Abstract
Genome alignment of a macrolide, lincosamide, and streptogramin B (MLSB)-resistant Staphylococcus fleurettii strain with an MLSB-susceptible S. fleurettii strain revealed a novel 11,513-bp genomic island carrying the new erythromycin resistance methylase gene erm(45). This gene was shown to confer inducible MLSB resistance when cloned into Staphylococcus aureus. The erm(45)-containing island was integrated into the housekeeping gene guaA in S. fleurettii and was able to form a circular intermediate but was not transmissible to S. aureus.
TEXT
Staphylococcus fleurettii is a commensal bacterium of various animal species and an occasional cause of bovine mastitis (1–3). It naturally contains the methicillin resistance gene mecA within its chromosome and is therefore suspected to have been the source of the mecA gene found in staphylococcal cassette chromosome mec (SCCmec) of methicillin-resistant staphylococci, including methicillin-resistant Staphylococcus aureus (MRSA) (4).
Due to this intrinsic resistance to β-lactams, other antibiotic classes such as macrolides or lincosamides are being used for the treatment of mastitis caused by S. fleurettii (5, 6).
S. fleurettii strain JW205, recently isolated from bovine milk in Switzerland, exhibited resistance to erythromycin and inducible resistance to clindamycin (3). This suggested the presence of a macrolide, lincosamide, and streptogramin B (MLSB) resistance methylase (Erm) (7). However, none of the erm genes commonly occurring in staphylococci were detected by microarray analysis (8, 9). We therefore examined S. fleurettii strain JW205 for a novel MLSB resistance mechanism by genome comparison with MLSB-susceptible S. fleurettii strain JW404 (Table 1).
TABLE 1.
MICs of erythromycin, clindamycin, and pristinamycin IA for different S. aureus and S. fleurettii strains as determined by broth microdilution
| Strain | Origin and characteristic(s) | Reference(s) | Antibiotic resistance gene(s)a | MIC (μg/ml) for:b |
||||
|---|---|---|---|---|---|---|---|---|
| ERY | CLI | iCLI | PIA | iPIA | ||||
| S. aureus | ||||||||
| 80CR5 | Recipient strain for conjugation, plasmid free, Rifr Fusr | 26 | ≤0.5 | ≤0.5 | NAc | 4 | NA | |
| RN4220 | Recipient strain for electrotransformation, plasmid free | 27 | ≤0.5 | ≤0.5 | NA | 4 | NA | |
| RN4220/pBUS1-HCd | RN4220 with S. aureus-E. coli shuttle vector pBUS1-HC | 28 | tet(L) | ≤0.5 | ≤0.5 | NA | 4 | NA |
| RN4220/pBUS1-Pcap-HCd | RN4220 with S. aureus-E. coli shuttle vector pBUS1-Pcap-HC | 28 | tet(L) | ≤0.5 | ≤0.5 | NA | 4 | NA |
| RN4220/pBJW15 | RN4220 with erm(45) and its regulatory region cloned into pBUS1-HC | This study | tet(L), erm(45) | 32 | ≤0.5 | 256 | 4 | 8 |
| RN4220/pBJW16 | RN4220 with erm(45) cloned into pBUS1-Pcap-HC | This study | tet(L), erm(45) | >256 | >256 | >256 | 64 | 64 |
| S. fleurettii | ||||||||
| JW205 | Bovine milk | 3, this study | erm(45), mecA, tet(K) | 16 | 1 | 64 | 4 | 8 |
| JW404 | Bovine mastitis milk | 3, this study | mecA | ≤0.5 | ≤0.5 | NA | 2 | NA |
Antibiotic resistance genes and functions: tet(L) and tet(K), tetracycline efflux genes; mecA, penicillin binding protein 2A gene; erm(45), 23S rRNA methylase gene.
ERY, erythromycin; CLI, clindamycin; PIA, pristinamycin IA; iCLI and iPIA, 2 μg/ml erythromycin added to the broth for the detection of inducible resistance to clindamycin (iCLI) and pristinamycin IA (iPIA).
NA, not applicable.
pBUS1-HC is a promoterless cloning vector and pBUS1-Pcap-HC is an expression vector that harbors the strong cap 1A promoter of the S. aureus type 1 capsular polysaccharide biosynthesis gene cluster.
Detection and characterization of erm(45).
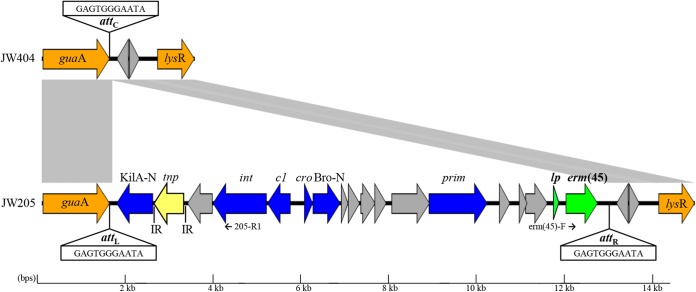
Genomes of strain JW205 and JW404 were sequenced using Ion Torrent (Life Technologies, Grand Island, NY) at the UZH/ETH Functional Genomics Center (Zurich, Switzerland) and Illumina MiSeq (Illumina, San Diego, CA) at the Department of Clinical Microbiology at Hvidovre Hospital (Hvidovre, Denmark), respectively. Contigs of the sequenced strains were aligned using the progressive Mauve algorithm (10). This revealed an 11,513-bp integrated genomic island in JW205, which was absent in strain JW404. The island contained 18 open reading frames (ORFs) (Fig. 1), which were identified by the Prokaryotic Dynamic Programming Genefinding Algorithm (Prodigal) (11) and compared to protein sequences and conserved domains in the BLASTp program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the Swiss Institute of Bioinformatics PROSITE database (http://prosite.expasy.org/). The rightmost ORF of the island encoded a 245-amino-acid (aa) protein, which contained the rRNA adenine dimethylase PROSITE signature PS01131, which is present in nearly all Erm 23S rRNA methylases (12). Of all 36 currently described Erm determinants, this methylase exhibited the highest similarity to Erm(B), with 64% aa and 67% nucleotide (nt) identity (Fig. 2). The novel gene was assigned the name erm(45) according to the established MLSB resistance gene nomenclature (http://faculty.washington.edu/marilynr/), which defines novel Erm determinants by amino acid sequence identities of ≤79% compared to those of their closest Erm protein (13). Like erm(B), the erm(45) gene was preceded by a leader region encoding a single 27-aa leader peptide (Lp) (14). The Lp of erm(B) and the Lp of erm(45) shared 81% aa and 81% nt identity and each harbored the MRNVD motif, which is crucial for inducible expression of erm methylases (14). The leader region of erm(45) also contained 2 different pairs of inverted repeats (IRs), identical to those found in erm(B), which have been shown to form stem-loops involved in translational attenuation (14).
FIG 1.
Schematic gene map showing the erm(45)-containing island and flanking region of S. fleurettii JW205 (ENA accession number LN680996), as well as the chromosomal glutamine amidotransferase gene (guaA) region of erm(45)-negative strain JW404. Gray areas represent regions showing high similarity at nucleotide level (>98%). Arrows represent position and orientation of open reading frames (ORFs). The erm(45) and its leader peptide (lp) gene are shown in green. The 11-bp (GAGTGGGAATA) chromosomal hot spot situated within the 3′ end of guaA is abbreviated as attC. This attachment site is illustrated as attL and attR at each extremity of the island. The IS431 transposase gene (tnp) illustrated in yellow is flanked by 16-bp inverted repeats (IR) (GGTTCTGTTGCAAAGT). Other putative genes and functions: KilA-N, KilA-N domain-carrying putative DNA-binding protein; c1/cro, repressors; Bro-N, Bro-N domain-carrying putative DNA-binding protein; lysR, transcriptional regulator; int, integrase; prim, primase. Color code: S. fleurettii core genome, orange; erm(45)-carrying island genes related to SaPI and bacteriophage genomes, blue; antibiotic resistance, green; no known function, significant similarity to proteins in the database used, or any known protein signatures, gray. Primers used to detect circular forms of the erm(45)-carrying island are indicated by small black arrows. The figure was generated using the program Easyfig (25).
FIG 2.
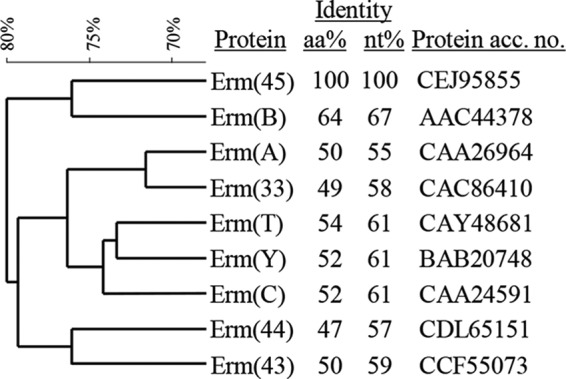
Relatedness of the novel erythromycin resistance methylase Erm(45) with other methylases commonly detected in Staphylococcus species (9, 12). Amino acid (aa) and nucleotide (nt) identity percentages were obtained by Clustal Omega sequence alignment (http://www.ebi.ac.uk/Tools/msa/clustalo/). Erm aa sequence clustering was performed by BioNumerics 7.1 (Applied Maths). Comparison settings used the standard algorithm for pairwise alignment, open gap penalty 100%, unit gap penalty 0%, and the unweighted-pair group method using average linkages.
Cloning and expression of erm(45).
To test functionality and inducibility of erm(45) a 1,098-bp region of strain JW205 including erm(45), its leader peptide and promoter (represented by green arrows in Fig. 1) was amplified by PCR (Pfu DNA Polymerase; Promega Corporation, Madison, WI) using primers erm(45)-Sal1-F2 (5′-cacagggtcgacATAAGTTGTTAGTAAATAGTATTCAAC) and erm(45)-XbaI-R (5′-cacaggtctagaCACCTATTTCAATACTAGG) (annealing temperatures, 50°C for cycles 1 to 3 and 56°C for cycles 4 to 24; elongation time, 2 min 30 s). The primers also contained polylinkers (lowercase) with restriction site sequences (underlined) to facilitate cloning. A 772-bp region containing erm(45) alone without its leader sequence was also amplified by PCR using primers erm(45)-NdeI-F2 (5′-cacacacggcatatgAATCAAAATATTAAGTTTACTC) and erm(45)-XbaI-R under the same conditions. The former amplicon was cloned into the SalI and XbaI restriction sites of pBUS1-HC, generating plasmid pBJW15, where erm(45) was under the control of its own promoter (Table 1). The latter amplicon was placed into the NdeI and XbaI restriction sites of pBUS1-Pcap-HC, generating plasmid pBJW16, where erm(45) was under the control of the strong S. aureus type I capsule gene 1A promoter provided by the vector (Table 1). Plasmids pBJW15 and pBJW16 were first transformed by heat shock into Escherichia coli DH5α and were subsequently electroporated into S. aureus RN4220 (15) (Table 1). All transformants were grown on LB agar plates containing 10 μg/ml tetracycline. MIC values of erythromycin, clindamycin (Sigma-Aldrich, St. Louis, MO), and pristinamycin IA (Molcan Corporation, Richmond Hill, ON, Canada) of S. fleurettii and S. aureus strains were determined by broth microdilution using Mueller-Hinton broth according to CLSI guidelines (16). MICs for inducible resistance were measured in the presence of 2 μg/ml erythromycin (16). When erm(45) was expressed from its own promoter in plasmid pBJW15 in S. aureus RN4220, the MIC of erythromycin increased at least 64-fold, while the MICs of clindamycin (>512-fold) and pristinamycin IA (2-fold) increased only after induction with erythromycin (Table 1). The RN4220 transformant carrying pBJW16 and expressing erm(45) constitutively exhibited a >512-fold increase of the MICs of erythromycin and clindamycin and a 16-fold increase of the MIC for pristinamycin IA (Table 1).
Characterization of the erm(45)-containing genomic island.
The 11,513-bp island containing erm(45) was flanked by 11-bp direct repeats (DRs) (GAGTGGGAATA) situated within the 3′ end of the glutamine amidotransferase (GMP) synthetase housekeeping gene guaA (Fig. 1), a known integration hot spot for genetic islands, transposons, and bacteriophages in different bacterial species (17, 18). For instance, identical DRs and guaA integration sites were associated with the pathogenicity islands of staphylococci (SaPIs) SaPIbov1 and SaPIbov2, identified in S. aureus from bovine mastitis milk (19). SaPIs are characterized by their specific set of bacteriophage-related genes and functions, the ability to exploit the life cycle of bacteriophages in favor of their own, and sizes ranging from 3 to 28 kb with most of them being 14 to 17 kb (19). Although the size of the element (11.5 kb) suggested a potential relation to SaPIs, the erm(45)-carrying island did not show significant DNA similarity to SaPI genomes in the GenBank and lacked the conserved genes rep, xis, pif, and terS, which are involved in the replication and excision of SaPIs, and in their interferences with phages (19). Nevertheless, six of the putative ORFs on the erm(45)-carrying island contained conserved domains related to SaPI proteins. In each element, these ORFs are structurally organized in a similar fashion at the 5′ end (Fig. 1). In the erm(45)-carrying island, they consisted of one putative integrase (Pfam signature cd00397), a putative cro repressor and c1 antirepressor (PROSITE signature PS50943), one putative primase (PS51206), and two ORFs containing the conserved DNA-binding domains KilA-N (PS51301) and Bro-N (SMART accession number 01040; no defined PS), domains which have been found in proteins of bacterial DNA viruses (20–23). The integrase, harboring the conserved similarity regions of tyrosine recombinases (24), was most likely responsible for integration into the S. fleurettii chromosome and the formation of a circular intermediate of the erm(45)-containing island. This intermediate form was detected by PCR (GoTaq Green master mix PCR; Promega, Madison, WI) using primers 205-R1 (5′-GTAACCCTATGGCTCTATCATC) and erm(45)-F (5′-CATAATTTATGAGGTTGGAACTGG), which read outwards from the island (annealing temperature, 55°C; elongation time, 5 min). Due to the structure and the ability to form a circular intermediate, the element carrying erm(45) was classified as a genomic island. Although a circular conformation was observed, transmission of erythromycin resistance was not observed either by electrotransformation into S. aureus RN4220 or by conjugation with S. aureus 80CR5 using conditions previously described (9). An IS431 flanked by 16-bp IRs (GGTTCTGTTGCAAAGT) was situated within the SaPI-like region between the KilA-N domain-containing ORF and the integrase (Fig. 1). The IRs flanking this transposase were the only repeats found that indicate integration of additional DNA into the island.
In conclusion, the detection of the new methylase gene erm(45) in S. fleurettii underlines the role of this bacterium as a reservoir of antibiotic resistance genes. The island containing erm(45) appears to have evolved from SaPIs and bacteriophages, emphasizing the potential of phage and phage-related structures to act as vehicles for antibiotic resistance.
Nucleotide sequence accession number.
The nucleotide sequence of the erm(45)-carrying island of S. fleurettii JW205 and its flanking region was deposited into the ENA database under accession number LN680996.
ACKNOWLEDGMENT
This study was financed by research grant 35-539 from the Institute of Veterinary Bacteriology, University of Bern, Switzerland.
REFERENCES
- 1.Vernozy-Rozand C, Mazuy C, Meugnier H, Bes M, Lasne Y, Fiedler F, Etienne J, Freney J. 2000. Staphylococcus fleurettii sp. nov., isolated from goat's milk cheeses. Int J Syst Evol Microbiol 50:1521–1527. doi: 10.1099/00207713-50-4-1521. [DOI] [PubMed] [Google Scholar]
- 2.Nemeghaire S, Argudín MA, Fessler AT, Hauschild T, Schwarz S, Butaye P. 2014. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet Microbiol 171:342–356. doi: 10.1016/j.vetmic.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Frey Y, Rodriguez JP, Thomann A, Schwendener S, Perreten V. 2013. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J Dairy Sci 96:2247–2257. doi: 10.3168/jds.2012-6091. [DOI] [PubMed] [Google Scholar]
- 4.Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. 2010. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother 54:4352–4359. doi: 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pyörälä S, Baptiste KE, Catry B, van Duijkeren E, Greko C, Moreno MA, Pomba MC, Rantala M, Ružauskas M, Sanders P, Threlfall EJ, Torren-Edo J, Törneke K. 2014. Macrolides and lincosamides in cattle and pigs: use and development of antimicrobial resistance. Vet J 200:230–239. doi: 10.1016/j.tvjl.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 6.Büttner S, Mehmann M, Müntener C, Torriani K, Overesch G. 2014. Bericht über den Vertrieb von Antibiotika in der Veterinärmedizin und das Antibiotikaresistenzmonitoring bei Nutztieren in der Schweiz (ARCH-VET 2013). Federal Food Safety and Veterinary Office and Swissmedic, Bern, Switzerland. [Google Scholar]
- 7.Roberts MC. 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett 282:147–159. doi: 10.1111/j.1574-6968.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 8.Strauss C, Endimiani A, Perreten V. 2015. A novel universal DNA labeling and amplification system for rapid microarray-based detection of 117 antibiotic resistance genes in Gram-positive bacteria. J Microbiol Methods 108:25–30. doi: 10.1016/j.mimet.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Wipf JRK, Schwendener S, Perreten V. 2014. The novel macrolide-lincosamide-streptogramin B resistance gene erm(44) is associated with a prophage in Staphylococcus xylosus. Antimicrob Agents Chemother 58:6133–6138. doi: 10.1128/AAC.02949-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwendener S, Perreten V. 2012. New MLSB resistance gene erm(43) in Staphylococcus lentus. Antimicrob Agents Chemother 56:4746–4752. doi: 10.1128/AAC.00627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppälä H. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother 43:2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min YH, Kwon AR, Yoon EJ, Shim MJ, Choi EC. 2008. Translational attenuation and mRNA stabilization as mechanisms of erm(B) induction by erythromycin. Antimicrob Agents Chemother 52:1782–1789. doi: 10.1128/AAC.01376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett 73:133–138. [DOI] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.Song L, Pan Y, Chen S, Zhang X. 2012. Structural characteristics of genomic islands associated with GMP synthases as integration hotspot among sequenced microbial genomes. Comput Biol Chem 36:62–70. doi: 10.1016/j.compbiolchem.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Mingoia M, Morici E, Tili E, Giovanetti E, Montanari MP, Varaldo PE. 2013. Characterization of Tn5801.Sag, a variant of Staphylococcus aureus Tn916 family transposon Tn5801 that is widespread in clinical isolates of Streptococcus agalactiae. Antimicrob Agents Chemother 57:4570–4574. doi: 10.1128/AAC.00521-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novick RP, Christie GE, Penadés JR. 2010. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol 8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ubeda C, Maiques E, Barry P, Matthews A, Tormo MA, Lasa I, Novick RP, Penadés JR. 2008. SaPI mutations affecting replication and transfer and enabling autonomous replication in the absence of helper phage. Mol Microbiol 67:493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 21.Iyer LM, Koonin E, Aravind VL. 2002. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol 3:research0012. doi: 10.1186/gb-2002-3-3-research0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato'o Y, Omoe K, Ono HK, Nakane A, Hu DL. 2013. A novel comprehensive analysis method for Staphylococcus aureus pathogenicity islands. Microbiol Immunol 57:91–99. doi: 10.1111/1348-0421.12007. [DOI] [PubMed] [Google Scholar]
- 23.Frick DN, Richardson CC. 2001. DNA primases. Annu Rev Biochem 70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 24.Esposito D, Scocca JJ. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res 25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engel HW, Soedirman N, Rost JA, van Leeuwen WJ, van Embden JD. 1980. Transferability of macrolide, lincomycin, and streptogramin resistances between group A, B, and D streptococci, Streptococcus pneumoniae, and Staphylococcus aureus. J Bacteriol 142:407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 28.Schwendener S, Perreten V. 6 March 2015. New shuttle vector-based expression system to generate polyhistidine-tagged fusion proteins in Staphylococcus aureus and Escherichia coli. Appl Environ Microbiol doi: 10.1128/AEM.03803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]