Abstract
Community-associated infections due to Escherichia coli producing CTX-M-type extended-spectrum β-lactamases are increasingly recognized in the United States. The blaCTX-M genes are frequently carried on IncF group plasmids. In this study, blaCTX-M-15-harboring plasmids pCA14 (sequence type 131 [ST131]) and pCA28 (ST44) and blaCTX-M-14-harboring plasmid pCA08 (ST131) were sequenced and characterized. The three plasmids were closely related to other IncFII plasmids from continents outside the United States in the conserved backbone region and multiresistance regions (MRRs). Each of the blaCTX-M-15-carrying plasmids pCA14 and pCA28 belonged to F31:A4:B1 (FAB [FII, FIA, FIB] formula) and showed a high level of similarity (92% coverage of pCA14 and 99% to 100% nucleotide identity), suggesting a possible common origin. The blaCTX-M-14-carrying plasmid pCA08 belonged to F2:A2:B20 and was highly similar to pKF3-140 from China (88% coverage of pCA08 and 99% to 100% nucleotide identity). All three plasmids carried multiple antimicrobial resistance genes and modules associated with virulence and biochemical pathways, which likely confer selective advantages for their host strains. The blaCTX-M-carrying IncFII-IA-IB plasmids implicated in community-associated infections in the United States shared key structural features with those identified from other continents, underscoring the global nature of this plasmid epidemic.
INTRODUCTION
CTX-M-producing Escherichia coli has become widespread in hospital and community settings in recent years (1–4). In particular, CTX-M-15, belonging to the CTX-M-1 group, appears to be the most common extended-spectrum β-lactamase (ESBL) globally, followed by CTX-M-14, another common variant of the CTX-M enzymes often reported in East Asia and parts of Europe (1, 2, 4, 5). The successful dissemination of blaCTX-M-15, the gene encoding CTX-M-15, has been mainly associated with the high-risk E. coli sequence type 131 (ST131) clonal group, as well as the epidemic multidrug-resistant IncF plasmids (6). Moreover, the ISEcp1 element, which can capture and mobilize blaCTX-M-15 and blaCTX-M-14 to IncF plasmids, has greatly facilitated their spread (6). In our previous study that identified 107 community-associated ESBL-producing E. coli isolates from five states in the United States, 54% belonged to ST131, and 91% of the 104 confirmed ESBL producers carried blaCTX-M (78 blaCTX-M-15, 12 blaCTX-M-14, and 5 other blaCTX-M variants) (2). In a follow-up study elucidating the molecular epidemiology of a subset of the isolates, 15 of the 24 blaCTX-M-carrying plasmids belonged to the IncF group by PCR-based replicon typing (PBRT), including IncFII-FIA-FIB, IncFII-FIA, IncFII, IncFIA-FIB, and IncFII-FIB (7). Many of these plasmids shared similar restriction profiles, and they occurred in ST131 and various non-ST131 isolates, suggesting the role of plasmid-mediated dissemination of blaCTX-M across different sequence types of ESBL-producing E. coli. Whole-plasmid sequencing and comparative analyses have facilitated our understanding of the process involved in the emergence and spread of these plasmids. To date, a few IncF plasmids carrying blaCTX-M-15 in E. coli isolates from different continents have been fully sequenced, which has demonstrated backbone regions and multiresistance regions (MRRs) that are related to various degrees (8, 9). However, IncF plasmids carrying blaCTX-M-15 in E. coli isolates from the United States have not been characterized. In addition, analysis of blaCTX-M-14-carrying plasmids has been limited in spite of their global spread. In this study, we present the complete nucleotide sequences of three representative blaCTX-M-carrying IncFII-IA-IB plasmids previously identified from E. coli isolates causing community-associated infections in the United States.
MATERIALS AND METHODS
Study strains.
E. coli isolates CA08, CA14, and CA28 were selected from our previous study in order to represent different states of origin, blaCTX-M types, and ST131 status (Table 1) (7). Isolates CA14 and CA28 carried blaCTX-M-15, while CA08 carried blaCTX-M-14. Isolates CA08 and CA14 belonged to ST131, while CA28 belonged to a non-ST131 sequence type (ST44). By PBRT, all three blaCTX-M-harboring plasmids belonged to IncFII-FIA-FIB, the dominant replicon type among the blaCTX-M-harboring plasmids in the United States (7). The antimicrobial susceptibilities of these isolates were reported previously (7).
TABLE 1.
Characteristics of three IncFII-IA-IB blaCTX-M-harboring plasmids from community-associated E. coli isolates in the United States
| Plasmid | Location | ST of host isolate | Size (bp) | No. of genes | FAB formula | Resistance genes | Putative virulence factor(s) |
|---|---|---|---|---|---|---|---|
| pCA08 | Michigan | 131 | 154,789 | 186 | F2:A2:B20 | blaCTX-M-14, tetA(A)-tetR, strAB, sul2, mph(A)-mrx-mphR(A), sul1-qacEΔ-aadA5-dfrA17 | ABC transporter system |
| pCA14 | Texas | 131 | 155,454 | 181 | F31:A4:B1 | blaCTX-M-15, ΔcatB3-blaOXA-1-aac(6′)-Ib-cr, tetA(B)-tetR-ΔtetC, mph(A)-mrx-mphR(A), dfrA17-aadA5-qacEΔ-sul1, ΔcatA1 | ABC transporter system, iucABCD-iutA, vagAB |
| pCA28 | Iowa | 44 | 172,280 | 203 | F31:A4:B1 | blaCTX-M-15, ΔcatB3-blaOXA-1-aac(6′)-Ib-cr, mph(A)-mrx-mphR(A), tetA(B)-tetR-tetC-tetD, dfrA17-aadA5-qacEΔ-sul1, aacC3 | ABC transporter system, iucABCD-iutA, sitABCD, vagAB |
Plasmid sequencing and bioinformatics.
A broth mating assay was performed using azide-resistant E. coli J53 as the recipient to test their self-transferability (10). The blaCTX-M-harboring plasmids of isolates CA08, CA14, and CA28 were extracted from the E. coli TOP10 transformants harboring them using the Qiagen Plasmid Maxi kit (Qiagen, Valencia, CA). Sequencing of the blaCTX-M-harboring plasmids was performed on a PacBio RSII single-molecule real-time (SMRT) sequencing instrument (Pacific Biosciences, Menlo Park, CA) at the Yale Center for Genome Analysis. The assembly process was described previously (11). In brief, the first-pass reads were assembled de novo using the hierarchical genome assembly process (HGAP) with SMRT Analysis v2.1 (Pacific Biosciences) (12). The single contigs representing the plasmids were circularized and used as references to reassemble the first-pass reads with Quiver v1 in SMRT Analysis v2.1. The plasmid sequences were initially annotated with the Rapid Annotations using Subsystem Technology (RAST) server (http://rast.nmpdr.org) and the Prokaryotic Genome Annotation Pipeline (PGAP) available through the NCBI and then curated manually using the BLASTn and BLASTp algorithms (http://blast.ncbi.nlm.nih.gov/blast). Easyfig 2.0 was used to map whole plasmids and to conduct a limited comparison. The MRRs in each plasmid were mapped with reference to MRRs of pEC958 (HG941719), pIP1206 (AM886293), pKF3-140 (FJ876827), and pETN48 (FQ482074). A phylogenetic tree based on the alignment of full sequences of each plasmid was constructed by the neighbor-joining method and bootstrap analysis with 100 replicates using the CLC Genomic Workbench version 7 (Qiagen).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the plasmids reported here are CP009233 (pCA08), CP009231 (pCA14), and CP009232 (pCA28).
RESULTS AND DISCUSSION
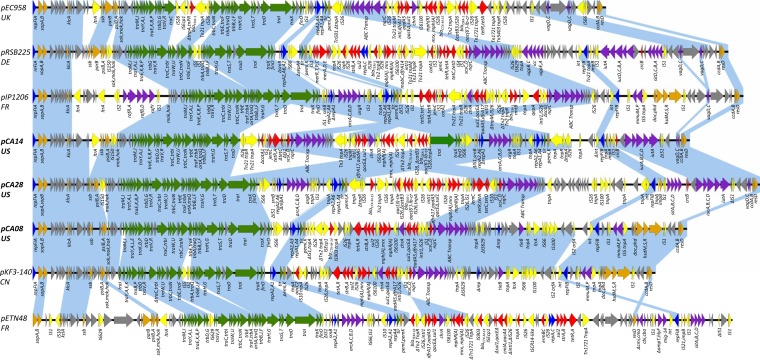
In this study, we determined the complete nucleotide sequences of two plasmids (pCA14 and pCA28) harboring blaCTX-M-15 and a plasmid (pCA08) harboring blaCTX-M-14 from community-associated E. coli isolates in the United States. Characteristics of the three plasmids are summarized in Table 1. Plasmids pCA08, pCA14, and pCA28 are circular molecules 154,789 bp, 155,454 bp, and 172,280 bp in size, with average GC contents of 50.94%, 51.54%, and 51.55% in their conserved backbone regions, respectively. They harbor 186, 181, and 203 predicted genes, respectively. Detailed structural features and comparisons with related IncF plasmids, including blaCTX-M-carrying plasmids, are shown in Fig. 1. All three plasmids have a typical IncFII-IA-IB plasmid scaffold and can be divided into two regions: the IncFII-IA-IB backbone and variable MRRs. The backbone region includes conserved genes for plasmid maintenance and conjugation, relatively variable genes for plasmid replication, and highly variable modules associated with virulence and biochemical pathways. The MRRs include several genes conferring antimicrobial resistance and associated mobile elements and are highly variable in composition and arrangement.
FIG 1.
Major structural features of plasmids pCA08 (ST131, blaCTX-M-14), pCA14 (ST131, blaCTX-M-15), and pCA28 (ST44, blaCTX-M-15) indicated with bold type compared with IncF plasmids pIP1206 (AM886293), pRSB225 (JX127248), and pKF3-140 (FJ876827), the IncF blaCTX-M-15-harboring plasmid pEC958 (HG941719), and the IncF blaCTX-M-14-harboring plasmid pETN48 (FQ482074). Light blue shades mainly indicate shared backbone regions and virulence factors with a high degree of similarity (>90% identity of nucleotide sequence). Open reading frames (ORFs) are portrayed by arrows and colored according to their putative functions. Dark-blue arrows indicate replication-associated genes. Genes associated with plasmid conjugal transfer are indicated by green arrows, and genes involved with plasmid stability are indicated by brown arrows. Red and yellow arrows indicate antimicrobial resistance genes and mobile element genes, respectively. Genes involved with virulence and biochemical pathways are indicated by dark-purple arrows. Gray arrows indicate genes for hypothetical proteins, as well as proteins of unknown function. UK, United Kingdom; DE, Germany; FR, France; CN, China.
According to the IncF replicon sequence typing (RST) scheme, which demonstrates a higher discriminatory power than PBRT (13), blaCTX-M-15-carrying plasmids pCA14 and pCA28 are assigned to FII, FIA, FIB (FAB) formula F31:A4:B1, whereas the blaCTX-M-14-carrying plasmid pCA08 is assigned to F2:A2:B20. These findings are consistent with the BLAST analysis, which indicates that pCA14 is closely related to pCA28, with 99% to 100% identity and 92% coverage, including the conserved backbone region and MRRs. pCA14 shares all backbone genes with pCA28, including genes for replication, plasmid maintenance, and conjugal transfer, as well as modules associated with virulence and biochemical pathways. A total of 20 nucleotide differences are observed between their backbone regions. A one-nucleotide difference and a two-nucleotide difference are found in the genes traE and iucC, respectively, with the remainder observed in genes of hypothetical proteins or intergenic regions. In addition, pCA14 shares all resistance genes with pCA28 except for the truncated catA1 gene. In contrast, the blaCTX-M-14-carrying plasmid pCA08 only shares 99% to 100% identity and 63% coverage with pCA28. Notably, a second copy of the FII allele is observed in each of the plasmids pCA14 and pCA28, which is composed of repA2 and repA6 genes and a 3′-end-truncated repA1 gene. This FII allele belongs to F36 according to the RST scheme and is almost identical to the FII-a replicon in pIP1206, with only 3 nucleotide differences. The multiple replication regions in IncF plasmids may have provided them with a broader host range and promoted their spread. F31:A4:B1 is a formula that includes a number of blaCTX-M-15-carrying plasmids and was reported in E. coli isolates from humans in the United Kingdom, Italy, and Australia, as well as in those from cattle and pets in France (9, 13–15). The plasmids from cattle and pets were identified from E. coli isolates in various non-ST131 sequence types, suggesting plasmid-mediated diffusion of blaCTX-M-15 genes across various E. coli lineages. F2:A2:B20 was also reported in E. coli ST131 from a pet cat in France, which harbored blaCTX-M-14 as in pCA08 (15).
pCA28 and pCA14 each contain a complete transfer region consisting of 24 tra genes, 9 trb genes, artA, and finO. In contrast, trbG and artA genes are absent in pCA08. However, all three plasmids are self-transmissible despite the incompleteness of the tra region in pCA08; this was supported by observations of their successful conjugal transfers to E. coli J53. pCA14 and pCA28 harbor several modules common across IncF plasmids that ensure stable plasmid inheritance and postsegregation killing: the sopABC operon for active plasmid partition, the postsegregation killing protein hok-mok system, the toxin-antitoxin systems pemKI and ccdAB, and the vagCD virulence-associated genes. The pCA08 plasmid also contains all these modules except for the vagCD virulence-associated genes. However, it possesses a restriction-modification (R-M) system (hsdMSR) that mediates plasmid maintenance. Another type I R-M system encoded by doc-phd is located upstream of the hsdMSR genes on pCA08 and is also present in the genome of bacteriophage P1 and encodes an addiction system stabilizing the P1 prophage (16). The doc-phd and hsdMSR in pCA08 exhibit high degrees of identity with their counterparts on pIP1206 (99% to 100%), which originated from an E. coli clinical isolate in France and carried rmtB, qepA, and an array of other antimicrobial resistance genes commonly found on IncF plasmids (16). Note that the parB gene mediating plasmid partition is absent in all three plasmids in our study. This gene is located upstream of the psiAB genes and is commonly observed in IncF plasmids.
Several known virulence determinants, as well as several modules associated with biochemical pathways, are identified on these plasmids. The module that encodes iron permease and ATP binding proteins of the ABC transporter family is present in all three plasmids. In addition, pCA28 contains two additional iron acquisition systems, IucABCD IutA and SitABCD. This region from sitABCD to iucABCD iutA on pCA28 shares high-level identity (with 10 nucleotide differences) with that on pSRB225, which originated from an unknown bacterium isolated from a municipal sewage treatment plant in Germany (17). In contrast, pCA14 only contains the IucABCD IutA system. Although these two gene clusters are rather common in IncF plasmids, they are rarely identified in blaCTX-M-15-carrying IncF plasmids (8, 18, 19). pCA14 and pCA28 have the arcACBD-argR gene cluster encoding proteins involved in the arginine deiminase pathway, which was almost identical to that on pIP1206, with only a two-nucleotide difference (16). pCA08 also carries an S-methylmethionine metabolism operon composed of mmuP and mmuM, which encode identical proteins as their counterparts on pIP1206 (16).
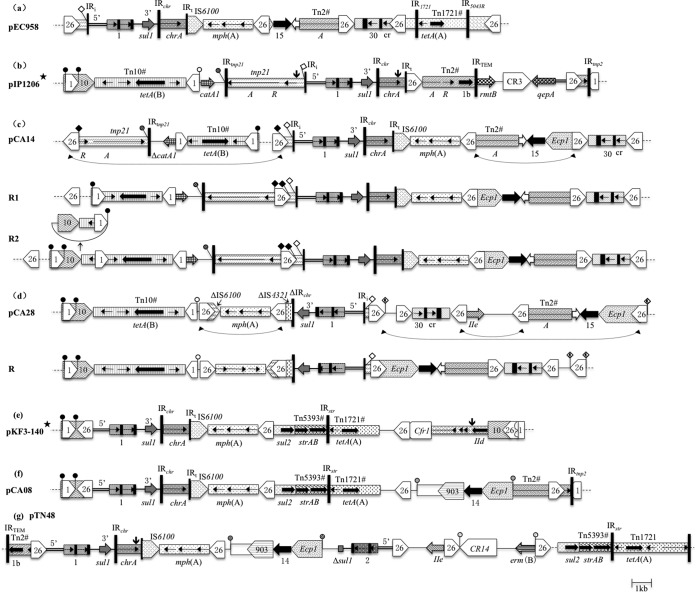
blaCTX-M-15 has always been found in association with ISEcp1 and is generally located 48 bp beyond its right-hand inverted repeat (IRR) (9). In our study, blaCTX-M-15 is also located 48 bp beyond the IRR of ISEcp1 in both pCA14 and pCA28. However, the ISEcp1 gene is truncated by IS26 at different locations, leaving ISEcp1 remnants of different lengths in pCA14 (545 bp) and pCA28 (1,224 bp), which correspond to the relatively common blaCTX-M-15 genetic environments 2b and 2c, respectively, found in E. coli strains isolated from travelers returning to the United Kingdom (20). Truncations of ISEcp1 by IS26 were also observed in other fully sequenced blaCTX-M-15-harboring plasmids, such as pEK499, pEC958, and pJIE286, underscoring the significant role of ISEcp1 in the initial spread of blaCTX-M-15 and the role of IS26 in the subsequent rearrangement of MRRs (8, 9). In addition to blaCTX-M-15, pCA14 and pCA28 also share other antimicrobial resistance genes, including the tetA(B)-tetR (tetracyclines) in a truncated Tn10 derivative, sul1 (sulfonamides)-qacEΔ (ammonium antiseptics)-aadA5 (spectinomycin and streptomycin)-dfrA17 (trimethoprim) cassette array carried by a class 1 integron, the mph(A)-mrx-mphR cluster (macrolides) following the class 1 integron, and the blaOXA-1 (penicillins/oxacillin)-ΔcatB3 (chloramphenicol)-aac(6′)-Ib-cr (aminoglycosides and fluoroquinolones) cassette array flanked by two copies of IS26 (Fig. 2). The tet cluster in pCA14 is truncated, probably as a result of insertion of an extra copy of IS1 followed by homologous recombination (HR) between directly oriented IS1 elements, deleting a circular molecule containing the intervening region and one copy of IS1. The gentamicin resistance gene aac(3)-IIe is only present in pCA28 between directly oriented copies of IS26, and its absence in pCA14 may be readily explained by IS26-mediated deletion (9). The missing chrA gene and truncated IS6100 in pCA28 is probably due to insertion of IS26 elements and IS26-mediated deletion between directly oriented copies of IS26. Furthermore, it appears that IS4321 was inserted into the chrA gene first and then truncated by IS26. pCA08 also encodes a class 1 integron with an sul1-qacEΔ-aadA5-dfrA17 cluster and the chrA-mph(A) module. However, it carries tetA(A) instead and harbors additional common MRR components sul2 (sulfonamides) and strAB (streptomycin) genes. The missing truncated Tn10 with tetA(B) and truncation of the integron may have been caused by insertion of two copies of directly oriented IS26 and homologous recombination. The blaTEM-1 gene, which is commonly found on IncF plasmids, especially blaCTX-M-carrying IncF plasmids, is noticeably absent in all three plasmids in our study. The compositions of MRRs on the three plasmids are in agreement with the susceptibility profiles of the E. coli TOP10 transformants harboring them (7). These MRR components have been commonly found in IncF plasmids, but they are in different combinations and arrangement with certain common boundaries (the catA1-IRtnp21 boundary and the tnp21-IRi boundary) in the plasmids in our study, highlighting the modular and mosaic characteristics of MRRs (21). The abundance of mobile elements, especially IS26, may have greatly facilitated the process of gene rearrangement. However, the limited number of target duplication repeats observed also suggests that homologous recombination (HR) may have played a key role in the gene rearrangement process. Moreover, a recent study reported that IS26 can mobilize adjacent DNA segments that carry genes encoding antimicrobial resistance or other functions via a translocatable unit that includes a single copy of IS26 and insert next to another IS26, with no additional IS26 or target duplications generated, which is quite similar to the process of HR between two copies of IS26 in direct orientation (9, 21, 22).
FIG 2.
MRRs of plasmids pCA14 (ST131, blaCTX-M-15), pCA28 (ST44, blaCTX-M-15), and pCA08 (ST131, blaCTX-M-14), compared with those of related IncF plasmids pIP1206 (AM886293) and pKF3-140 (FJ876827), the IncF blaCTX-M-15-harboring plasmid pEC958 (HG941719), and the IncF blaCTX-M-14-harboring plasmid pETN48 (FQ482074). Various transposons and other modules have different shading and are generally labeled once for each plasmid. # indicates that a transposon is incomplete. ISs are pointed boxes labeled with their number/name. Tall bars represent the 38-bp IR of transposons, as indicated. Positions/orientations of selected resistance and other genes are indicated by arrows, generally labeled once for each plasmid. Abbreviations: A, tnpA; R, tnpR; 30, blaOXA-30; cr, aac(6′)-Ib-cr; IIe, aac(3)-IIe; 15, blaCTX-M-15; 1b, blaTEM-1b; 14, blaCTX-M-14; IId, aac(3)-IId. Class 1 integron components are indicated as follows: 5′, 5′-CS; 3′, 3′-CS; small black boxes, attC sites; narrow box 1, dfrA17-aadA5 cassette array; narrow box 2, aacA4-cmlA1 cassette array; IRi, 25-bp IR at intI1 end; IRt, 25-bp IR at tni end. The chrA-mph(A) module is usually located after the 3′-CS, which consists of part of a chromate resistance transposon (IRchr-chrA), the 123-bp IRt end of tni402, IS6100, and the mph(A)-mrx-mphR(A) macrolide resistance gene cluster (9). Paired filled circles represent direct repeats (DRs), and paired squares represent flanking sequences that are reverse complementary to one another. An unpaired circle or square indicates the same boundary between regions present in another of the structures shown. Asterisks against plasmid names indicate that the structures shown have been rearranged to emphasize relationship, usually by inverting regions flanked by IS26 (21). Vertical arrows indicate the positions of IS26 elements with DRs that have been removed for ease of comparison. Dashed lines represent the IncFII backbone. Structures named R1, R2 below CA14, and R below CA28 are hypothetical and are intended to facilitate comparison between related MRRs. Dashed arrowed lines indicate where homologous recombination (HR) happens to invert or delete the regions between two IS26 elements. (c) pCA14 MRR. R1, two inversion events by homologous recombination between IS26 elements in opposite orientations may have occurred (shown by the dashed arrowed lines) in hypothetical structure R1 to yield pCA14 MRR; R2, the circular molecule above R2 may have been deleted in R2 by recombination between IS1 leading to the generation of R1. (d) pCA28 MRR. R, two inversion events by HR between IS26 elements in opposite orientations and a deletion event by recombination between IS26 in the same orientation (shown by the dashed arrowed lines) may have occurred and resulted in hypothetical structure R.
pCA08 is highly similar to pKF3-140, described in a Klebsiella pneumoniae clinical strain from China (23). They share almost all genes for the plasmid scaffold and most resistance genes. A major difference between pCA08 and pKF3-140 is that, in the region located downstream of the pemKI genes, blaCTX-M-14 is identified on pCA08, while aacC2 resides instead on pKF3-140. blaCTX-M-14 and its environment (ISEcp1 and IS903) together with the iroN gene located downstream of IS903 constitute a functional transposition unit typical for the blaCTX-M-9 group genes and have been reported mostly in blaCTX-M-14-carrying plasmids such as pETN48 (FQ482074.1) and pE66An (NC_020086.1), as well as in plasmids harboring blaCTX-M-17 (pIP843, AY033516.1), blaCTX-M-19 (pILT-3, AF458080.1), and blaCTX-M-24 (pKP96, EU195449.1) (24–28). This module is flanked by direct repeats TAAAA in pCA08. Interestingly, the K. pneumoniae strain from which pKF3-140 originated also contained another plasmid, pKF3-70, harboring blaCTX-M-14 (23). Therefore, acquisition of the module containing blaCTX-M-14 by pKF3-140 from pKF3-70 might have generated a pCA08-like plasmid.
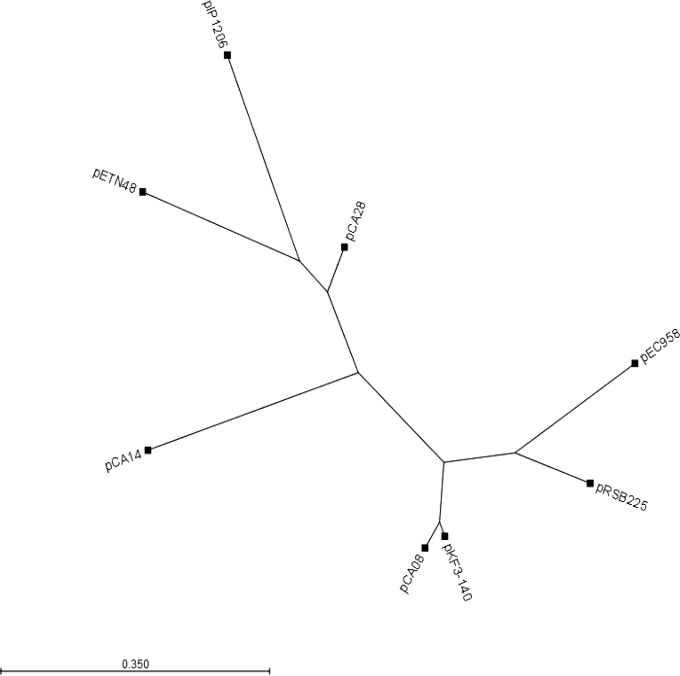
The phylogenetic tree generated from the nucleotide alignment of all of the plasmids shows that pCA08 clusters with the plasmid pKF3-140, which coincides with the above-described BLAST findings and further suggests a possible common origin (Fig. 3). pIP1206 and the blaCTX-M-14-harboring plasmid pETN48 originated from France and also cluster together. pCA14 and pCA28, which are closely related based on the BLAST search and FAB (FII, FIA, FIB) formula, do not form a single cluster. This distance is likely due to the large rearrangements present between the regions encoding the ABC transporter and arginine deiminase pathway. pCA28 is also related to the above-named plasmids from France, suggesting a shared evolutionary relationship between these blaCTX-M-harboring plasmids.
FIG 3.
Phylogenetic tree of plasmids pCA08 (ST131, blaCTX-M-14), pCA14 (ST131, blaCTX-M-15), pCA28 (ST44, blaCTX-M-15), and related IncF plasmids pIP1206 (AM886293), pRSB225 (JX127248), pKF3-140 (FJ876827), pEC958 (HG941719), and pETN48 (FQ482074).
In summary, we described the complete nucleotide sequences of three blaCTX-M-carrying IncFII-IA-IB plasmids from community-associated E. coli strains in the United States. blaCTX-M-15-harboring plasmids pCA14 and pCA28 are closely related to other IncFII plasmids from other continents in the backbone region and MRRs, especially pETN48 and pIP1206 from France. They belong to F31:A4:B1 and share a high degree of identity, suggesting a possible common origin. The blaCTX-M-14-harboring plasmid pCA08 is very similar to pKF3-140 from China. All three plasmids carry multiple antimicrobial resistance genes, as well as modules associated with virulence and biochemical pathways, which together likely confer significant selective advantages for their host strains.
ACKNOWLEDGMENTS
Y.D. was supported by the National Institutes of Health (research grants R21AI107302 and R01AI104895). J.-J.L. was supported by the National Scientific and Technological Major Projects of China (grants 2013ZX10004-904 and 2014ZX10004-008).
Y.D. has served on an advisory board for Shionogi, Inc., consulted for Melinta Therapeutics, and received research funding from Merck & Co. for a study unrelated to this work. The other authors declare no potential conflicts of interest.
REFERENCES
- 1.Pitout JD, Laupland KB. 2008. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 2.Doi Y, Park YS, Rivera JI, Adams-Haduch JM, Hingwe A, Sordillo EM, Lewis JS II, Howard WJ, Johnson LE, Polsky B, Jorgensen JH, Richter SS, Shutt KA, Paterson DL. 2013. Community-associated extended-spectrum β-lactamase-producing Escherichia coli infection in the United States. Clin Infect Dis 56:641–648. doi: 10.1093/cid/cis942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seiffert SN, Hilty M, Kronenberg A, Droz S, Perreten V, Endimiani A. 2013. Extended-spectrum cephalosporin-resistant Escherichia coli in community, specialized outpatient clinic and hospital settings in Switzerland. J Antimicrob Chemother 68:2249–2254. doi: 10.1093/jac/dkt208. [DOI] [PubMed] [Google Scholar]
- 4.Zong Z, Partridge SR, Thomas L, Iredell JR. 2008. Dominance of blaCTX-M within an Australian extended-spectrum β-lactamase gene pool. Antimicrob Agents Chemother 52:4198–4202. doi: 10.1128/AAC.00107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peirano G, Costello M, Pitout JD. 2010. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int J Antimicrob Agents 36:19–23. doi: 10.1016/j.ijantimicag.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu F, O'Hara JA, Rivera JI, Doi Y. 2014. Molecular features of community-associated extended-spectrum-β-lactamase-producing Escherichia coli strains in the United States. Antimicrob Agents Chemother 58:6953–6957. doi: 10.1128/AAC.03321-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother 53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Partridge SR, Zong Z, Iredell JR. 2011. Recombination in IS26 and Tn2 in the evolution of multiresistance regions carrying blaCTX-M-15 on conjugative IncF plasmids from Escherichia coli. Antimicrob Agents Chemother 55:4971–4978. doi: 10.1128/AAC.00025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M, Sahm DF, Jacoby GA, Hooper DC. 2004. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob Agents Chemother 48:1295–1299. doi: 10.1128/AAC.48.4.1295-1299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li JJ, Lee CS, Sheng J F, Doi Y. 2014. Complete sequence of a conjugative IncN plasmid harboring blaKPC-2, blaSHV-12, and qnrS1 from an Escherichia coli sequence type 648 strain. Antimicrob Agents Chemother 58:6974–6977. doi: 10.1128/AAC.03632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 13.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 14.Madec JY, Poirel L, Saras E, Gourguechon A, Girlich D, Nordmann P, Haenni M. 2012. Non-ST131 Escherichia coli from cattle harbouring human-like blaCTX-M-15-carrying plasmids. J Antimicrob Chemother 67:578–581. doi: 10.1093/jac/dkr542. [DOI] [PubMed] [Google Scholar]
- 15.Dahmen S, Haenni M, Chatre P, Madec JY. 2013. Characterization of blaCTX-M IncFII plasmids and clones of Escherichia coli from pets in France. J Antimicrob Chemother 68:2797–2801. doi: 10.1093/jac/dkt291. [DOI] [PubMed] [Google Scholar]
- 16.Perichon B, Bogaerts P, Lambert T, Frangeul L, Courvalin P, Galimand M. 2008. Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob Agents Chemother 52:2581–2592. doi: 10.1128/AAC.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wibberg D, Szczepanowski R, Eikmeyer F, Puhler A, Schluter A. 2013. The IncF plasmid pRSB225 isolated from a municipal wastewater treatment plant's on-site preflooder combining antibiotic resistance and putative virulence functions is highly related to virulence plasmids identified in pathogenic E. coli isolates. Plasmid 69:127–137. doi: 10.1016/j.plasmid.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum β-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother 48:3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smet A, Van Nieuwerburgh F, Vandekerckhove TT, Martel A, Deforce D, Butaye P, Haesebrouck F. 2010. Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences. PLoS One 5:e11202. doi: 10.1371/journal.pone.0011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhanji H, Patel R, Wall R, Doumith M, Patel B, Hope R, Livermore DM, Woodford N. 2011. Variation in the genetic environments of blaCTX-M-15 in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother 66:1005–1012. doi: 10.1093/jac/dkr041. [DOI] [PubMed] [Google Scholar]
- 21.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 22.Harmer CJ, Moran RA, Hall RM. 2014. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai J, Liu Q, Yang Y, Wang J, Yang Y, Li J, Li P, Li X, Xi Y, Ying J, Ren P, Yang L, Ni L, Wu J, Bao Q, Zhou T. 2013. Insights into the evolution of gene organization and multidrug resistance from Klebsiella pneumoniae plasmid pKF3-140. Gene 519:60–66. doi: 10.1016/j.gene.2013.01.050. [DOI] [PubMed] [Google Scholar]
- 24.Billard-Pomares T, Tenaillon O, Le Nagard H, Rouy Z, Cruveiller S, Medigue C, Arlet G, Denamur E, Branger C. 2011. Complete nucleotide sequence of plasmid pTN48, encoding the CTX-M-14 extended-spectrum β-lactamase from an Escherichia coli O102-ST405 strain. Antimicrob Agents Chemother 55:1270–1273. doi: 10.1128/AAC.01108-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao V, Lambert T, Courvalin P. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum β-lactamase CTX-M-17. Antimicrob Agents Chemother 46:1212–1217. doi: 10.1128/AAC.46.5.1212-1217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel L, Decousser JW, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother 47:2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen P, Jiang Y, Zhou Z, Zhang J, Yu Y, Li L. 2008. Complete nucleotide sequence of pKP96, a 67 850 bp multiresistance plasmid encoding qnrA1, aac(6′)-Ib-cr and blaCTX-M-24 from Klebsiella pneumoniae. J Antimicrob Chemother 62:1252–1256. doi: 10.1093/jac/dkn397. [DOI] [PubMed] [Google Scholar]
- 28.He L, Partridge SR, Yang X, Hou J, Deng Y, Yao Q, Zeng Z, Chen Z, Liu JH. 2013. Complete nucleotide sequence of pHN7A8, an F33:A-:B-type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. J Antimicrob Chemother 68:46–50. doi: 10.1093/jac/dks369. [DOI] [PubMed] [Google Scholar]