Abstract
Topical mupirocin is used widely to treat skin and soft tissue infections and to eradicate nasal carriage of methicillin-resistant Staphylococcus aureus (MRSA). Few studies to date have characterized the rates of S. aureus mupirocin resistance in pediatric populations. We retrospectively studied 358 unique S. aureus isolates obtained from 249 children seen in a predominantly outpatient setting by the Division of Pediatric Dermatology at a major academic center in New York City between 1 May 2012 and 17 September 2013. Mupirocin resistance rates and the associated risk factors were determined using a logistic regression analysis. In our patient population, 19.3% of patients had mupirocin-resistant S. aureus isolates at the time of their first culture, and 22.1% of patients with S. aureus infection had a mupirocin-resistant isolate at some time during the study period. Overall, 31.3% of all S. aureus isolates collected during the study period were resistant to mupirocin. Prior mupirocin use was strongly correlated (odds ratio [OR] = 26.5; P = <0.001) with mupirocin resistance. Additional risk factors for mupirocin resistance included methicillin resistance, atopic dermatitis (AD), epidermolysis bullosa (EB), immunosuppression, and residence in northern Manhattan and the Bronx. Resistance to mupirocin is widespread in children with dermatologic complaints in the New York City area, and given the strong association with mupirocin exposure, it is likely that mupirocin use contributes to the increased resistance. Routine mupirocin testing may be important for MRSA decolonization strategies or the treatment of minor skin infections in children.
INTRODUCTION
Mupirocin is a topical antibiotic widely used to treat skin and soft tissue infections and to eliminate nasal carriage of methicillin-resistant Staphylococcus aureus (MRSA) (1). Mupirocin was introduced into clinical practice in 1985, with mupirocin-resistant S. aureus (MupRSA) first reported in 1987 (2, 3). Resistance is classified into two categories: low-level resistance, with MICs ranging from 8 to 256 μg/ml, and high-level resistance, with MICs of ≥512 μg/ml. High-level resistance is in most cases conferred by the plasmid-borne gene mupA, which produces a “eukaryotic-like” tRNA synthetase with no affinity for mupirocin (4). A related gene, mupB, has also been shown to confer high-level resistance (5). Carriage of a high-level-resistant MupRSA strain has been shown to predict decolonization failure after treatment with mupirocin (6, 7). Low-level mupirocin resistance is due to point mutations in the native isoleucyl-tRNA synthetase gene (ileS), most commonly V588F (8), and may be associated with higher rates of recolonization after efforts to eradicate S. aureus carriage (9).
Mupirocin susceptibility often is not tested as part of routine clinical care because high-level mupirocin resistance has been reported to be relatively rare, ranging from 1% to 5% of MRSA isolates from hospitalized adult populations in North America and Europe (9–12). However, prevalences of 13% of MRSA isolates (13) and 45% of S. aureus isolates have been reported in single-center studies (9, 14). In the few studies that have examined rates of mupirocin resistance in children, the prevalence has ranged from 2% to 15% (15–17).
Several studies have linked mupirocin resistance to mupirocin use. A 20-year analysis of MRSA blood culture isolates in Europe found mupirocin resistance to be associated with increased use (18). In another study, decreased clinical usage of mupirocin over 5 years at a French hospital mirrored a decrease in resistance rates (19). A case-control study by Caffrey et al. revealed a strong association between previous mupirocin exposure and subsequent mupirocin resistance in MRSA (20).
Mupirocin resistance may also aid in the spread of multidrug resistance through coselection with other resistance genes. For instance, high rates of clindamycin resistance have been observed in MupRSA isolates (7, 16). Cadilla et al. found an association between strains of mupirocin-resistant MRSA and resistance to three or more non-beta-lactam antimicrobial classes (21). Mupirocin-resistant MRSA strains isolated from several hospitals in Korea were also resistant to ciprofloxacin, clindamycin, and tetracycline (22).
To determine the prevalence of mupirocin resistance and its associated risk factors in our pediatric population, we retrospectively reviewed all skin cultures from the Division of Pediatric Dermatology that were positive for S. aureus at our center over a 16-month period.
MATERIALS AND METHODS
Following institutional review board approval, the computer database of the Clinical Microbiology Laboratory at Columbia University Medical Center was queried for all culture results positive for S. aureus and tested for susceptibility to mupirocin during the 16-month period from 1 May 2012 to 17 September 2013. In our pediatric dermatology practice, mupirocin sensitivity testing of S. aureus isolates is routinely requested, and 91.4% of S. aureus cultures obtained during the study period were tested for mupirocin susceptibility. Outside our dermatology clinic, mupirocin susceptibility testing was not routine, which limited our study population to children seen by pediatric dermatologists.
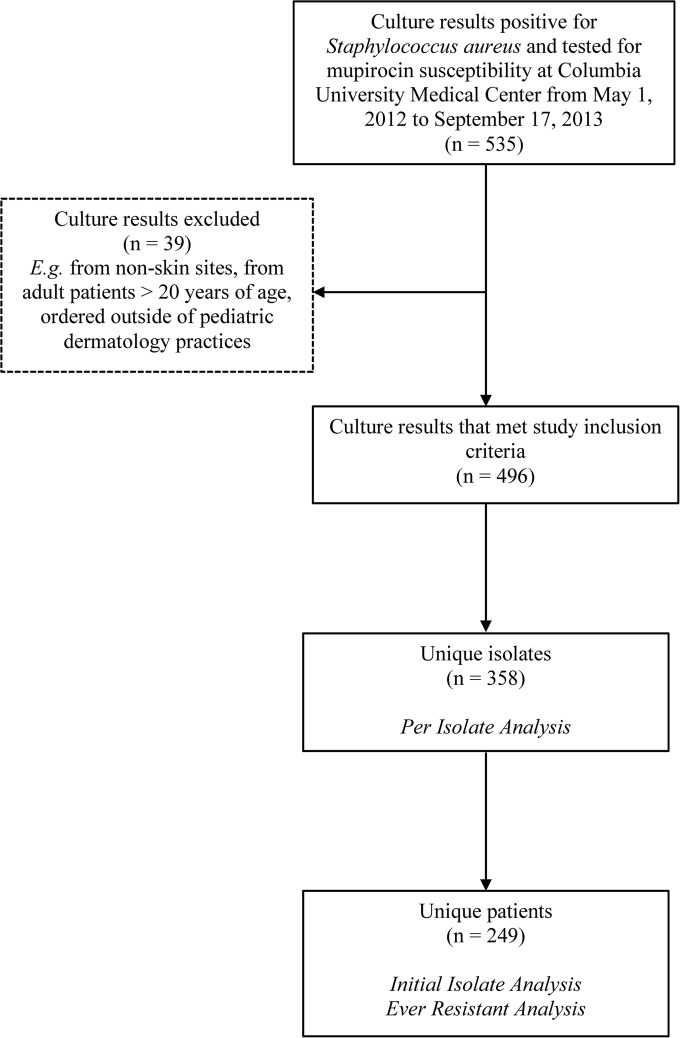
A list of 535 individual culture results (isolates) was generated. We excluded culture results from nonskin sites (e.g., blood culture) or adults (>20 years old) and cultures not sent by physicians from our division. Isolates were obtained from sites of suspected infection and colonization. Thirty-nine culture results were excluded, resulting in 496 specimens from 249 patients (Fig. 1). Isolates obtained from the same patient on the same date with equivalent antibiotic susceptibility profiles (beta-lactams, clindamycin, daptomycin, erythromycin, levofloxacin, linezolid, trimethoprim-sulfamethoxazole, rifampin, tetracycline, vancomycin, and mupirocin) were assumed to be the same isolate. Isolates from different dates and/or isolates showing variability in susceptibility to one or more of the routinely tested antibiotics were regarded, for our analyses, as different or unique isolates. The 496 skin culture isolates were grouped together by these criteria, resulting in 358 unique isolates from 249 patients. Fifty-seven patients had more than one unique isolate identified at the time of initial culture or on different visit dates; 35 patients had 2 unique isolates, and 22 patients had 3 or more unique isolates. Forty-two patients were cultured on more than one visit date.
FIG 1.
Flowchart summarizing the study design.
The clinical records of all patients whose cultures met the inclusion criteria were reviewed for demographic and clinical information, including patient age, gender, zip code, primary dermatologic diagnosis, additional dermatologic diagnoses if applicable, immunosuppression, visit status, and documented history and timing of mupirocin use. Among the subset of immunosuppressed patients, conditions included solid-organ transplant, leukemia, graft-versus-host disease, severe atopic dermatitis (AD), severe granulomatous colitis, and primary immune deficiency; 7 patients were receiving therapy with a cytotoxic agent (e.g., mycophenolate mofetil, cyclosporine, tacrolimus, sirolimus, or mercaptopurine) and/or systemic steroids. Cultures obtained from patients in the emergency department were considered outpatient. Patient zip codes were grouped into five geographic regions: Mid- and Lower Manhattan (10002 to 10025; 10065 to 10282); Brooklyn, Queens, Long Island, and Staten Island (10306 to 10309; 11040 to 11787); Upper Manhattan and the Bronx (10026 to 10040; 10451 to 10472); Connecticut and Northern New York State (06831 to 06870; 10512 to 10977; 12508 to 12589); and New Jersey (07003 to 08854). Three zip codes were outside this area and were excluded from the statistical analysis of geography. Groups were selected based on borough and state boundaries; however, Upper Manhattan was included with the Bronx due to prior work suggesting these areas may be demographically and epidemiologically linked (23). Primary dermatologic diagnoses, defined as the reason the culture was sent and determined by chart review, were classified into six categories: atopic dermatitis (group 1); dermatitis not otherwise specified (NOS) (group 2); impetigo (group 3); folliculitis, pustulosis, furunculosis, or abscess (group 4); epidermolysis bullosa (EB) (group 5); and other (group 6). Group 6 included other diagnoses, such as molluscum, skin ulcer, erythema, and paronychia.
Culture result details were recorded, including body site(s), quantity of bacterial growth (classified as few, moderate, or many), presence of additional bacterial species other than S. aureus on the culture result (additional strains), susceptibility to a panel of routinely tested antibiotics, and mupirocin susceptibility, including the quantitative MIC. Body sites were grouped into five broad categories: head, nares, trunk, extremity (shoulders, arms, hands, hips, buttocks, legs, and feet), and skin fold (neck, axilla, perianal, inguinal and genital areas, umbilicus, popliteal fossa, and antecubital fossa). A unique isolate may have been obtained from multiple body sites from a patient during a clinic visit, and as such, body site categories were not mutually exclusive in our analysis. Mupirocin resistance was characterized as a binary outcome variable, with a MIC of <8 μg/ml regarded as susceptible and a MIC of ≥8 μg/ml as resistant.
We organized the data in three different ways for statistical analysis. First, the “per-isolate” analysis included all 358 unique isolates. In this analysis, each patient may have been represented more than once due to inclusion of isolates from more than one visit. Second, the “initial-isolate” analysis included a single isolate from the first culture date collected from each patient during the study time period. Third, the “ever-resistant” analysis included a single isolate for each patient on the visit when mupirocin resistance was first, if ever, recorded or from the initial culture date if mupirocin resistance was never recorded. The initial-isolate and ever-resistant analyses included 249 separate observations, 1 for each individual patient.
For all analyses, both simple and multiple logistic regression models were fitted. Logistic regression models for per-isolate analysis included a correction for the effect of subjects having multiple samples. Briefly, if the correlation between the isolates from the same patient is r and that patient has m isolates, then the effective sample size from that patient that contributes to the final estimates is m/(1 + r). To perform variable selection for the multiple logistic regression model, a backward elimination approach was used, where 0.1 was the threshold for keeping a variable in the model. Statistical analysis was performed using SAS 9.3 and R.
RESULTS
A total of 358 unique isolates and 249 individual patients were included in this retrospective study, with an average initial age of 5.4 (range, 0 to 19.2) years; 53.8% were male, and 46.2% were female, with the largest percentage of patients residing in Upper Manhattan and the Bronx (30.1% of patients) (see Table S1 in the supplemental material).
At the time of initial culture, 35.3% of the patients with S. aureus infection had a documented clinical history of prior mupirocin use. A large proportion (55.4%) of patients carried a diagnosis of atopic dermatitis. The most common primary dermatologic diagnosis (i.e., the clinical reason a skin culture was obtained) on the day of culture was atopic dermatitis, accounting for 36.6% of patients at initial culture, followed by dermatitis NOS (16.5%), impetigo (14.9%), folliculitis/pustulosis and abscess (14.1%), and EB (6.4%). The majority of initial cultures were performed in the outpatient setting (96.0%) in a non-immune-suppressed (96.4%) group of patients (see Table S1 in the supplemental material).
For all 358 unique isolates, the overall prevalence of mupirocin resistance was 31.3%. The majority of isolates (96 of 112; 85.7%) carried high-level resistance, with MICs of ≥1,024 μg/ml. Sixteen isolates (14.3%) were classified as having low-level resistance, with MICs ranging from 8 to 64 μg/ml. Two hundred ninety-three isolates (81.8%) were methicillin-susceptible S. aureus (MSSA), and 65 (18.2%) were MRSA, in line with MRSA rates from other pediatric populations (24, 25); 19.3% of patients had a mupirocin-resistant isolate on their first culture in the study period, and 22.1% had at least one culture that was mupirocin resistant over the study period.
The results of univariate logistic regression of the per-isolate, initial-isolate, and ever-resistant analyses identified many of the same risk factors, including prior mupirocin use, atopic dermatitis, immunosuppression, and MRSA, among which only a history of prior mupirocin use was found to be a significant risk factor for all three in multivariate analysis (Tables 1 and 2; see Table S1 in the supplemental material). However, atopic dermatitis, immunosuppression, and inpatient status were found to be significant in 2 out of 3 multivariate analyses.
TABLE 1.
Per-isolate analysis
| Parameter | Total [n (%)] | No. (%) mupirocin susceptible (n = 246)a | No. (%) mupirocin resistant (n = 112) | Univariate OR (95% CIb) | P value | Multivariate ORc (95% CI) | P value |
|---|---|---|---|---|---|---|---|
| Demographic factorsa | |||||||
| Female sex | 170 (47.49) | 109 (44.31) | 61 (54.46) | 1.503 (0.727–3.111) | 0.272 | ||
| Additional dermatologic diagnosis | 126 (35.20) | 82 (33.33) | 44 (39.29) | 1.294 (0.661–2.533) | 0.452 | ||
| Atopic dermatitis | 225 (62.85) | 139 (56.50) | 86 (76.79) | 2.546 (1.274–5.088) | 0.008 | 4.906 (1.435–16.778) | 0.011 |
| Immunosuppression | 30 (8.40) | 8 (3.27) | 22 (19.64) | 7.242 (2.899–18.089) | <0.001 | 7.166 (2.613–19.651) | <0.001 |
| History of mupirocin use | 185 (51.68) | 81 (32.93) | 104 (92.86) | 26.482 (11.197–62.631) | <0.001 | 29.128 (10.819–78.421) | <0.001 |
| Outpatient | 339 (94.69) | 237 (96.34) | 102 (91.07) | 0.387 (0.156–0.961) | 0.041 | ||
| Zip code group | |||||||
| Mid- and Lower Manhattan (ZCG 1) | 60 (16.76) | 52 (21.14) | 8 (7.14) | 1 | |||
| Brooklyn, Queens, Long Island, and Staten Island (ZCG 2) | 69 (19.27) | 40 (16.26) | 29 (25.89) | 4.713 (1.073–20.701) | 0.04 | ||
| Upper Manhattan and the Bronx (ZCG 3) | 124 (34.64) | 75 (30.49) | 49 (43.75) | 4.247 (1.158–15.579) | 0.029 | ||
| Connecticut and northern New York State (ZCG 4) | 47 (13.13) | 35 (14.23) | 12 (10.71) | 2.229 (0.557–8.916) | 0.257 | ||
| New Jersey (ZCG 5) | 55 (15.36) | 41 (16.67) | 14 (12.50) | 2.220 (0.543–9.068) | 0.267 | ||
| Primary diagnosis | |||||||
| Atopic dermatitis (group 1) | 144 (40.22) | 88 (35.77) | 56 (50.00) | 1 | 1 | ||
| Dermatitis NOS (group 2) | 46 (12.85) | 43 (17.48) | 3 (2.68) | 0.110 (0.023–0.529) | 0.006 | 0.283 (0.067–1.206) | 0.088 |
| Impetigo (group 3) | 48 (13.41) | 36 (14.63) | 12 (10.71) | 0.524 (0.237–1.159) | 0.111 | 1.059 (0.295–3.795) | 0.93 |
| Folliculitis, pustulosis, abscess (group 4) | 58 (16.20) | 38 (15.45) | 20 (17.68) | 0.827 (0.372–1.837) | 0.641 | 0.576 (0.204–1.625) | 0.298 |
| Epidermolysis bullosa (group 5) | 28 (7.82) | 10 (4.07) | 18 (16.07) | 2.829 (1.051–7.611) | 0.04 | 5.567 (1.088–28.482) | 0.039 |
| Other (group 6) | 34 (9.50) | 31 (12.60) | 3 (2.68) | 0.152 (0.046–0.505) | 0.002 | 0.376 (0.095–1.493) | 0.164 |
| Culture site | |||||||
| Head | 94 (26.40) | 69 (28.05) | 25 (22.73) | 0.755 (0.432–1.318) | 0.322 | ||
| Nares | 49 (13.76) | 27 (10.98) | 22 (20.00) | 2.028 (1.098–3.744) | 0.024 | ||
| Trunk | 30 (8.43) | 19 (7.72) | 11 (10.00) | 1.328 (0.602–2.928) | 0.483 | ||
| Extremity | 165 (46.35) | 103 (41.87) | 62 (56.36) | 1.793 (1.129–2.848) | 0.013 | ||
| Skin fold | 112 (31.46) | 80 (32.52) | 32 (29.09) | 0.851 (0.501–1.446) | 0.552 | ||
| Unspecified | 6 (1.69) | 5 (2.03) | 1 (0.91) | 0.442 (0.050–3.898) | 0.462 | ||
| Strain characteristics | |||||||
| MRSA | 65 (18.16) | 29 (11.79) | 36 (32.14) | 3.545 (1.576–7.970) | 0.002 | ||
| Additional strain(s) | 41 (11.45) | 28 (11.38) | 13 (11.61) | 1.022 (0.441–2.369) | 0.959 | ||
| Antibiotic resistance | |||||||
| Amoxicillin | 3 (1.02) | 1 (0.46) | 2 (2.60) | 5.760 (0.524–63.367) | 0.152 | ||
| Clindamycin | 90 (25.49) | 60 (24.80) | 30 (27.03) | 1.124 (0.625–2.020) | 0.697 | ||
| Erythromycin | 153 (42.86) | 97 (39.43) | 56 (50.45) | 1.564 (0.818–2.991) | 0.176 | 2.054 (0.938–4.499) | 0.072 |
| Levofloxacin | 51 (14.24) | 22 (8.95) | 29 (25.90) | 3.558 (1.433–8.833) | 0.006 | ||
| Oxacillin | 65 (18.16) | 29 (11.79) | 36 (32.14) | 3.545 (1.576–7.970) | 0.002 | ||
| Rifampin | 6 (1.68) | 3 (1.22) | 3 (2.68) | 2.229 (0.498–9.989) | 0.295 | ||
| Trimethoprim-sulfamethoxazole | 4 (1.12) | 1 (0.41) | 3 (2.68) | 6.743 (0.576–78.942) | 0.128 | ||
| Tetracycline | 44 (12.29) | 30 (12.20) | 14 (12.50) | 1.029 (0.300–3.527) | 0.964 | ||
| Amt of bacterial growth | |||||||
| Few | 98 (28.32) | 80 (33.33) | 18 (16.98) | 1 | 1 | ||
| Moderate | 157 (45.38) | 102 (42.50) | 55 (51.89) | 2.397 (1.334–4.304) | 0.003 | 3.327 (1.486–7.446) | 0.004 |
| Many | 91 (26.30) | 58 (24.17) | 33 (31.13) | 2.529 (1.190–5.376) | 0.016 | 2.271 (0.932–5.534) | 0.071 |
The mean ages were 5.00 years (mupirocin-susceptible group) and 8.21 years (mupirocin-resistant group); univariate OR (95% CI), 1.136 (1.066–1.210); P < 0.001.
CI, confidence interval.
For the multivariate analysis, variables trimmed from the backward model selection are excluded, and only variables selected for the final model are shown.
TABLE 2.
Ever-resistant analysis
| Parameter | Total [n (%)] | No. (%) mupirocin susceptible (n = 194) | No. (%) mupirocin resistant (n = 55) | Univariate OR (95% CIb) | P value | Multivariate ORc (95% CI) | P value |
|---|---|---|---|---|---|---|---|
| Demographic factorsa | |||||||
| Female sex | 115 (46.18) | 88 (45.36) | 27 (49.09) | 1.162 (0.638–2.115) | 0.624 | ||
| Additional dermatologic diagnosis | 79 (31.73) | 60 (30.93) | 19 (34.55) | 1.179 (0.625–2.221) | 0.611 | ||
| Atopic dermatitis | 138 (55.42) | 100 (51.55) | 38 (69.09) | 2.101 (1.111–3.975) | 0.022 | 11.454 (1.986–66.066) | 0.006 |
| Immunosuppression | 9 (3.63) | 4 (2.07) | 5 (9.09) | 4.725 (1.223–18.249) | 0.024 | 17.142 (1.302–225.727) | 0.031 |
| History of mupirocin use | 89 (35.74) | 41 (21.13) | 48 (87.27) | 25.589 (10.778–60.752) | <0.001 | 51.571 (13.720–193.841) | <0.001 |
| Outpatient | 238 (95.58) | 188 (96.91) | 50 (90.91) | 0.319 (0.094–1.089) | 0.068 | 0.087 (0.011–0.663) | 0.018 |
| Zip code group | |||||||
| Mid- and Lower Manhattan (ZCG 1) | 51 (20.48) | 47 (24.23) | 4 (7.27) | 1 | 1 | ||
| Brooklyn, Queens, Long Island, and Staten Island (ZCG 2) | 42 (16.87) | 32 (16.49) | 10 (18.18) | 3.672 (1.059–12.733) | 0.04 | 1.277 (0.218–7.490) | 0.787 |
| Upper Manhattan and the Bronx (ZCG 3) | 75 (30.12) | 53 (27.32) | 22 (40.00) | 4.877 (1.567–15.181) | 0.006 | 7.971 (1.555–40.851) | 0.013 |
| Connecticut and Northern New York State (ZCG 4) | 37 (14.86) | 27 (13.92) | 10 (18.18) | 4.352 (1.244–15.226) | 0.021 | 4.201 (0.756–23.343) | 0.101 |
| New Jersey (ZCG 5) | 41 (16.47) | 32 (16.49) | 9 (16.36) | 3.305 (0.937–11.657) | 0.063 | 3.065 (0.494–19.030) | 0.229 |
| Primary diagnosis | |||||||
| Atopic dermatitis (group 1) | 92 (36.95) | 66 (34.02) | 26 (47.27) | 1 | 1 | ||
| Dermatitis NOS (group 2) | 42 (16.87) | 40 (20.62) | 2 (3.64) | 0.127 (0.029–0.564) | 0.007 | 0.506 (0.049–5.201) | 0.567 |
| Impetigo (group 3) | 36 (14.46) | 30 (15.46) | 6 (10.91) | 0.508 (0.189–1.362) | 0.178 | 1.844 (0.336–10.115) | 0.481 |
| Folliculitis, pustulosis, abscess (group 4) | 34 (13.65) | 26 (13.40) | 8 (14.55) | 0.781 (0.313–1.947) | 0.596 | 1.035 (0.216–4.961) | 0.966 |
| Epidermolysis bullosa (group 5) | 16 (6.43) | 5 (2.58) | 11 (20.00) | 5.585 (1.768–17.645) | 0.003 | 13.037 (1.468–115.739) | 0.021 |
| Other (group 6) | 29 (11.65) | 27 (13.92) | 2 (3.64) | 0.188 (0.042–0.848) | 0.03 | 0.170 (0.010–2.849) | 0.218 |
| Culture site | |||||||
| Head | 69 (27.71) | 58 (29.90) | 11 (20.00) | 0.586 (0.283–1.215) | 0.151 | ||
| Nares | 36 (14.46) | 22 (11.34) | 14 (25.45) | 2.670 (1.259–5.661) | 0.01 | ||
| Trunk | 19 (7.63) | 14 (7.22) | 5 (9.09) | 1.286(0.442–3.741) | 0.645 | ||
| Extremity | 106 (42.57) | 74 (38.14) | 32 (58.18) | 2.256 (1.227–4.149) | 0.009 | 2.782 (0.967–8.003) | 0.058 |
| Skin fold | 90 (36.14) | 69 (35.57) | 21 (38.18) | 1.119 (0.603–2.077) | 0.722 | ||
| Unspecified | 5 (2.01) | 4 (2.06) | 1 (1.82) | 0.880 (0.096–8.035) | 0.91 | ||
| Strain characteristics | |||||||
| MRSA | 36 | 21 (10.82) | 15 (27.27) | 3.089 (1.464–6.517) | 0.003 | ||
| Additional strain(s) | 31 (12.45) | 23 (11.86) | 8 (14.55) | 1.265 (0.532–3.011) | 0.594 | ||
| Amt of bacterial growth | |||||||
| Few | 72 (29.88) | 63 (33.33) | 9 (17.31) | 1 | 1 | ||
| Moderate | 109 (45.23) | 82 (43.39) | 27 (51.92) | 2.305 (1.012–5.248) | 0.047 | 4.030 (1.122–14.475) | 0.033 |
| Many | 60 (24.90) | 44 (23.28) | 16 (30.77) | 2.545 (1.032–6.279) | 0.043 | 1.699 (0.430–6.721) | 0.45 |
The mean ages were 4.77 years (mupirocin-susceptible group) and 7.86 years (mupirocin-resistant group); univariate OR (95% CI), 1.123 (1.060–1.190; P < 0.001; n = 249.
CI, confidence interval.
For the multivariate analysis, variables trimmed from the backward model selection are excluded, and only variables selected for the final model are shown.
There were also significant differences between zip code groups (ZCG). ZCG 1, which included Mid- and Lower Manhattan, had the least mupirocin resistance (8.3%) and was used as the reference group. In per-isolate and ever-resistant univariate analyses, ZCG 3, which comprises Upper Manhattan and the Bronx, had significantly more mupirocin resistance (Tables 1 and 2). Multivariate analysis of the ever-resistant data set also highlighted ZCG 3 (Table 2).
Significant differences were also identified between primary-diagnosis groups. For all three data analyses, patients with a diagnosis of dermatitis NOS were less likely than patients with atopic dermatitis to have mupirocin-resistant isolates, whereas those of patients with EB were more likely to be mupirocin resistant (Tables 1 and 2; see Table S1 in the supplemental material).
Isolates from patients with AD were more likely to be oxacillin resistant (i.e., MRSA) and mupirocin resistant than isolates from nonatopic children. Of the MRSA isolates in this study, 67.7% were found in patients with AD. The majority of MRSA isolates (68.2%) in patients with AD were resistant to mupirocin compared to only 28.6% of MRSA isolates in nonatopic patients.
Forty-two subjects had cultures obtained on more than one date during the study period. Not unexpectedly, in 19 subjects with MupRSA isolates (7 of which were MRSA), colonization persisted despite the use of mupirocin. In six subjects initially found to have mupirocin-sensitive MSSA on culture, a subsequent isolate was found to be mupirocin resistant; all 6 patients had a history of mupirocin use prior to repeat cultures. In contrast, we never observed conversion to mupirocin resistance in the absence of mupirocin use. There were no instances where exposure to mupirocin preceded the acquisition of MRSA.
DISCUSSION
A study period prevalence of 31.3% MupRSA is one of the highest reported to date, and it is alarming that 22.1% of the children had a mupirocin-resistant isolate at some time during the study period. Other studies in pediatric populations have found various rates of mupirocin resistance, including 1.8% in initial MRSA isolates from a predominantly outpatient setting in the northwest United States (15), 14.7% in S. aureus isolates from children with recurrent skin and soft tissue infections seen at a tertiary care center in Houston, TX (16), and 9.8% in S. aureus isolates from a mixed group of children with single or recurrent skin and soft tissue infections from the same center in Houston (17). In adult populations, the prevalence of mupirocin resistance reported is low, generally ranging from 1 to 5% (7, 9–12, 26). The highest rate reported was 45% in clinical staphylococcal isolates from patients at a Turkish hospital (14).
We found a strong association between mupirocin resistance and prior mupirocin use in all three analyses (odds ratio [OR] = 19.2 to 26.5; P < 0.001), supporting the notion that use of mupirocin may be the primary driver of resistance. This is consistent with previous reports, including a case-control study revealing a strong association (OR = 9.8) between previous mupirocin exposure and subsequent resistance in MRSA (20) and several observational studies (18, 19). A recent study in a pediatric population in Texas also reported an association between previous mupirocin use and mupirocin resistance (17).
Mupirocin resistance was highly prevalent in our MRSA isolates (55.4%), and MRSA was a strong risk factor for resistance to mupirocin. This finding agrees with studies in adult populations (27, 28), where mupirocin resistance was found more frequently in MRSA than MSSA, but contrasts with a recent study in children (8.3% and 21.4% [MRSA:MSSA]) (16). The association between levofloxacin and mupirocin resistance (OR = 3.6; P = 0.006) in univariate but not in multivariate analysis may be related to the fact that levofloxacin resistance is more common in MRSA (29). Over three-quarters (39/51) of levofloxacin-resistant isolates in our study were MRSA. We did not see an association between mupirocin resistance and clindamycin resistance, as has been reported previously (7).
Although our study was not designed to examine longitudinal factors influencing the emergence and persistence of mupirocin resistance, the patterns observed in patients with multiple isolates may give some insight into this process. There were multiple instances where mupirocin treatment failed to eradicate MupRSA isolates and where a MupRSA isolate replaced a mupirocin-sensitive S. aureus (MupSSA) isolate after treatment with mupirocin, showing that mupirocin eradication strategies may fail in multiple ways.
Place of residence was associated with mupirocin resistance, suggesting that geographic or socioeconomic factors may contribute to the development of resistance. Compared to the reference ZCG 1 (Mid- and Lower Manhattan), patients from ZCG 3 (Harlem, Washington Heights, Inwood, and the Bronx) were more likely to carry MupRSA, indicating that there may be geographic pockets of higher/lower mupirocin resistance in the greater New York City area. This pattern may be due to factors such as transmission rates, population density, prescribing patterns, or higher MRSA prevalence. Indeed, we found higher rates of MRSA in ZCG 3 (18.5%) than in ZCG 1 (11.7%). A recent study found a high incidence of intrahousehold S. aureus transmission in northern Manhattan (similar to ZCG 3) and demonstrated that environmental contamination with a colonizing or clinical infection strain was associated with transmission (30). Mupirocin resistance may also vary on a broader geographic scale. A large multicenter study found generally low rates of mupirocin resistance in S. aureus isolates from outpatient dermatology centers across the United States, with the exception of 33.9% at a center in Florida (31).
The strong association between MupRSA and diseases such as AD and EB is in agreement with prior studies (17, 32) and is likely due at least in part to the increased use of antibiotics in these patients. It is also likely that high cutaneous bacterial burdens (33–35), more frequent infection, and the inability to eradicate S. aureus lead to a higher probability of acquiring a persistent MupRSA strain.
There are several limitations to this study. It is likely that patients presenting to dermatology at a tertiary care center represent a more severely and/or chronically infected population. These patients may be more likely to be exposed to antibiotics and less likely to clear infection, resulting in higher rates of MupRSA than in the general population. More severely infected patients may have been overrepresented because they were seen more often in follow-up. Indeed, culture results from patients with MupRSA do appear to be overrepresented in our per-isolate data, but this did not seem to significantly affect our analysis of risk factors, which were similar in all three analyses. A review of pediatric-dermatology cultures obtained over the study period revealed that 8.6% of the cultures were not tested for mupirocin resistance for a variety of logistical reasons, and as cultures are not routinely held after processing, we were unable to retrospectively obtain mupirocin sensitivity for these isolates. While we cannot rule out a bias in isolate selection, the small percentage of untested isolates suggests that this may not have had a significant impact on our analysis.
Mupirocin is an important component of antimicrobial therapy that is recommended by the Infectious Disease Society of America (IDSA) for treatment of children with minor skin infections caused by MRSA or neonatal pustulosis and for decolonization of patients with recurrent MRSA skin and soft tissue infections (36). Prophylactic intranasal mupirocin has been shown to decrease rates of nosocomial S. aureus infection in carriers (37). A recent multicenter study found a significant decrease in intensive care unit bloodstream infections when universal intranasal mupirocin and chlorhexidine washes were employed as a decolonization strategy (38). It is unclear what the impact of very high rates of MupRSA would have been in these studies, but one may speculate that the strategies may have been less successful. Furthermore, in situations where rates of mupirocin resistance in MRSA are high, it is possible that indiscriminate mupirocin use could lead to coselection of methicillin-resistant strains. Presently, it seems prudent to test for mupirocin susceptibility in patients with a history of mupirocin use, culture-positive MRSA, or atopic dermatitis and in those patients who are immune suppressed or hospitalized. Judicious clinical use of mupirocin, particularly in high-risk populations, may prevent the development of additional and widespread resistance. Going forward, it will be critical to identify and validate the efficacies of alternative topical strategies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jimmy Duong, Ying Wei, and Yuan Zhang of the Department of Biostatistics at the Mailman School of Public Health of Columbia University for their consultation and assistance with statistical analysis of data.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00079-15.
REFERENCES
- 1.Coia JE, Duckworth GJ, Edwards DI, Farrington M, Fry C, Humphreys H, Mallaghan C, Tucker DR, Joint Working Party of the British Society of Antimicrobial Chemotherapy, Hospital Infection Society, Infection Control Nurses Association. 2006. Guidelines for the control and prevention of meticillin-resistant Staphylococcus aureus (MRSA) in healthcare facilities. J Hosp Infect 63(Suppl 1):S1–S44. doi: 10.1016/j.jhin.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Rahman M, Noble WC, Cookson B. 1987. Mupirocin-resistant Staphylococcus aureus. Lancet ii:387–388. [DOI] [PubMed] [Google Scholar]
- 3.Kavi J, Andrews JM, Wise R, Smith MD, Sanghrajka M, Lock S. 1987. Mupirocin-resistant Staphylococcus aureus. Lancet ii:1472–1473. [PubMed] [Google Scholar]
- 4.Hodgson JE, Curnock SP, Dyke KG, Morris R, Sylvester DR, Gross MS. 1994. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob Agents Chemother 38:1205–1208. doi: 10.1128/AAC.38.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seah C, Alexander DC, Louie L, Simor A, Low DE, Longtin J, Melano RG. 2012. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob Agents Chemother 56:1916–1920. doi: 10.1128/AAC.05325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker ES, Vasquez JE, Dula R, Bullock H, Sarubbi FA. 2003. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus: does mupirocin remain effective? Infect Control Hosp Epidemiol 24:342–346. doi: 10.1086/502218. [DOI] [PubMed] [Google Scholar]
- 7.Fritz SA, Hogan PG, Camins BC, Ainsworth AJ, Patrick C, Martin MS, Krauss MJ, Rodriguez M, Burnham CA. 2013. Mupirocin and chlorhexidine resistance in Staphylococcus aureus in patients with community-onset skin and soft tissue infections. Antimicrob Agents Chemother 57:559–568. doi: 10.1128/AAC.01633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonio M, McFerran N, Pallen MJ. 2002. Mutations affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 46:438–442. doi: 10.1128/AAC.46.2.438-442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hetem DJ, Bonten MJ. 2013. Clinical relevance of mupirocin resistance in Staphylococcus aureus. J Hosp Infect 85:249–256. doi: 10.1016/j.jhin.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Simor AE, Stuart TL, Louie L, Watt C, Ofner-Agostini M, Gravel D, Mulvey M, Loeb M, McGeer A, Bryce E, Matlow A, Canadian Nosocomial Infection Surveillance Program. 2007. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus strains in Canadian hospitals. Antimicrob Agents Chemother 51:3880–3886. doi: 10.1128/AAC.00846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossney A, O'Connell S. 2008. Emerging high-level mupirocin resistance among MRSA isolates in Ireland. Euro Surveill 13:8084. [PubMed] [Google Scholar]
- 12.Desroches M, Potier J, Laurent F, Bourrel AS, Doucet-Populaire F, Decousser JW, Microbs Study Group. 2013. Prevalence of mupirocin resistance among invasive coagulase-negative staphylococci and methicillin-resistant Staphylococcus aureus (MRSA) in France: emergence of a mupirocin-resistant MRSA clone harbouring mupA. J Antimicrob Chemother 68:1714–1717. doi: 10.1093/jac/dkt085. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Roth E, Lopez-Aguilar C, Alcoba-Florez J, Mendez-Alvarez S. 2006. High-level mupirocin resistance within methicillin-resistant Staphylococcus aureus pandemic lineages. Antimicrob Agents Chemother 50:3207–3211. doi: 10.1128/AAC.00059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sareyyupoglu B, Ozyurt M, Haznedaroglu T, Ardic N. 2008. Detection of methicillin and mupirocin resistance in staphylococcal hospital isolates with a touchdown multiplex polymerase chain reaction. Folia Microbiol (Praha) 53:363–367. doi: 10.1007/s12223-008-0056-4. [DOI] [PubMed] [Google Scholar]
- 15.Hogue JS, Buttke P, Braun LE, Fairchok MP. 2010. Mupirocin resistance related to increasing mupirocin use in clinical isolates of methicillin-resistant Staphylococcus aureus in a pediatric population. J Clin Microbiol 48:2599–2600. doi: 10.1128/JCM.02118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNeil JC, Hulten KG, Kaplan SL, Mason EO. 2011. Mupirocin resistance in Staphylococcus aureus causing recurrent skin and soft tissue infections in children. Antimicrob Agents Chemother 55:2431–2433. doi: 10.1128/AAC.01587-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNeil JC, Hulten KG, Kaplan SL, Mason EO. 2014. Decreased susceptibilities to Retapamulin, Mupirocin, and Chlorhexidine among Staphylococcus aureus isolates causing skin and soft tissue infections in otherwise healthy children. Antimicrob Agents Chemother 58:2878–2883. doi: 10.1128/AAC.02707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee AS, Macedo-Vinas M, François P, Renzi G, Vernaz N, Schrenzel J, Pittet D, Harbarth S. 2011. Trends in mupirocin resistance in meticillin-resistant Staphylococcus aureus and mupirocin consumption at a tertiary care hospital. J Hosp Infect 77:360–362. doi: 10.1016/j.jhin.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Talon D, Marion C, Thouverez M, Bertrand X. 2011. Mupirocin resistance is not an inevitable consequence of mupirocin use. J Hosp Infect 79:366–367. doi: 10.1016/j.jhin.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Caffrey AR, Quilliam BJ, LaPlante KL. 2010. Risk factors associated with mupirocin resistance in meticillin-resistant Staphylococcus aureus. J Hosp Infect 76:206–210. doi: 10.1016/j.jhin.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 21.Cadilla A, David MZ, Daum RS, Boyle-Vavra S. 2011. Association of high-level mupirocin resistance and multidrug-resistant methicillin-resistant Staphylococcus aureus at an academic center in the midwestern United States. J Clin Microbiol 49:95–100. doi: 10.1128/JCM.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SY, Kim SM, Park SD. 2012. The prevalence, genotype and antimicrobial susceptibility of high- and low-level mupirocin resistant methicillin-resistant Staphylococcus aureus. Ann Dermatol 24:32–38. doi: 10.5021/ad.2012.24.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlemann AC, Dordel J, Knox JR, Raven KE, Parkhill J, Holden MT, Peacock SJ, Lowy FD. 2014. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci U S A 111:6738–6743. doi: 10.1073/pnas.1401006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suh L, Coffin S, Leckerman KH, Gelfand JM, Honig PJ, Yan AC. 2008. Methicillin-resistant Staphylococcus aureus colonization in children with atopic dermatitis. Pediatr Dermatol 25:528–534. doi: 10.1111/j.1525-1470.2008.00768.x. [DOI] [PubMed] [Google Scholar]
- 25.Matiz C, Tom WL, Eichenfield LF, Pong A, Friedlander SF. 2011. Children with atopic dermatitis appear less likely to be infected with community acquired methicillin-resistant Staphylococcus aureus: the San Diego experience. Pediatr Dermatol 28:6–11. doi: 10.1111/j.1525-1470.2010.01293.x. [DOI] [PubMed] [Google Scholar]
- 26.Perkins D, Hogue JS, Fairchok M, Braun L, Viscount HB. 2008. Mupirocin resistance screening of methicillin-resistant Staphylococcus aureus isolates at Madigan Army Medical Center. Mil Med 173:604–608. doi: 10.7205/MILMED.173.6.604. [DOI] [PubMed] [Google Scholar]
- 27.Chaves F, Garcia-Martinez J, de Miguel S, Otero JR. 2004. Molecular characterization of resistance to mupirocin in methicillin-susceptible and -resistant isolates of Staphylococcus aureus from nasal samples. J Clin Microbiol 42:822–824. doi: 10.1128/JCM.42.2.822-824.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rotger M, Trampuz A, Piper KE, Steckelberg JM, Patel R. 2005. Phenotypic and genotypic mupirocin resistance among staphylococci causing prosthetic joint infection. J Clin Microbiol 43:4266–4268. doi: 10.1128/JCM.43.8.4266-4268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limbago B, Fosheim GE, Schoonover V, Crane CE, Nadle J, Petit S, Heltzel D, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Mu Y, Fridkin SK, Active Bacterial Core Surveillance Investigators MRSA. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J Clin Microbiol 47:1344–1351. doi: 10.1128/JCM.02264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knox J, Uhlemann AC, Miller M, Hafer C, Vasquez G, Vavagiakis P, Shi Q, Lowy FD. 2012. Environmental contamination as a risk factor for intra-household Staphylococcus aureus transmission. PLoS One 7:e49900. doi: 10.1371/journal.pone.0049900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biedenbach DJ, Bouchillon SK, Johnson SA, Hoban DJ, Hackel M. 2014. Susceptibility of Staphylococcus aureus to topical agents in the United States: a sentinel study. Clin Ther 36:953–960. doi: 10.1016/j.clinthera.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Moy JA, Caldwell-Brown D, Lin AN, Pappa KA, Carter DM. 1990. Mupirocin-resistant Staphylococcus aureus after long-term treatment of patients with epidermolysis bullosa. J Am Acad Dermatol 22:893–895. doi: 10.1016/0190-9622(90)70120-7. [DOI] [PubMed] [Google Scholar]
- 33.van der Kooi-Pol MM, Veenstra-Kyuchukova YK, Duipmans JC, Pluister GN, Schouls LM, de Neeling AJ, Grundmann H, Jonkman MF, van Dijl JM. 2012. High genetic diversity of Staphylococcus aureus strains colonizing patients with epidermolysis bullosa. Exp Dermatol 21:463–466. doi: 10.1111/j.1600-0625.2012.01502.x. [DOI] [PubMed] [Google Scholar]
- 34.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Comparative Sequence Program NISC, Murray PR, Turner ML, Segre JA. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jinnestål CL, Belfrage E, Bäck O, Schmidtchen A, Sonesson A. 2014. Skin barrier impairment correlates with cutaneous Staphylococcus aureus colonization and sensitization to skin-associated microbial antigens in adult patients with atopic dermatitis. Int J Dermatol 53:27–33. doi: 10.1111/ijd.12198. [DOI] [PubMed] [Google Scholar]
- 36.Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, Rybak JM, Talan DA, Chambers HF. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin Infect Dis 52:285–292. doi: 10.1093/cid/cir034. [DOI] [PubMed] [Google Scholar]
- 37.Perl TM, Cullen JJ, Wenzel RP, Zimmerman MB, Pfaller MA, Sheppard D, Twombley J, French PP, Herwaldt LA, Mupirocin and the Risk Of Staphylococcus Aureus Study Team. 2002. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N Engl J Med 346:1871–1877. doi: 10.1056/NEJMoa003069. [DOI] [PubMed] [Google Scholar]
- 38.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, Lankiewicz J, Gombosev A, Terpstra L, Hartford F, Hayden MK, Jernigan JA, Weinstein RA, Fraser VJ, Haffenreffer K, Cui E, Kaganov RE, Lolans K, Perlin JB, Platt R, CDC Prevention Epicenters Program, AHRQ DECIDE Network and Healthcare-Associated Infections Program. 2013. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med 368:2255–2265. doi: 10.1056/NEJMoa1207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.