Abstract
Streptococcus pneumoniae is a leading cause of pneumonia, meningitis, septicemia, and middle ear infections. The incidence of S. pneumoniae isolates that are not susceptible to penicillin has risen worldwide and may be above 20% in some countries. Beta-lactam antibiotic resistance in pneumococci is associated with significant sequence polymorphism in penicillin-binding proteins (PBPs). Commensal streptococci, especially S. mitis and S. oralis, have been identified as putative donors of mutated gene fragments. However, no studies have compared sequences of the involved pbp genes in large collections of commensal streptococci with those of S. pneumoniae. We therefore investigated the sequence diversity of the transpeptidase region of the three pbp genes, pbp2x, pbp2b, and pbp1a in 107, 96, and 88 susceptible and nonsusceptible strains of commensal streptococci, respectively, at the nucleotide and amino acid levels to determine to what extent homologous recombination between commensal streptococci and S. pneumoniae plays a role in the development of beta-lactam resistance in S. pneumoniae. In contrast to pneumococci, extensive sequence variation in the transpeptidase region of pbp2x, pbp2b, and pbp1a was observed in both susceptible and nonsusceptible strains of commensal streptococci, conceivably reflecting the genetic diversity of the many evolutionary lineages of commensal streptococci combined with the recombination events occurring with intra- and interspecies homologues. Our data support the notion that resistance to beta-lactam antibiotics in pneumococci is due to sequences acquired from commensal Mitis group streptococci, especially S. mitis. However, several amino acid alterations previously linked to beta-lactam resistance in pneumococci appear to represent species signatures of the donor strain rather than being causal of resistance.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) is a leading cause of pneumonia, meningitis, septicemia, and middle ear infections (1). According to data from the World Health Organization, S. pneumoniae is the fourth-most-frequent cause of fatal infections worldwide (2). For many years, S. pneumoniae was considered to always be susceptible to beta-lactam antibiotics, and since the first reports in the late 1970s on beta-lactam antibiotic resistance in clinical pneumococcal isolates, the incidence of nonsusceptible S. pneumoniae isolates has risen worldwide and may be above 20% in some countries, especially in southern Europe (3). However, the introduction of the protein conjugate pneumococcal vaccines in treatment of children has reduced the rates of beta-lactam resistance in some countries, e.g., in the United States (4). Generally, the rate of nonsusceptible S. pneumoniae isolates correlates with the level of usage of beta-lactam antibiotics (5). In such countries, a high occurrence of nonsusceptible strains of the closely related commensal streptococci, in particular, S. mitis and S. oralis, has also been observed (6, 7).
Beta-lactam antibiotic resistance in pneumococci and commensal streptococci develops upon the accumulation of mutations in three of the six transpeptidases involved in cell wall biosynthesis, i.e., the so-called penicillin-binding proteins PBP2x, PBP2b, and PBP1a (8). These sequence alterations reduce the affinity of the transpeptidases for the drugs while leaving the enzyme function unaffected and confer an advantage for the mutated bacteria in the presence of antibiotics. While the pbp genes of susceptible pneumococci are well conserved, the sequences of the pbp genes in nonsusceptible clinical strains of pneumococci show a mosaic structure, with blocks of sequences of different lengths that may differ by up to 20% at the DNA level and by up to 10% at the amino acid level compared to the corresponding regions in susceptible pneumococci (9–11). Although one paper by Dowson et al. (12) concluded that viridans streptococci (S. oralis) have obtained altered penicillin-binding protein genes from penicillin-resistant strains of S. pneumoniae, commensal streptococci, especially S. mitis and S. oralis, have been identified as putative donors to the mosaic structure of the pbp genes found in resistant pneumococci (13–16). The evolution of resistance in pneumococci and commensal streptococci has been hypothesized to involve at least two steps (13): first, selection of resistant commensal streptococci with point mutations in their pbp genes; second, transfer of parts of these resistance genes to competent pneumococci through homologous recombination. This is in agreement with the generally one-directional transfer of genes from S. mitis and, to a lesser extent, S. oralis to S. pneumoniae (17). In addition, mosaic structures have been observed in resistant commensal streptococci (18–20). Comprehensive studies have identified specific mutations in pneumococci that are associated with beta-lactam resistance (21, 22). However, no studies supported the conclusions with reference to pbp gene sequences in resistant and susceptible members of the large populations of potential donor species. Therefore, the purpose of the present study was to investigate the sequence diversity at the nucleotide and amino acid level of the transpeptidase region of the three pbp genes, pbp2x, pbp2b, and ppb1a, in 107, 96, and 88 strains, respectively, of susceptible and nonsusceptible species of the Mitis group of streptococci in order to map the natural history of beta-lactam resistance in these streptococci.
MATERIALS AND METHODS
Strains—collection and identification.
A total of 63 commensal streptococcal strains belonging to the Mitis group of streptococci were included the study. Strains were randomly picked from several strain collections, with no prior knowledge of their antibiotic susceptibility (23–25). They were isolated from the upper respiratory tract of healthy subjects or from blood of patients with bacteremia. The strains were identified by multilocus sequence analysis (MLSA) either as described by Bishop et al. (26) or using a combination of at least three of the following genes: gdh, rpoB, dnaJ, ddl, pdgA, recP, and aroE (27). Forty-six strains were identified as S. mitis, 10 as S. oralis, 6 as S. infantis, and 1 as S. pseudopneumoniae. Twenty-eight isolates were from the United States, 13 from Denmark, 11 from Greece, 5 from Sweden, 4 from Switzerland, and 2 from the United Kingdom. A complete list of the strains is shown in Table S1 in the supplemental material. In addition to the sequences of the 63 strains generated during this study, sequences available in GenBank of pbp1a, pbp2b, and pbp2x from strains of S. mitis and S. oralis with at least one known MIC value of beta-lactam antibiotics were included in the analysis (see Table S1). Ten sequences from the work of Chi et al. (13) that have not been deposited in GenBank were kindly supplied by Regine Hakenbeck. Furthermore, a total of 20 sequences from susceptible and nonsusceptible strains of S. pneumoniae were included for comparison (see Table S1).
Antibiotic susceptibility testing.
MICs were determined by the agar dilution method as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Strains were grown in Todd-Hewitt broth supplemented with 0.1% pyruvate and incubated at 37°C in a 5% CO2-enriched atmosphere to a turbidity of a 0.5 McFarland standard (∼108 CFU/ml). The suspension for each strain was then transferred to a single well on a sterile 96-well microtiter plate. By using an in-house multi-inoculator and applying 1 μl of the suspension, duplicate sets of Mueller-Hinton agar plates supplemented with 5% defibrinated horse blood and appropriate concentrations of antibiotics in 2-fold dilutions were inoculated with up to 96 different strains. MICs were determined after incubation at 37°C in a 5% CO2-enriched atmosphere for 48 h. S. pneumoniae R6 and S. pneumoniae ATCC 49619 were used for quality control. The MIC was read as the lowest concentration of antibiotic inhibiting growth of the bacteria. MIC interpretation was done according to the guidelines from EUCAST (28) using the MIC values for “viridans streptococci” or for S. pneumoniae when no value for the relevant antibiotic was available for viridans streptococci. The following beta-lactam antibiotics were used for MIC determination: benzylpenicillin, oxacillin, piperacillin, cefotaxime, cefuroxime, and cephalothin.
DNA extraction, PCR amplification, and DNA sequencing.
DNA was extracted as previously described (29). Novel primer pairs for the pbp1a, pbp2b, and pbp2x genes were developed by using sequences extracted from publicly available genomes of Mitis group streptococci as the templates (Table 1). All PCRs were carried out in a 25-μl volume containing 10 μl of 5′-Prime Hotmastermix (5 Prime, Hamburg, Germany), 3 μl of each primer (10 μM), 4 μl of PCR-grade H2O, and 5 μl of DNA. Amplification was achieved with an initial cycle of 2 min of denaturation at 94°C and 30 cycles of 30 s at 94°C for denaturation, 60 s at 52°C (pbp1a) or 55°C (pbp2b and pbp2x) for annealing, and 60 s at 72°C for extension with a final extension step at 72°C for 5 min. Purified PCR products were sequenced on both strands at GATC Biotech, Constance, Germany.
TABLE 1.
List of primers used in this study and sequence information
| Gene(s) | Primer name | Primer sequence (5′ to 3′) | Position(s)a | Trimmed amino acid sequence (positions)b | No. (%) of variable nucleotide sites/total no. of nucleotide sitsf | No. (%) of variable amino acid sites/total no. of amino acid sitesf |
|---|---|---|---|---|---|---|
| pbp1a | pbp1a-f | ACIACDGGDATGGAHGTHTA | 877–896 | 318–577 | 371/780 (48) | 98/263(37%) |
| pbp1a-r | GTCCADACRGCCATWGMATA | 1790–1771 | ||||
| pbp2b | pbp2b-f | AATGAYCGHGTBGGDACYTC | 793–812 | 440–643 | 294/612 (48) | 79/204 (39) |
| pbp2b-r | TTGATRATRTCRCGHGCAAT | 2027–2008 | ||||
| pbp2x | pbp2x-f | AAGTAYATGACIGCDACCTT | 862–881 | 308–554 | 320/741 (43) | 79/247 (32) |
| pbp2x-r | TGVAGRTTRAGRGADTCTTT | 1865–1846 | ||||
| murM and murN | murMN-f | CAYGARTGGTAYTAYTGGGAA | 694–714c | |||
| murMN-r | TGAATRTARCAWCCAGGRCT | 62–43d | ||||
| Ortholog of murM | murM_uo5-f | TTGCARAGTAGTGATTGGKCCA | 76–97e | |||
| murM_uo5-r | TCCTTTATTYYTTGCAGTTCGGA | 556–534e |
Nucleotide positions according to the S. pneumoniae R6 sequence.
Amino acid positions in S. pneumoniae R6. Active sites range from residue 337 to residue 549 (pbp2x), 386 to 617 (pbp2b), and 370 to 557 (pbp1a).
Nucleotide position in the flanking SP_0614 gene in S. pneumoniae R6 of the mugsy cluster of murM and murN.
Nucleotide position in the flanking SP_0618 gene in S. pneumoniae R6 of the mugsy cluster of murM and murN.
Nucleotide position in the ortholog of the murM gene in S. oralis uo5.
Numbers of all variable sites for all strains included in the sequence analysis.
A novel primer pair designed according to an alignment of the flanking regions of the murM and murN genes extracted from available genomes of S. mitis and S. oralis in the GenBank database was used to amplify the murM and murN genes (Table 1). The amplicon resulted in a 3.3-kb band on an agarose gel when both genes were present in a strain. In some strains, however, an ortholog of the murM gene, but not the murN gene, was located at a different position in the genome and was detected using a novel primer pair that amplifies a 500-bp internal fragment of the murM gene (Table 1). Reactions were carried out in a 25-μl volume containing 10 μl of 5′-Prime Hotmastermix (5 Prime, Hamburg, Germany), 3 μl of each primer (10 μM), 7 μl of PCR-grade H2O, and 2 μl of DNA. Amplification was achieved with an initial cycle of 2 min of denaturation at 94°C and 34 cycles of 30 s at 94°C for denaturation, 40 s at 60°C for annealing, and 90 s (murM) or 3 min (murM and murN) at 72°C for extension with a final extension step at 72°C for 7 min. PCR products were visualized on a 1% agarose gel by ethidium bromide staining.
Sequence analysis.
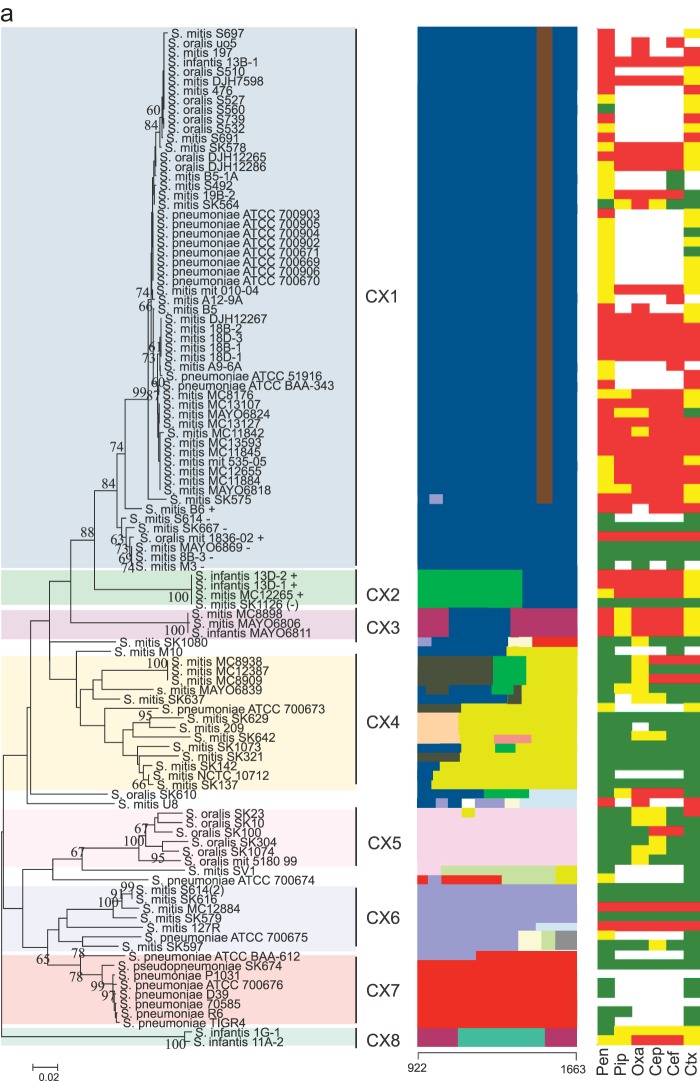
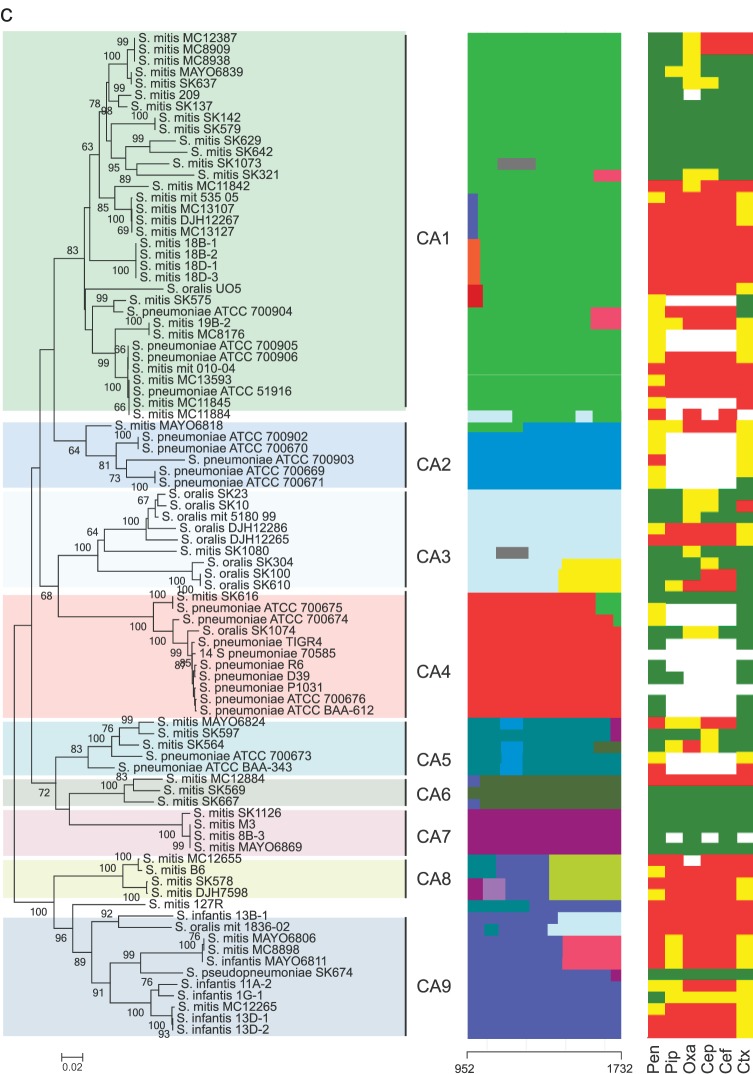
Sequences of the pbp genes were edited, aligned, and subjected to nucleotide and protein analysis using MEGA version 6.0 (30). Sequences were trimmed to cover the active-site motifs of the transpeptidase region, except for the pbp2b gene, in which the first active motif, SVVK, is outside the sequence alignment (Table 1). The trimmed sequences were subjected to phylogenetic analysis using the minimum evolution algorithm. Bootstrap tests were performed with 500 replicates. The mosaic structures of the partial sequences of the pbp1a, pbp2b, and pbp2x genes were detected using a modified Linux version of BRATNextGen software (31). Using a window size of 40 bp and an alpha value of 1, the pbp1a, pbp2b, and pbp2x genes were divided into 9, 5, and 8 subclusters which were consistent with the phylogenetic clusters identified by the minimum evolution algorithm (Fig. 1). Segments of sequences potentially originating from recombination events were identified using a P value of 0.05 and calculated through 100 replicates of 10 iterations. The possible origin of the resulting recombinational segments was then potentially identified by phylogenetic analyses of the segments and manual sequence comparison. Segments with the possible same origin are unicolored in Fig. 1.
FIG 1.
(a to c) Comparative analyses of pbp genes in Mitis group streptococci. Left columns show minimum evolution trees generated in MEGA of the transpeptidase region of pbp2x (a), pbp2b (b), and pbp1a (c) genes of susceptible and nonsusceptible strains of commensal streptococci and S. pneumoniae. Bootstrap values of 500 replications are shown. Clusters CX1 to CX8 (pbp2x), CB1 to CB5 (pbp2B), and CA1 to CA9 (pbp1a) identified by the BratNextgen software for detecting recombination are highlighted. In the pbp2x tree, +, −, or (−) after a strain name denotes the presence of, absence of, or a nonfunctional murM gene. The middle columns show recombination segments identified by the BratNextgen software over the length of the sequenced part of the transpeptidase region of the pbp genes. Blocks with the same color are hypothesized to be of the same origin. The numbers at the bottom of the middle columns indicate the region of the genes used for the analysis according to the positions in sequences of S. pneumoniae R6. Right columns show the susceptibility of six beta-lactam antibiotics (benzylpenicillin [pen], piperacillin [pip], oxacillin [oxa], cephalothin [cep], cefuroxime [cef], and cefotaxime [ctx]) indicated as susceptible (green), intermediate (yellow), and resistant (red) according the thresholds given by EUCAST (28) and shown in Table 2.
Nucleotide sequence accession numbers.
Sequences of the three genes were deposited at GenBank with the following accession numbers: for pbp2x, KP732106 to KP732168 and KP984507 to KP984516; for pbp2b, KP732232 to KP732294; and for pbp1a, KP732169 to KP732231.
RESULTS
Antibiotic susceptibility.
The data corresponding to susceptibility to six different beta-lactam antibiotics determined with reference to the MIC cutoff values of EUCAST are shown in Table 2 (see also Table S1 in the supplemental material). Among the commensal streptococcal strains, resistance to the three penicillins tested ranged from 35% for penicillin to 52% for oxacillin, with the median MIC value of oxacillin being 4-fold higher (4 μg/ml) than those for penicillin (1 μg/ml) and piperacillin (1 μg/ml). The same was observed for the three cephalosporins, with the median MIC value for cephalothin (8 μg/ml) being higher than those for cefuroxime (4 μg/ml) and cefotaxime (2 μg/ml) (Table 2). Only a few strains showed marked variation in susceptibility to the beta-lactam antibiotics tested, most strains showing either resistance or intermediate or full susceptibility to all six beta-lactam antibiotics. Few strains were susceptible to penicillins but resistant to cephalosporins (e.g., MC8909 and MC8938), while some strains also showed considerable variation in susceptibility to each of the three penicillins (e.g., S. mitis SK564 and S. oralis SK610) or to each of the three cephalosporins (e.g., S. mitis 19B-2 and S. oralis SK23/ATCC 35037T).
TABLE 2.
MICs of 63 strains of commensal streptococci divided by species designation
| Antibiotic and MIC (μg/ml) | No. (%) of strains | No. (%) of strains of species: |
|||
|---|---|---|---|---|---|
| S. mitis (n = 46) | S. oralis (n = 10) | S. infantis (n = 6) | S. pseudopneumoniae (n = 1) | ||
| Benzylpenicillin | |||||
| ≤0.25 | 30 (48) | 21 (46) | 7 (70) | 1 (17) | 1 |
| >0.25–2 | 11 (17) | 9 (19) | 1 (10) | 1 (17) | |
| >2 | 22 (35) | 16 (35) | 2 (20) | 4 (66) | |
| Median | 1.0 | 1.0 | 0.1875 | 4.0 | |
| Piperacillina | |||||
| ≤0.25 | 22 (35) | 15 (33) | 6 (60) | 1 | |
| >0.25–2 | 15 (24) | 11 (24) | 1 (10) | 3 (50) | |
| >2 | 26 (41) | 20 (43) | 3 (30) | 3 (50) | |
| Mean | 1.0 | 1.0 | 0.125 | 2.5 | |
| Oxacillina | |||||
| ≤0.25 | 15 (24) | 12 (26) | 2 (20) | 1 | |
| >0.25–2 | 15 (24) | 10 (22) | 4 (40) | 1 (16) | |
| >2 | 33 (52) | 24 (52) | 4 (40) | 5 (84) | |
| Median | 4.0 | 4.0 | 1.25 | 12.0 | |
| Cephalothinb | |||||
| ≤0.5 | 16 (25) | 14 (30) | 1 (10) | 1 | |
| >0.5–2 | 10 (16) | 5 (11) | 4 (40) | 1 (16) | |
| >2 | 37 (59) | 27 (59) | 5 (50) | 5 (84) | |
| Median | 8.0 | 4.0 | 3.0 | 8.0 | |
| Cefuroxime | |||||
| ≤0.5 | 24 (39) | 18 (40) | 5 (50) | 1 | |
| >0.5–1 | 1 (2) | 1 (16) | |||
| >1 | 37 (59) | 27 (60) | 5 (50) | 5 (84) | |
| Median | 4.0 | 4.0 | 1.25 | 4.0 | |
| Cefotaxime | |||||
| ≤0.5 | 27 (44) | 20 (44) | 6 (60) | 1 | |
| >0.5–2 | 18 (29) | 11 (25) | 2 (20) | 5 (84) | |
| >2 | 17 (27) | 14 (31) | 2 (20) | 1 (16) | |
| Median | 2.0 | 2.0 | 0.282 | 2.0 | |
No resistant breakpoints for the mitis group of streptococci are given by EUCAST. Breakpoints for benzylpenicillin are used for comparison.
No resistant breakpoints for the mitis group of streptococci are given by EUCAST. Breakpoints for cefotaxime are used for comparison.
Although direct comparison of the different countries is not possible due to the nonsystematic collection of strains, isolates from the United States and Greece were clearly more resistant to most of the antibiotics compared to isolates from Denmark, presumably reflecting different policies for antibiotic usage.
Sequence analyses.
The molecular mechanisms of beta-lactam resistance were studied by partial sequencing of the transpeptidase region of the pbp1a, pbp2b, and pbp2x genes for all the included strains (see Table S1 in the supplemental material). Combined with GenBank-derived sequences from relevant S. pneumoniae strains and strains of S. oralis and S. mitis with known beta-lactam susceptibility, a total of 107, 96, and 88 sequences were included in the analyses of pbp2x, pbp2b, and pbp1a, respectively.
All three genes showed remarkable nucleotide variation within the sequenced part of the transpeptidase region with 43% (pbp2x), 48% (pbp2b), and 48% (pbp1a) polymorphic sites, which resulted in 32% (pbp2x), 39% (pbp2b), and 37% (pbp1a) variation at the amino acid level (Table 1).
Phylogenetic analyses.
The data for the deep-rooted branches in the phylogenetic trees based on the sequenced part of the transpeptidase regions of the pbp2x, pbp2b, and pbp1a genes (Fig. 1) support the notion of extensive sequence polymorphism. With some exceptions, the sequences clustered according to the antibiotic susceptibility of the respective strains. Susceptible strains of S. mitis and S. oralis showed remarkable sequence variation and formed several separate clusters in all three genes, in contrast to susceptible strains of S. pneumoniae, where all three genes were highly conserved. This is in agreement with the general differences in sequence polymorphisms among S. pneumoniae, S. mitis, and S. oralis genomes (17). Below, the three pbp genes are discussed in more detail.
pbp2x.
The sequenced part of the transpeptidase region of the pbp2x gene showed remarkable sequence variation, with several distinct clusters (Fig. 1a). Generally, strains clustered according to antibiotic susceptibility but not according to species affiliation. A large cluster, designated CX1, consisted mostly of resistant strains of S. oralis, S. mitis, and S. pneumoniae, including the multidrug-resistant S. pneumoniae ATCC 700669 strain (Spain 23F-1; PMEN1). Interestingly, the same cluster also contained a subcluster of susceptible S. mitis strains, but our mosaic structure analysis performed using BRATNextGen software revealed a small sequence segment located just upstream of the K547SG active motif specific to all resistant strains of the large subcluster of CX1. Amino acid alterations within this small fragment may contribute to the reduced susceptibility among the Mitis group streptococci, including S. pneumoniae, in the CX1 cluster. While S. pneumoniae strains with reduced susceptibility were found in several clusters, susceptible strains of S. pneumoniae were found only in cluster CX7.
pbp2b.
Minimum evolution analysis of the pbp2b gene revealed several distinct clusters that included strains of S. mitis, S. oralis, S. infantis, and S. pneumoniae (Fig. 1b). In contrast to the pbp2x gene, susceptible strains of S. pneumoniae were found in the same cluster (CB3) as nonsusceptible strains of S. pneumoniae and susceptible as well as nonsusceptible strains of S. mitis. Although this might indicate that the pbp2b gene is less involved in beta-lactam resistance, our mosaic structure analysis revealed a sequence segment at the end of the alignment that was shared by most of the nonsusceptible strains of CB3 and all strains located in clusters CB1 and CB2. These two clusters contained most nonsusceptible strains of S. mitis and S. oralis. Although no S. pneumoniae strains included in this study were found in CB1 and CB2, a BLAST search in GenBank revealed that the pbp2 genes of several strains of S. pneumoniae in the NCBI database do belong to these clusters. Interestingly, all such S. pneumoniae strains for which susceptibility data were available in the GenBank database had elevated MIC values against beta-lactam antibiotics (data not shown).
pbp1a.
Several distinct clusters, many of which included strains of S. mitis, S. oralis, S. infantis, and S. pneumoniae, were identified in the minimum evolution tree of the pbp1a gene (Fig. 1c). Most strains of S. mitis, S. oralis, and S. infantis with reduced susceptibility were found in CA1, CA8, and CA9, whereas CA8 and CA9 consisted almost exclusively of nonsusceptible strains. The clustering of many susceptible strains of, in particular, S. mitis in CA1 and S. pseudopneumoniae SK674 in CA9 suggests that pbp1a has an effect on beta-lactam resistance in Mitis group streptococci only when altered alleles of pbp2x or pbp2b are present too.
Mosaic structure of the pbp genes.
Analysis of recombination in the three pbp genes using BRATNextGen software revealed extensive inter- and intraspecies recombination resulting in a mosaic structure in all three pbp genes (Fig. 1). Interestingly, mosaic structures in the pbp genes were found both in susceptible and in nonsusceptible strains of S. mitis, S. oralis, and S. infantis, indicating that such pbp alleles resulting from random recombination, in these three species, were maintained in the absence of selection pressure from antibiotic use. In the pbp2x gene, extensive recombination was observed, particularly in cluster CX4, but all of the pbp2x genes displayed a mosaic structure. In the CX1 cluster, a small segment of 73 bp near the end of our alignment was observed in all but one strain with reduced susceptibility. Furthermore, phylogenetic and recombination analysis of available nearly full-length pbp2x gene sequences revealed that nonsusceptible strains of S. pneumoniae in cluster CX1 had similar sequence segments and clustered with susceptible strains of S. pneumoniae when the sequence of the pbp2x gene outside the transpeptidase region was used for analysis (data not shown). This may indicate that most of the transpeptidase region of S. pneumoniae strains found within the CX1 cluster originated in susceptible strains of S. mitis and S. oralis and had been transferred intact by one or several homologous recombination events (Fig. 1a). A 211-bp sequence fragment in the pbp2x gene of S. mitis SK1080 that was identical to that in susceptible strains of S. pneumoniae in cluster CX7 may represent a recombination event involving transfer of gene fragments from susceptible strains of S. pneumoniae to S. mitis or remnants of the ancestral allele.
The pbp2b gene recombination analysis revealed that strains of S. mitis, S. oralis, and S. infantis with reduced susceptibility in the CB3 cluster harbored a 165-bp segment at the end of the alignment that was similar to segments found in strains with reduced susceptibility in CB1 and CB2 (Fig. 1b). Amino acid alterations within this segment may play an important role in beta-lactam resistance in the Mitis group of streptococci, and a BLAST search using this segment as a query revealed that all strains of S. pneumoniae harboring this segment for which susceptibility data were available are nonsusceptible (data not shown). In the pbp1a gene, we detected several smaller fragments that were a result of recombination (Fig. 1c), as was also the case for the pbp2x and pbp2b genes. Whether these segments are due to a single recombination event involving very small DNA fragments or are a result of overlapping recombination events involving larger fragments is unknown. Unlike the pbp2x and pbp2b genes, no segments in the pbp1a gene were related to reduced susceptibility to beta-lactam antibiotics.
Analysis of the amino acid sequence of the transpeptidase region of the PBPs.
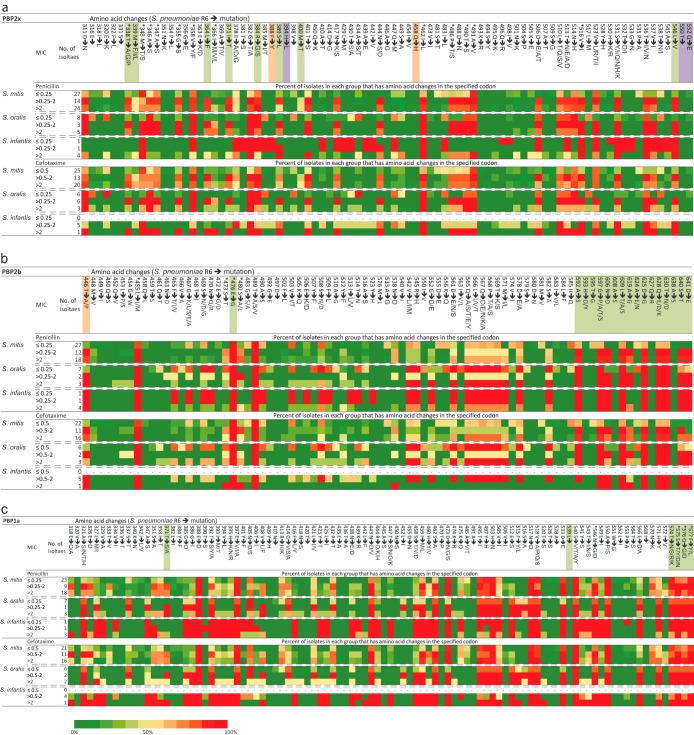
Although extensive recombination resulting in the characteristic mosaic structure of the pbp genes is the basis of reduced susceptibility in pneumococci, it is the amino acid changes that are responsible for the altered susceptibility. Of the many amino acid substitutions detected among our sequences compared to the susceptible S. pneumoniae R6 strain, none were universally found in all strains with reduced susceptibility for beta-lactam antibiotics as illustrated for penicillin and cefotaxime in Fig. 2 (see also Table S2 to Table S4 in the supplemental material). Yet several substitutions in all three pbp genes were associated with reduced susceptibility (discussed in detail later) (Fig. 2). In addition to substitutions, we also found an insertion of an alanine after I366 (according to S. pneumoniae R6 amino acid coordinates) in the pbp2x gene of the resistant S. mitis 127R. Likewise, D325 was deleted in the resistant S. mitis SK575 strain and G545 and R546 were deleted in the resistant S. infantis 13D-1 and S. infantis 13D-2 strains in pbp1a. As these indels were found only in nonsusceptible strains, these insertions and deletions may be involved in beta-lactam resistance.
FIG 2.
(a to c) Codons with amino acid changes in the transpeptidase region of PBP2x (a), PBP2b (b), and PBP1a (c) compared to the S. pneumoniae R6 reference strain. Only amino acids that differ from the reference sequence of the R6 strain are shown. Percentages of isolates in each group that have amino acid changes in the specified position are indicated. Amino acid positions further marked with colors or an asterisk indicate sites previously associated with resistance in S. pneumoniae and commensal streptococci (see reference 8). Orange, laboratory mutants; green, clinical isolates; purple, both laboratory mutants and clinical isolates. Amino acid alterations linked to reduced beta-lactam susceptibility in a study by Chewapreecha et al. (21) are marked with an asterisk. The color scale bar represents the percentage of isolates with altered amino acids compared to S. pneumoniae R6 in each position.
Previously, mutations in the pbp2x gene corresponding to the S337TMK active-site motif were linked to beta-lactam resistance in pneumococci (22, 32–36). We found that substitutions T338G and M339F were indeed associated with reduced susceptibility in our strain collection and were limited to nonsusceptible strains of S. oralis, S. mitis, and S. pneumoniae from the large CX1 cluster (Fig. 2a; see also Table S2 in the supplemental material). In contrast, T338A and M339L were also found in susceptible strains of S. mitis. Furthermore, the M400T mutation, which also has been associated with resistance in clinical isolates of pneumococci (37) and is located in close proximity to the S396SN active-site motif, was in our sequences always found together with the M339F mutations in S. mitis strains but not in S. oralis strains. Combined, these two mutations may play a crucial role in beta-lactam resistance in S. mitis and S. pneumoniae. The T338P mutation was found in only two strains, i.e., the S. mitis U8 and S. mitis 127R singletons located in CX6. The MIC values of all tested beta-lactam antibiotics for these two strains were very high, indicating that this substitution may play an important role in beta-lactam resistance in some strains. The amino acid sequence segment unique to nonsusceptible strains in CX1 included two unique substitutions, L510T and T513N, that had never previously been linked to resistance in S. pneumoniae. To what extent these substitutions may contribute to resistance has yet to be evaluated.
The highly resistant S. mitis B6 strain (38) located between the resistant and susceptible subclusters in CX1 may reveal important substitutions involved in beta-lactam resistance. The N417K and N444S substitutions were found in the resistant subcluster and in S. mitis B6 but not in the susceptible subcluster of CX1 or in any other of the included susceptible strains. However, none of these substitutions were previously associated with resistance in S. pneumoniae. The multiresistant S. mitis MC12884 strain clustered very closely with susceptible strains in cluster CX6. However, this strain harbors two amino acid substitutions, A369T and I454V, which are not found in the susceptible strains in cluster CX7. While A369T is found also in the resistant S. mitis 127R strain, I454V is unique to S. mitis MC12284. Whether or not these mutations are contributing to the observed resistance has to be determined. Interestingly, the Q552E substitution, which was previously associated with reduced susceptibility in S. pneumoniae (39), was found only in susceptible strains of S. mitis and S. oralis.
All nonsusceptible commensal streptococci in CB3 (except for S. mitis MC12884) harbored a sequence segment in the pbp2b gene similar to that of strains in CB1 and CB2. This was characterized by several amino acid substitutions located close to the active-site motif K615TG, including G597P (unique), N606D (unique), L609T (unique), and A619G, which was found in most of these strains but also in S. pneumoniae ATCC 51916 from CB7 (Fig. 2b; see also Table S3 in the supplemental material). The nonsusceptible S. pneumoniae ATCC 51916 strain is particularly informative, as this strain had the same amino acid sequence as that found in the susceptible strains of S. pneumoniae, except for six amino acid substitutions from A619G to D641E. These six substitutions are identical to the substitutions found in the resistant strains of CB1 and CB2 as well as in some strains in CB3. Although S. pneumoniae ATCC 51916 also differs from the susceptible strains of S. pneumoniae in the other two pbp genes, this may indicate that these substitutions collectively play a role in the resistance against penicillins as also previously suggested (40–42). The T446A and E476Q substitutions, which have been found in most clinical isolates of S. pneumoniae with reduced susceptibility (43, 44), were both found in susceptible strains of S. mitis and S. oralis (Fig. 2b; see also Table S3). Interestingly, the E476Q substitution was found in all strains of S. mitis and S. oralis regardless of their susceptibility but not in any susceptible strains of S. pneumoniae.
The alteration of T371 (A/S) in the pbp1a gene in the S370TMK active-site motif was found only in strains with reduced susceptibility (Fig. 2c; see also Table S4 in the supplemental material), in agreement with previous observations on clinical isolates of pneumococci (22, 45). Likewise, the V408L substitution was associated with reduced susceptibility although this substitution was found also in a single susceptible strain, S. mitis SK616. The four consecutive residues of T574SQF to NTGY have been linked to resistance in S. pneumoniae (45) but were found in many susceptible strains of S. mitis and S. oralis.
Impact of the murM gene on beta-lactam susceptibility.
In the cluster analysis of the pbp2x gene, we observed that some strains clustered very closely independently of their susceptibility to beta-lactams. This was the case for resistant S. oralis mit_1836_02, which was very closely related to five susceptible strains of S. mitis in a subcluster of CX1 (subcluster a). Another example was observed in CX2, where the susceptible S. mitis SK1126 strain in CX2 was genetically identical in the pbp2x gene to the resistant S. mitis MC12265, S. infantis 13D-1, and S. infantis 13D-2. Although these strains were dissimilar in the two other genes, this probably does not explain all the observed differences in the beta-lactam susceptibility. On the other hand, it was previously shown that a characteristic mosaic structure in the murM gene may contribute to beta-lactam resistance in S. pneumoniae strains and that a deletion of this gene in resistant strains of S. pneumoniae would change the phenotype of resistant strains to susceptible (46, 47). Whole-genome comparisons of S. mitis and S. oralis strains revealed that some S. mitis and S. oralis strains lacked a murM gene similar to that found in S. pneumoniae (data not shown). Using two specific PCR assays, we found that S. oralis mit_1836_02 did harbor the murM gene whereas the closely related S. mitis M3, 8B-3, SK667, and MAYO 6869 strains in CX2 did not, which perfectly correlates with the susceptibility of these strains (Fig. 1a). Likewise, S. mitis MC12265, S. infantis 13D-1, and S. infantis 13D-2 did harbor a murM gene, while sequence analysis of the murM gene in S. mitis SK1126 derived from genome sequences revealed a stop codon in the murM gene of this strain. This pseudogene may be responsible for the susceptible phenotype of SK1126 in spite of the presence of the altered pbp genes. Other strains were not tested for the presence of the murM gene.
DISCUSSION
Understanding the mechanisms of the evolution of antibiotic resistance in bacteria is crucial. It is generally recognized that resistance to beta-lactam antibiotics in pneumococci is due to alterations in the amino acid sequence of the PBP2x, PBP2b, and PBP1a penicillin-binding proteins (8) driven by selection in the presence of excessive use of antibiotics. PBP alterations are reflected in a mosaic structure in the proteins and the corresponding genes as a result of homologous recombination with other pneumococci and closely related commensal streptococci, most notably, S. mitis and S. oralis, which share the pharynx as an ecological habitat (9, 10). Although previous studies have shown that S. mitis is the main donor of genes acquired by S. pneumoniae in homologous recombination events, our data were unable to distinguish between S. mitis and S. oralis as the main donor, presumably due to prior recombination between the two species (17, 21, 48). To fully understand this interspecies relationship, it is necessary to identify the sequence diversity of the pbp genes in commensal streptococci. To our knowledge, this study is the first to identify the sequence diversity of the three major pbp genes, pbp2x, pbp2b, and pbp1a, in a large collection of susceptible and nonsusceptible strains of commensal streptococci, in particular, S. mitis. We found that the overall number of polymorphic sites generated by spontaneous mutations in all three genes was considerable (up to 39% of all sites) for both susceptible and nonsusceptible strains of S. mitis, S. oralis, and S. infantis. This contrasts with the situation among pneumococci, where only nonsusceptible strains display extensive sequence variations as a consequence of the import of sequences previously diversified in commensal streptococci. This is in agreement with the general population structure of Mitis group streptococci, where pneumococci have a clonal origin, while both S. mitis and S. oralis consist of multiple evolutionary lineages that serve as a gene reservoir for pneumococci (17, 26, 48). Our phylogenetic analysis of the pbp genes revealed that the distances between lineages of S. mitis were greater than between those of susceptible pneumococci and S. mitis, supporting the hypothesis that S. pneumoniae is a successful pathogenic lineage within the S. mitis cluster (17, 48).
In agreement with a previous report (8), homologous recombination affecting pbp genes was detected almost exclusively in nonsusceptible strains of pneumococci. In contrast, commensal streptococci showed evidence of homologous inter- and intraspecies recombination in both susceptible and nonsusceptible strains. The finding of interspecies recombination affecting pbp genes of S. pneumoniae, S. mitis, and S. oralis is in agreement with previous reports (9, 13–16). The predominantly unidirectional gene transfer from S. mitis to S. pneumoniae is in agreement with results of our recent comparative analyses of S. mitis and S. pneumoniae genomes (17). By this process, pneumococci have exploited the more extensive diversification that already occurs in S. mitis lineages as a result of accumulation of mutations and intraspecies recombination. In a few cases, in this study, we observed potential homologous sequence transfer from S. pneumoniae to S. mitis and S. oralis. It is conceivable that these potential exceptions were facilitated by the extensive selective pressure on the pbp genes exerted by beta-lactam antibiotic usage (49).
The absence of interspecies recombination affecting pbp genes in susceptible strains of pneumococci may be due to reduced fitness of strains displaying mosaic structure in the absence of antibiotic selection pressure. In support of this, Albarracín Orio et al. (50) observed that incorporation of pbp2b mutant alleles from commensal streptococci resulted in fitness costs. Incorporation of pbp1a and pbp2x mutant alleles, however, had a compensatory effect, which may explain why most resistant pneumococci are altered in all three pbp genes.
Our recombination analysis suggests that very small (i.e., below 100-bp) sequence fragments were involved in recombination, which would be in agreement with a recent conclusion reported by Mostowy et al. (51) that recombination involving small sequence fragments, termed microrecombination, has an important impact on pneumococcal evolution. Nevertheless, Sauerbier et al. (52) showed that, under in vitro conditions, random transformation events occurring between S. mitis B6 and S. pneumoniae R6 involved from 160 to 23,000 nucleotides. Therefore, a potential explanation is that the small mosaic units observed by Mostowy et al. (51) and us are a result of several independent but partially overlapping recombination events. Either way, these small diverse fragments generated by mutations play a crucial role in shaping the pbp genes and, thus, the evolution of beta-lactam resistance in commensal streptococci and S. pneumoniae.
Although the collection of strains for this study was not systematic, our data imply that the occurrence of commensal Mitis group streptococci with reduced susceptibility is more common in countries with high-level usage of antibiotics such as the United States and Greece than in countries with a more restricted use of antibiotics such as Denmark. This is in agreement with studies that have shown an increase of the carriage of resistant nonpneumococcal alpha-hemolytic streptococci after antibiotic therapy (53, 54). Apparently, these commensal streptococci may serve as a reservoir long after the occurrence of selective pressure of beta-lactam antibiotic usage because the fitness cost of the resistant phenotype appears to be lower in S. mitis and S. oralis strains than in S. pneumoniae strains.
A recent genome-wide association study by Chewapreecha et al. (21) involving 4,001 pneumococcal isolates aimed at identifying single nucleotide polymorphisms (SNPs) that could confer beta-lactam nonsusceptibility. The study identified 301 SNPs (207 in pbp genes), of which 71 (52 in pbp genes) were nonsynonymous and associated with beta-lactam nonsusceptibility. Interestingly, of the 20 amino acid alterations shared with our study, all but 1 were detected in susceptible S. mitis or S. oralis strains (MIC values for penicillin, below 0.032 μg/ml) (Fig. 2). Furthermore, 15 of these substitutions were found in more than 75% of all the S. mitis and S. oralis strains in our study (Fig. 2). Similar correlations were observed for many of the amino acid alterations associated with beta-lactam resistance in 80 pneumococcal strains found in the study of Hsieh et al. (22). These included several alterations associated with pneumococcal resistance also reported in other studies (8) such as R384G in the pbp2x gene, T574SQF to NTGY in the pbp1a gene, and E476G in the pbp2b gene. These observations clearly suggest that many of the amino acid alterations previously found to be associated with resistance in pneumococci more likely are species signatures of the donor of the sequence rather than the specific alteration causing resistance. Our findings highlight the difficulties in detecting true causal amino acid alterations even in large genome-wide association studies without reference to the large population of closely related Mitis group streptococci.
Our results support the hypothesis that the murM gene may contribute to beta-lactam resistance in commensal streptococci, as is the case for S. pneumoniae (47, 55). We found that the presence of the murM gene was necessary for commensal streptococci to develop a resistant phenotype. In pneumococci, the murM gene is always present (46), indicating that the murM gene is indispensable in pneumococci but not in commensal streptococci as long as they remain susceptible to beta-lactam antibiotics. To what degree the murM gene and, potentially, other genes may contribute to beta-lactam resistance in commensal streptococci needs to be evaluated in more detail.
In conclusion, the transpeptidase regions of pbp2x, pbp2b, and pbp1a in commensal Mitis group streptococci display considerable sequence variation at both the nucleotide and the amino acid levels. In contrast to pneumococci, sequence variation was observed in both susceptible and nonsusceptible strains, reflecting the level of genetic diversification within the species, including consequences of prior intra- and interspecies homologous recombination events. Furthermore, our data support the hypothesis that resistance to beta-lactam antibiotics in pneumococci is due to sequences acquired from commensal Mitis group streptococci, especially S. mitis. Several amino acid alterations found to be associated with beta-lactam resistance in pneumococci were also found to be associated with reduced susceptibility in the commensal streptococci. However, other amino acid alterations previously linked to beta-lactam resistance in pneumococci appear to represent species signatures of the donor strain rather than being causal of resistance. This report clearly demonstrates the importance of the commensal Mitis group streptococci as a reservoir of beta-lactam resistance exploited by pneumococci. Further insight into this relationship obtained by whole-genome sequencing of a comprehensive collection of susceptible and resistant strains of Mitis group streptococci will facilitate the exact identification of structural changes in penicillin-binding proteins responsible for resistance to beta-lactam antibiotics. Such information may eventually enable the design of modified antibiotics that circumvent these resistance mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
Strains with names with the prefixes MC and DJH used in this study were received from Michael F. Cole at Georgetown and Dwight J. Hardy at the University of Rochester, respectively. Those with the prefixes mit_ and Mayo were from within the collections of U.S. strains held at the University of Alabama in Birmingham. We gratefully acknowledge the assistance of Pat Coan in strain isolation and maintenance at UAB and Sotos Kalfas for collecting oral samples in Greece.
This work was supported by grant 10-083748 from the Danish Research Council for Health and Disease.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00429-15.
REFERENCES
- 1.Mitchell TJ. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat Rev Microbiol 1:219–230. doi: 10.1038/nrmicro771. [DOI] [PubMed] [Google Scholar]
- 2.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 3.Henriques-Normark B. 2007. Molecular epidemiology and mechanisms for antibiotic resistance in Streptococcus pneumoniae, p 269–290. In Hakenbeck R, Chhatwal GS (ed), Molecular biology of streptococci. Horizon Press, Wymondham, Norfolk, United Kingdom. [Google Scholar]
- 4.Anonymous. 2013. Centers for Disease Control and Prevention. Streptococcus pneumoniae. Active Bacterial Core Surveillance report, Emerging Infections Program Network 2013. http://www.cdc.gov/abcs/reports-findings/survreports/spneu13.pdf.
- 5.Granizo JJ, Aguilar L, Casal J, García-Rey C, Dal-Ré R, Baquero F. 2000. Streptococcus pneumoniae resistance to erythromycin and penicillin in relation to macrolide and beta-lactam consumption in Spain (1979–1997). J Antimicrob Chemother 46:767–773. doi: 10.1093/jac/46.5.767. [DOI] [PubMed] [Google Scholar]
- 6.Carratalá J, Alcaide F, Fernández-Sevilla A, Corbella X, Lińares J, Gudiol F. 1995. Bacteremia due to viridans streptococci that are highly resistant to penicillin: increase among neutropenic patients with cancer. Clin Infect Dis 20:1169–1173. doi: 10.1093/clinids/20.5.1169. [DOI] [PubMed] [Google Scholar]
- 7.Doern GV, Ferraro MJ, Brueggemann AB, Ruoff KL. 1996. Emergence of high rates of antimicrobial resistance among viridans group streptococci in the United States. Antimicrob Agents Chemother 40:891–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakenbeck R, Bruckner R, Denapaite D, Maurer P. 2012. Molecular mechanisms of beta-lactam resistance in Streptococcus pneumoniae. Future Microbiol 7:395–410. doi: 10.2217/fmb.12.2. [DOI] [PubMed] [Google Scholar]
- 9.Dowson CG, Hutchison A, Brannigan JA, George RC, Hansman D, Linares J, Tomasz A, Smith JM, Spratt BG. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci U S A 86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laible G, Spratt BG, Hakenbeck R. 1991. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol 5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 11.Martin C, Sibold C, Hakenbeck R. 1992. Relatedness of penicillin-binding protein 1a genes from different clones of penicillin-resistant Streptococcus pneumoniae isolated in South Africa and Spain. EMBO J 11:3831–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowson CG, Hutchison A, Woodford N, Johnson AP, George RC, Spratt BG. 1990. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc Natl Acad Sci U S A 87:5858–5862. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi F, Nolte O, Bergmann C, Ip M, Hakenbeck R. 2007. Crossing the barrier: evolution and spread of a major class of mosaic pbp2x in Streptococcus pneumoniae, S. mitis and S. oralis. Int J Med Microbiol 297:503–512. doi: 10.1016/j.ijmm.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Dowson CG, Coffey TJ, Kell C, Whiley RA. 1993. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol Microbiol 9:635–643. doi: 10.1111/j.1365-2958.1993.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 15.Reichmann P, Konig A, Linares J, Alcaide F, Tenover FC, McDougal L, Swidsinski S, Hakenbeck R. 1997. A global gene pool for high-level cephalosporin resistance in commensal Streptococcus species and Streptococcus pneumoniae. J Infect Dis 176:1001–1012. doi: 10.1086/516532. [DOI] [PubMed] [Google Scholar]
- 16.Sibold C, Henrichsen J, Konig A, Martin C, Chalkley L, Hakenbeck R. 1994. Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol Microbiol 12:1013–1023. doi: 10.1111/j.1365-2958.1994.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 17.Kilian M, Riley DR, Jensen A, Bruggemann H, Tettelin H. 2014. Parallel evolution of Streptococcus pneumoniae and Streptococcus mitis to pathogenic and mutualistic lifestyles. mBio 5:e01490-14. doi: 10.1128/mBio.01490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amoroso A, Demares D, Mollerach M, Gutkind G, Coyette J. 2001. All detectable high-molecular-mass penicillin-binding proteins are modified in a high-level beta-lactam-resistant clinical isolate of Streptococcus mitis. Antimicrob Agents Chemother 45:2075–2081. doi: 10.1128/AAC.45.7.2075-2081.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.König A, Reinert RR, Hakenbeck R. 1998. Streptococcus mitis with unusually high level resistance to beta-lactam antibiotics. Microb Drug Resist 4:45–49. doi: 10.1089/mdr.1998.4.45. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama A, Takao A. 2003. Beta-lactam resistance in Streptococcus mitis isolated from saliva of healthy subjects. J Infect Chemother 9:321–327. doi: 10.1007/s10156-003-0286-Y. [DOI] [PubMed] [Google Scholar]
- 21.Chewapreecha C, Marttinen P, Croucher NJ, Salter SJ, Harris SR, Mather AE, Hanage WP, Goldblatt D, Nosten FH, Turner C, Turner P, Bentley SD, Parkhill J. 2014. Comprehensive identification of single nucleotide polymorphisms associated with beta-lactam resistance within pneumococcal mosaic genes. PLoS Genet 10:e1004547. doi: 10.1371/journal.pgen.1004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh YC, Su LH, Hsu MH, Chiu CH. 2013. Alterations of penicillin-binding proteins in pneumococci with stepwise increase in beta-lactam resistance. Pathog Dis 67:84–88. doi: 10.1111/2049-632x.12018. [DOI] [PubMed] [Google Scholar]
- 23.Bruckner LB, Korones DN, Karnauchow T, Hardy DJ, Gigliotti F. 2002. High incidence of penicillin resistance among alpha-hemolytic streptococci isolated from the blood of children with cancer. J Pediatr 140:20–26. doi: 10.1067/mpd.2002.118886. [DOI] [PubMed] [Google Scholar]
- 24.Cole MF, Bryan S, Evans MK, Pearce CL, Sheridan MJ, Sura PA, Wientzen RL, Bowden GH. 1999. Humoral immunity to commensal oral bacteria in human infants: salivary secretory immunoglobulin A antibodies reactive with Streptococcus mitis biovar 1, Streptococcus oralis, Streptococcus mutans, and Enterococcus faecalis during the first two years of life. Infect Immun 67:1878–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Razonable RR, Litzow MR, Khaliq Y, Piper KE, Rouse MS, Patel R. 2002. Bacteremia due to viridans group Streptococci with diminished susceptibility to levofloxacin among neutropenic patients receiving levofloxacin prophylaxis. Clin Infect Dis 34:1469–1474. doi: 10.1086/340352. [DOI] [PubMed] [Google Scholar]
- 26.Bishop CJ, Aanensen DM, Jordan GE, Kilian M, Hanage WP, Spratt BG. 2009. Assigning strains to bacterial species via the internet. BMC Biol 7:3. doi: 10.1186/1741-7007-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoshino T, Fujiwara T, Kilian M. 2005. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J Clin Microbiol 43:6073–6085. doi: 10.1128/JCM.43.12.6073-6085.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anonymous. 2014. Breakpoint tables for interpretation of MICs and zone diameters, version 4.0. The European Committee on Antimicrobial Susceptibility Testing. http://www.eucast.org.
- 29.Jensen A, Kilian M. 2012. Delineation of Streptococcus dysgalactiae, its subspecies, and its clinical and phylogenetic relationship to Streptococcus pyogenes. J Clin Microbiol 50:113–126. doi: 10.1128/JCM.05900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J. 2012. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res 40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asahi Y, Takeuchi Y, Ubukata K. 1999. Diversity of substitutions within or adjacent to conserved amino acid motifs of penicillin-binding protein 2X in cephalosporin-resistant Streptococcus pneumoniae isolates. Antimicrob Agents Chemother 43:1252–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chesnel L, Pernot L, Lemaire D, Champelovier D, Croize J, Dideberg O, Vernet T, Zapun A. 2003. The structural modifications induced by the M339F substitution in PBP2x from Streptococcus pneumoniae further decreases the susceptibility to beta-lactams of resistant strains. J Biol Chem 278:44448–44456. doi: 10.1074/jbc.M305948200. [DOI] [PubMed] [Google Scholar]
- 34.Granger D, Boily-Larouche G, Turgeon P, Weiss K, Roger M. 2005. Genetic analysis of pbp2x in clinical Streptococcus pneumoniae isolates in Quebec, Canada. J Antimicrob Chemother 55:832–839. doi: 10.1093/jac/dki118. [DOI] [PubMed] [Google Scholar]
- 35.Mouz N, Di Guilmi AM, Gordon E, Hakenbeck R, Dideberg O, Vernet T. 1999. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumoniae. Role in the specificity for beta-lactam antibiotics. J Biol Chem 274:19175–19180. [DOI] [PubMed] [Google Scholar]
- 36.Nichol KA, Zhanel GG, Hoban DJ. 2002. Penicillin-binding protein 1A, 2B, and 2X alterations in Canadian isolates of penicillin-resistant Streptococcus pneumoniae. Antimicrob Agents Chemother 46:3261–3264. doi: 10.1128/AAC.46.10.3261-3264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carapito R, Chesnel L, Vernet T, Zapun A. 2006. Pneumococcal beta-lactam resistance due to a conformational change in penicillin-binding protein 2x. J Biol Chem 281:1771–1777. doi: 10.1074/jbc.M511506200. [DOI] [PubMed] [Google Scholar]
- 38.Denapaite D, Bruckner R, Nuhn M, Reichmann P, Henrich B, Maurer P, Schahle Y, Selbmann P, Zimmermann W, Wambutt R, Hakenbeck R. 2010. The genome of Streptococcus mitis B6—what is a commensal? PLoS One 5:e9426. doi: 10.1371/journal.pone.0009426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maurer P, Koch B, Zerfass I, Krauss J, van der Linden M, Frere JM, Contreras-Martel C, Hakenbeck R. 2008. Penicillin-binding protein 2x of Streptococcus pneumoniae: three new mutational pathways for remodelling an essential enzyme into a resistance determinant. J Mol Biol 376:1403–1416. doi: 10.1016/j.jmb.2007.12.058. [DOI] [PubMed] [Google Scholar]
- 40.Cafini F, del Campo R, Alou L, Sevillano D, Morosini MI, Baquero F, Prieto J, Spanish Pneumococcal Network (G03/103) . 2006. Alterations of the penicillin-binding proteins and murM alleles of clinical Streptococcus pneumoniae isolates with high-level resistance to amoxicillin in Spain. J Antimicrob Chemother 57:224–229. doi: 10.1093/jac/dki442. [DOI] [PubMed] [Google Scholar]
- 41.du Plessis M, Bingen E, Klugman KP. 2002. Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob Agents Chemother 46:2349–2357. doi: 10.1128/AAC.46.8.2349-2357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kosowska K, Jacobs MR, Bajaksouzian S, Koeth L, Appelbaum PC. 2004. Alterations of penicillin-binding proteins 1A, 2X, and 2B in Streptococcus pneumoniae isolates for which amoxicillin MICs are higher than penicillin MICs. Antimicrob Agents Chemother 48:4020–4022. doi: 10.1128/AAC.48.10.4020-4022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanbongi Y, Ida T, Ishikawa M, Osaki Y, Kataoka H, Suzuki T, Kondo K, Ohsawa F, Yonezawa M. 2004. Complete sequences of six penicillin-binding protein genes from 40 Streptococcus pneumoniae clinical isolates collected in Japan. Antimicrob Agents Chemother 48:2244–2250. doi: 10.1128/AAC.48.6.2244-2250.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith AM, Klugman KP. 1995. Alterations in penicillin-binding protein 2b from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob Agents Chemother 39:859–867. doi: 10.1128/AAC.39.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith AM, Klugman KP. 1998. Alterations in PBP 1A essential for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother 42:1329–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filipe SR, Tomasz A. 2000. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc Natl Acad Sci U S A 97:4891–4896. doi: 10.1073/pnas.080067697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber B, Ehlert K, Diehl A, Reichmann P, Labischinski H, Hakenbeck R. 2000. The fib locus in Streptococcus pneumoniae is required for peptidoglycan crosslinking and PBP-mediated beta-lactam resistance. Fems Microbiology Lett 188:81–85. [DOI] [PubMed] [Google Scholar]
- 48.Kilian M, Poulsen K, Blomqvist T, Havarstein LS, Bek-Thomsen M, Tettelin H, Sorensen UBS. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3:e2683. doi: 10.1371/journal.pone.0002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, Pessia A, Aanensen DM, Mather AE, Page AJ, Salter SJ, Harris D, Nosten F, Goldblatt D, Corander J, Parkhill J, Turner P, Bentley SD. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet 46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albarracín Orio AG, Piñas GE, Cortes PR, Cian MB, Echenique J. 2011. Compensatory evolution of pbp mutations restores the fitness cost imposed by beta-lactam resistance in Streptococcus pneumoniae. PLoS Pathog 7:e1002000. doi: 10.1371/journal.ppat.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mostowy R, Croucher NJ, Hanage WP, Harris SR, Bentley S, Fraser C. 2014. Heterogeneity in the frequency and characteristics of homologous recombination in pneumococcal evolution. PLoS Genet 10:e1004300. doi: 10.1371/journal.pgen.1004300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sauerbier J, Maurer P, Rieger M, Hakenbeck R. 2012. Streptococcus pneumoniae R6 interspecies transformation: genetic analysis of penicillin resistance determinants and genome-wide recombination events. Mol Microbiol 86:692–706. doi: 10.1111/mmi.12009. [DOI] [PubMed] [Google Scholar]
- 53.Ghaffar F, Friedland IR, Katz K, Muniz LS, Smith JL, Davis P, Reynolds J, McCracken GH Jr. 1999. Increased carriage of resistant non-pneumococcal alpha-hemolytic streptococci after antibiotic therapy. J Pediatr 135:618–623. doi: 10.1016/S0022-3476(99)70061-2. [DOI] [PubMed] [Google Scholar]
- 54.Ghaffar F, Muniz LS, Katz K, Smith JL, Shouse T, Davis P, McCracken GH Jr. 2002. Effects of large dosages of amoxicillin/clavulanate or azithromycin on nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, nonpneumococcal alpha-hemolytic streptococci, and Staphylococcus aureus in children with acute otitis media. Clin Infect Dis 34:1301–1309. doi: 10.1086/340054. [DOI] [PubMed] [Google Scholar]
- 55.Filipe SR, Severina E, Tomasz A. 2000. Distribution of the mosaic structured murM genes among natural populations of Streptococcus pneumoniae. J Bacteriol 182:6798–6805. doi: 10.1128/JB.182.23.6798-6805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.