 72-h-old P. fluorescens biofilms exposed to glutaraldehyde and PBS. The 72-h time point was chosen because it reflected an intermediate time point between susceptible and resistant phenotypes. Four biological processes were induced following glutaraldehyde exposure: phosphonate degradation, polyamine biosynthesis, lipid metabolism, and efflux pumps (see Table S3 in the supplemental material). Phosphonate degradation, lipid metabolism, and polyamine biosynthesis have been reported to influence biofilm formation. For instance, antibodies against PhnD, the phosphonate ABC transporter substrate-binding protein, were reported to block initial attachment and aggregation (induced 2.5-fold in our study) (47). Similarly, a defect in polyamine biosynthesis has been shown to reduce planktonic and biofilm growth in several species that may be rescued with exogenous polyamine supplementation (48–52). Moreover, spermidine and putrescine were shown to stabilize the outer membrane and protect P. aeruginosa from antibiotic and oxidative stress by binding to lipopolysaccharides (53). Lipid metabolism contributes to the production of fatty acids, phospholipid, lipopolysaccharides, and acyl-homoserine lactone biosynthesis, which play critical roles in biofilm formation (54). It is possible that phosphonate degradation, lipid metabolism, and polyamine biosynthesis can help in recovery and regrowth of the biofilm following disinfection; however, the roles of these processes need to be evaluated further to determine their contribution to biofilm formation.
Two genes (PFLU2929 and PFLU3876) related to multidrug efflux activity were induced in our RNA-seq data. Independent measurement using qRT-PCR on a different set of samples confirmed the results observed in the RNA-seq data. The expression of PFLU2929 and PFLU3876 appeared to be greatest at 62.5 mg/liter glutaraldehyde. Efflux pumps significantly contribute to antimicrobial resistance, including biocide resistance in bacteria (7), and have been shown to confer resistance to triclosan and chlorheximide in P. aeruginosa and other bacterial species (55–57). Chlorheximide treatment strongly induces the MexCD-OprJ efflux pump in P. aeruginosa that contributes to resistance against chlorheximide and various antimicrobials (57). It is likely that the induction of efflux pumps can contribute to increased glutaraldehyde resistance. This conjecture is supported by the fact that EPIs potentiate the activity of glutaraldehyde.
PFLU2929 and PFLU3876 appear to be orthologs of P. aeruginosa oprN and PA5159, respectively. In P. aeruginosa, OprN is an outer membrane channel-forming protein in the MexEF-OprN efflux pump (58). It is likely that P. aeruginosa demonstrates glutaraldehyde resistance in a similar fashion, by active efflux of glutaraldehyde. Potentiation of glutaraldehyde activity in P. aeruginosa PAO1 and PAmuc by EPIs further suggests that efflux pumps contribute to glutaraldehyde resistance in P. aeruginosa.
MexEF-OprN and its homologue PFLU2929-PFLU2931 encode a resistance-nodulation-cell division (RND) pump. Three of the four EPIs in our study are RND pump inhibitors. Specifically, NMP, PAβN, and Cpz (a phenothiazine) modulate activity of RND pumps (38, 39, 41). The fourth EPI, CCCP, is an energy uncoupler (40). Three of the EPIs affect RND pump activity and CCCP influences most of the energy-dependent efflux pumps, and we observed potentiation of glutaraldehyde activity by all four EPIs but to different levels. Potentiation of glutaraldehyde activity by NMP, PAβN, and Cpz also suggests that the RND pump encoded by PFLU2929-PFLU2931 and MexEF-OprN may be involved in glutaraldehyde resistance. Taken together, these data suggest that efflux pumps contribute to glutaraldehyde resistance, and their inhibition potentiates the biocidal activity of glutaraldehyde in P. fluorescens and P. aeruginosa. Additionally, due to the nonspecific nature of these EPIs, other mechanisms of glutaraldehyde activity potentiation cannot be excluded, including inhibition of constitutively induced efflux pumps. In conclusion, this study provides new insight into the mechanism by which biofilms demonstrate glutaraldehyde resistance and suggests a mechanism of post-glutaraldehyde exposure biofilm recovery. Altogether, our study suggests that efflux pumps, lipid biosynthesis, polyamine biosynthesis, and phosphonate degradation contribute to glutaraldehyde resistance and biofilm resilience in pseudomonad biofilms. This study further indicates that combining EPIs with glutaraldehyde improves glutaraldehyde effectiveness and may serve to improve the effectiveness of other biocides as well.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the startup funds provided by the University of Pittsburgh to K.J.B. and by R00HL098342 to J.M.B.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.05152-14.
REFERENCES
- 1.Meyer B, Cookson B. 2010. Does microbial resistance or adaptation to biocides create a hazard in infection prevention and control? J Hosp Infect 76:200–205. doi: 10.1016/j.jhin.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 2.Maillard JY. 2007. Bacterial resistance to biocides in the healthcare environment: should it be of genuine concern? J Hosp Infect 65(Suppl 2):60–72. doi: 10.1016/S0195-6701(07)60018-8. [DOI] [PubMed] [Google Scholar]
- 3.McDonnell G, Burke P. 2011. Disinfection: is it time to reconsider Spaulding? J Hosp Infect 78:163–170. doi: 10.1016/j.jhin.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Russell AD. 2002. Mechanisms of antimicrobial action of antiseptics and disinfectants: an increasingly important area of investigation. J Antimicrob Chemother 49:597–599. doi: 10.1093/jac/49.4.597. [DOI] [PubMed] [Google Scholar]
- 5.Bryers JD. 2008. Medical biofilms. Biotechnol Bioeng 100:1–18. doi: 10.1002/bit.21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevenson BS, Drilling HS, Lawson PA, Duncan KE, Parisi VA, Suflita JM. 2011. Microbial communities in bulk fluids and biofilms of an oil facility have similar composition but different structure. Environ Microbiol 13:1078–1090. doi: 10.1111/j.1462-2920.2010.02413.x. [DOI] [PubMed] [Google Scholar]
- 7.Gnanadhas DP, Marathe SA, Chakravortty D. 2013. Biocides—resistance, cross-resistance mechanisms and assessment. Expert Opin Invest Drugs 22:191–206. doi: 10.1517/13543784.2013.748035. [DOI] [PubMed] [Google Scholar]
- 8.Bridier A, Dubois-Brissonnet F, Greub G, Thomas V, Briandet R. 2011. Dynamics of the action of biocides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 55:2648–2654. doi: 10.1128/AAC.01760-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davison WM, Pitts B, Stewart PS. 2010. Spatial and temporal patterns of biocide action against Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 54:2920–2927. doi: 10.1128/AAC.01734-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maseda H, Hashida Y, Konaka R, Shirai A, Kourai H. 2009. Mutational upregulation of a resistance-nodulation-cell division-type multidrug efflux pump, SdeAB, upon exposure to a biocide, cetylpyridinium chloride, and antibiotic resistance in Serratia marcescens. Antimicrob Agents Chemother 53:5230–5235. doi: 10.1128/AAC.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez P, Moreno E, Martinez JL. 2005. The biocide triclosan selects Stenotrophomonas maltophilia mutants that overproduce the SmeDEF multidrug efflux pump. Antimicrob Agents Chemother 49:781–782. doi: 10.1128/AAC.49.2.781-782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith K, Hunter IS. 2008. Efficacy of common hospital biocides with biofilms of multi-drug resistant clinical isolates. J Med Microbiol 57:966–973. doi: 10.1099/jmm.0.47668-0. [DOI] [PubMed] [Google Scholar]
- 13.Van Acker H, Van Dijck P, Coenye T. 2014. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol 22:326–333. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Bridier A, Briandet R, Thomas V, Dubois-Brissonnet F. 2011. Resistance of bacterial biofilms to disinfectants: a review. Biofouling 27:1017–1032. doi: 10.1080/08927014.2011.626899. [DOI] [PubMed] [Google Scholar]
- 15.Poole K. 2002. Mechanisms of bacterial biocide and antibiotic resistance. J Appl Microbiol 92:55S–64S. doi: 10.1046/j.1365-2672.92.5s1.8.x. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira C, Pereira AM, Pereira MC, Melo LF, Simões M. 2011. Physiological changes induced by the quaternary ammonium compound benzyldimethyldodecylammonium chloride on Pseudomonas fluorescens. J Antimicrob Chemother 66:1036–1043. doi: 10.1093/jac/dkr028. [DOI] [PubMed] [Google Scholar]
- 17.McDonnell G, Russell AD. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 12:147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tschudin-Sutter S, Frei R, Kampf G, Tamm M, Pflimlin E, Battegay M, Widmer AF. 2011. Emergence of glutaraldehyde-resistant Pseudomonas aeruginosa. Infect Control Hosp Epidemiol 32:1173–1178. doi: 10.1086/662624. [DOI] [PubMed] [Google Scholar]
- 19.Kampf G, Ostermeyer C, Tschudin-Sutter S, Widmer AF. 2013. Resistance or adaptation? How susceptible is a ‘glutaraldehyde-resistant’ Pseudomonas aeruginosa isolate in the absence of selection pressure? J Hosp Infect 84:316–318. doi: 10.1016/j.jhin.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Svetlíková Z, Škovierová H, Niederweis M, Gaillard J-L, McDonnell G, Jackson M. 2009. Role of porins in the susceptibility of Mycobacterium smegmatis and Mycobacterium chelonae to aldehyde-based disinfectants and drugs. Antimicrob Agents Chemother 53:4015–4018. doi: 10.1128/AAC.00590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simões LC, Lemos M, Araújo P, Pereira AM, Simões M. 2011. The effects of glutaraldehyde on the control of single and dual biofilms of Bacillus cereus and Pseudomonas fluorescens. Biofouling 27:337–346. doi: 10.1080/08927014.2011.575935. [DOI] [PubMed] [Google Scholar]
- 22.Simões M, Pereira MO, Machado I, Simões LC, Vieira MJ. 2006. Comparative antibacterial potential of selected aldehyde-based biocides and surfactants against planktonic Pseudomonas fluorescens. J Ind Microbiol Biotechnol 33:741–749. doi: 10.1007/s10295-006-0120-5. [DOI] [PubMed] [Google Scholar]
- 23.Kirschke DL, Jones TF, Craig AS, Chu PS, Mayernick GG, Patel JA, Schaffner W. 2003. Pseudomonas aeruginosa and Serratia marcescens contamination associated with a manufacturing defect in bronchoscopes. N Engl J Med 348:214–220. doi: 10.1056/NEJMoa021791. [DOI] [PubMed] [Google Scholar]
- 24.Vikram A, Lipus D, Bibby K. 2014. Produced water exposure alters bacterial response to biocides. Environ Sci Technol 48:13001–13009. doi: 10.1021/es5036915. [DOI] [PubMed] [Google Scholar]
- 25.Vikram A, Jesudhasan P, Pillai S, Patil B. 2012. Isolimonic acid interferes with Escherichia coli O157:H7 biofilm and TTSS in QseBC and QseA dependent fashion. BMC Microbiol 12:261. doi: 10.1186/1471-2180-12-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chitwood JL, Rincon G, Kaiser GG, Medrano JF, Ross PJ. 2013. RNA-seq analysis of single bovine blastocysts. BMC Genomics 14:350. doi: 10.1186/1471-2164-14-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CLC bio. 2013. Application note: expression analysis of mRNA-Seq reads from Arabidopsis tissues. CLC bio, Aarhus, Denmark: http://www.clcbio.com/files/appnotes/mRNA-Seq.pdf. [Google Scholar]
- 28.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 29.Robinson M, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baggerly KA, Deng L, Morris JS, Aldaz CM. 2003. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 19:1477–1483. doi: 10.1093/bioinformatics/btg173. [DOI] [PubMed] [Google Scholar]
- 31.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PubMed] [Google Scholar]
- 32.Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers, p 365–386. In Krawetz S, Misener S (ed), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 33.Vikram A, Jesudhasan PR, Jayaprakasha GK, Pillai SD, Patil BS. 2010. Grapefruit bioactive limonoids modulate E. coli O157:H7 TTSS and biofilm. Int J Food Microbiol 140:109–116. doi: 10.1016/j.ijfoodmicro.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Kulakova AN, Kulakov LA, Akulenko NV, Ksenzenko VN, Hamilton JTG, Quinn JP. 2001. Structural and functional analysis of the phosphonoacetate hydrolase (phnA) gene region in Pseudomonas fluorescens 23F. J Bacteriol 183:3268–3275. doi: 10.1128/JB.183.11.3268-3275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jochimsen B, Lolle S, McSorley FR, Nabi M, Stougaard J, Zechel DL, Hove-Jensen B. 2011. Five phosphonate operon gene products as components of a multi-subunit complex of the carbon-phosphorus lyase pathway. Proc Natl Acad Sci U S A 108:11393–11398. doi: 10.1073/pnas.1104922108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y, Cronan JE. 2011. Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol Microbiol 80:195–218. doi: 10.1111/j.1365-2958.2011.07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohnert JA, Kern WV. 2005. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother 49:849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pages JM, Amaral L, Fanning S. 2011. An original deal for new molecule: reversal of efflux pump activity, a rational strategy to combat Gram-negative resistant bacteria. Curr Med Chem 18:2969–2980. doi: 10.2174/092986711796150469. [DOI] [PubMed] [Google Scholar]
- 41.Kristiansen MM, Leandro C, Ordway D, Martins M, Viveiros M, Pacheco T, Molnar J, Kristiansen JE, Amaral L. 2006. Thioridazine reduces resistance of methicillin-resistant Staphylococcus aureus by inhibiting a reserpine-sensitive efflux pump. In Vivo 20:361–366. [PubMed] [Google Scholar]
- 42.Anwar H, Strap JL, Chen K, Costerton JW. 1992. Dynamic interactions of biofilms of mucoid Pseudomonas aeruginosa with tobramycin and piperacillin. Antimicrob Agents Chemother 36:1208–1214. doi: 10.1128/AAC.36.6.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grobe KJ, Zahller J, Stewart PS. 2002. Role of dose concentration in biocide efficacy against Pseudomonas aeruginosa biofilms. J Ind Microbiol Biotechnol 29:10–15. doi: 10.1038/sj.jim.7000256. [DOI] [PubMed] [Google Scholar]
- 44.O'Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp 30:pii=2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomes LC, Moreira JMR, Simões M, Melo LF, Mergulhão FJ. 2014. Biofilm localization in the vertical wall of shaking 96-well plates. Scientifica 2014:231083. doi: 10.1155/2014/231083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campanac C, Pineau L, Payard A, Baziard-Mouysset G, Roques C. 2002. Interactions between biocide cationic agents and bacterial biofilms. Antimicrob Agents Chemother 46:1469–1474. doi: 10.1128/AAC.46.5.1469-1474.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam H, Kesselly A, Stegalkina S, Kleanthous H, Yethon JA. 2014. Antibodies to PhnD inhibit Staphylococcal biofilms. Infect Immun 82:3764–3774. doi: 10.1128/IAI.02168-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel CN, Wortham BW, Lines JL, Fetherston JD, Perry RD, Oliveira MA. 2006. Polyamines are essential for the formation of plague biofilm. J Bacteriol 188:2355–2363. doi: 10.1128/JB.188.7.2355-2363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wortham BW, Oliveira MA, Fetherston JD, Perry RD. 2010. Polyamines are required for the expression of key Hms proteins important for Yersinia pestis biofilm formation. Environ Microbiol 12:2034–2047. doi: 10.1111/j.1462-2920.2010.02219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J, Sperandio V, Frantz DE, Longgood J, Camilli A, Phillips MA, Michael AJ. 2009. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J Biol Chem 284:9899–9907. doi: 10.1074/jbc.M900110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakada Y, Itoh Y. 2003. Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology 149:707–714. doi: 10.1099/mic.0.26009-0. [DOI] [PubMed] [Google Scholar]
- 52.Burrell M, Hanfrey CC, Murray EJ, Stanley-Wall NR, Michael AJ. 2010. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J Biol Chem 285:39224–39238. doi: 10.1074/jbc.M110.163154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson L, Mulcahy H, Kanevets U, Shi Y, Lewenza S. 2012. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J Bacteriol 194:813–826. doi: 10.1128/JB.05230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernando DM, Xu W, Loewen PC, Zhanel GG, Kumar A. 2014. Triclosan can select for an AdeIJK-overexpressing mutant of Acinetobacter baumannii ATCC 17978 that displays reduced susceptibility to multiple antibiotics. Antimicrob Agents Chemother 58:6424–6431. doi: 10.1128/AAC.03074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chuanchuen R, Beinlich K, Hoang TT, Becher A, Karkhoff-Schweizer RR, Schweizer HP. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob Agents Chemother 45:428–432. doi: 10.1128/AAC.45.2.428-432.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fraud S, Campigotto AJ, Chen Z, Poole K. 2008. MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa: involvement in chlorhexidine resistance and induction by membrane-damaging agents dependent upon the AlgU stress response sigma factor. Antimicrob Agents Chemother 52:4478–4482. doi: 10.1128/AAC.01072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poole K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J Mol Microbiol Biotechnol 3:255–264. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.