Abstract
Enterobacteriaceae strains producing the Klebsiella pneumoniae carbapenemase (KPC) have disseminated worldwide, causing an urgent threat to public health. KPC-producing strains often exhibit low-level carbapenem resistance, which may be missed by automated clinical detection systems. In this study, eight Klebsiella pneumoniae strains with heterogeneous resistance to imipenem were used to elucidate the factors leading from imipenem susceptibility to high-level resistance as defined by clinical laboratory testing standards. Time-kill analysis with an inoculum as low as 3 × 106 CFU/ml and concentrations of imipenem 8- and 16-fold higher than the MIC resulted in the initial killing of 99.9% of the population. However, full recovery of the population occurred by 20 h of incubation in the same drug concentrations. Population profiles showed that recovery was mediated by a heteroresistant subpopulation at a frequency of 2 × 10−7 to 3 × 10−6. Samples selected 2 h after exposure to imipenem were as susceptible as the unexposed parental strain and produced the major outer membrane porin OmpK36. However, between 4 to 8 h after exposure, OmpK36 became absent, and the imipenem MIC increased at least 32-fold. Individual colonies isolated from cultures after 20 h of exposure revealed both susceptible and resistant subpopulations. Once induced, however, the high-level imipenem resistance was maintained, and OmpK36 remained unexpressed even without continued carbapenem exposure. This study demonstrates the essential coordination between blaKPC and ompK36 expression mediating high-level imipenem resistance from a population of bacteria that initially exhibits a carbapenem-susceptibility phenotype.
INTRODUCTION
The widespread dissemination of carbapenem-resistant Enterobacteriaceae (CRE) has reached a state of urgency in the United States and abroad, greatly diminishing the ability to rely on carbapenems as the drugs of last resort to treat multidrug-resistant CRE infections (1, 2). Strains that produce Klebsiella pneumoniae carbapenemase (KPC), encoded by the blaKPC gene, first emerged with large-scale outbreaks in U.S. hospitals and are now some of the most important contributors to carbapenem resistance worldwide among Gram-negative bacteria (GNB) (2–6). KPC-producing strains coharbor numerous drug resistance determinants, making clinical management of infections caused by such strains very complicated. Mortality exceeds 40% in patients infected with KPC-producing strains, especially when the infection results in bacteremia (2, 4, 7–11).
The failure to detect carbapenem resistance in a timely manner is a major contributor to the high rates of mortality in infections caused by KPC-producing GNB strains. Indeed, strains that harbor blaKPC commonly exhibit low-level resistance to carbapenem drugs and are frequently missed due to inconsistencies across various automated detection systems (7, 11–16). Moreover, carbapenemase-producing organisms are often detected only after therapy in patients fails (10, 14).
These strains often exhibit full or reduced susceptibility to a carbapenem according to standard laboratory testing (1 to 2 μg/ml), but, upon single exposure to a carbapenem, generate subpopulations with MICs of >64 μg/ml. Such strains are said to exhibit heteroresistance. The factors that determine carbapenem heteroresistance are unknown. Here we show how such conversion occurs through coordinated expression of blaKPC and decreased production of the major outer membrane porin OmpK36.
MATERIALS AND METHODS
Strains and susceptibility testing.
Antimicrobial susceptibility testing was performed by broth microdilution in accordance with the standards set by the Clinical and Laboratory Standards Institute (CLSI) and Etest (bioMérieux, Marcy l'Etoile, France). Imipenem (Sigma-Aldrich, St. Louis, MO) was used as the representative carbapenem drug in all experiments. Phenylboronic acid (PBA) (Sigma-Aldrich), an inhibitor of KPC hydrolysis, was used to analyze its effect on imipenem MICs. K. pneumoniae strains were obtained from rectal swabs and bloodstream and urinary tract infection samples collected by hospitals in Brazil and San Francisco. Eight KPC-producing K. pneumoniae strains with clinically relevant imipenem-heteroresistant phenotypes and three KPC-producing K. pneumoniae strains with high-level imipenem resistance were chosen from this set for our analysis (Table 1). Four non-KPC-producing K. pneumoniae clinical strains were chosen as controls. The KPC-producing strains belonged to three different multilocus sequence type (MLST) clonal groups. Strains were considered heteroresistant if colonies grew within the zone of inhibition with an imipenem Etest. Heteroresistant strains were considered clinically relevant if their reference standard broth microdilution imipenem MIC was ≤2 μg/ml. All experiments were prepared with one isolated colony from a freshly streaked Mueller-Hinton (MH) agar plate, which was grown overnight in MH broth at 37°C with shaking. Samples were tested in triplicate, and experiments were performed at least three times.
TABLE 1.
K. pneumoniae strains used in this studya
| Strainb | ST | β-Lactamase gene(s) | IPM MIC (μg/ml) with inoculum of: |
Etest zonec,d | |||
|---|---|---|---|---|---|---|---|
| 5 × 105 (ref)e | 5 × 106 | 5 × 107 | 5 × 108 | ||||
| BR6 (HET) | 437 | blaKPC-2, blaCTX-M (NTf) | 1–2 | 16 | 64 | >64 | 1+ |
| BR7 (HET) | 437 | blaKPC-2, blaCTX-M-1, blaTEM-1 | 1–2 | 16 | 64 | >64 | 1+ |
| BR14 (HET) | 437 | blaKPC-2, blaCTX-M-9, blaTEM-1, blaSHV-11 | 2 | 16 | 64 | >64 | 1+ |
| BR19 (HET) | 437 | blaKPC-2, blaCTX-M-1, blaTEM-1, blaSHV-11 | 2 | 16 | 64 | >64 | 1+ |
| BR21 (HET) | 437 | blaKPC-2, blaCTX-M-1, blaSHV-11, blaOXA-1 | 1–2 | 16 | 64 | >64 | 1+ |
| BR23 (HET) | 437 | blaKPC-2, blaCTX-M-1, blaTEM-1, blaSHV-11 | 2 | 16 | 64 | >64 | 1+ |
| BR26 (HET) | 437 | blaKPC-2, blaCTX-M (NT) | 2 | 16 | 64 | >64 | 1+ |
| BR28 (HET) | 483 | blaKPC-2, blaSHV-11 | 2 | 16 | 64 | >64 | 1+ |
| BR1 (RES) | 340 | blaKPC-2, blaCTX-M (NT) | 16 | >32 | >64 | >64 | 2+ |
| BR20 (RES) | 437 | blaKPC-2, blaCTX-M (NT) | 16 | 32 | >64 | >64 | 2+ |
| BR3 (RES) | 340 | blaKPC-2, blaSHV-11 | >64 | >64 | >64 | >64 | 3+ |
| SF701 (SUSC) | 514 | None | 0.25 | 0.5 | 1 | 4 | 0 |
| SF705 (SUSC) | 1248 | None | 0.25 | 0.25 | 0.5 | 4 | 0 |
| SF519 (SUSC) | 66 | None | 0.25 | 0.25 | 0.5 | 4 | 0 |
| SF681 (SUSC) | 392 | blaCTX-M-15 | 0.25 | 0.5 | 1 | 4 | 0 |
Coharbored β-lactamase genes, the change in imipenem (IPM) susceptibility due to increased inoculum, and the multilocus sequence type (ST) are shown.
Study strain sources: BR, 6 hospitals, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil; SF, San Francisco General Hospital, San Francisco, CA, USA; HET, IPM-heteroresistant phenotype; RES, high-level IPM resistance; SUSC, IPM-susceptible control strains.
Number of colonies within the zone of inhibition with an Etest: 0, no colonies; 1+, <50 colonies (within the lower region of the zone); 2+, >50 colonies (within the entire zone); 3+, no zone of inhibition.
The ertapenem Etest MIC was similar to that for imipenem, with colonies growing within the zone of inhibition.
CLSI reference standard inoculum.
NT, not typed.
Inoculum effect analysis.
Inoculum-dependent increases in the MICs for imipenem were determined based on the CLSI reference standard starting inoculum of 5 × 105 CFU/ml compared to those for inocula of 5 × 106, 5 × 107, and 5 × 108 CFU/ml. An inoculum effect was considered positive if the higher test inocula resulted in a ≥8-fold increase in the imipenem MIC.
Population analysis.
Population analysis was performed with 106 and 107 bacterial CFU spread on imipenem-containing MH agar plates (0.25 to 64 μg/ml). We calculated the frequency of heteroresistant subpopulations at the highest drug concentrations after 24 h of growth by dividing the number of colonies grown on imipenem-containing plates by the colony counts from the same bacterial inoculum plated on drug-free MH agar plates (17).
Time-kill analysis.
The frequency of survival in bactericidal concentrations of imipenem was quantified with starting inocula of 5 × 105 and 1 to 9 × 106 CFU/ml in a total volume of 3 ml of MH broth and with concentrations of imipenem 4- to 16-fold above the reference MIC. The starting inoculum was prepared from appropriate dilutions of overnight cultures standardized by optical density at 600 nm (OD600). Starting inocula were enumerated on drug-free agar plates. At 2, 4, 6, 8, and 20 h after imipenem exposure, 50-μl aliquots were serially diluted in 0.85% saline and plated on drug-free agar for enumeration. Control samples of the strains were grown in MH broth without drug and enumerated at the same time points. Population recovery was considered achieved if, after 20 h of drug exposure, enumeration yielded at least 109 CFU/ml or if the OD600 of the cultures was >1. The 20-hour endpoint was determined based on results of imipenem stability experiments (described below). Aliquots removed from the wells at 2, 4, 6, 8, and 20 h after imipenem exposure were also plated on MH agar containing the same concentration of imipenem used in the time-kill analysis.
Bioassay for imipenem hydrolysis.
Inocula of 5 × 105 and 5 × 106 CFU/ml of heteroresistant KPC-producing strains were incubated in the same imipenem concentration as that used in the time-kill experiments for 2, 8, and 20 h. Triplicate samples were used for each time point. At each time point, the cells were spun down, and the supernatant was passed through a 0.2-μm filter and frozen at −80°C. Aliquots were plated on LB agar to ensure that they were cell free. An Escherichia coli ATCC 25922 reference strain was then used to test the residual imipenem concentrations in these filtrates. Spontaneous imipenem hydrolysis was assessed by incubation of MH broth with the appropriate concentrations of imipenem for 4, 6, 12, 18, and 24 h. The E. coli ATCC 25922 reference strain was then inoculated into tubes of these preparations to perform standard imipenem broth microdilution testing. Fresh imipenem in MH broth was prepared as a control.
PCR and sequencing of blaKPC structural region and outer membrane porin genes.
We conducted PCR analysis of the Tn4401 regions upstream and downstream of the blaKPC open reading frame with primers based on a report by Naas et al. (18) and with primers designed within this study by Primer-BLAST (National Center for Biotechnology Information [NCBI]) (Table 2). PCR analysis of the coding regions of ompK35 and ompK36 was performed with primers designed by Primer-BLAST. Sequencing was performed on an Applied Biosystems 3730 DNA analyzer (Applied Biosystems, Foster City, CA) at the University of California (UC) Berkeley DNA Sequencing Facility. We visually inspected, edited, and assembled the DNA sequences with BioEdit (version 7.0.1) and then used ClustalW to perform multiple alignment analyses of the sequences. Sequences were analyzed for single nucleotide polymorphisms (SNPs) between the time-kill survivor strains and unexposed parental strains. Sequences were compared to those of the Tn4401 structural genes, ompK35, ompK36, and ompK37, deposited in the NCBI database by an updated version of the BLAST program.
TABLE 2.
PCR primers used in this study
| Primer target | Primer name | Sequence (5′ to 3′) | Expected amplicon size (bp) | Reference or study |
|---|---|---|---|---|
| blaKPC promoter region | Naas1 | ACCCTTGCCATCCCGTGTGC | 1,659 | 18 |
| Naas11 | AATTGGCGGCGGCGTTATCA | |||
| blaKPC | Naas3 | CTTCAAACAAGGAATATCGTTG | 1,040 | 18 |
| Naas2 | ATGCGCCATCGTCAGTGCTCTAC | |||
| ompK36 | ompK36-5F | AACTGGTAAACCAGGCCCAG | 829 | This study |
| ompK36-834R | CGTTCAGGCGAACAACACTG | |||
| ompK36-782F | AATTTCAGACCTGCGAATGC | 213 | ||
| ompK36-995R | ACCTGTACGGCAAAATCGAC | |||
| ompK35 | ompK35-83 | AAAACGGCAACAAACTGGAC | 971 | This study |
| ompK35-1054 | TGGTAAACGATACCCACGGC |
Real-time RT-PCR analysis.
We performed real-time reverse transcription-PCR (RT-PCR) of blaKPC gene expression for time-kill survivor samples of four heteroresistant K. pneumoniae strains (BR6, BR7, BR14, BR21) according to previously published protocols with modifications for comparative quantification by the standard curve method (19). Expression was compared between unexposed samples and those exposed to imipenem for 2, 4, 6, 8, or 20 h. The rpoB gene was used as an endogenous reference. An untreated wild-type sample of each strain was used as a calibrator gene standard. Total RNA was extracted with the RNeasy minikit (Qiagen, Valencia, CA) at each of the experimental time points. cDNA was generated by reverse transcription with random hexamer primers and SuperScript III according to the manufacturer's instructions (Life Technologies/Thermo Fisher Scientific, Waltham, MA). Samples were prepared with Maxima SYBR Green/Rox qPCR master mix (Thermo Fisher Scientific) and procedures were performed on an AB7300 real-time PCR system (Applied Biosystems). All samples were amplified in triplicate. Comparative quantification (fold change) of gene expression between samples was analyzed with the equation 2−ΔΔCT, where ΔΔCT = ΔCT KPC − ΔCT rpoB.
Analysis of outer membrane proteins.
Outer membrane proteins were isolated according to the method of Carlone et al. (20). Briefly, samples were grown in nutrient broth or MH broth at an OD600 of 0.6, centrifuged at 5,000 × g for 10 min, washed and resuspended in 10 mM HEPES buffer (pH 7.4), and sonicated. The sodium N-lauroyl sarcosinate insoluble outer membrane porins were selectively obtained by incubation in 10 mM HEPES buffer with 2% Sarkosyl, followed by a 30-min centrifugation at 15,600 × g. Samples were boiled and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10% polyacrylamide gels (Bio-Rad, Hercules, CA). Controls included drug-susceptible Klebsiella pneumoniae strains.
ESI-MS.
Electrospray ionization mass spectrometry (ESI-MS) of the outer membrane proteins was performed on a Thermo LTQ-Orbitrap-XL mass spectrometer at the QB3/Chemistry Mass Spectrometry Facility at UC Berkeley. Samples were prepared by excising the band of interest from SDS-PAGE gels, followed by in-gel tryptic digestion according to the facility protocol. Data analysis was performed with Thermo Scientific Proteome Discoverer (version 1.3) software.
Efflux pump analysis.
We used 100 μM concentrations of the efflux pump inhibitor, Phe-Arg β-naphthylamide dihydrochloride (PaβN), in conjunction with imipenem broth microdilution to assess efflux activity. Both unexposed parental-type and 8-h imipenem-exposed samples were tested in triplicate against three concentrations of the inhibitor. MgSO4 was used in a separate set of experiments to ensure that membrane permeability was not contributing to MIC differences. Experiments were repeated twice. Efflux activity was considered significant if there was a ≥2-fold difference in the imipenem MIC in the presence of the inhibitor (21).
Statistical analysis.
Categorical variables were compared by a chi-square or Fisher exact test (2-tailed). Differences in means or proportions were compared with Student's t test. Differences were considered statistically significant at a P value of ≤0.05.
RESULTS
Pronounced inoculum effect in heteroresistant strains.
Imipenem MICs of the eight heteroresistant KPC-producing K. pneumoniae strains were in the range of 1 to 2 μg/ml. They increased to 16 μg/ml with the 106-CFU/ml inoculum and were ≥64 μg/ml (the maximum of this test) with the 107-CFU/ml inoculum, a 32-fold increase (Table 1). These strains all produced colonies within the zone of inhibition of the imipenem Etest. Two strains (K. pneumoniae BR1 and BR20) with high-level imipenem resistance at the reference standard (16 μg/ml) showed a 4-fold MIC increase with the higher inocula. The Etest results for these strains showed dense growth of colonies throughout the zone of inhibition. An inoculum effect was not observed among the non-KPC-producing K. pneumoniae control strains (SF701, SF705, SF519, SF681).
A minor subpopulation of survivors mediates population recovery after lethal imipenem exposure.
Bactericidal levels were achieved for all study strains in the first 2 h of exposure. The mean numbers of colonies enumerated 2 h after exposure were 1.6 × 103 CFU/ml (95% confidence interval [CI], 1.2 × 103 to 2.1 × 103) for higher-inoculum samples and 1.7 × 103 CFU/ml (95% CI, 7.4 × 102 to 2.6 × 103) for standard-inoculum samples (P > 0.05). The mean number of colonies for non-KPC-producing strains 2 h after exposure was 1.9 × 104 CFU/ml (95% CI, 1.1 × 104 to 2.7 × 104).
For KPC-producing strains, 32 (76%) of 42 time-kill samples at a starting inoculum of >3.3 × 106 CFU/ml yielded >109 CFU/ml by 20 h postexposure (recovery), whereas only 4 (11%) of 36 samples below this starting inoculum recovered (P < 0.0001). None (n = 30) of the non-KPC-producing strain samples recovered even at concentrations of imipenem at the MIC.
The numbers of colonies enumerated 8 h after exposure ranged from 102 to 104 CFU/ml, with more survivors enumerated in higher- versus standard-inoculum experiments (P = 0.005). No significant difference was found in the number of survivors after 8 h of imipenem exposure between non-KPC-producing strains with higher inocula and KPC-producing strains with standard inocula (P > 0.05). However, 11% of the latter and none of the former group recovered.
At 2 h after imipenem exposure for all inocula tested, the survivors were as imipenem-susceptible as their parental strain and did not produce any colonies on imipenem agar plates. At 8 h of exposure, survivors had severalfold-higher imipenem MICs among groups that exhibited recovery at 20 h, while survivors that showed no recovery had MICs that were not different from those of the parental strain (Table 3). Population profiles revealed that an even smaller proportion of the initial 2-hour imipenem exposure survivors recovered (Table 4). For the heteroresistant K. pneumoniae strains (BR6, BR7, BR21, BR23, BR28), the frequencies of colonies that grew on imipenem agar in concentrations 8-fold higher (16 μg/ml) than the reference standard MIC were similar for both the 107- and 106-CFU-inoculum samples, with a range of 2 × 10−7 to 3 × 10−6, relative to those for samples grown on drug-free agar. The frequencies of colonies that grew on concentrations of 32 μg/ml were 2 × 10−7 and 0 to 3 × 10−7 for the 107- and 106-CFU-inoculum samples, respectively. Non-KPC-producing strains grew on imipenem agar at a maximum of 4-fold above the MIC at frequencies of 7 × 10−6 to 4 × 10−6 of the original inoculum.
TABLE 3.
Klebsiella pneumoniae OmpK36 porin analysis and imipenem susceptibility of unexposed and time-kill survivor samples
| Strain(s)a | IPM (μg/ml) | IPM expos time (h) | IPM MIC (μg/ml) |
ompK36 SNPb | OmpK36 statusc | ||
|---|---|---|---|---|---|---|---|
| STDd | HId | PBA (STD, HI)e | |||||
| BR6 (HET) | None | 1–2 | 16 | 0.5, 2 | WTf | + | |
| 16 | 2 | 1–2 | 16 | WT | + | ||
| 16 | 8 | >32 | >32 | 8, 8 | WT | − | |
| BR6, passaged | 16g | 8 | >32 | >32 | WT | − | |
| BR7 (HET) | None | 1–2 | 16 | 1, 4 | WT | + | |
| 16 | 2 | 1–2 | 16 | WT | + | ||
| 16 | 8 | >32 | >32 | 1, 4 | WT | + | |
| BR7, passaged | 16g | 8 | 2 | 16 | WT | + | |
| BR14 (HET) | None | 2 | 16 | 1, 4 | WT | + | |
| 16 | 2 | 2 | 16 | WT | + | ||
| 16 | 4 | >32 | >32 | C430T | − | ||
| 16 | 8 | >32 | >32 | 16, 16 | C731T, G374A | − | |
| BR14, passaged | 16g | 8 | >32 | >32 | ompK37h | − | |
| BR19 (HET) | None | 4 | 16 | 2, 2 | WT | + | |
| 16 | 2 | 2 | 16 | WT | + | ||
| 16 | 8 | >32 | >32 | 16, 16 | WT | − | |
| BR19, passaged | 16 | 8 | >32 | >32 | WT | − | |
| BR21 (HET) | None | 1–2 | 16 | 1, 4 | WT | + | |
| 16 | 2 | 1–2 | 16 | WT | + | ||
| 16 | 4 | >32 | >32 | WT | − | ||
| 16 | 8 | >32 | >32 | 16, 16 | ompK37h | − | |
| BR21, passaged | 16g | 8 | >32 | >32 | ompK37h | − | |
| BR21, standard inoculum | None | 8 | 1–2 | 8 | NDi | + | |
| BR21, standard inoculum, no recovery | 8 | 8 | 2 | 8 | WT | + | |
| BR21, standard inoculum, recovery | 8 | 8 | >32 | >32 | ND | − | |
| BR23 (HET) | None | 2 | 16 | 1, 4 | WT | + | |
| 16 | 2 | 2 | 16 | WT | + | ||
| 16 | 8 | >32 | >32 | 1, 4 | WT | − | |
| BR23, passaged | 16g | 8 | 2 | 16 | WT | + | |
| BR26 (HET) | None | 2 | 16 | 0.5, 2 | WT | + | |
| 16 | 8 | >32 | >32 | 4, 8 | WT | − | |
| BR26, passaged | 16g | 8 | >32 | >32 | ND | ND | |
| BR28 (HET) | None | 2 | 16 | 1, 4 | WT | + | |
| 16 | 8 | >32 | >32 | 16, 16 | WT | − | |
| BR28, passaged | 16g | 8 | >32 | >32 | ompK37h | − | |
| BR1 (RES) | None | 16 | >32 | 4, 4 | WT | − | |
| 32 | 8 | >64 | >64 | 16, 8 | WT | − | |
| BR3 (RES) | None | >64 | >64 | 16, >16 | WT | − | |
| BR20 (RES) | None | 16 | 32 | 2, 4 | ins: 403 GACGGCj | + | |
| 64 | 2 | 16 | 32 | ins: 403 GACGGC | + | ||
| BR20, no recovery | 64 | 8 | 16 | 32 | ins: 403 GACGGC | + | |
| BR20, recovery | 64 | 8 | 16 | 32 | ins: 403 GACGGC | + | |
| SF701, SF705 (CTL) | None | 0.5 | 0.5 | WT | + | ||
| 2, 1, 0.5 | 8, 24 | 0.5 | 0.5 | WT | + | ||
All samples were selected from higher-inoculum time-kill experiments unless otherwise noted. Passaged, 8-h imipenem (IPM)-exposed samples were passaged daily on drug-free MH plates for at least 7 days; HET, IPM-heteroresistant phenotype; RES, high-level IPM resistance; CTL, IPM susceptible control strains.
SNP, single nucleotide polymorphisms detected by PCR analysis.
SDS-PAGE analysis of bands corresponding to OmpK36 porin. +, present; −, not present.
IPM MIC results for starting inocula, 5 × 105 CFU/ml (standard [STD]) and 5 × 106 CFU/ml (high [HI]).
IPM MIC results (μg/ml) for starting inocula 5 × 105 (STD)/5 × 106 (HI) CFU/ml in the presence of 100 μM phenylboronic acid (PBA).
WT, wild-type sequence (GenBank accession no. JX310551).
Original IPM exposure concentration prior to drug-free passage.
ompK37 gene product (100% identity to that of GenBank accession no. KC534871) obtained by PCR with ompK36 primers.
ND, not determined.
100% identity to that of GenBank accession no. HM769261.
TABLE 4.
Frequency of heteroresistant subcolonies for select heteroresistant KPC-producing K. pneumoniae strains
| Strain(s) | Highest IPMa concn (μg/ml) | Frequency |
Fold-increase in IPM MICb | |
|---|---|---|---|---|
| 107-CFU inoculum | 106-CFU inoculum | |||
| BR6, BR7, BR21, BR23, BR28c | 16 | 2 × 10−7–3 × 10−6 | 3 × 10−7–1 × 10−6 | 8 |
| 32 | 2 × 10−7 | 3 × 10−7 | 16 | |
| 2-h-exposure samples, BR6, BR7, BR21d | 16 | 1 × 10−6−2 × 10−6 | 3 × 10−7–1 × 10−6 | 8 |
| 32 | 2 × 10−7–1 × 10−6 | 3 × 10−7 | 16 | |
| SF519, SF701e | 1 (SF519); 2 (SF701) | 4 × 10−6–7 × 10−6 | 4 × 10−6–7 × 10−6 | 4 |
| SF681f | 1 | 2 × 10−6−3 × 10−6 | 2 × 10−6−3 × 10−6 | 4 |
IPM, imipenem.
CLSI reference standard IPM MIC.
Heteroresistant KPC-producing K. pneumoniae strains.
Heteroresistant KPC-producing K. pneumoniae strains, exposed for 2 h to imipenem.
Non-KPC-producing K. pneumoniae strains.
CTX-M-producing K. pneumoniae strain.
KPC enzyme from lysed cells during imipenem exposure does not contribute to population survival.
The imipenem MICs for E. coli ATCC 25922 were 0.125 to 0.25 μg/ml in all of the KPC-producing K. pneumoniae culture filtrates from all incubation time samples, with the exception of supernatant removed from samples 20 h after exposure to imipenem in a population that recovered; these six samples grew in wells with imipenem concentrations of 8 μg/ml, which was the maximum concentration of the test (data not shown). Spontaneous degradation of imipenem was not observed in the test samples until 24 h of incubation (data not shown).
Increased expression of the blaKPC gene does not contribute to high-level resistance in heteroresistant strains.
All KPC-producing study strains contained blaKPC-2. The transcription start site region was 100% identical at the nucleotide level among all strains. The sequence upstream of blaKPC in all other strains was 100% identical to the region mapped by Naas et al. to contain three transcription start sites (18).
When blaKPC expression was normalized to that for unexposed samples, changes in expression for heteroresistant K. pneumoniae strains (BR6, BR7, BR14, BR21) ranged between 0.5-fold and 0.7-fold lower for the 2-h and 8-h imipenem-exposed samples. The four strains had similar expression levels, with the highest expression 2.4-fold higher than the lowest expression (data not shown).
Efflux pump activity does not contribute to survival in lethal doses of imipenem.
Imipenem broth microdilution with the efflux pump inhibitor, PaβN, showed no imipenem MIC reduction in any of the unexposed or 8-h-exposed heteroresistant K. pneumoniae samples (data not shown).
Porin expression changes contribute to high-level imipenem resistance.
The non-KPC-producing K. pneumoniae control strain SF519, but none of the KPC-producing K. pneumoniae strains, expressed OmpK35. By SDS-PAGE, all heteroresistant K. pneumoniae strains exposed to imipenem for 2 h, as well as their unexposed parental types, showed two bands, which were confirmed by ESI-MS as OmpA and OmpK36 (Table 3; Fig. 1). As early as 4 h postexposure, OmpK36 porin disappeared in some strains. In all 8-h exposure samples that subsequently recovered, OmpK36 was absent. The imipenem MICs for all such samples were >32 μg/ml. The OmpK36 band was present, however, in 8-h exposure samples of cultures that did not recover, as well as in the non-KPC-producing K. pneumoniae control strains. The OmpK36 band was also present in all samples at 2 and 8 h of drug-free growth. By PCR, in most cases, the ompK35 PCR product was not obtained, and evidence of insertions was seen in the ompK35 coding region in the sequenced PCR products. PCR results for the ompK36 gene agreed with SDS-PAGE and ESI-MS results. That is, unexposed and 2-h exposure samples with the OmpK36 protein band yielded an ompK36 PCR product with 100% nucleotide identity to that of the NCBI reference sequence (GenBank accession no. JX310551) (Table 3). For the 4- and 8-h exposure samples of one heteroresistant K. pneumoniae strain (BR14), the ompK36 sequence had mutations predicted to encode premature stop codons. In 8-h exposure samples of several heteroresistant K. pneumoniae strains (BR14, BR21, BR28), no ompK36 product was obtained by PCR, but an ompK37 PCR product with 100% nucleotide identity to that of the NCBI reference sequence (GenBank accession no. KC534871) was obtained. Wild-type ompK36 sequences were obtained for 8-h exposure samples of several heteroresistant K. pneumoniae strains (BR6, BR7, BR19, BR23), even though the OmpK36 protein band was absent. We did not analyze the region upstream of the open reading frame of the porin genes, so we cannot rule out mutations in the promoter or ribosome binding sites, which have been noted by others (19, 22).
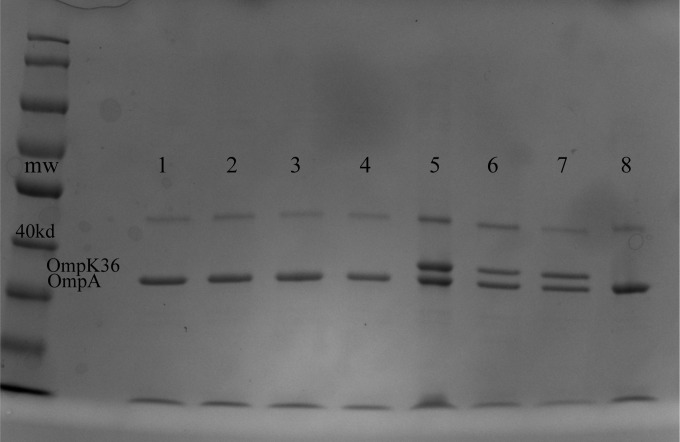
FIG 1.
Outer membrane fractions of 8-h imipenem-exposed OmpK36-deficient heteroresistant K. pneumoniae strain BR7 samples analyzed by SDS-PAGE. The identities of OmpA and OmpK36 were confirmed by ESI-MS with locations marked on the image; a 170-kDa molecular mass ladder is shown on the left. Lanes 1 to 4, samples from 2-μg/ml imipenem wells; lanes 5 to 7, samples from 2-μg/ml imipenem and 100 μM KPC enzyme inhibitor (PBA) wells (maximum concentration of sample growth); lane 8, BR7 wild-type control, initially expressing OmpK36, exposed to 16 μg/ml imipenem. Note that strain BR7 loses expression of OmpK36 with lethal imipenem exposure, but regains expression of OmpK36 upon drug-free passage (refer to text).
The imipenem-resistant K. pneumoniae strain BR20 demonstrated a 2- to 4-fold increase in imipenem resistance when tested at higher inocula. In contrast to the porin profiles of the heteroresistant K. pneumoniae strains, a 2-codon nucleotide insertion was found in all imipenem-exposed and unexposed samples, and OmpK36 was present by SDS-PAGE analysis in both exposed and unexposed samples. The insertion of GACGGC at position 403 of the NCBI reference sequence (GenBank accession no. HM769261) generates Asp135, Gly136 insertions in the L3 channel eyelet loop region described by others (23, 24). Mutations in this region have been predicted to reduce the uptake of carbapenems and cephalosporins, thus increasing the MICs against these drugs through selective restriction without abolishing expression of the porin (25).
In contrast, SDS-PAGE analysis of the constitutively highly resistant K. pneumoniae strain BR3 (with no observed inoculum effect) showed no OmpK36 protein even in the unexposed samples.
Stability of high-level resistance is associated with OmpK36 porin loss.
Heteroresistant strains were passaged daily on drug-free medium and then retested for their imipenem MICs (Table 3). After 8 h of imipenem exposure, six of the eight strains (BR6, BR14, BR19, BR21, BR26, BR28) showed no decreases in their MICs and had Etests with no zone of inhibition. Two strains (BR7, BR23) reverted to the heteroresistant phenotype, showing colonies in the zone of inhibition of the imipenem Etest. Imipenem broth microdilution MIC results showed reversion to the pronounced inoculum effect of the original unexposed strain. SDS-PAGE analysis showed that the strains with no reversion were still missing OmpK36, while the strains that reverted to heteroresistance regained the presence of OmpK36. By PCR, the nonrevertant strains did not yield ompK36 amplification products, while the revertant strain yielded a sequence with 100% identity to that of the wild-type coding region.
KPC enzyme activity is necessary for expression of imipenem heteroresistance.
In 100 μM concentrations of PBA (an inhibitor of KPC hydrolysis), growth in imipenem of heteroresistant KPC-producing K. pneumoniae strains expressing ompK36 was reduced 2- and 4-fold for standard and higher inocula, respectively (Table 3). The highly resistant K. pneumoniae strain BR20 (with the OmpK36 channel mutation) grew only in a maximum of 1 to 2 μg/ml imipenem in the presence of PBA, regardless of inocula. In the presence of PBA, the imipenem MICs of the OmpK36-deficient, nonrevertant strains BR14, BR19, BR21, and BR28 decreased only 2- to 4-fold, while MICs of the nonrevertant strains BR6 and BR26 decreased 4- to 8-fold. However, the OmpK36-deficient revertant strains BR7 and BR23 became as susceptible to imipenem as their OmpK36-expressing counterparts (a 16- to 32-fold decrease relative to the test maximum of 32 μg/ml imipenem). OmpK36 was expressed by individual colonies of strain BR7 after dual exposure to PBA and imipenem (at the highest concentration it grew, 2 μg/ml), but the porin was still absent in comparison samples exposed to the same dose of imipenem without PBA (Fig. 1).
Porin loss does not confer increased resistance to non-β-lactam drugs.
Five heteroresistant K. pneumoniae strains (BR7, BR14, BR21, BR23, BR28) were tested for resistance to other β-lactam drugs and to unrelated classes of antimicrobial agents to assess the potential contribution of efflux pumps or AmpC-type mechanisms to imipenem resistance (data not shown). A pronounced inoculum effect was observed with cefotaxime (8-fold difference), but not with ceftazidime, two extended-spectrum β-lactam drugs. The 2-h imipenem-exposed samples showed the same MICs as their nonexposed counterparts for all other drugs tested, while the 8-h imipenem-exposed samples showed a 4-fold increase against cefotaxime and a 2-fold increase against aztreonam. No increased MICs were observed for 8-hour imipenem-exposed samples against levofloxacin, gentamicin, or trimethoprim-sulfamethoxazole.
Recovered populations comprise subpopulations with heterogeneous imipenem resistance.
We analyzed five heteroresistant K. pneumoniae strains (BR6, BR14, BR21, BR23, BR28) by serial dilution and plating them after 20 h of incubation with imipenem, as well as by direct imipenem MIC testing (Table 5). Six to 12 individual colonies/strains were selected for imipenem MIC analysis. Interestingly, most aliquots of these imipenem-exposed total cultures had imipenem MICs in the highly resistant range (>32 μg/ml), while the isolated colonies had mixed results with MICs and OmpK36 porin profiles similar to those of the unexposed parental strains.
TABLE 5.
Imipenem MICs of individual colonies selected from 20-h imipenem-exposed cultures reveal the presence of heteroresistant subpopulations
| IPMa | No. of individual colonies for strain (revertant typeb) |
OmpK36 proteinc | ||||
|---|---|---|---|---|---|---|
| BR14 (NRv) | BR21 (NRv) | BR6 (NRv) | BR23 (Rv) | BR28 (NRv) | ||
| Highest concn grown (μg/ml)d | ||||||
| 2 | 18 | 13 | 17 | 6 | 0 | + |
| 4 | 2 | 1 | 1 | 0 | 0 | + |
| 8 | 1 | 1 | 0 | 0 | 0 | + |
| 16 | 3 | 4 | 0 | 0 | 0 | − |
| 32 | 16 | 18 | 0 | 12 | 24 | − |
| MIC, 20-h total culture | >32 | 1−>32 | 2−>32 | >32 | >32 | Variablee |
| MIC as above, >7 days drug-free passage | >32 | >32 | NDf | >32 | >32 | ND |
IPM, imipenem.
The revertant type is defined as nonrevertant (NRv) if conversion to high-level IPM resistance upon IPM exposure was retained or revertant (Rv) if the original IPM-heteroresistant phenotype was observed after 1 week of daily drug-free passage.
The presence of OmpK36 was determined by SDS-PAGE: +, present; −, not present.
Tests were performed with CLSI reference standard inocula (5 × 105 CFU/ml).
OmpK36 was present in all except one of the whole culture samples tested (BR21 strain).
ND, not determined.
DISCUSSION
There is no unified definition for heteroresistance. It is most commonly defined as a characteristic of a bacterial strain population susceptible to a drug according to clinical standards, but that contains subpopulations of much higher resistance. It commonly involves nonheritable phenotypic variability in a genetically homogeneous population (26–29). Heteroresistance was first reported in Staphylococcus aureus (methicillin, vancomycin) (30, 31), followed by reports in Acinetobacter baumannii (carbapenems, colistin, cephalosporins, penicillins) (32–34), Pseudomonas aeruginosa (carbapenems) (35, 36), Streptococcus pneumoniae (penicillin) (29), and Klebsiella pneumoniae (carbapenems, colistin, chlorhexidine) (37–39). For most of these, the mechanisms mediating heteroresistance remain elusive or suggest multiple pathways (32, 40–44).
In this study, we analyzed the phenotypic heteroresistance of KPC-producing K. pneumoniae strains to a carbapenem, imipenem. We showed that heteroresistant KPC-producing K. pneumoniae strains survive bactericidal concentrations of imipenem from 8- to 32-fold higher than their reference standard MICs. This survival was associated with (i) an inoculum density of at least 3 × 106 CFU/ml, (ii) carriage of the blaKPC gene, and (iii) the imipenem-induced generation of a subpopulation of cells with decreased expression of the major outer membrane porin, OmpK36. The survival was not related to other factors such as imipenem degradation or hydrolysis of the drug or increased expression of blaKPC.
OmpK36 porin loss by KPC-producing strains greatly increased the imipenem MIC. Landman et al. found by real-time RT-PCR analysis that even for K. pneumoniae strains with relatively low expression of blaKPC, decreased expression of ompK36 results in substantially higher imipenem MICs (16). Similar quantitative ompK36 expression studies should be performed with our heteroresistant K. pneumoniae strains. Tsai et al. also showed that loss of OmpK36 on its own increased imipenem MICs (45, 46). One expects OmpK36 loss to be detrimental for bacterial nutrient uptake, but this sacrifice of a subpopulation may have a beneficial outcome for the population as a whole in its defense against antimicrobial stress.
Porin loss in Enterobacteriaceae organisms is commonly reported in clinical treatment cases and has been shown to occur during the course of carbapenem treatment (47–51). Carbapenem resistance can develop in strains with OmpK36 loss in the absence of a carbapenemase (16, 19, 45, 52). Such strains usually express plasmid-mediated AmpC type β-lactamases or extended-spectrum β-lactamases (ESBLs) such as CTX-M types. Our KPC-producing K. pneumoniae strains nearly all coharbored blaCTX-M-1- or blaCTX-M-9-type ESBLs, and many coharbored blaTEM-1- and blaSHV-11-type β-lactamases. While it is possible that these enzymes contribute to the heteroresistant phenotype, our findings indicate that coordination of blaKPC and OmpK36 expression are key components of this phenotype. PBA-mediated inhibition of KPC enzyme activity prevented loss of OmpK36 and population recovery. Moreover, none of the four control strains in this study lacking blaKPC (one harbored blaCTX-M-15) achieved such abrupt imipenem MIC increases with such minor changes in inoculum, and no OmpK36 porin loss was observed under any of the experimental conditions.
There is evidence that carbapenem monotherapy for infections caused by strains with low-level resistance leads to high rates of clinical treatment failure (7, 10, 11, 14, 53, 54). There is debate over whether heteroresistant strains are associated with treatment failure (34, 55–59). Nevertheless, our experimental data suggest that the use of carbapenem monotherapy for heteroresistant strains, especially at infection sites where bacterial density may be high and drug penetration suboptimal, may unintentionally lead to induction of higher-level resistance and treatment failure.
The limitation of our study in extrapolating to clinical relevance is that our study is based on in vitro data and for a limited number of strains. However, it does provide some clue on the physiology and importance of resistant subpopulations generated by strains with apparent carbapenem susceptibility upon exposure to bactericidal doses of imipenem. Development of new therapeutic targets, such as those regulating porin expression, for carbapenemase-producing strains is urgently needed, especially for heteroresistant strains, which most likely contribute to the urgent threat of CRE infections.
ACKNOWLEDGMENTS
Study strains were kindly provided by Li Basuino (San Francisco General Hospital, San Francisco, CA), with coordination by Binh An Diep (Department of Medicine, University of California, San Francisco). We thank Anthony Iavarone for assistance with ESI-MS (QB3/Chemistry Mass Spectrometry Facility, University of California, Berkeley, CA). We thank Sangwei Lu for help with protocols. We thank Melaine Delcroix and Nicole Tarlton for critical review of the manuscript.
This study was supported in part by the NIH Fogarty International Center (grant D43 TW006563) and the RB Roberts Bacterial Drug-Resistant Infection Research Fund.
We declare no conflicts of interest.
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 3.Bratu S, Tolaney P, Karumudi U, Quale J, Mooty M, Nichani S, Landman D. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J Antimicrob Chemother 56:128–132. doi: 10.1093/jac/dki175. [DOI] [PubMed] [Google Scholar]
- 4.Woodford N, Tierno PM Jr, Young K, Tysall L, Palepou MF, Ward E, Painter RE, Suber DF, Shungu D, Silver LL, Inglima K, Kornblum J, Livermore DM. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York medical center. Antimicrob Agents Chemother 48:4793–4799. doi: 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, Rahal JJ, Brooks S, Cebular S, Quale J. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin Infect Dis 39:55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 7.Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B, Adams-Haduch JM, Doi Y. 2012. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother 56:2108–2113. doi: 10.1128/AAC.06268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, Prekates A, Themeli-Digalaki K, Tsakris A. 2011. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin Microbiol Infect 17:1798–1803. doi: 10.1111/j.1469-0691.2011.03514.x. [DOI] [PubMed] [Google Scholar]
- 9.Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch Intern Med 165:1430–1435. doi: 10.1001/archinte.165.12.1430. [DOI] [PubMed] [Google Scholar]
- 10.Weisenberg SA, Morgan DJ, Espinal-Witter R, Larone DH. 2009. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn Microbiol Infect Dis 64:233–235. doi: 10.1016/j.diagmicrobio.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, Morgan DJ. 2011. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J 104:40–45. doi: 10.1097/SMJ.0b013e3181fd7d5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith Moland E, Hanson ND, Herrera VL, Black JA, Lockhart TJ, Hossain A, Johnson JA, Goering RV, Thomson KS. 2003. Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J Antimicrob Chemother 51:711–714. doi: 10.1093/jac/dkg124. [DOI] [PubMed] [Google Scholar]
- 13.Tenover FC, Kalsi RK, Williams PP, Carey RB, Stocker S, Lonsway D, Rasheed JK, Biddle JW, McGowan JE Jr, Hanna B. 2006. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg Infect Dis 12:1209–1213. doi: 10.3201/eid1208.0602910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirsch EB, Tam VH. 2010. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J Antimicrob Chemother 65:1119–1125. doi: 10.1093/jac/dkq108. [DOI] [PubMed] [Google Scholar]
- 15.Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 16.Landman D, Bratu S, Quale J. 2009. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J Med Microbiol 58:1303–1308. doi: 10.1099/jmm.0.012575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wootton M, Howe RA, Hillman R, Walsh TR, Bennett PM, MacGowan AP. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother 47:399–403. doi: 10.1093/jac/47.4.399. [DOI] [PubMed] [Google Scholar]
- 18.Naas T, Cuzon G, Truong HV, Nordmann P. 2012. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother 56:4753–4759. doi: 10.1128/AAC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 20.Carlone GM, Thomas ML, Rumschlag HS, Sottnek FO. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol 24:330–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamers RP, Cavallari JF, Burrows LL. 2013. The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAbetaN) permeabilizes the outer membrane of gram-negative bacteria. PLoS One 8:e60666. doi: 10.1371/journal.pone.0060666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai JC, Zhou HW, Zhang R, Chen GX. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother 52:2014–2018. doi: 10.1128/AAC.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Fernández A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, Carattoli A. 2010. An ertapenem-resistant extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother 54:4178–4184. doi: 10.1128/AAC.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García-Fernández A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albertí S, Rodriquez-Quinones F, Schirmer T, Rummel G, Tomas JM, Rosenbusch JP, Benedi VJ. 1995. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect Immun 63:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rinder H. 2001. Hetero-resistance: an under-recognised confounder in diagnosis and therapy? J Med Microbiol 50:1018–1020. [DOI] [PubMed] [Google Scholar]
- 27.Tomasz A, Nachman S, Leaf H. 1991. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother 35:124–129. doi: 10.1128/AAC.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeltz RF, Schmidt JL, Wilkinson BJ. 2001. A microdilution plating method for population analysis of antibiotic-resistant staphylococci. Microb Drug Resist 7:289–295. doi: 10.1089/10766290152652846. [DOI] [PubMed] [Google Scholar]
- 29.Morand B, Muhlemann K. 2007. Heteroresistance to penicillin in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 104:14098–14103. doi: 10.1073/pnas.0702377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harigaya Y, Ngo D, Lesse AJ, Huang V, Tsuji BT. 2011. Characterization of heterogeneous vancomycin-intermediate resistance, MIC and accessory gene regulator (agr) dysfunction among clinical bloodstream isolates of Staphyloccocus aureus. BMC Infect Dis 11:287. doi: 10.1186/1471-2334-11-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu C, Chambers HF. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob Agents Chemother 47:3040–3045. doi: 10.1128/AAC.47.10.3040-3045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai Y, Chai D, Wang R, Liang B, Bai N. 2012. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 33.Hung KH, Wang MC, Huang AH, Yan JJ, Wu JJ. 2012. Heteroresistance to cephalosporins and penicillins in Acinetobacter baumannii. J Clin Microbiol 50:721–726. doi: 10.1128/JCM.05085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HY, Chen CL, Wang SB, Su LH, Chen SH, Liu SY, Wu TL, Lin TY, Chiu CH. 2011. Imipenem heteroresistance induced by imipenem in multidrug-resistant Acinetobacter baumannii: mechanism and clinical implications. Int J Antimicrob Agents 37:302–308. doi: 10.1016/j.ijantimicag.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Oikonomou O, Panopoulou M, Ikonomidis A. 2011. Investigation of carbapenem heteroresistance among different sequence types of Pseudomonas aeruginosa clinical isolates reveals further diversity. J Med Microbiol 60:1556–1558. doi: 10.1099/jmm.0.032276-0. [DOI] [PubMed] [Google Scholar]
- 36.Pournaras S, Ikonomidis A, Markogiannakis A, Spanakis N, Maniatis AN, Tsakris A. 2007. Characterization of clinical isolates of Pseudomonas aeruginosa heterogeneously resistant to carbapenems. J Med Microbiol 56:66–70. doi: 10.1099/jmm.0.46816-0. [DOI] [PubMed] [Google Scholar]
- 37.Meletis G, Tzampaz E, Sianou E, Tzavaras I, Sofianou D. 2011. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J Antimicrob Chemother 66:946–947. doi: 10.1093/jac/dkr007. [DOI] [PubMed] [Google Scholar]
- 38.Naparstek L, Carmeli Y, Chmelnitsky I, Banin E, Navon-Venezia S. 2012. Reduced susceptibility to chlorhexidine among extremely-drug-resistant strains of Klebsiella pneumoniae. J Hosp Infect 81:15–19. doi: 10.1016/j.jhin.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Pournaras S, Kristo I, Vrioni G, Ikonomidis A, Poulou A, Petropoulou D, Tsakris A. 2010. Characteristics of meropenem heteroresistance in Klebsiella pneumoniae carbapenemase (KPC)-producing clinical isolates of K. pneumoniae. J Clin Microbiol 48:2601–2604. doi: 10.1128/JCM.02134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohrer S, Maki H, Berger-Bachi B. 2003. What makes resistance to methicillin heterogeneous? J Med Microbiol 52:605–607. doi: 10.1099/jmm.0.05176-0. [DOI] [PubMed] [Google Scholar]
- 41.Deresinski S. 2013. The multiple paths to heteroresistance and intermediate resistance to vancomycin in Staphylococcus aureus. J Infect Dis 208:7–9. doi: 10.1093/infdis/jit136. [DOI] [PubMed] [Google Scholar]
- 42.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maor Y, Lago L, Zlotkin A, Nitzan Y, Belausov N, Ben-David D, Keller N, Rahav G. 2009. Molecular features of heterogeneous vancomycin-intermediate Staphylococcus aureus strains isolated from bacteremic patients. BMC Microbiol 9:189. doi: 10.1186/1471-2180-9-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mwangi MM, Kim C, Chung M, Tsai J, Vijayadamodar G, Benitez M, Jarvie TP, Du L, Tomasz A. 2013. Whole-genome sequencing reveals a link between beta-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb Drug Resist 19:153–159. doi: 10.1089/mdr.2013.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother 55:1485–1493. doi: 10.1128/AAC.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai YK, Liou CH, Fung CP, Lin JC, Siu LK. 2013. Single or in combination antimicrobial resistance mechanisms of Klebsiella pneumoniae contribute to varied susceptibility to different carbapenems. PLoS One 8:e79640. doi: 10.1371/journal.pone.0079640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elliott E, Brink AJ, van Greune J, Els Z, Woodford N, Turton J, Warner M, Livermore DM. 2006. In vivo development of ertapenem resistance in a patient with pneumonia caused by Klebsiella pneumoniae with an extended-spectrum beta-lactamase. Clin Infect Dis 42:e95–98. doi: 10.1086/503264. [DOI] [PubMed] [Google Scholar]
- 48.Mena A, Plasencia V, Garcia L, Hidalgo O, Ayestaran JI, Alberti S, Borrell N, Perez JL, Oliver A. 2006. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J Clin Microbiol 44:2831–2837. doi: 10.1128/JCM.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poirel L, Heritier C, Spicq C, Nordmann P. 2004. In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J Clin Microbiol 42:3831–3833. doi: 10.1128/JCM.42.8.3831-3833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song W, Suh B, Choi JY, Jeong SH, Jeon EH, Lee YK, Hong SG, Lee K. 2009. In vivo selection of carbapenem-resistant Klebsiella pneumoniae by OmpK36 loss during meropenem treatment. Diagn Microbiol Infect Dis 65:447–449. doi: 10.1016/j.diagmicrobio.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 51.Thiolas A, Bollet C, La Scola B, Raoult D, Pages JM. 2005. Successive emergence of Enterobacter aerogenes strains resistant to imipenem and colistin in a patient. Antimicrob Agents Chemother 49:1354–1358. doi: 10.1128/AAC.49.4.1354-1358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martínez-Martínez L, Pascual A, Hernandez-Alles S, Alvarez-Diaz D, Suarez AI, Tran J, Benedi VJ, Jacoby GA. 1999. Roles of beta-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob Agents Chemother 43:1669–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paterson DL, Ko WC, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, Bonomo RA, Rice LB, Wagener MM, McCormack JG, Yu VL. 2004. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum beta-lactamases. Clin Infect Dis 39:31–37. doi: 10.1086/420816. [DOI] [PubMed] [Google Scholar]
- 54.Lee GC, Burgess DS. 2012. Treatment of Klebsiella pneumoniae carbapenemase (KPC) infections: a review of published case series and case reports. Ann Clin Microbiol Antimicrob 11:32. doi: 10.1186/1476-0711-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deresinski S. 2009. Vancomycin heteroresistance and methicillin-resistant Staphylococcus aureus. J Infect Dis 199:605–609. doi: 10.1086/596630. [DOI] [PubMed] [Google Scholar]
- 56.Falagas ME, Makris GC, Dimopoulos G, Matthaiou DK. 2008. Heteroresistance: a concern of increasing clinical significance? Clin Microbiol Infect 14:101–104. doi: 10.1111/j.1469-0691.2007.01912.x. [DOI] [PubMed] [Google Scholar]
- 57.Ikonomidis A, Neou E, Gogou V, Vrioni G, Tsakris A, Pournaras S. 2009. Heteroresistance to meropenem in carbapenem-susceptible Acinetobacter baumannii. J Clin Microbiol 47:4055–4059. doi: 10.1128/JCM.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moore MR, Perdreau-Remington F, Chambers HF. 2003. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob Agents Chemother 47:1262–1266. doi: 10.1128/AAC.47.4.1262-1266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satola SW, Farley MM, Anderson KF, Patel JB. 2011. Comparison of detection methods for heteroresistant vancomycin-intermediate Staphylococcus aureus, with the population analysis profile method as the reference method. J Clin Microbiol 49:177–183. doi: 10.1128/JCM.01128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]