Abstract
We examined whether Escherichia coli isolates that produce CTX-M-9-type extended-spectrum β-lactamases (ESBL) are transferred between humans and chickens in a Vietnamese community. The phylogenetic group compositions, sequence types, antimicrobial resistance profiles, the prevalence of plasmid antibiotic resistance genes, and the plasmid replicon types generally differed between the human and chicken E. coli isolates. Our results suggest that transmission of the blaCTX-M-9-positive E. coli between humans and poultry was limited.
TEXT
Extended-spectrum-β-lactamase (ESBL)-producing bacteria have been rapidly spreading worldwide over the last 30 years (1, 2). ESBL hydrolyzes third-generation cephalosporins and is produced in bacteria, such as Escherichia coli and Klebsiella spp. There are three main groups of ESBL genes: blaTEM, blaSHV, and blaCTX-M. Recently, the CTX-M type of ESBL has become the most prevalent (1). So far, more than 100 genetic variants of blaCTX-M have been confirmed, and the genetic variants were categorized by their amino acids sequence into five main groups: CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25 types (2, 3).
The plasmids harboring most blaCTX-M and other antibiotics genes, such as aac (3)-II, aac(6′)-Ib, and aph(3′)-I (4), are transferable among bacterial cells. Therefore, it has been regarded as one of the major contributing factors to the rapid dissemination of multidrug-resistant bacteria.
Previous reports have indicated that the prevalence of ESBL-producing bacteria in healthy individuals is highest in China (50.5%) (5) and Thailand (65.7%) (6–8). Transmission mechanisms of these antibiotic-resistant bacteria are still unclear. Several previous reports suggest that companion animals might be reservoirs of the antibiotic-resistant bacteria in the community (9–12). However, direct transmission of antibiotic-resistant bacteria between humans and other animals has not been fully proved. Considering food-producing animals as one source of extraintestinal ESBL-producing bacteria to human has been a point of controversy (13). Therefore, in this study, we compared the antimicrobial susceptibility and the underlying genetic elements in the genomes of blaCTX-M-9-positive E. coli isolated from stool samples provided by asymptomatic human individuals and obtained from poultry in a small Vietnamese village. We assessed whether the blaCTX-M-9-positive E. coli was transferred between humans and poultry.
In this study, 199 fecal samples were collected from asymptomatic participants belonging to 47 households in a rural area of Vietnam (within a radius of approximately 150 m) from June 1 to 6 2013. At the same time, a rectal swab from a backyard chicken of each household was collected (n = 47). Each fecal sample was directly inoculated on a MacConkey Agar plate containing 1 μg/ml cefotaxime. After an overnight incubation at 37°C, a typical single red colony of each sample was isolated and subjected to bacterial species determination and ESBL phenotype confirmation. The bacterial species were identified by the VITEK2 system (bioMérieux, Marcy l'Etoile, France). The ESBL phenotypes were confirmed by disc diffusion tests according to the Clinical and Laboratory Standards Institute standard M100-S23. Consequently, 93 (46.7%) and 20 (42.6%) E. coli isolates from 199 human and 47 chicken fecal samples, respectively, were confirmed as ESBL-phenotype-positive E. coli isolates. In this area, the CTX-M-9-type ESBL gene, blaCTX-M-9 group, was most prevalent among CTX-M main groups (preliminary observation). Therefore, we confirmed CTX-M-9 group genes by PCR with TaKaRa EX Taq DNA polymerase (TaKaRa Bio Inc., Shiga, Japan) and primers targeting the blaCTX-M-9 group (CTX-M-IV-SeqF [5′-TGTAACACGGATTGACCGTAT-3′] and CTX-M-IV-SeqR-IS903 [5′-ACTCAGCAAAAGTTCGATTTATTC-3′]) according to the manufacturer's instruction. Consequently, the blaCTX-M-9 type was detected in 64 (32.2%) and 15 (31.9%) of the 93 human and 20 chicken ESBL-phenotype-positive E. coli isolates, respectively. E. coli isolates other than the 64 and 15 blaCTX-M-9-type-positive E. coli isolates possessed other blaCTX-M types than the blaCTX-M-9 type (B. Ngan et al., unpublished data). In this study, the 93 human and 20 chicken blaCTX-M-9-type-positive E. coli isolates were examined. The genes aac (3)-II, aac(6′)-Ib, aadA, aph(3′)-I, cat1, and cmlA were detected by PCR with heat-extracted bacterial DNA as the template (8, 14–16). Phylogenetic grouping of the E. coli isolates was determined by the protocol described by Clermont et al., using the three genetic markers chuA, yjaA, and TspE4.C2 (17). Plasmid replicons of plasmids harbored by the E. coli isolates were determined by following the original protocol described by Carattoli et al. (18). All isolates were typed by using a conventional multilocus sequence typing method described by Wirth et al. (19).
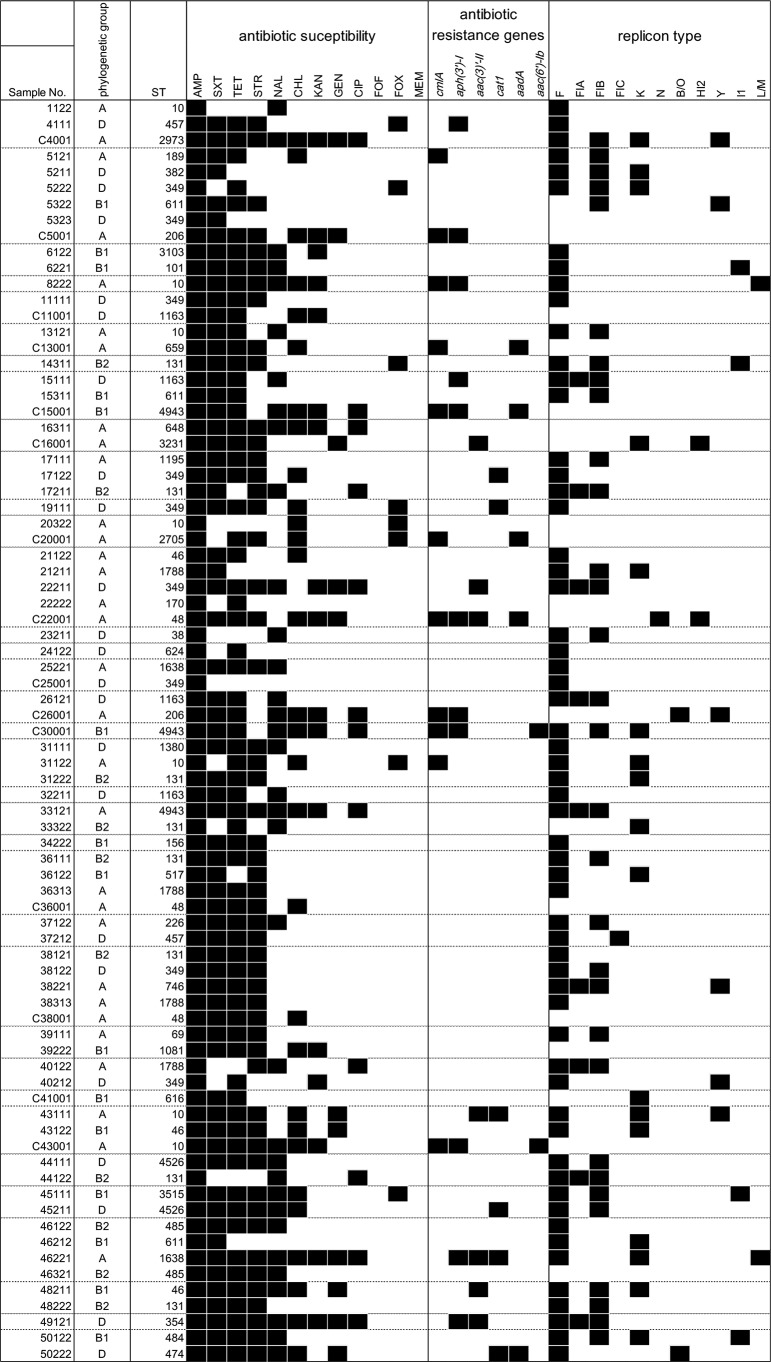
For nosocomial infections, clonal or oligoclonal distributions of antibiotic-resistant bacteria have often been observed (20). If the E. coli isolates obtained from the asymptomatic participants and their own domestic chickens were consistent with a clonal or oligoclonal distribution of a particular ancestral E. coli clone(s), similar phylogenetic relationships among the E. coli isolates would be observed for humans and chickens. The E. coli isolates from human were classified as groups A (32.8%), B1 (18.8%), B2 (15.6%), and D (32.8%). The isolates from chickens were primarily classified as group A (66.7%), and B1 (20.0%) and D (13.3%) were less frequent. Phylogenetic group B2, which has been regarded as including extraintestinal virulent strains, was not detected in the chicken isolates. Several sequence types (STs) were detected in the chicken and human isolates (Fig. 1). ST10 and ST349 were detected in the chickens and human isolates; however, antibiotic susceptibility, antibiotic resistance genes, and replicon type were not completely identical among the isolates with same STs (Table 1). There was no isolate set with the identical genomic characters of human and chicken isolates belonging to same household (Fig. 1).
FIG 1.
Antibiotic susceptibility, antibiotic-related genes, and plasmid replicon type of the blaCTX-M-9-positive E. coli. Black squares, resistant to antibiotic or detected each gene and replicon type.
TABLE 1.
Summary of MLSTa analysis
| ST |
E. coli isolates from: |
|||||
|---|---|---|---|---|---|---|
| All sources |
Human |
Chicken |
||||
| No. | % | No. | % | No. | % | |
| 349 | 9 | 11.4 | 8 | 12.5 | 1 | 6.7 |
| 131 | 8 | 10.1 | 8 | 12.5 | 0 | 0.0 |
| 10 | 7 | 8.9 | 6 | 9.4 | 1 | 6.7 |
| 1788 | 4 | 5.1 | 4 | 6.3 | 0 | 0.0 |
| 1163 | 3 | 3.8 | 2 | 3.1 | 1 | 6.7 |
| 46 | 3 | 3.8 | 3 | 4.7 | 0 | 0.0 |
| 611 | 3 | 3.8 | 3 | 4.7 | 0 | 0.0 |
| 48 | 3 | 3.8 | 0 | 0.0 | 3 | 20.0 |
| Other | 39 | 49.4 | 30 | 46.9 | 9 | 60.0 |
| Total | 79 | 100.0 | 64 | 100.0 | 15 | 100.0 |
MLST, multilocus sequence typing.
We assessed the susceptibility of the blaCTX-M-9-positive E. coli isolates by the standard disc diffusion test using Mueller-Hinton agar and 12 kinds of antibiotics discs (Eiken Chemical Co., Tokyo, Japan), including ampicillin (AMP), ciprofloxacin (CIP), chloramphenicol (CHL), fosfomycin (FOF), cefoxitin (FOX), gentamicin (GEN), kanamycin (KAN), meropenem (MEM), nalidixic acid (NAL), streptomycin (STR), trimethoprim-sulfamethoxazole (SXT), and tetracycline (TET) (Fig. 1). The proportions of E. coli isolates resistant to CHL and KAN were higher in chickens than in humans. None of the isolates was resistant to FOF or MEM. More than 80% of the isolates from humans and chickens were resistant to STR.
The genes cmlA and cat1 confer resistance to CHL; aac (3)-II and aac(6′)-Ib, to GEN; aph(3′)-I, to KAN; and aadA, to STR (4). The prevalence of the E. coli isolates possessing at least one plasmid antibiotic resistance gene, such as cmlA, cat1, aac (3)-II, aac(6′)-Ib, aph(3′)-I, or aadA, was 21.9% in humans and 60.0% in chickens. The prevalence of each gene differed between human and chicken isolates. For example, 33.3% and 0.0% of the E. coli isolates from humans and chickens, respectively, possessed cat1. The CHL resistance gene cmlA was detected in both human and chicken isolates; however, cat1 was detected only in human isolates (Fig. 1). In addition, the plasmid replicon types differed between the human and chicken E. coli isolates. Replicon type F was more prevalent in human isolates (87.5%) than in chicken isolates (20.0%). The replicons detected in chicken isolates included K, N, HI2, and Y. Furthermore, common combinations of replicon type and antibiotic resistance genes were not observed for human and chicken isolates.
Our results thus suggest that the transfer of the blaCTX-M-9-type-positive E. coli isolate was very limited between humans and chickens in the rural Vietnamese community. However, our sample size was insufficient to exclude the possibility of transfer between human and poultry. Further studies with larger sample sizes will be important to address these issues.
ACKNOWLEDGMENTS
This work was supported by the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA) as part of the Science and Technology Research Partnership for Sustainable Development (SATREPS).
We thank the staff (N. T. A. Tuyet, H. T. T. Van, and P. T. T. Ha) at the Food Microbiology Department of the National Institute of Nutrition for their excellent technical assistance.
REFERENCES
- 1.Cantón R, Gonzalez-Alba JM, Galan JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao WH, Hu ZQ. 2013. Epidemiology and genetics of CTX-M extended-spectrum beta-lactamases in Gram-negative bacteria. Crit Rev Microbiol 39:79–101. doi: 10.3109/1040841X.2012.691460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Andrea MM, Arena F, Pallecchi L, Rossolini GM. 2013. CTX-M-type beta-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol 303:305–317. doi: 10.1016/j.ijmm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 4.van Hoek AH, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJ. 2011. Acquired antibiotic resistance genes: an overview. Front Microbiol 2:203. doi: 10.3389/fmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Sun JY, Liu QZ, Han LZ, Huang XH, Ni YX. 2011. High prevalence of CTX-M beta-lactamases in faecal Escherichia coli strains from healthy humans in Fuzhou, China. Scand J Infect Dis 43:170–174. doi: 10.3109/00365548.2010.538856. [DOI] [PubMed] [Google Scholar]
- 6.Luvsansharav UO, Hirai I, Nakata A, Imura K, Yamauchi K, Niki M, Komalamisra C, Kusolsuk T, Yamamoto Y. 2012. Prevalence of and risk factors associated with faecal carriage of CTX-M beta-lactamase-producing Enterobacteriaceae in rural Thai communities. J Antimicrob Chemother 67:1769–1774. doi: 10.1093/jac/dks118. [DOI] [PubMed] [Google Scholar]
- 7.Luvsansharav UO, Hirai I, Niki M, Sasaki T, Makimoto K, Komalamisra C, Maipanich W, Kusolsuk T, Sa-Nguankiat S, Pubampen S, Yamamoto Y. 2011. Analysis of risk factors for a high prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in asymptomatic individuals in rural Thailand. J Med Microbiol 60:619–624. doi: 10.1099/jmm.0.026955-0. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki T, Hirai I, Niki M, Nakamura T, Komalamisra C, Maipanich W, Kusolsuk T, Sa-Nguankiat S, Pubampen S, Yamamoto Y. 2010. High prevalence of CTX-M beta-lactamase-producing Enterobacteriaceae in stool specimens obtained from healthy individuals in Thailand. J Antimicrob Chemother 65:666–668. doi: 10.1093/jac/dkq008. [DOI] [PubMed] [Google Scholar]
- 9.Blanc V, Mesa R, Saco M, Lavilla S, Prats G, Miro E, Navarro F, Cortes P, Llagostera M. 2006. ESBL- and plasmidic class C beta-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet Microbiol 118:299–304. doi: 10.1016/j.vetmic.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. 2012. Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect 18:646–655. doi: 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- 11.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ. 2011. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect 17:873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 12.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X, Kluytmans J. 2011. Extended-spectrum beta-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg Infect Dis 17:1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazarus B, Paterson DL, Mollinger JL, Rogers BA. 2015. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis 60:439–452. doi: 10.1093/cid/ciu785. [DOI] [PubMed] [Google Scholar]
- 14.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosengren LB, Waldner CL, Reid-Smith RJ. 2009. Associations between antimicrobial resistance phenotypes, antimicrobial resistance genes, and virulence genes of fecal Escherichia coli isolates from healthy grow-finish pigs. Appl Environ Microbiol 75:1373–1380. doi: 10.1128/AEM.01253-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van TT, Chin J, Chapman T, Tran LT, Coloe PJ. 2008. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int J Food Microbiol 124:217–223. doi: 10.1016/j.ijfoodmicro.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falagas ME, Karageorgopoulos DE. 2009. Extended-spectrum beta-lactamase-producing organisms. J Hosp Infect 73:345–354. doi: 10.1016/j.jhin.2009.02.021. [DOI] [PubMed] [Google Scholar]