Abstract
Selective fluorescent β-lactam chemical probes enable the visualization of the transpeptidase activity of penicillin-binding proteins (PBPs) at different stages of bacterial cell division. To facilitate the development of new fluorescent probes for PBP imaging, we evaluated 20 commercially available β-lactams for selective PBP inhibition in an unencapsulated derivative of the D39 strain of Streptococcus pneumoniae. Live cells were treated with β-lactam antibiotics at different concentrations and subsequently incubated with Bocillin FL (Boc-FL; fluorescent penicillin) to saturate uninhibited PBPs. Fluorophore-labeled PBPs were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorescence scanning. Among 20 compounds tested, carbapenems (doripenem and meropenem) were coselective for PBP1a, PBP2x, and PBP3, while six of the nine penicillin compounds were coselective for PBP2x and PBP3. In contrast, the seven cephalosporin compounds tested display variability in their PBP-binding profiles. Three cephalosporin compounds (cefoxitin, cephalexin, and cefsulodin) and the monobactam aztreonam exhibited selectivity for PBP3, while only cefuroxime (a cephalosporin) was selective for PBP2x. Treatment of S. pneumoniae cultures with a sublethal concentration of cefuroxime that inhibited 60% of PBP2x activity and less than 20% of the activity of other PBPs resulted in formation of elongated cells. In contrast, treatment of S. pneumoniae cultures with concentrations of aztreonam and cefoxitin that inhibited up to 70% of PBP3 activity and less than 30% of other PBPs resulted in no discernible morphological changes. Additionally, correlation of the MIC and IC50s for each PBP, with the exception of faropenem, amdinocillin (mecillinam), and 6-APA, suggests that pneumococcal growth inhibition is primarily due to the inhibition of PBP2x.
INTRODUCTION
Penicillin-binding proteins (PBPs) are membrane-anchored enzymes involved in the final step of bacterial cell wall synthesis and are targets for β-lactam antibiotics (1). These proteins are classified based upon their molecular weights and conserved domain structures. Class A high-molecular-weight (HMW) PBPs are bifunctional proteins with transglycosylase (TG) and transpeptidase (TP) activities. Class B HMW PBPs are monofunctional TPs. Lastly, class C, or low-molecular-weight (LMW), PBPs are d,d-carboxypeptidases or d,d-endopeptidases (2). Streptococcus pneumoniae, an opportunistic pathogen that causes serious infectious diseases, such as pneumonia, meningitis, otitis media, and bacteremia (3–5), possesses three class A PBPs (PBP1a, PBP1b, and PBP2a), two class B PBPs (PBP2x and PBP2b), and one class C PBP (PBP3) with d,d-carboxypeptidase activity (Fig. 1) (6–8). In addition, PBP2x possesses a C-terminal extension consisting of two PASTA (PBP- and serine/threonine kinase-associated) domains (Fig. 1), each containing one α-helix and three β-strands (8, 9).
FIG 1.
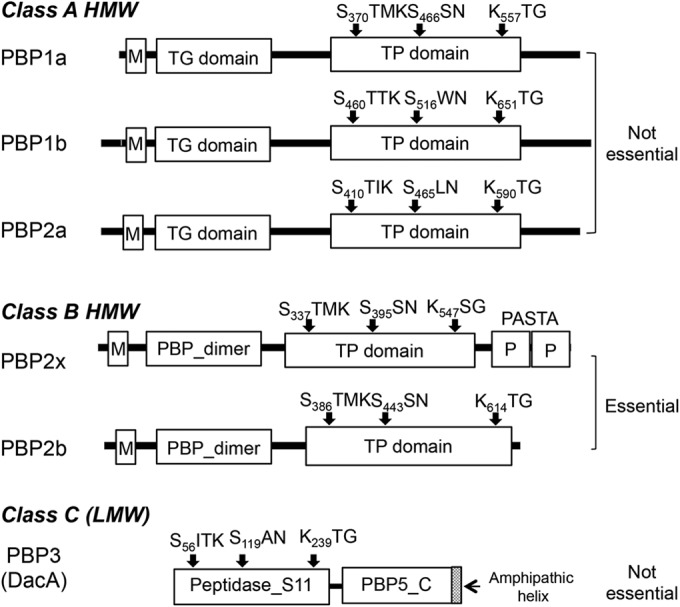
Domain architecture and conserved motifs in penicillin-binding proteins (PBPs) of Streptococcus pneumoniae. S. pneumoniae contains six PBPs: the high-molecular-weight (HMW) class A members PBP1a, PBP1b, and PBP2a; the HMW class B members PBP2x and PBP2b; and the low-molecular-weight (LMW) protein PBP3 (1, 8). The topology of the three class A PBPs consists of a cytoplasmic tail, a transmembrane (M) anchor, and extracellular transglycosylase (TG) and transpeptidase (TP) domains (8). The two class B PBPs consist of a cytoplasmic tail, a transmembrane anchor, an extracellular N-terminal domain (PBP_dimer) that is implicated in PBP polymerization, and a TP domain. In addition, PBP2x contains a C-terminal extension consisting of two penicillin-binding protein and serine/threonine kinase-associated (PASTA [P]) domains (15, 16). LMW PBP3 is a d-alanyl–d-alanine carboxypeptidase that contains an N-terminal catalytic peptidase S11 domain and a C-terminal PBP5 C domain, followed by an amphipathic helix (39). The TP and peptidase S11 domains contain the conserved PBP-binding motifs: SXXK with the active-site serine, S/YXN, and K/H(S/T)G. PBP2x and PBP2b are essential while the other four PBPs are not required for growth of S. pneumoniae (8).
The two class B PBPs (PBP2x and PBP2b) are individually essential in S. pneumoniae, performing critical roles in septal and peripheral peptidoglycan synthesis, respectively (10–13), and they are therefore likely critical targets for the β-lactam antibiotics. PBP2x is the functional equivalent of PBP3 in Escherichia coli, as both inactivation of PBP2x of S. pneumoniae by methicillin (6) and inactivation or depletion of PBP3 of E. coli result in filamentous cells (1). Similarly, depletion of PBP2b of S. pneumoniae (10, 12) and inactivation or depletion of PBP2 of E. coli result in spherical cells (1). Point and mosaic mutations in the conserved motifs of PBP2x and PBP2b result in decreased β-lactam affinity, which is associated with resistance to these compounds (8, 14, 15). In particular, the PASTA domains of PBP2x were shown to be essential for the binding of Bocillin FL (Boc-FL; fluorescent penicillin) (15), in addition to providing stability to PBP2x (16). Consistent with the role of the PASTA domains in β-lactam binding, the X-ray structure of an acylated PBP2x crystallized in the presence of a high concentration (14 mg/ml) of cefuroxime revealed the presence of two cefuroxime molecules. One was covalently bound to the active-site serine (Ser337), and the second was noncovalently sandwiched between the TP and the first PASTA domain (17). In contrast to the class B PBPs, each of the class A PBPs and PBP3 can be deleted individually in S. pneumoniae (18, 19), but pbp1a pbp2a double mutants are not viable (7, 19).
Historically, the earliest strategy for detection of PBP activity was tagging of these proteins with radiolabeled penicillin to examine the affinity of the β-lactams for each PBP either from membrane preparations or in live cells (20–22). More recently, nonradioactive β-lactams, such as fluorescent derivatives, have been used to detect PBPs in both gel-based analyses (23, 24) and localization studies in live cells (25, 26). Previous PBP profiling with β-lactams was performed with the laboratory strains R36A and R6 (10, 22, 27, 28). Compared to their D39 progenitor, these strains have accrued over 80 mutations that affect metabolism and peptidoglycan-modifying enzymes (29, 30). In particular, R6 protein PBP1a (PBP1aR6) contains two amino acid differences (Thr124→Ala and Asp388→Gln) compared to its D39 progenitor (30) or strains of other serotypes, such as TIGR4 (31). Our previous study on suppression of the ΔmreC essentiality phenotype suggested that PBP1aR6 is only partially active compared to PBP1aD39 (32). We used an unencapsulated (Δcps) D39 strain in previous pneumococcal cell division studies, because the presence of capsule masks or alters some cell division defects in S. pneumoniae D39 strains (18) and the absence of cell chains in the unencapsulated strain allowed us to readily classify the divisional stage of each cell (6, 12). Hence, an unencapsulated D39 strain that has been characterized extensively with respect to the function and localization of the PBPs was used in this study to provide an understanding of antibiotic-PBP interactions in the genetic background of the virulent serotype 2 progenitor strain.
PBP2x (initially named PBP2′) from S. pneumoniae was not identified until 1980 (33) and was therefore not described in an early PBP selectivity study that included 18 β-lactam antibiotics (22). In addition, comigration of PBP2x, PBP2a, and PBP2b during SDS-PAGE gel analysis has been problematic in some investigations, making accurate gel band quantitation difficult (10, 11, 27, 34). In this investigation, these proteins were distinguished from one another as Boc-FL-labeled conjugates. Our recent study using superresolution microscopy revealed that PBP2x of S. pneumoniae is directed to a discrete location separate from PBP2b and PBP1a within the septal aperture during the later stages of cell division (12). Selective fluorescent β-lactam chemical probes would provide further insight by visualization of the TP activity of PBPs at different stages of bacterial cell division (25, 26).
To facilitate the development of probes that target single PBPs, we evaluated 20 commercially available β-lactams from five different subclasses (monobactam, penem, carbapenem, penicillin, and cephalosporin) for selective PBP inhibition in an unencapsulated derivative of the D39 strain of S. pneumoniae. We identified the cephalosporin antibiotic cefuroxime as PBP2x selective and the monobactam aztreonam and three cephalosporins (cefoxitin, cephalexin, and cefsulodin) as PBP3 selective. In addition, comparison of MICs of the examined compounds with the 50% inhibitory concentrations (IC50s) for each PBP suggests that pneumococcal killing is often associated with PBP2x inhibition, with the exception of faropenem, (+)-6-aminopenicillanic acid (6-APA), and amdinocillin (mecillinam). Overall, we identify candidate β-lactam compounds for designing next-generation probes for specific pneumococcal PBP labeling.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and antibiotics.
S. pneumoniae strains IU1945, an unencapsulated derivative of D39 (Δcps) (30), and IU6647 (IU1945 Δpbp1a::P-erm), an isogenic reconstructed strain of E177 (32), were cultured statically in Becton-Dickinson brain heart infusion (BHI) broth at 37°C in an atmosphere of 5% CO2, and growth was monitored by determining the optical density at 620 nm (OD620), as described before (6). For SDS-PAGE gel analysis and morphological studies, bacteria were inoculated into BHI broth from glycerol stocks, serially diluted into the same medium, and propagated overnight. An overnight culture that was still in exponential phase (OD620 = 0.1 to 0.4) was diluted back to an OD620 of ∼0.005 to start final cultures.
Faropenem, doripenem, meropenem, (+)-6-aminopenicillanic acid (6-APA), penicillin V, penicillin G, ampicillin, methicillin, amdinocillin, oxacillin, piperacillin, cephalexin, cefsulodin, cefoxitin, cephalothin, cefuroxime, and ceftriaxone were purchased from Sigma-Aldrich (St. Louis, MO). Cefotaxime was purchased from Calbiochem (Billerica, MA). Aztreonam and amoxicillin were obtained from MP Biomedicals (Solon, OH). Bocillin FL (Boc-FL) was purchased from Life Technologies (Grand Island, NY).
All antibiotics were stored as solids and were dissolved in Milli-Q purified water (unless noted otherwise) at 10 mg/ml immediately before each experiment. 6-APA, amoxicillin, and cefuroxime were not soluble in water. Thus, 6-APA was dissolved in phosphate-buffered saline (PBS), pH 7.4, at a 5-mg/ml concentration. Amoxicillin and cefuroxime were dissolved in dimethyl sulfoxide (DMSO) at a 100-mg/ml stock concentration (further dilution in PBS decreases the final DMSO concentration to 1% or lower). All stock solutions were serially diluted (10-fold) in PBS to make 0.0001- to 1,000-μg/ml working solutions. For morphological studies, antibiotics were dissolved and serially diluted in BHI.
β-Lactam titration and detection of PBPs.
S. pneumoniae IU1945 cells from 1.5 ml of an exponential culture at an OD620 of ∼0.18 to 0.20 were harvested by centrifugation (16,000 × g for 2 min at room temperature). The cell pellets were washed in 1 ml PBS, pH 7.4, by pipetting up and down a few times. Then cells were pelleted using the same centrifugation conditions. The cells were resuspended in 50 μl PBS containing 0.0001 to 1,000 μg/ml β-lactam antibiotics. A reference sample was resuspended in 50 μl PBS without any antibiotics. After 30 min of incubation at room temperature, the cells were pelleted, washed in 1 ml PBS, and resuspended in 50 μl PBS containing 5 μg/ml Boc-FL (0.1% DMSO). After 10 min of incubation at room temperature, the cells were pelleted and washed in 1 ml PBS. Next, the cells were resuspended in 100 μl PBS containing 1 mg/ml lysozyme and incubated for 30 min at 37°C. The cells were sonicated using a Branson Sonifier 250 (Branson Ultrasonic, Danbury, CT) on ice (power setting 3, 30% duty cycle for three 10-s intervals with a 10-s cooling time between rounds). The membrane proteome was isolated by centrifugation at 21,000 × g for 15 min at 4°C. The supernatant was discarded, the membrane was resuspended in 100 μl PBS, and the samples were homogenized by sonication (power setting 1, 10% duty cycle for 1 s). The protein concentration was measured with a NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE). The protein concentration was adjusted to 2.5 mg/ml by diluting with PBS.
Fifty-one microliters of proteome sample was dispensed into a clean 1.5-ml microcentrifuge tube, and 17 μl of 4× SDS-PAGE loading buffer was added to each sample. The samples were heated for 5 min at 90°C to denature the proteins, then cooled down to room temperature. Ten microliters of sample was loaded onto a 10% SDS-PAGE gel. The protein bands were separated by gel electrophoresis for 1.5 h at 180 V, 400 mA, and 60 W. The gel was rinsed with distilled water three times and then scanned using a Typhoon 9210 gel scanner (Amersham Biosciences, Pittsburgh, PA) with a 526-nm short-pass filter at a 50-μm resolution. The gel images were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD). The background signal of the gel images was subtracted, and the brightness and contrast adjusted to optimize the signal-to-noise ratio (all operations were performed over the entire gel uniformly). Integrated density values were measured for gel band quantitation, and Boc-FL labeling of each PBP in antibiotic-treated samples is shown relative to samples not treated with antibiotics. Integrated density values were inserted into GraphPad Prism (GraphPad Software, La Jolla, CA) to create graphs showing relative percentages of Boc-FL labeling versus inhibitor concentration. IC50 determinations were performed using the same software. For all dose-response curves, data were fitted to a four-parameter logistic equation. If one of the two runs yielded an ambiguous curve fit but the obtained IC50 was ≤4-fold higher than the value from the duplicate run, it was considered valid, and the two values were averaged.
Antimicrobial susceptibility assay for S. pneumoniae IU1945.
S. pneumoniae strain IU1945 (D39 Δcps) was streaked from glycerol stock to plates containing Trypticase soy agar II (modified; Becton-Dickinson) and 5% (vol/vol) defibrinated sheep blood (TSAIIBA) and incubated at 37°C in an atmosphere of 5% CO2 for 14 to 16 h. Four colonies were inoculated into 3 to 5 ml of BHI and grown to an OD620 of ∼0.085 (2.2 × 105 CFU/ml). Antimicrobial susceptibility assays were performed as described previously (35). Briefly, cultures at an OD620 of ∼0.085 were diluted 100-fold with BHI. β-Lactam antibiotic stocks were serially diluted 2-fold in BHI. In 96-well round-bottom plates, 50 μl diluted culture was added to 50 μl of serially diluted antibiotics in each well (final number of CFU per well was 1.1 × 104). Duplicates of each antibiotic concentration were performed on a different plate. After incubation at 37°C in an atmosphere of 5% CO2 for 18 to 20 h, plates were examined by eye and with an inverted enlargement mirror. The lowest concentration that inhibited cell growth was noted as the MIC.
Morphological studies.
Cefuroxime at a final concentration of 0.006 μg/ml (twice the IC50 of PBP2x) was added to a 5-ml culture of exponentially growing IU1945 (OD620 ≈ 0.085), and 1 μl of the culture was examined with phase-contrast microscopy for 15 min to 5.5 h. Aztreonam at a final concentration of 0.2, 0.4, or 1 μg/ml or cefoxitin at 0.008, 0.016, or 0.04 μg/ml was added to 5-ml cultures of IU1945 at an OD620 of ∼0.003, and the cultures were examined between 3 h and 4.5 h after addition of antibiotics. In addition, aztreonam at a final concentration of 0.2 μg/ml was added to 5-ml cultures of IU1945 at an OD620 of ∼0.09, and the cultures were examined between 21 and 81 min after addition of antibiotics.
RESULTS AND DISCUSSION
It is well established that penicillins and cephalosporins inhibit the growth of S. pneumoniae by interacting with the TP domain of PBPs (1, 8, 34, 36). β-Lactams act as suicide inhibitors, acylating these proteins and yielding a covalent small-molecule–enzyme complex (36). Because the hydrolysis of this complex is slow, β-lactams occupy the active site of the PBPs for extended periods and prevent them from catalyzing further reactions, often resulting in cell lysis, except for treatment with cephalosporins (e.g., cefotaxime) that do not inhibit PBP2b (8, 27, 37). We assessed the selectivity of 20 β-lactams for the PBPs in live S. pneumoniae using fluorescent penicillin, Boc-FL, as the readout probe (Table 1; also, see Table S1 and Fig. S2 in the supplemental material) with an unencapsulated derivative of the D39 strain. Earlier work used the laboratory strains R36A and R6 (22, 27, 28), which harbor mutations in many genes involved in peptidoglycan synthesis, including pbp1a, compared to the progenitor D39 strain. Our study provides an up-to-date assessment, as we were able to resolve all six PBPs as their Boc-FL conjugates by SDS-PAGE analysis, while in previous studies PBP2x had either not yet been identified or comigrated with PBP2a and/or PBP2b, making assignment of selectivity difficult.
TABLE 1.
IC50s of β-lactams for S. pneumoniae IU1945
| β-Lactam class and drug | MIC (μg/ml) | IC50 (μg/ml)a |
PBP selectivity | |||||
|---|---|---|---|---|---|---|---|---|
| PBP1a | PBP1b | PBP2x | PBP2a | PBP2b | PBP3 | |||
| Monobactams | ||||||||
| Aztreonam | 64 | ND | ND | 67 ± 1 | 100 ± 1 | >1,000 | 0.051 ± 0.003 | 3c |
| Penems | ||||||||
| Faropenem | 0.016 | 0.0024 ± 0.0002 | 0.011 ± 0.001 | 0.007* ± 0.002 | 0.0089* ± 0.0004 | 0.0038* ± 0.0005 | 0.0035* ± 0.0008 | NSe |
| Carbapenems | ||||||||
| Doripenem | 0.008 | 0.0085* ± 0.0013 | 0.035 ± 0.018 | 0.0082* ± 0.0009 | 0.082 ± 0.041 | 0.029 ± 0.011 | 0.0042 ± 0.0001 | 1a, 2x, 3 |
| Meropenem | 0.016 | 0.0054* ± 0.0006 | 0.059 ± 0.012 | 0.011* ± 0.002 | 1.6 ± 0.8 | 0.027 ± 0.001 | 0.0031 ± 0.0005 | 1a, 2x, 3 |
| Penicillins | ||||||||
| Penicillin V | 0.008 | 0.022 ± 0.001 | 0.036 ± 0.005 | 0.0074* ± 0.0009 | 0.14 ± 0.03 | 0.15 ± 0.05 | 0.0046 ± 0.0003 | 2x, 3 |
| Penicillin G | 0.008 | 0.020 ± 0.001 | 0.028 ± 0.002 | 0.0059* ± 0.0000 | 0.038 ± 0.002 | 0.13 ± 0.01 | 0.0024 ± 0.0001 | 2x, 3 |
| Ampicillin | 0.016 | 0.077* ± 0.031 | 0.24 ± 0.11 | 0.046* ± 0.031b | 0.14 ± 0.09b | 0.12 ± 0.05 | 0.022 ± 0.013b | 1a, 2x, 3d |
| Amoxicillin | 0.016 | 0.069 ± 0.029 | 0.17 ± 0.12b | 0.025* ± 0.014 | 0.12 ± 0.05 | 0.078 ± 0.027 | 0.012 ± 0.005 | 2x, 3 |
| Oxacillin | 0.016 | 1.5 ± 0.3 | 1.3 ± 0.2 | 0.038 ± 0.013 | 3.7 ± 0.6 | 1.4 ± 0.3 | 0.071* ± 0.006 | 2x, 3 |
| Piperacillin | 0.016 | 4.8 ± 4.0b | 4.1 ± 3.5b | 0.020 ± 0.008 | 0.94 ± 0.07 | 0.18 ± 0.11b | 0.056* ± 0.000 | 2x, 3d |
| Methicillin | 0.125 | 9.9 ± 0.4 | 4.9 ± 0.5 | 0.083 ± 0.012 | 4.3 ± 0.9 | 2.5 ± 0.4 | 0.16* ± 0.00 | 2x, 3 |
| Amdinocillin | 4 | 73* ± 2 | 48 ± 4 | >1,000 | >1,000 | >1,000 | >1,000 | 1a, 1b |
| 6-APA | 64 | >1,000 | >1,000 | >1,000 | >1,000 | >1,000 | >1,000 | NS |
| Cephalosporins | ||||||||
| Ceftriaxone | 0.008 | 0.15 ± 0.05 | 0.064 ± 0.006 | 0.012* ± 0.002 | 0.21 ± 0.03 | >1,000 | 0.011 ± 0.009b | 2x, 3d |
| Cefuroxime | 0.016 | 0.22 ± 0.01 | 0.10 ± 0.02 | 0.0031 ± 0.0007 | 0.053 ± 0.005 | 64 ± 5 | 0.020 ± 0.000 | 2x |
| Cefotaxime | 0.016 | 0.33 ± 0.01 | 0.22 ± 0.01 | 0.0078* ± 0.0002 | 0.17 ± 0.00 | >1,000 | 0.0031 ± 0.0001 | 2x, 3 |
| Cephalothin | 0.25 | 0.018* ± 0.010 | 0.013* ± 0.005 | 0.043 ± 0.005 | 0.13 ± 0.02 | 1.9 ± 0.2 | 0.0060 ± 0.0002 | 1a, 1b, 3 |
| Cefoxitin | 0.5 | 0.77 ± 0.36 | 0.14 ± 0.04 | 0.81 ± 0.24 | 0.42 ± 0.07 | 1.6 ± 0.5 | 0.0020 ± 0.0005 | 3 |
| Cephalexin | 1 | >1,000 | >1,000 | 0.67 ± 0.06 | >1,000 | 45 ± 2 | 0.035 ± 0.001 | 3 |
| Cefsulodin | 4 | 16 ± 4 | 2.4 ± 0.2 | 1.7 ± 0.6 | 4.3 ± 0.7 | >1,000 | 0.33 ± 0.17 | 3 |
IC50s were generated as described in Materials and Methods using a log(inhibitor)-versus-response-variable slope (four parameters) and least square (ordinary) fit with GraphPad Prism software. Data from two experiments with the standard error are provided. An IC50 of >1,000 was assigned when GraphPad Prism reported an ambiguous number and significant inhibition was not seen at 1,000 μg/ml. Underlining indicates the lowest IC50 for each compound. Asterisks indicate IC50s within 4-fold of the lowest value. ND, not determined (GraphPad Prism reported an ambiguous number and/or an extremely wide confidence interval for IC50 determination [>106] for both data sets).
IC50s obtained from duplicate data sets are ≥4-fold different.
Selectivity assignment was possible due to apparent IC50s of PBP1a and PBP1b (≥4-fold above that of PBP3).
Selectivity assignment is an estimate due to a high standard error value(s) in IC50 determination.
NS, not selective.
To demonstrate that accurate band quantitation could be achieved for PBP1b despite its close migration with the much more prominently labeled PBP1a in the SDS-PAGE gels, we performed titration experiments with amoxicillin, doripenem, and meropenem on IU6647 (IU1945 Δpbp1a::P-erm), an isogenic reconstructed strain of E177 (see Fig. S3 in the supplemental material) (32). The IC50s of these compounds with PBP1b in the Δpbp1a strain are consistent with data obtained from the wild-type organism (average of 2-fold difference [see Table S2 in the supplemental material]).
Cefuroxime is selective for PBP2x, and aztreonam, cefoxitin, cephalexin, and cefsulodin are selective for PBP3.
Representative gel images and associated gel band quantitation graphs are shown in Fig. 2 and 3 and in Fig. S2 and S4 in the supplemental material. IC50s for each compound with every PBP (Table 1) were obtained as described in Materials and Methods. To facilitate analysis, we set criteria of selectivity and coselectivity. A compound was considered selective for a single PBP if its IC50 was at least 4-fold lower than that of the next most inhibited PBP. We used a 4-fold difference to define selectivity because this range is much larger than the standard error of our IC50 determinations, with the majority of the standard error values being less than 25% of the corresponding IC50 (>70% of the compounds). When a second or third PBP fell below this 4-fold threshold, the compound was considered coselective for two or three PBPs. Following these criteria, a trend between compound class and PBP selectivity profiles can be seen (Table 1). Only five molecules demonstrated selective inhibition of a single PBP: cefoxitin, cephalexin, and cefsulodin (cephalosporins) and aztreonam (monobactam) showed selectivity for PBP3, while only cefuroxime (a cephalosporin) selectively blocked PBP2x. Although the IC50s of aztreonam for PBP1a and PBP1b could not be determined in our assay (Table 1), they are clearly substantially higher (>4-fold) than that of PBP3, enabling assignment of this selectivity profile (Fig. 2B and C). Six of the nine penicillin compounds were coselective for PBP2x and PBP3. Finally, both carbapenem compounds (doripenem and meropenem) were coselective for PBP1a, PBP2x, and PBP3, while seven additional cephalosporins showed variability in their PBP-binding profiles. The selective inhibition profiles of PBP3 by aztreonam, cefoxitin, cephalexin, and cefsulodin in our study (Fig. 2 and 3A; also, see Fig. S4S in the supplemental material) are similar to those in previous studies with R6 strains (22, 27).
FIG 2.
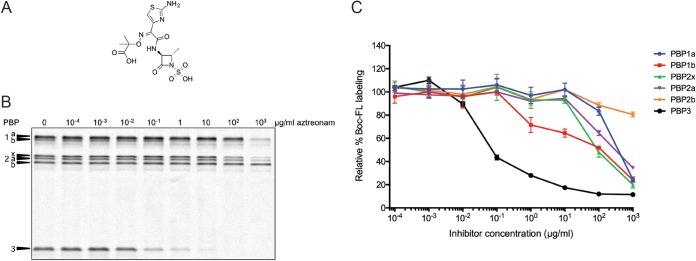
Aztreonam titration of PBPs in S. pneumoniae. Whole cells were treated with various concentrations of aztreonam and subsequently labeled with Boc-FL. (A) Structure of aztreonam; (B) representative SDS-PAGE gel image for aztreonam inhibition of the PBPs in S. pneumoniae over a range of drug concentrations; (C) gel band quantitation for the gel shown in panel A plotted with the standard deviation for each inhibitor concentration obtained from two independent experiments.
FIG 3.
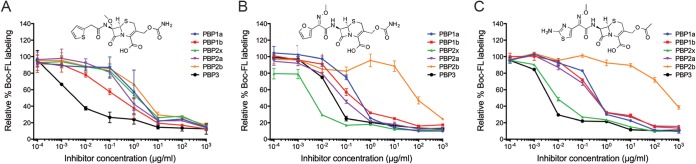
Representative graphs showing the inhibition of PBPs with cefoxitin (A), cefuroxime (B), and cefotaxime (C). Standard deviations for each inhibitor concentration obtained from two independent experiments are plotted. Despite these three molecules' belonging to the same β-lactam class, their selectivity profiles differed substantially, with cefoxitin being selective for PBP3, cefuroxime demonstrating selective inhibition of PBP2x, and cefotaxime inhibiting both proteins (0.01 μg/ml).
We found that cefuroxime, a member of the cephalosporin class, selectively targets PBP2x (Fig. 3B). This compound is a therapeutically relevant antibiotic and was used in a cocrystallization analysis with purified PBP2x (17), although its selectivity profile in S. pneumoniae has not been reported. PBP2x is the functional equivalent of PBP3 in E. coli (1, 6); however, the inhibition profiles obtained using the same assay in these two organisms showed substantial variation between S. pneumoniae PBP2x and E. coli PBP3 (38). One compound, cefuroxime, was found to be selective for both proteins. Four additional compounds demonstrated selective inhibition of E. coli PBP3 (aztreonam, piperacillin, cefotaxime, and ceftriaxone), but three of these molecules (piperacillin, cefotaxime, and ceftriaxone) were coselective for PBP2x and PBP3 in S. pneumoniae, while aztreonam primarily targeted PBP3 in this organism. Intriguingly, not all compounds that exhibited preference for S. pneumoniae PBP2x and PBP3 showed preferential binding of E. coli PBP3 (penicillin V, penicillin G, amoxicillin, methicillin), suggesting that the functional relationship between protein homologs is highly complex.
Methicillin is considered coselective for PBP2x and PBP3 in this study because the IC50s for PBP2x and PBP3 are closer than our 4-fold criteria using the described protocol. In a previous study, we identified an experimental condition under which the addition of 0.1 μg/ml methicillin resulted in an ∼80% reduction in Boc-FL labeling of PBP2x and ∼20% reduction in Boc-FL labeling of the other five PBPs (6). There are several differences in the two protocols used in the previous and the current studies. First, the earlier study was performed on cells growing in BHI at 37°C during methicillin titration, whereas the current investigation was performed in PBS buffer at room temperature. Second, the previous study titrated methicillin in BHI with a bacterial concentration of 2 × 107 CFU/ml, while here, methicillin titration was done in PBS with bacterial cells at a higher concentration of ∼8 × 108 CFU/ml. We performed a parallel experiment using either the BHI or PBS incubation protocol and found IC50 ratios for PBP2x and PBP3 of 6-fold and 3-fold, respectively. As a result, in this work we report methicillin to be coselective for PBP2x and PBP3, while our previous result of methicillin being PBP2x selective under BHI incubation conditions was also confirmed.
The majority of β-lactams exhibit most potent inhibition of PBP3 or PBP2x.
Among the 20 compounds tested, 13 molecules have the lowest IC50 for PBP3 and four compounds have the lowest IC50 for PBP2x. The three exceptions are faropenem, amdinocillin, and 6-APA (which have the lowest IC50 for PBP1a, the lowest IC50 for PBP1b, and poor affinity for all PBPs, respectively). These results are consistent with measured kcat/Km values of the different purified pneumococcal PBPs. Comparison of the catalytic efficiency of a soluble form of PBP3 to those of PBP2a, -2x, -2b, and -1a indicated that PBP3 hydrolyzes a synthetic peptide substrate dramatically faster than any of the other family members (39). This protein was found to be >600 times more efficient than PBP2b, the homolog with the lowest kcat/Km value. Given that β-lactam antibiotic interactions with the PBPs are activity dependent, it could be postulated that the catalytic efficiency of PBP3 contributes to the facile inhibition of this protein. Interestingly, PBP2x was found to be the next most catalytically competent (>30-fold more efficient than PBP2b) (39, 40) and is the only other protein that was specifically inhibited in our studies.
The remainder of the PBPs in S. pneumoniae were inhibited only in combination with one another or in some cases were poorly targeted by the examined antibiotics. We found that PBP1a was inactivated concurrently with PBP1b, PBP2x, and/or PBP3. Overall, none of the examined β-lactams had high affinity for PBP2a or PBP2b in our assay. This was expected, as PBP2a was previously shown to have poor affinity for β-lactam antibiotics in comparison to other PBPs in S. pneumoniae (28). In general, the lowest affinity observed among cephalosporins was for PBP2b. Finally, the penem antibiotic faropenem showed minimal PBP selectivity, which is consistent with data obtained in clinical isolates, with the exception that the affinity of PBP2x for this compound was much more similar to the binding seen with other PBPs in our studies than in previous work (e.g., the difference between the lowest IC50 and IC50PBP2x is 2-fold versus 126-fold) (41).
Cephalothin inhibited PBP1a, PBP1b, and PBP3 in our assay in which the IC50 for PBP3 was 2-fold lower than the IC50 of PBP1a and PBP1b. This antibiotic exhibited maximal potency against PBP1a and PBP1b in previous work performed in the R6 strain (22), which has subsequently been shown to possess mutations that result in decreased PBP1aR6 activity (30, 32). The penicillin structure amdinocillin showed the greatest affinity for PBP1a and PBP1b (<100 μg/ml). In the study by Williamson et al., the IC50s for PBP1a and PBP1b with this drug were significantly higher than those for other PBPs, indicating that these proteins had poor affinity for amdinocillin in their assay (22).
Selectivity profiles of cefotaxime and piperacillin.
Cefotaxime and piperacillin are classified as coselective for PBP2x and PBP3 in this study. The selectivity profiles for each compound were similar when the assays were performed with the current protocol in PBS and with the previous protocol in BHI (6) and are in accordance with inhibition patterns reported by Williamson et al. (22), taking into account that the reported PBP2a inhibition in the previous study corresponded to that of both PBP2x and PBP2a. Our selectivity results are also consistent with later studies by Grebe and Hakenbeck (34) and Laible and Hakenbeck (42). The selectivity profiles of these compounds are of particular interest because PBP2x and PBP2b were described as the primary targets of cefotaxime and piperacillin, respectively, based on reduced affinity to PBP2x or PBP2b seen in a radiolabeled β-lactam binding assay with strains containing pbp2x or pbp2b alleles, which conferred resistance to these molecules (34). We report here that direct inhibition studies confirm previous findings that cefotaxime and piperacillin are coselective for PBP3 and PBP2x and that piperacillin does possess a partial relative affinity for PBP2b in our assay. Importantly, a recent study confirmed this piperacillin inhibition profile and also provided compelling evidence that PBP2b is a critical target of this drug and that combined inhibition of PBP2x and PBP2b may be key to the unusual resistance pattern seen with this molecule (43).
Pneumococcal growth inhibition is correlated with PBP2x inhibition.
PBP2x, PBP2b, and PBP1a are the primary contributors to the development of resistance in S. pneumoniae. PBP2x and PBP2b are known to be essential for S. pneumoniae, and mutations in these PBPs result in low-level resistance in this organism (8). PBP1a is a class A HMW PBP that is not essential individually, but the absence of pbp1a together with pbp2a is lethal to pneumococci. A PBP2x and/or PBP2b mutation combined with that of PBP1a gives rise to high-level resistance (19). Studies previously demonstrated that an altered PBP2x active site leads to reduced β-lactam affinity in resistant clinical isolates and laboratory mutants (8). However, recent evidence showed that alteration in other domains could also affect the β-lactam affinity. PASTA domains were shown to be not only essential for functionality and localization of PBP2x but also important for β-lactam binding. Deletion of these domains from PBP2x results in >90% loss of β-lactam binding (15).
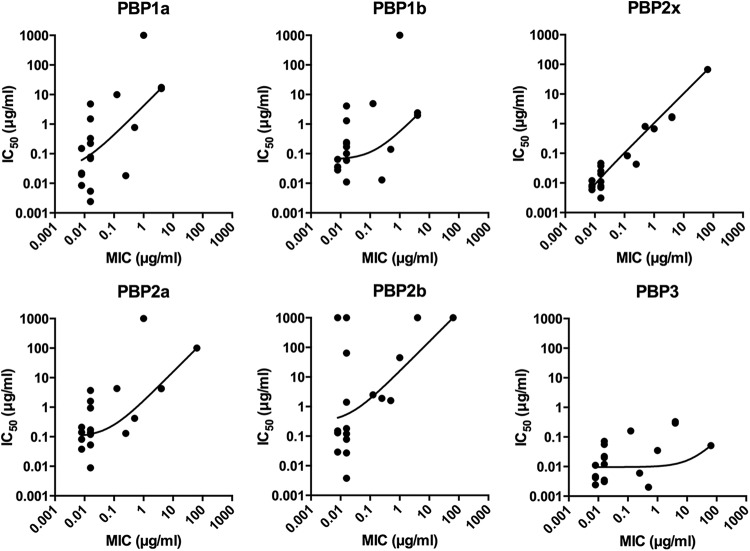
To elucidate the PBP(s) that is most responsible for cell death, we generated scatter plots to relate PBP IC50s and S. pneumoniae MICs (Fig. 4). The MICs of 18 compounds correlated well with the PBP2x IC50s. Furthermore, only PBP2x has a nearly linear correlation between MICs and IC50s, indicating that inhibition of this protein is the primary cause of growth inhibition by the majority of β-lactams (17 out of 20 tested). Previously, it was generally thought that both PBP2x and PBP2b are primary targets for the β-lactams (8). The PBP inhibition profiles for amdinocillin and 6-APA do not correlate with the observed MICs (the MICs are 12- and 16-fold less than the lowest IC50, respectively). As confirmation, the selectivity profiles for amdinocillin and 6-APA were assessed in BHI and found to be similar (see Fig. S5A and D in the supplemental material for growth inhibition and Fig. S5B, C, E, and F for Boc-FL labeling). As noted above, Williamson et al. observed different inhibition profiles with these two molecules in the R6 strain, with the lowest IC50s being for PBP2a (combination of PBP2a and PBP2x) and correlated to the measured MICs (22). Instead, we determined the IC50s for PBP2a, PBP2b, and PBP2x to be >1,000 μg/ml and poorly correlated with the MIC of amdinocillin in the D39 strain (4 μg/ml). We found the IC50s for all PBPs to be >1,000 μg/ml with 6-APA, yet the MIC of this compound is 64 μg/ml, suggesting that although it contains the motif required for PBP inhibition (β-lactam), the target of this compound is not the PBPs. This is consistent with recent studies performed in E. coli that also showed no correlation between the IC50s with 6-APA (all >1,000 μg/ml) and the MIC of this compound (32 μg/ml) (38). The mechanism underlying killing of S. pneumoniae D39 by amdinocillin and 6-APA in the absence of significant PBP inhibition remains to be determined.
FIG 4.
Scatter plots of IC50s of each PBP versus MICs. The data points are plotted in log-log scale and fitted with a straight line and robust fit using GraphPad Prism. Amdinocillin, 6-APA, and the two undetermined IC50s for aztreonam (PBP1a and PBP1b) are not included.
The more dispersed distributions in other PBP plots suggest a weak correlation between individual PBP inhibition and bacterial growth. Thus, we conclude that pneumococcal growth inhibition can, in general, be attributed primarily to the inhibition of PBP2x. This is consistent with the finding that the IC50s for PBP2b are higher than those for PBP2x for all compounds except for faropenem, amdinocillin, and 6-APA. Both PBP2x and PBP2b are essential class B HMW PBPs, but the main distinction between them is the presence of the PASTA domains in PBP2x. Because none of the tested molecules were selective for PBP2b, we hypothesize that the PASTA domains may play a critical role in enhancing antibiotic specificity for PBP2x.
The other PBP that was selectively inhibited by four compounds (aztreonam, cefoxitin, cephalexin, and cefsulodin) from our library was PBP3. Interestingly, the IC50s for PBP3 are much lower than the associated MICs for these compounds, implying that inhibition of PBP3 does not stop growth. This result is consistent with previous data showing that PBP3 is not essential for pneumococcal cell survival (18).
Treatment of S. pneumoniae cells with cefuroxime at a PBP2x-selective concentration results in elongated cells.
We previously reported that treatment of methicillin at a concentration that selectively inhibits PBP2x resulted in elongated cells, suggesting that PBP2x plays a role in septal ring closure (6). This function of PBP2x is consistent with the observation by superresolution microscopy that PBP2x is directed to a discrete location within the septal aperture during the later stages of cell division (6, 12). In this study, we treated exponentially growing IU1945 cells with 0.006 μg/ml of cefuroxime, a concentration that is below the MIC and two times higher than the IC50 of PBP2x. This concentration inhibits 60% of PBP2x activity and less than 20% of the activity of other PBPs. Starting at 30 min after cefuroxime addition, about 30% of the cells appeared elongated (see Fig. S6 in the supplemental material). This result further confirms that inhibition of PBP2x results in the inability for the cells to form division septa.
PBP3, also known as DacA, does not contain a transpeptidase domain shared by the other five pneumococcal PBPs. It is a d,d-carboxypeptidase that cleaves the ultimate d-Ala from peptidoglycan peptides. Δpbp3 strains are viable but show severe growth defects and have been characterized as having decreased growth and yield rounded cell shape, division asymmetry, and an aberrant fluorescent vancomycin (FL-V) staining pattern (18). To investigate whether inhibition of PBP3 activity would result in the ΔdacA phenotype, we examined S. pneumoniae cells treated with the PBP3-specific compounds aztreonam at concentrations of 0.2 to 1 μg/ml or cefoxitin at concentrations of 0.008 to 0.04 μg/ml. At 1 μg/ml of aztreonam and 0.04 μg/ml of cefoxitin, 70% of PBP3 activity and less than 30% of other PBP activities were inhibited. No alteration of growth rate, growth yield, or cell morphology was observed for up to 4.5 h of treatment. These results suggest that PBP3 activity is robust in a wild-type pbp3+ genetic background, and a reduction of 70% activity does not produce any growth defect. Alternatively, Δpbp3 phenotypes can be mainly due to other nonenzymatic functions of PBP3. The absence of morphological changes in S. pneumoniae associated with aztreonam treatment is in contrast with filament formation of E. coli upon treatment with aztreonam at 0.2 μg/ml, which specifically inhibits PBP3 of E. coli (44).
In this study, we have reevaluated PBP selectivity in an unencapsulated derivative of the D39 strain of S. pneumoniae for a variety of β-lactam antibiotics using Boc-FL labeling as the readout method. The specificity data reported here will be valuable for interpreting β-lactam sensitivity patterns and are the foundation for designing future PBP-specific β-lactam probes by conjugation of selective antibiotics with readout tags such as fluorophores. These fluorescent probes will be important tools for correlating individual PBPs with physiological and microscopic studies. Because preliminary genetic modification is not required and these probes mimic the natural substrate of PBPs, they can be utilized to visualize tagged PBPs in their native environment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karen Bush for helpful discussions and careful reading of the manuscript. We also thank the Indiana University Physical Biochemistry Instrumentation Facility for use of a fluorescence gel scanner.
This work was supported by NIH DP2OD008592 (E.E.C.), a Pew Biomedical Scholar Award (E.E.C.), a Sloan Research Fellow Award (E.E.C.), start-up funds and a Dean's Fellow Award from the Indiana University-Bloomington, Department of Chemistry, and NIH grants AI107075 (M.E.W.) and GM113172 (M.E.W.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.05142-14.
REFERENCES
- 1.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 2.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev 30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 3.Donkor ES. 2013. Understanding the pneumococcus: transmission and evolution. Front Cell Infect Microbiol 3:7. doi: 10.3389/fcimb.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henriques-Normark B, Tuomanen EI. 2013. The pneumococcus: epidemiology, microbiology, and pathogenesis. Cold Spring Harb Perspect Med 3:a010215. doi: 10.1101/cshperspect.a010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sham LT, Tsui HC, Land AD, Barendt SM, Winkler ME. 2012. Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Curr Opin Microbiol 15:194–203. doi: 10.1016/j.mib.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Land AD, Tsui HC, Kocaoglu O, Vella SA, Shaw SL, Keen SK, Sham LT, Carlson EE, Winkler ME. 2013. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol Microbiol 90:939–955. doi: 10.1111/mmi.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massidda O, Novakova L, Vollmer W. 2013. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division? Environ Microbiol 15:3133–3157. doi: 10.1111/1462-2920.12189. [DOI] [PubMed] [Google Scholar]
- 8.Hakenbeck R, Bruckner R, Denapaite D, Maurer P. 2012. Molecular mechanisms of beta-lactam resistance in Streptococcus pneumoniae. Future Microbiol 7:395–410. doi: 10.2217/fmb.12.2. [DOI] [PubMed] [Google Scholar]
- 9.Yeats C, Finn RD, Bateman A. 2002. The PASTA domain: a beta-lactam-binding domain. Trends Biochem Sci 27:438. doi: 10.1016/S0968-0004(02)02164-3. [DOI] [PubMed] [Google Scholar]
- 10.Berg KH, Stamsas GA, Straume D, Havarstein LS. 2013. Effects of low PBP2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. J Bacteriol 195:4342–4354. doi: 10.1128/JB.00184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters K, Schweizer I, Beilharz K, Stahlmann C, Veening JW, Hakenbeck R, Denapaite D. 2014. Streptococcus pneumoniae PBP2x mid-cell localization requires the C-terminal PASTA domains and is essential for cell shape maintenance. Mol Microbiol 92:733–755. doi: 10.1111/mmi.12588. [DOI] [PubMed] [Google Scholar]
- 12.Tsui HC, Boersma MJ, Vella SA, Kocaoglu O, Kuru E, Peceny JK, Carlson EE, VanNieuwenhze MS, Brun YV, Shaw SL, Winkler ME. 2014. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol Microbiol 94:21–40. doi: 10.1111/mmi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philippe J, Vernet T, Zapun A. 2014. The elongation of ovococci. Microb Drug Resist 20:215–221. doi: 10.1089/mdr.2014.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagai K, Davies TA, Jacobs MR, Appelbaum PC. 2002. Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob Agents Chemother 46:1273–1280. doi: 10.1128/AAC.46.5.1273-1280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurer P, Todorova K, Sauerbier J, Hakenbeck R. 2012. Mutations in Streptococcus pneumoniae penicillin-binding protein 2x: importance of the C-terminal penicillin-binding protein and serine/threonine kinase-associated domains for beta-lactam binding. Microb Drug Resist 18:314–321. doi: 10.1089/mdr.2012.0022. [DOI] [PubMed] [Google Scholar]
- 16.Schweizer I, Peters K, Stahlmann C, Hakenbeck R, Denapaite D. 2014. Penicillin-binding protein 2x of Streptococcus pneumoniae: the mutation Ala707Asp within the C-terminal PASTA2 domain leads to destabilization. Microb Drug Resist 20:250–257. doi: 10.1089/mdr.2014.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon E, Mouz N, Duee E, Dideberg O. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J Mol Biol 299:477–485. doi: 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 18.Barendt SM, Land AD, Sham LT, Ng WL, Tsui HC, Arnold RJ, Winkler ME. 2009. Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J Bacteriol 191:3024–3040. doi: 10.1128/JB.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paik J, Kern I, Lurz R, Hakenbeck R. 1999. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J Bacteriol 181:3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spratt BG, Pardee AB. 1975. Penicillin-binding proteins and cell shape in E. coli. Nature 254:516–517. doi: 10.1038/254516a0. [DOI] [PubMed] [Google Scholar]
- 21.Curtis NA, Orr D, Ross GW, Boulton MG. 1979. Affinities of penicillins and cephalosporins for the penicillin-binding proteins of Escherichia coli K-12 and their antibacterial activity. Antimicrob Agents Chemother 16:533–539. doi: 10.1128/AAC.16.5.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williamson R, Hakenbeck R, Tomasz A. 1980. In vivo interaction of beta-lactam antibiotics with the penicillin-binding proteins of Streptococcus pneumoniae. Antimicrob Agents Chemother 18:629–637. doi: 10.1128/AAC.18.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dargis M, Malouin F. 1994. Use of biotinylated beta-lactams and chemiluminescence for study and purification of penicillin-binding proteins in bacteria. Antimicrob Agents Chemother 38:973–980. doi: 10.1128/AAC.38.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. 1999. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother 43:1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocaoglu O, Calvo RA, Sham LT, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, Carlson EE. 2012. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem Biol 7:1746–1753. doi: 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kocaoglu O, Carlson EE. 2013. Penicillin-binding protein imaging probes. Curr Protoc Chem Biol 5:239–250. doi: 10.1002/9780470559277.ch130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hakenbeck R, Tornette S, Adkinson NF. 1987. Interaction of non-lytic beta-lactams with penicillin-binding proteins in Streptococcus pneumoniae. J Gen Microbiol 133:755–760. [DOI] [PubMed] [Google Scholar]
- 28.Zhao G, Meier TI, Hoskins J, McAllister KA. 2000. Identification and characterization of the penicillin-binding protein 2a of Streptococcus pneumoniae and its possible role in resistance to beta-lactam antibiotics. Antimicrob Agents Chemother 44:1745–1748. doi: 10.1128/AAC.44.6.1745-1748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvalho SM, Kuipers OP, Neves AR. 2013. Environmental and nutritional factors that affect growth and metabolism of the pneumococcal serotype 2 strain D39 and its nonencapsulated derivative strain R6. PLoS One 8:e58492. doi: 10.1371/journal.pone.0058492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 32.Land AD, Winkler ME. 2011. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J Bacteriol 193:4166–4179. doi: 10.1128/JB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hakenbeck R, Tarpay M, Tomasz A. 1980. Multiple changes of penicillin-binding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 17:364–371. doi: 10.1128/AAC.17.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grebe T, Hakenbeck R. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of beta-lactam antibiotics. Antimicrob Agents Chemother 40:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 36.Waxman DJ, Strominger JL. 1983. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu Rev Biochem 52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 37.Ghuysen JM. 1994. Molecular structures of penicillin-binding proteins and beta-lactamases. Trends Microbiol 2:372–380. doi: 10.1016/0966-842X(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 38.Kocaoglu O, Carlson EE. 2 March 2015. Profiling of β-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob Agents Chemother 59:2785–2790. doi: 10.1128/AAC.04552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morlot C, Pernot L, Le Gouellec A, Di Guilmi AM, Vernet T, Dideberg O, Dessen A. 2005. Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae. J Biol Chem 280:15984–15991. doi: 10.1074/jbc.M408446200. [DOI] [PubMed] [Google Scholar]
- 40.Mouz N, Gordon E, Di Guilmi AM, Petit I, Petillot Y, Dupont Y, Hakenbeck R, Vernet T, Dideberg O. 1998. Identification of a structural determinant for resistance to beta-lactam antibiotics in Gram-positive bacteria. Proc Natl Acad Sci U S A 95:13403–13406. doi: 10.1073/pnas.95.23.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosowska-Shick K, McGhee P, Appelbaum PC. 2009. Binding of faropenem and other beta-lactam agents to penicillin-binding proteins of pneumococci with various beta-lactam susceptibilities. Antimicrob Agents Chemother 53:2176–2180. doi: 10.1128/AAC.01566-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laible G, Hakenbeck R. 1987. Penicillin-binding proteins in beta-lactam-resistant laboratory mutants of Streptococcus pneumoniae. Mol Microbiol 1:355–363. doi: 10.1111/j.1365-2958.1987.tb01942.x. [DOI] [PubMed] [Google Scholar]
- 43.Philippe J, Gallet B, Morlot C, Denapaite D, Hakenbeck R, Chen Y, Vernet T, Zapun A. 2015. Mechanism of β-lactam action in Streptococcus pneumoniae: the piperacillin paradox. Antimicrob Agents Chemother 59:609–621. doi: 10.1128/AAC.04283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georgopapadakou NH, Smith SA, Sykes RB. 1982. Mode of action of azthreonam. Antimicrob Agents Chemother 21:950–956. doi: 10.1128/AAC.21.6.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.