Abstract
Prostate cancer is the second leading cause of cancer-related death for men in the United States. The TMPRSS2/ERG (T/E) fusion gene is present in approximately 50% of prostate cancers and promotes tumor progression in vivo. The presence of the T/E fusion gene is strongly associated with ERG protein expression but emerging evidence suggests significant interfocal and intrafocal variability in levels of ERG protein expression. We therefore analyzed ERG protein expression by image analysis to objectively quantitate the extent of such heterogeneity and confirmed significant interfocal and intrafocal variability of ERG protein expression levels in cancer expressing ERG. To define the pathways associated with ERG and its variable expression in prostate cancer we have analyzed correlations of ERG expression, as evaluated by immunohistochemistry, with 46 key proteins associated with signal transduction, transcriptional control and other processes using a large tissue microarray with more than 500 prostate cancers. We found significant correlation of ERG expression with markers of activation of the PI3K, MYC and NFkB pathways, which had previously been linked directly or indirectly to ERG expression. We have also identified significant correlations with novel proteins not previously linked to ERG expression including serum response factor, the p160 coactivator SRC1 and Sprouty1. Of note, SKP2 is only correlated with high level ERG protein expression. Thus ERG expression is variable in prostate cancer and is associated with activation of multiple pathways and proteins including several potentially targetable pathways.
Keywords: prostate cancer, TMPRSS2/ERG, MYC, PTEN, NFkB
Introduction
Prostate cancer remains the most common malignancy affecting men and the second leading cause of cancer-related death of men in the United States. It is a heterogeneous disease and the biology of various subtypes is still poorly understood. The pathways altered at high frequency in specific patient tumor types need to be better defined to optimize individually targeted therapy. Over the past 8 years important progress has been made in the subclassification of prostate cancer, in particular the finding that the TMPRSS2/ERG (T/E) fusion gene is present in approximately 50% of prostate cancers. Experiments in prostate cancer cells containing the T/E fusion (Tomlins, et al. 2005) indicate that the TMPRSS2 promoter, which contains androgen receptor (AR)-responsive promoter elements (Lin, et al. 1999), increases ERG expression in response to androgens. The ubiquitous activity of AR in prostate cancer cells thus results in the constitutive expression of ERG fusion transcripts and ERG protein. We have demonstrated that the T/E fusion gene can enhance proliferation, invasion and motility of prostate epithelial cells (Wang, et al. 2008). More importantly, stable knockdown of the T/E fusion mRNA in VCaP cells inhibits tumor growth in vivo, indicating that the T/E fusion gene is a potential therapeutic target which is present in the majority of prostate cancers (Wang et al. 2008). More recently we have shown that highly specific knockdown of the T/E fusion gene with siRNAS delivered via nanoliposomal vectors substantially decreases growth of established tumors, confirming the importance of the T/E fusion gene in cancer progression in vivo (Shao, et al. 2012).
The pathways and proteins implicated in the ability of the ERG oncoprotein to promote prostate cancer progression has been a focus of active investigation. Prostate cancers with ERG expression have been shown to have activation of multiple proteins and pathways such as Ezh2, Wnts, TGFB and Sox9 (Brase, et al. 2011; Cai, et al. 2013; Gupta, et al. 2010; Magistroni, et al. 2011; Massoner, et al. 2013; Sun, et al. 2008; Tian, et al. 2013; Tomlins, et al. 2008; Vainio, et al. 2011; Wu, et al. 2013; Yu, et al. 2010). We have shown that the T/E fusion gene increases NFkB mediated transcription via increased phosphorylation of NFkB p65 on Ser536 (Wang, et al. 2011). Correlative studies in human prostate cancers reveal a strong association of ERG expression with loss of PTEN and studies in mouse models reveal that ERG expression and PTEN loss synergistically promote prostate cancer progression (Chen, et al. 2013; Han, et al. 2009; King, et al. 2009; Leinonen, et al. 2013). Thus alterations in a number of key pathways and proteins important in tumor progression have been linked to the presence of the ERG expression
One aspect of the biology of the T/E fusion gene that has received limited attention to date is the substantial variability of ERG protein expression levels in cancers containing the T/E fusion. The presence of the T/E fusion gene in a focus of prostate cancer by immunohistochemistry (IHC) is strongly associated with the presence of the T/E fusion gene in that focus, although less common variant translocations driving ERG expression have been identified. However, there can be substantial intrafocal variability in the levels of ERG expression (Mertz, et al. 2013; Minner, et al. 2013). To evaluate objectively the variability of ERG protein expression in prostate cancers we have carried out a multispectral image analysis of ERG expression in a cohort of more than 500 radical prostatectomies that we have studied extensively over the last 10 years. In addition, we have examined the correlation of 46 proteins and tumor specific factors in this same cohort with ERG expression. Our studies confirm the extent of variability of ERG expression in prostate cancer. The correlative studies have confirmed known or inferred associations with several important pathways/proteins which had previously been linked directly or indirectly to ERG expression, including the PI3K, MYC and NFkB pathways. We have also identified significant correlations with novel proteins not previously linked to ERG expression including serum response factor, the p160 coactivator SRC1 and Sprouty1. Importantly, we have found differential correlations based on the level of ERG expression consistent with a significant biological impact of ERG expression levels. In particular, SKP2 is only correlated with high level ERG protein expression. Thus ERG expression is variable in prostate cancer and is associated with activation of multiple pathways and proteins including several potentially targetable pathways.
Materials and Methods
Immunohistochemical evaluation
Tissues for analysis were collected with informed consent under and Institutional Review Board approved protocol. Immunohistochemistry (IHC) for ERG expression in the tissue microarray has been described previously (Wang et al. 2011). Briefly, antigen retrieval was carried out with heat using high pH Tris/borate/EDTA. Slides were incubated with a 1:100 dilution of rabbit anti-ERG monoclonal antibody (clone EPR3864, Abcam, Cambridge, MA, USA). The anti-rabbit HRP antibody was applied for 16 min at room temperature and detected using a ChromoMap DAB detection system (Ventana, Tucson, AZ, USA). Slides were then counterstained with hematoxylin. The same primary antibody was used in other studies as well. In our initial study ERG staining was scored visually as positive or negative, with any staining of cancer nuclei called positive regardless of intensity or extent based on the finding that even weak expression was correlated with the presence of the T/E fusion gene (Park, et al. 2010). To more quantitatively evaluate ERG protein expression each tissue microarray (TMA) slide was imaged with the Vectra™-Inform™ image analysis system (Cambridge Research & Instrumentation, Inc., Hopkinton, MA), an automated multispectral slide analysis system. 4× scans were obtained to detect the TMA, and 20× multispectral images were acquired from each core for IHC scoring of ERG with inform™. The inForm™ software supports image analysis projects by combining automated image processing with advanced object recognition and data analysis tools. Using image segmentation technology we trained the system to recognize and distinguish significant histological features including cancerous epithelial cells, stroma and luminal spaces in entire images. Cellular segmentation was used to detect cellular components such as nuclei and cytoplasm. After segmenting tissues and nuclei, we used the IHC scoring tool to score intensity of ERG staining in cancer nuclei. We selected 4 bins, ranging from 0-3+, to score ERG in cancerous epithelium with ERG staining. This was then applied to all images, yielding, for each case, a “distribution” of staining intensity represented as the fraction of cancer nuclei in each bin.
Over the last 10 years we have carried out numerous studies of protein expression and biological alterations in prostate cancer using a large TMA of radical prostatectomy specimens containing more than 500 prostate cancers. Most of these studies have been published (Agoulnik, et al. 2003; Agoulnik, et al. 2005; Agoulnik, et al. 2006; Ayala, et al. 2013a; Ayala, et al. 2013b; Ayala, et al. 2004; Ayala, et al. 2003a; Ayala, et al. 2003b; Cordon-Cardo, et al. 2007; Dai, et al. 2005; Ding, et al. 2013; Haqq, et al. 2005; Hodgson, et al. 2011; Knudsen, et al. 2005; Kwabi-Addo, et al. 2004; Li, et al. 2009a; Li, et al. 2009b; Li, et al. 2007; Li, et al. 2004a; Li, et al. 2006; Li, et al. 2003; Li, et al. 2004b; McAlhany, et al. 2004; Qin, et al. 2013; Yang, et al. 2002; Yu, et al. 2011; Zhou, et al. 2005) and the 46 biomarkers evaluated in this study are summarized in Table 1. For most markers staining was quantitated in tumor cells in the appropriate subcellular compartment (nuclear/cytoplasmic) after IHC using a multiplicative index of staining intensity (0-3) and extent (0-3) yielding a 10 point staining index (0-9). For other markers, such as Ki67 and TUNEL, stained cancer cells in each core were counted while for CD34 and c-KIT positive cells in the stroma were counted. Not every tumor was scored on every marker, in part due to exhaustion of tumor, folds, other artifacts etc.
Table 1. Markers evaluated for association with ERG expression.
| Transcription | Signaling | Microenvironment | Others |
|---|---|---|---|
|
| |||
| AR | PTEN | CD34 | ADRB2 |
| SRC1 | INPP4b | Semaphorin 4F | ASCT2 |
| SRC2 | AKT | c-KIT | CAS |
| SRC3 | P-AKT | CAV1 | |
|
|
|||
| COUPTFII | FKHR | Cell Cycle | FGFR4 |
|
|
|||
| ER | P-FKHR | Cyclin D1 | HAI1 |
| DAX1 | GSK3B | p27 | VEGFR3 |
| NCoR | P-GSK3B | SKP2 | SPINK1 |
| REGG | GGAP2 | GLIPR1 | |
|
|
|||
| SRF | NFkB p65 | Proliferation/death | PIN1 |
|
|
|||
| MYC | NFkB p65 P-276 | Ki67 | WFDC1 |
| p53 | NFkB p65 P-536 | TUNEL | |
| PIM2 | PKC-E | ||
| Sprouty1 | |||
Statistical Analysis
ERG expression was summarized in three ways – by visual assessment as a dichotomous variable, as a distribution with 4 levels, and as the Mean Bin, computed as the frequency weighted average of the bin intensities. Distributional patterns of ERG expression were visualized using a heatmap, with additional side bars to display corresponding values for visually assessed ERG status, and Mean Bin ERG. Agglomerative hierarchical clustering with complete linkage was used to cluster cases with similar distributional patterns. The Kolmogorov-Smirnov (KS) statistic, which quantifies the distance between two empirical distribution functions and is sensitive to differences in both location and spread, was used as the distance metric to compute the ‘distance’ between each case and every other case. Clustering and heat mapping were performed using R. The Mann-Whitney U test was used to test for differences in levels of studied markers between visually ERG positive and negative cancers. The association of average ERG expression (Mean Bin) with clinical and pathological variables as well as the panel of 46 previously studied TMA biomarkers was evaluated using Spearman rank correlation. There was no formal adjustment for multiple comparisons although a more stringent alpha=0.01 was used. Kaplan-Meier survival curves and log rank tests were used to evaluate the effect of ERG positivity on biochemical recurrence free survival and the effect of other biomarkers on RFS within each ERG visual expression group model. Associations were summarized by hazard ratios (HR) and 95% confidence intervals (95%CI), estimated by Cox proportional hazards regression. Candidate biomarkers were dichotomized as “low” or “high” using previously determined cutpoints. Unless otherwise specified, P values of ≤0.05 were considered statistically significant. Correlations and statistical comparisons were performed using the SPSS software package. (IBM SPSS Statistics, Version 20.0. Armonk, NY, USA)
Results
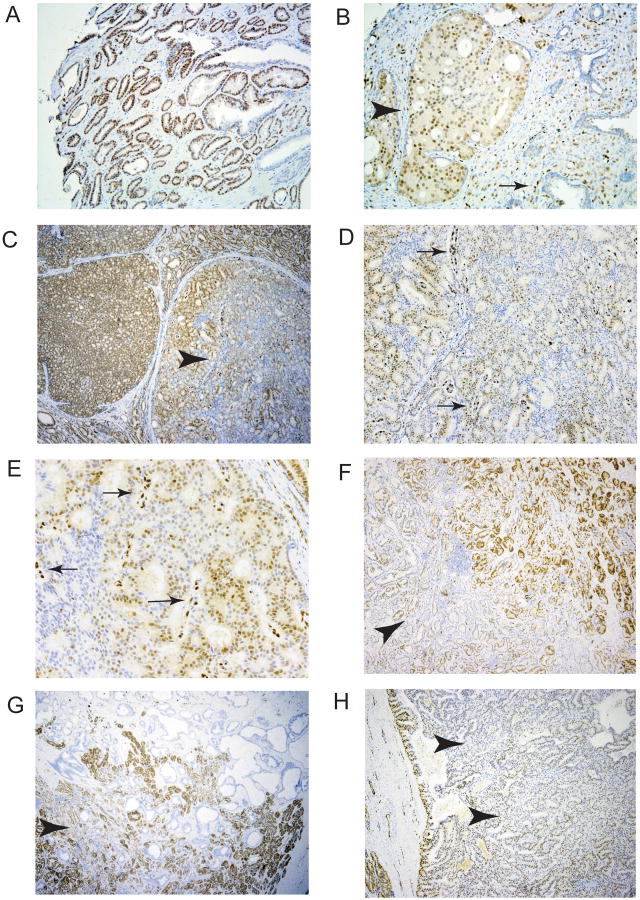
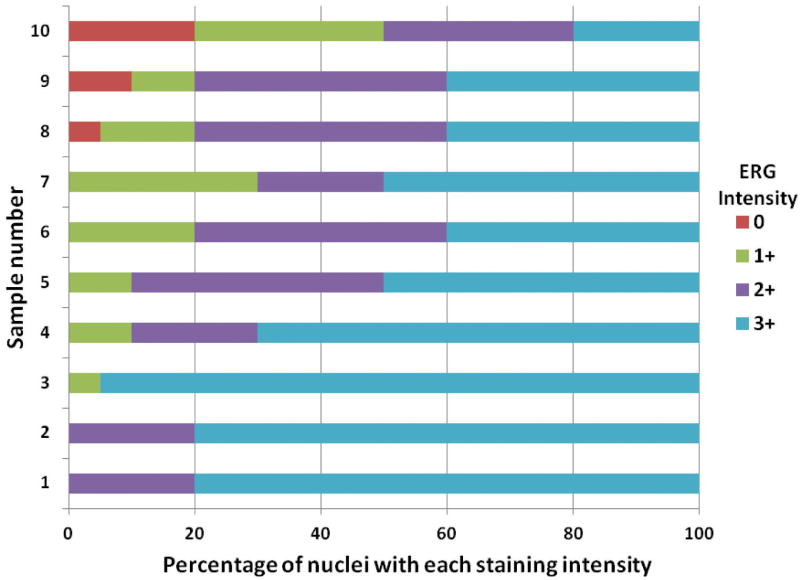
Variability of ERG expression levels in prostate cancer expressing the ERG oncoprotein. In prior studies we have carried out immunohistochemistry (IHC) for ERG expression using a large tissue microarray of cancers from radical prostatectomy specimens performed to treat clinically localized prostate cancer (Wang et al. 2011). As in most studies in the literature, prostate cancer cases were classified as positive for ERG expression if distinct staining was seen in cancer cell nuclei of any core since even weak expression is correlated with the presence of the T/E fusion gene (Park et al. 2010). However, we noted differences in staining intensity between cases stained on the same tissue microarray slide (Fig 1A and 1B). It was also noted that in some cases there was substantial variability of expression between tumor nuclei within a single cancer (intrafocal variability; Fig 1B). To better understand the extent of such variability we evaluated ERG expression in 16 consecutive radical prostatectomy specimens by IHC. In each case, the section with the largest percentage of tumor from the radical prostatectomy specimen was used for IHC. Ten cases stained positively for ERG. As in the tissue microarray we noted substantial variability in staining intensity between different cancers and within a single cancer focus. We quantitated the expression of ERG protein in cancer nuclei visually as 0, 1+, 2+ or 3+ and estimated the percentage of cancer cells with each staining intensity throughout the section. As shown in Figure 2 there was substantial variability in staining between the 10 cases. Examples of variable staining are shown in Fig 1C-1H. We noted that in several cases there was a tendency to stain more strongly at the edge of tumors (Fig 1F-H). This is unlikely to be an artifact of fixation since higher staining was seen both on the inner and outer (capsular) sides of the tumor, areas of strong staining often surrounded areas with weak or no staining and staining of ERG in endothelial cells remained quite strong adjacent to cancer cells with deceased staining.
Figure 1. Immunohistochemical analysis of ERG expression in prostate cancers.
A, B. ERG expression in a tissue microarray. Some cancers showed uniform strong expression (A, 40×) while other cancers on the same tissue microarray slide showed distinctly lower overall staining, and in some cases substantial intrafocal variability (B, 100×), Arrowhead indicates a cribiform tumor nodule with variable ERG staining. Arrow shows ERG staining in endothelial cells which are uniformly strong. C-H. ERG staining in cancer foci in radical prostatectomy specimens. C-E. C (40×). Heterogeneous staining in a cribiform nodule of cancer (arrowhead). Note strong uniform staining in adjacent tumor nodule. D (100×) and E (400×) show heterogeneous staining with retained strong staining of endothelial cells (arrows). F-H. Cancers with weaker staining in center of tumor (arrowheads).
Figure 2. Analysis of ERG expression in prostate cancers by visual quantitation.
Ten tumors from radical prostatectomy specimens with ERG staining by IHC were scored for staining intensity in cancer nuclei (0, 1+, 2+ or 3+). The percentage of nuclei with each staining intensity was estimated to the nearest 5%.
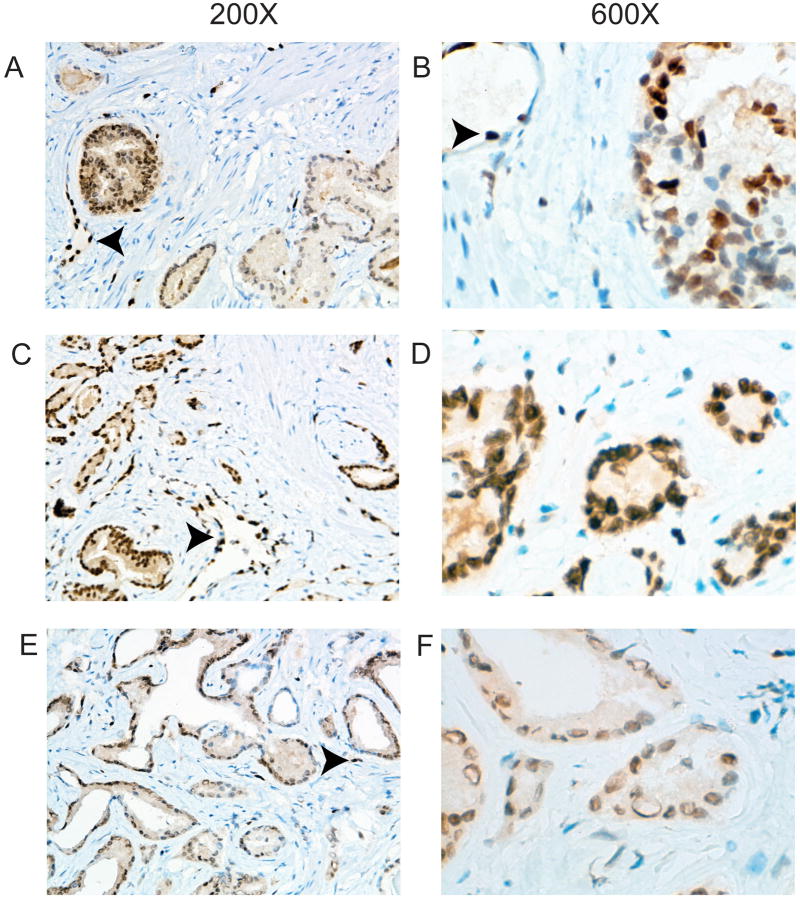
To confirm that the variable protein levels were not an artifact of delayed or poor fixation we took prostate cancer tissues that were snap frozen in liquid nitrogen within 15 minutes of surgery and took a thin slice that was then fixed overnight in formalin, embedded and ERG IHC performed. As seen in Figure 3, we still observed both interfocal and intrafocal variability of ERG expression in these rapidly fixed tissues. It should be noted that if immunohistochemistry protocols are optimized to give a maximal ERG signal, as is desirable for clinical use, the variable expression may be far less apparent than reported here.
Figure 3. Immunohistochemistry with anti-ERG antibody using rapidly fixed prostatectomy tissues.
Prostate cancer tissues were snap frozen in liquid nitrogen within 15 minutes of surgery and a thin slice taken that was then fixed overnight in formalin, embedded and ERG IHC performed. A. Intrafocal variability. Weakly staining tumor is on the right with strongly staining tumor on the left. A blood vessel is indicated by the arrowhead. B. Intrafocal variability. A tumor with variable staining within a single focus ranging from negative to strong. An endothelial cell with strong staining is indicated by the arrowhead. C-F. Interfocal variability. C and D show medium and high power views of a cancer with uniform strong staining similar in intensity to endothelial cells. E and F show a cancer with medium staining. Endothelial cells are indicated by the arrowheads. Magnification is shown at top.
Image analysis of ERG expression in prostate cancer
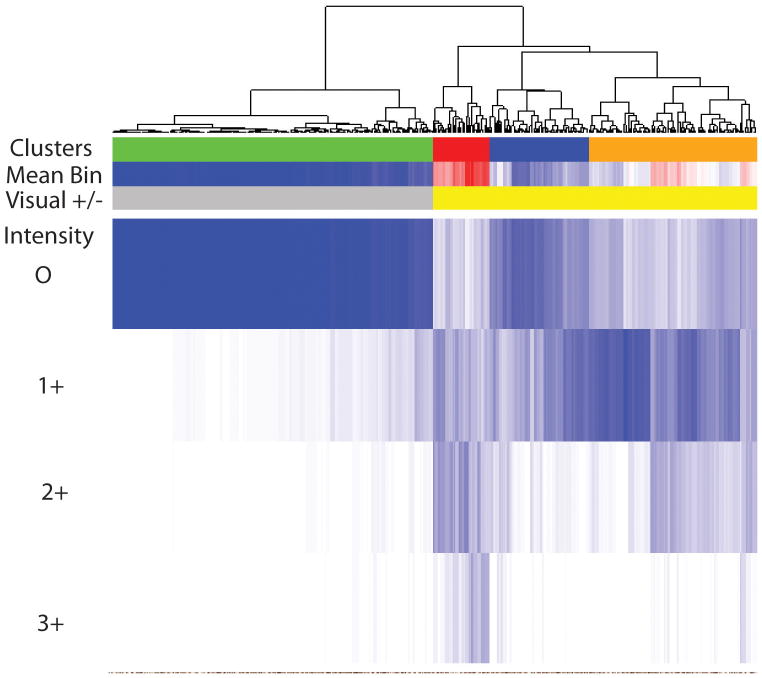
Given the substantial variability in staining we carried out image analysis using multispectral imaging to quantify the extent of variable staining objectively. This analysis allowed us to determine the intensity of staining in cancer nuclei on a cell-by-cell basis, with each nucleus binned into one of 4 expression levels (0, 1+, 2+ or 3+). The mean intensity of ERG staining was then determined for each cancer (ERG Mean Bin) based on the mean of all cancer nuclei. This analysis is shown in Figure 4 in the Mean Bin row, with the lowest mean intensity shown in blue and the highest in red. The visual image quantitation data for each case is also shown. There was a very strong, although not perfect correlation, correlation between the visual classification and the Mean Bin scores (r=0.797, p<0.001). We then carried out hierarchical clustering of the distributions of staining, using the Kolmogorov-Smirnov statistic as the distance metric. The dendogram is shown in Figure 4 with cases as columns showing the proportion of nuclei in each staining intensity (white=0, dark blue=1.0). The largest cluster (negative; green) corresponds perfectly with the cases evaluated as negative by visual inspection. The visually positively staining cases are clustered into three groups, corresponding to low (blue), intermediate (orange) and strong (red) average staining. Most cases showed variable levels of staining within the tumor as well as between tumors when analyzed by image analysis. No strong association with pathological variables or biochemical recurrence was noted based on either the visual presence or absence of ERG, Mean Bin values or cluster group (data not shown).
Figure 4. Analysis of ERG expression in prostate cancers by image analysis.
ERG expression was evaluated by immunohistochemistry using a prostate cancer tissue microarray as described previously. Expression was evaluated visually and classified as positive if any staining was seen in cancer nuclei. Expression was also quantitated by image analysis of all cancer nuclei. We selected four bins from 0-3+ to score ERG in cancerous epithelia with ERG staining which was then applied to all images. Among cases with any visually positive staining there was substantial variability between tumors and within each tumor. The mean staining (Mean Bin) was calculated for each case. Cluster analysis of quantitative expression was then carried out identifying four cluster groups indicated by green (negative), blue (low) orange (medium), and red (strong).
Correlation of ERG expression with other proteins and biological processes in prostate cancer
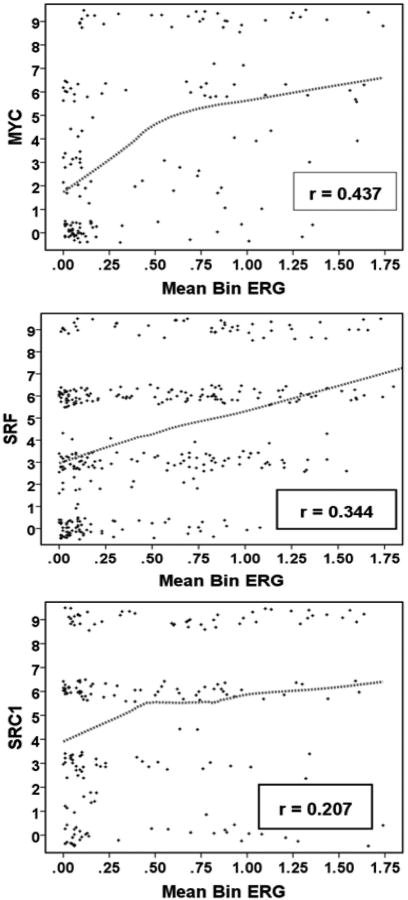
To better understand the biological impact of ERG expression and its variability in prostate cancer we examined the correlation of ERG expression with expression of proteins and biological factors in this same tissue microarray in Table 1. We compared marker levels between visually graded ERG groups and evaluated correlations of these markers with Mean Bin ERG expression. The proteins that showed highly significant associations with ERG expression by both visual and quantitative methods of analysis are shown in Table 2. To be included in this table, associations with both methods of analysis had to be statistically significant (p≤0.05) with at least one method being significant at p≤0.01. Scatter plots demonstrating some of these associations are shown in Figure 5.
Table 2. Markers associated with ERG expression.
| Group | Protein or factor | Binary ERG | Mean Bin ERG | |
|---|---|---|---|---|
|
| ||||
| M-W p-value# | r | p-value$ | ||
| MYC | MYC | <.001 | 0.437 | <.001 |
| PIM2 | 0.001 | 0.231 | 0.002 | |
|
| ||||
| PI3K/AKT | PTEN | 0.004 | -0.224 | 0.007 |
| INPP4B | 0.002 | -0.140 | 0.011 | |
| P-AKT | 0.005 | 0.195 | 0.003 | |
| P-GSK3B | 0.022 | 0.244 | <.001 | |
|
| ||||
| NFkB | NFkB p65 | 0.001 | 0.303 | <.001 |
| NFkB P-S536 | <.001 | 0.316 | <.001 | |
|
| ||||
| Transcription | p53 | 0.013 | 0.257 | <.001 |
| SRF | <.001 | 0.344 | <.001 | |
| SRC1 | 0.008 | 0.207 | 0.004 | |
|
| ||||
| Signal transduction | Sprouty1 | 0.002 | 0.244 | 0.001 |
Mann-Whitney U test comparing expression in ERG negative versus ERG positive.
Spearman rank correlation and associated p-value.
Figure 5. Relationship between expression of selected proteins and ERG.
Scatter plots with conditional uniform jitter and Loess fit curve (75% of points) to illustrate the relationship between MYC, SRF and SRC1. The correlation coefficient r is Spearman rank.
The strongest correlation is with MYC expression (r=0.437; Figure 5). Of note, MYC has been shown to be a direct transcriptional target of ERG (Sun et al. 2008) and our data are consistent with a significant role for ERG in driving MYC expression in primary prostate cancers. PIM2 is also correlated with ERG expression. While PIM2 has pleiotrophic effects on tumor progression, it can synergize with MYC overexpression (Allen, et al. 1997) and this may be at least one of its important roles in prostate cancer, where MYC overexpression is common (Antonarakis, et al. 2012).
As described previously, there is a significant positive correlation of ERG with increased levels of NFkB p65 phospho-Ser536, which is an indirect target of the ERG oncoprotein via its induction of TLR4. In addition, nuclear NFkB p65 is correlated with ERG expression. This may be in part via increased PIM1 kinase, which has been reported to be increased by ERG (Magistroni et al. 2011) and is known to activate the NFkB pathway. PIM2 has also been linked to NFkB pathway activation (Dai et al. 2005).
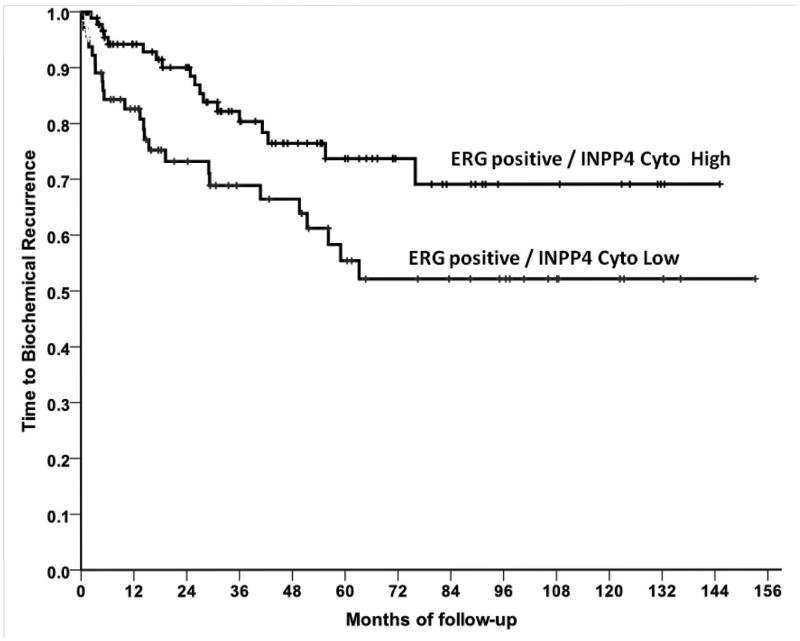
ERG is negatively correlated with both PTEN and INPP4B (Table 2). Loss of the INPP4B phosphatase also activates the AKT pathway and is an alternative mode of activating this pathway in prostate cancer (Agoulnik, et al. 2011). The association of ERG expression with loss of PTEN at the genomic level has been described previously and loss of PTEN and expression of ERG synergize in prostate cancer induction in genetically engineered mouse models of prostate cancer. Our data confirm this association and indicate that there is also correlation with activation of the AKT pathway by alternative genomic alterations, and thus the observed association with PTEN loss is unlikely to be due to loss of other activities of PTEN (Bassi, et al. 2013). Of note, in those cancers with ERG expression, low INPP4B levels were associated with a significantly decreased time to biochemical recurrence (p=0.023, Hazard ratio=2.01, 95% Confidence Interval (1.09, 3.70), Figure 6). There was no difference in ERG negative cases (P=0.31). We did not see a similar association with PTEN levels (data not shown). Loss of PTEN has been associated with worse outcome in T/E fusion positive prostate cancer in some but not all studies (Leinonen et al. 2013; Reid, et al. 2010). Consistent with these findings, ERG is positively associated with phosphorylated AKT (Ser473) and phosphorylated GSK3B. Taken together, these data are mutually consistent and indicate that ERG expression is broadly associated with activation of the PI3-K pathway.
Figure 6. INPP4B loss decreases time to biochemical recurrence in prostate cancers expressing the TMPRSS2/ERG fusion gene.
Kaplan Meier analysis of time to biochemical recurrence following radical prostatectomy of ERG positive cancers with low (“0-5”, n=65) or high (“6-9”, n=103) INPP4B Index. The Log-rank Test p-value is 0.023.
We have previously shown that serum response factor (SRF) is increased in prostate cancer and higher expression is associated with biochemical recurrence after radical prostatectomy (Yu et al. 2011). Recent studies have shown that SRF is associated with aggressive metastatic disease as well (O'Hurley, et al. 2014). Unexpectedly, we have found that SRF is significantly positively associated with ERG expression and is the second strongest association (r=0.344; Figure 5) we identified (after MYC).
Expression of the p160 steroid receptor co-activator SRC1 is also positively correlated with ERG protein levels (Table 2 and Figure 5). This positive association is most likely due the known role of SRC1 in promoting TMPRSS2 (and presumably T/E fusion gene) expression (Nakka, et al. 2013).
The p53 pathway is among the most commonly altered pathways in cancer and although p53 mutation is uncommon in clinically localized cancer, we noted a positive correlation between ERG and p53. Increased p53 protein is generally seen when this pathway is inactivated by mutation or some other mechanisms. Others have previously noted a correlation between the presence of the T/E fusion gene and p53 protein expression (Kluth, et al. 2014; Minner, et al. 2011)
Increased Sprouty1 expression is associated with ERG expression. We have previously shown significant heterogeneity of Sprouty1 expression in prostate cancer with some cases being negative and other cases retaining staining or having increased staining compared to benign epithelium (Kwabi-Addo et al. 2004). Why ERG expressing cancers would express in Sprouty1 is not known, but ETS transcription factors are known to play an important role in Sprouty gene expression (Tsang and Dawid 2004) and so it may be a direct target for ERG. Sprouty1 is known to inhibit FGF receptor signaling but can enhance EGFR signaling (Tsang and Dawid 2004), so some ERG expressing cancers may have enhanced EGFR signaling via this mechanism.
Associations with ERG protein levels
To determine if there is unique biology associated with different levels of ERG protein as determined by image analysis we examined the association of ERG protein levels with the 46 proteins/phosphoproteins and biological processes in Table 1, excluding all cases that were ERG negative (green cluster, Figure 4). Thus all associations are driven by differences in ERG protein level between positive cases. Results of this analysis are shown in Table 3, which shows the factors strongly associated with higher ERG expression (p<.01). Of note, most of the factors in Table 2 are no longer strongly significant except for NFkB p65 phospho-Ser536, SRF and Sprouty1. SKP2 is identified as a novel factor associate with ERG protein levels. Thus differential ERG protein expression levels are associated with activation of distinct pathways and proteins that are distinct from the pathways linked to the presence of ERG per se.
Table 3. Markers associated with ERG expression levels.
| Protein or P-protein | r | p-value$ |
|---|---|---|
| SKP2 | 0.291 | 0.003 |
| NFkB P-S536 | 0.328 | <0.001 |
| Sprouty1 | 0.244 | 0.001 |
| SRF | 0.274 | 0.002 |
Spearman rank correlation and associated p-value.
Discussion
Prostate cancers expressing the TMPRSS2/ERG fusion gene is by far the most commonly identified prostate cancer variant and understanding how this fusion impacts carcinogenesis is critical in developing novel approaches to therapy in men with prostate cancers bearing this fusion gene. We have analyzed associations 46 proteins/phosphoproteins or biological process with the presence and level of ERG protein in clinically localized prostate cancer and identified multiple key proteins and pathways that are associated with expression and/or level of expression of ERG. Such associations may reflect different types of underlying biology. One potential cause of such association is if the identified protein drives expression of the fusion gene, as is probably the case for SRC1 (Nakka et al. 2013). Alternatively, the ERG fusion gene may directly drive transcription of a protein (such as MYC) or indirectly drive increased expression (NFkB p65 phospho-Ser536 via TLR4). In addition, such associations may be due to selection due to strong biological complementation of a pathway/protein with ERG expression as is apparently the case for the PI3K/AKT pathway. Finally, it is possible that some observed associations may be due to chance, as may occur with examination of multiple associations, although our stringent p values make this less likely. At present the exact mechanisms underlying many of the associations we have identified will require further study. However, we have confirmed in human tissues some previously inferred associations based on mechanistic data (MYC), identified new associations related to prior identified associations (INPP4B, NFkB, PIM2) and also identified novel associations (SRF, Sprouty1). These associations in human cancers provide a strong rationale for further mechanistic studies.
Our studies confirm, using quantitative image analysis, that there is interfocal and intrafocal variability in ERG protein levels in prostate cancer. Early studies had shown that different cancer foci in radical prostatectomy specimens were discordant for the presence of the fusion gene in some cases (Mehra, et al. 2007). This is well established and we have not examined this question in the current studies. On the contrary, our studies of both the radical prostatectomy specimens and in the TMAs examine only a single cancer focus in each radical prostatectomy. We now show objectively that there is heterogeneity in overall levels of ERG protein expression in different patients. Such heterogeneity may arise due to germline variability between patients and/or genomic variability between cancers. For example, germline variants in the TMPRSS2 enhancer affecting AR binding (Clinckemalie, et al. 2013), germline AR variants and or germline variants affecting androgen synthesis and/or metabolism may all potentially impact T/E fusion protein levels. Vitamin D can also increase transcription of the T/E fusion gene (Washington and Weigel 2010) and individual variation in vitamin D metabolism and/or intake may also potentially impact fusion gene expression. Variable levels of coactivators (such as SRC1 and potentially others) may also effect transcription. Differential miRNA expression between different cancers may also affect ERG levels since miRNAs targeting the T/E fusion have been identified (Hart, et al. 2013). In any case, we have previously shown marked variability in levels of fusion gene mRNA in different fusion gene expressing cancers (Wang, et al. 2006), so some of the variability between different patient's cancers is almost certainly due to differential levels of transcription and/or mRNA stability in those tumors. We have also described significant differences in alternative splicing in different cancers that can impact translational efficiency (Zammarchi, et al. 2013) so translational alterations must also be considered. Differences in protein stability or degradation based on differences in alternatively spliced isoforms or associated tumor alterations may also play a role.
We also observed significant differences in ERG levels within a single cancer lesion. The finding that expression was stronger at the edge of some tumors implies that invasion, proliferation or local microenvironmental factors may modulate ERG expression.
Alternatively, genomic heterogeneity within cancers, including prostate cancer, is well known, and other genomic alterations may be heterogeneous within a given cancer focus and this could impact ERG protein levels. Further studies needed to understand the basis of intrafocal heterogeneity in fusion gene protein levels in prostate cancer.
The impact of different levels of ERG protein on the biology of fusion gene expressing cancers is not completely clear. The core proteins/pathways identified in our study (Table 2) appear to be associated with any level of ERG protein expression since the associations are present even when tumors are classified based on visual detection of any ERG protein in cancer cells. However, the associations noted with variable ERG expression are different and only partially overlapping. Thus there is likely significant similarity in the biology in all ERG expressing prostate cancers but also some differences. The differences may be important is some contexts. For example, NFkB p65 phospho-Ser536 levels are positively correlated with levels of ERG protein, which indicates that cancers with high ERG protein may be more sensitive to therapies targeting the NFkB pathway. We did not observe any differences in clinical outcome based on levels of ERG expression. However, a recent study in a radical prostatectomy cohort enriched for highly aggressive disease have shown that higher levels of ERG protein are significantly associated with distant metastasis and death from prostate cancer (Spencer, et al. 2013). Of note, the presence of ERG protein per se in was not associated with aggressive prostate cancer in this study, concordant with most but not all studies of radical prostatectomy cohorts to date. It is possible that our study is underpowered to see such associations since the majority of our cases were not aggressive. Additional studies with larger cohorts with larger numbers of aggressive cancers will be needed to determine if levels of ERG expression impact clinical outcome.
Acknowledgments
The technical assistance of Billie Smith and the Human Tissue Acquisition and Pathology Core of the Dan L. Duncan Cancer Center is gratefully acknowledged.
Funding. This work was supported by grant R01CA140734 from the National Cancer Institute (GA), the Dan L. Duncan Cancer Center P30 Cancer Center support grant (P30 CA125123) supporting the Human Tissue Acquisition and Pathology Core and the Biostatistics and Bioinformatics Core, the Dept of Veterans Affairs Merit Review program (MI) and by the use of the facilities of the Michael E. DeBakey VAMC.
Footnotes
Declaration of Interest. There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Agoulnik IU, Hodgson MC, Bowden WA, Ittmann MM. INPP4B: the new kid on the PI3K block. Oncotarget. 2011;2:321–328. doi: 10.18632/oncotarget.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoulnik IU, Krause WC, Bingman WE, 3rd, Rahman HT, Amrikachi M, Ayala GE, Weigel NL. Repressors of androgen and progesterone receptor action. J Biol Chem. 2003;278:31136–31148. doi: 10.1074/jbc.M305153200. [DOI] [PubMed] [Google Scholar]
- Agoulnik IU, Vaid A, Bingman WE, 3rd, Erdeme H, Frolov A, Smith CL, Ayala G, Ittmann MM, Weigel NL. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65:7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- Agoulnik IU, Vaid A, Nakka M, Alvarado M, Bingman WE, 3rd, Erdem H, Frolov A, Smith CL, Ayala GE, Ittmann MM, et al. Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer Res. 2006;66:10594–10602. doi: 10.1158/0008-5472.CAN-06-1023. [DOI] [PubMed] [Google Scholar]
- Allen JD, Verhoeven E, Domen J, van der Valk M, Berns A. Pim-2 transgene induces lymphoid tumors, exhibiting potent synergy with c-myc. Oncogene. 1997;15:1133–1141. doi: 10.1038/sj.onc.1201288. [DOI] [PubMed] [Google Scholar]
- Antonarakis ES, Keizman D, Zhang Z, Gurel B, Lotan TL, Hicks JL, Fedor HL, Carducci MA, De Marzo AM, Eisenberger MA. An immunohistochemical signature comprising PTEN, MYC, and Ki67 predicts progression in prostate cancer patients receiving adjuvant docetaxel after prostatectomy. Cancer. 2012;118:6063–6071. doi: 10.1002/cncr.27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala G, Frolov A, Ittman M, Mohammed S, LeBlanc M, Falcon S, Rowley D, Etzioni R. Biological correlates of biochemical recurrence free survival using multiple markers in a large tissue microarray cohort. Annals of clinical and laboratory science. 2013a;43:11–21. [PubMed] [Google Scholar]
- Ayala G, Morello M, Frolov A, You S, Li R, Rosati F, Bartolucci G, Danza G, Adam RM, Thompson TC, et al. Loss of caveolin-1 in prostate cancer stroma correlates with reduced relapse-free survival and is functionally relevant to tumour progression. The Journal of pathology. 2013b;231:77–87. doi: 10.1002/path.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala G, Thompson T, Yang G, Frolov A, Li R, Scardino P, Ohori M, Wheeler T, Harper W. High levels of phosphorylated form of Akt-1 in prostate cancer and non-neoplastic prostate tissues are strong predictors of biochemical recurrence. Clin Cancer Res. 2004;10:6572–6578. doi: 10.1158/1078-0432.CCR-04-0477. [DOI] [PubMed] [Google Scholar]
- Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, Wheeler M, Spitler J, Rowley DR. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res. 2003a;9:4792–4801. [PubMed] [Google Scholar]
- Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J, Wheeler TM, Lu KP, Bao L. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 2003b;63:6244–6251. [PubMed] [Google Scholar]
- Bassi C, Ho J, Srikumar T, Dowling RJ, Gorrini C, Miller SJ, Mak TW, Neel BG, Raught B, Stambolic V. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341:395–399. doi: 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brase JC, Johannes M, Mannsperger H, Falth M, Metzger J, Kacprzyk LA, Andrasiuk T, Gade S, Meister M, Sirma H, et al. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011;11:507. doi: 10.1186/1471-2407-11-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Wang H, He HH, Chen S, He L, Ma F, Mucci L, Wang Q, Fiore C, Sowalsky AG, et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. The Journal of clinical investigation. 2013;123:1109–1122. doi: 10.1172/JCI66666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nature medicine. 2013 doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinckemalie L, Spans L, Dubois V, Laurent M, Helsen C, Joniau S, Claessens F. Androgen regulation of the TMPRSS2 gene and the effect of a SNP in an androgen response element. Molecular endocrinology. 2013;27:2028–2040. doi: 10.1210/me.2013-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordon-Cardo C, Kotsianti A, Verbel DA, Teverovskiy M, Capodieci P, Hamann S, Jeffers Y, Clayton M, Elkhettabi F, Khan FM, et al. Improved prediction of prostate cancer recurrence through systems pathology. J Clin Invest. 2007;117:1876–1883. doi: 10.1172/JCI31399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Li R, Wheeler T, Diaz de Vivar A, Frolov A, Tahir S, Agoulnik I, Thompson T, Rowley D, Ayala G. Pim-2 upregulation: biological implications associated with disease progression and perinueral invasion in prostate cancer. Prostate. 2005;65:276–286. doi: 10.1002/pros.20294. [DOI] [PubMed] [Google Scholar]
- Ding Y, He D, Florentin D, Frolov A, Hilsenbeck S, Ittmann M, Kadmon D, Miles B, Rowley D, Ayala G. Semaphorin 4F as a critical regulator of neuroepithelial interactions and a biomarker of aggressive prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:6101–6111. doi: 10.1158/1078-0432.CCR-12-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, Rantala J, Alanen K, Nees M, Kallioniemi O. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2009;22:1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqq C, Li R, Khodabakhsh D, Frolov A, Ginzinger D, Thompson T, Wheeler T, Carroll P, Ayala G. Ethnic and racial differences in prostate stromal estrogen receptor alpha. Prostate. 2005;65:101–109. doi: 10.1002/pros.20272. [DOI] [PubMed] [Google Scholar]
- Hart M, Wach S, Nolte E, Szczyrba J, Menon R, Taubert H, Hartmann A, Stoehr R, Wieland W, Grasser FA, et al. The proto-oncogene ERG is a target of microRNA miR-145 in prostate cancer. The FEBS journal. 2013;280:2105–2116. doi: 10.1111/febs.12236. [DOI] [PubMed] [Google Scholar]
- Hodgson MC, Shao LJ, Frolov A, Li R, Peterson LE, Ayala G, Ittmann MM, Weigel NL, Agoulnik IU. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer Res. 2011;71:572–582. doi: 10.1158/0008-5472.CAN-10-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nature genetics. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluth M, Harasimowicz S, Burkhardt L, Grupp K, Krohn A, Prien K, Gjoni J, Hass T, Galal R, Graefen M, et al. Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. International journal of cancer Journal international du cancer. 2014;135:1369–1380. doi: 10.1002/ijc.28784. [DOI] [PubMed] [Google Scholar]
- Knudsen BS, Lucas JM, Fazli L, Hawley S, Falcon S, Coleman IM, Martin DB, Xu C, True LD, Gleave ME, et al. Regulation of hepatocyte activator inhibitor-1 expression by androgen and oncogenic transformation in the prostate. Am J Pathol. 2005;167:255–266. doi: 10.1016/S0002-9440(10)62970-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwabi-Addo B, Wang J, Erdem H, Vaid A, Castro P, Ayala G, Ittmann M. The expression of Sprouty1, an inhibitor of fibroblast growth factor signal transduction, is decreased in human prostate cancer. Cancer Res. 2004;64:4728–4735. doi: 10.1158/0008-5472.CAN-03-3759. [DOI] [PubMed] [Google Scholar]
- Leinonen KA, Saramaki OR, Furusato B, Kimura T, Takahashi H, Egawa S, Suzuki H, Keiger K, Ho Hahm S, Isaacs WB, et al. Loss of PTEN is associated with aggressive behavior in ERG-positive prostate cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2013;22:2333–2344. doi: 10.1158/1055-9965.EPI-13-0333-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Dai H, Wheeler TM, Sayeeduddin M, Scardino PT, Frolov A, Ayala GE. Prognostic value of Akt-1 in human prostate cancer: a computerized quantitative assessment with quantum dot technology. Clin Cancer Res. 2009a;15:3568–3573. doi: 10.1158/1078-0432.CCR-08-0826. [DOI] [PubMed] [Google Scholar]
- Li R, Erdamar S, Dai H, Sayeeduddin M, Frolov A, Wheeler TM, Ayala GE. Cytoplasmic accumulation of glycogen synthase kinase-3beta is associated with aggressive clinicopathological features in human prostate cancer. Anticancer Res. 2009b;29:2077–2081. [PubMed] [Google Scholar]
- Li R, Erdamar S, Dai H, Wheeler TM, Frolov A, Scardino PT, Thompson TC, Ayala GE. Forkhead protein FKHR and its phosphorylated form p-FKHR in human prostate cancer. Hum Pathol. 2007;38:1501–1507. doi: 10.1016/j.humpath.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Wheeler T, Dai H, Frolov A, Thompson T, Ayala G. High level of androgen receptor is associated with aggressive clinicopathologic features and decreased biochemical recurrence-free survival in prostate: cancer patients treated with radical prostatectomy. Am J Surg Pathol. 2004a;28:928–934. doi: 10.1097/00000478-200407000-00013. [DOI] [PubMed] [Google Scholar]
- Li R, Wheeler TM, Dai H, Sayeeduddin M, Scardino PT, Frolov A, Ayala GE. Biological correlates of p27 compartmental expression in prostate cancer. J Urol. 2006;175:528–532. doi: 10.1016/S0022-5347(05)00151-5. [DOI] [PubMed] [Google Scholar]
- Li R, Younes M, Frolov A, Wheeler TM, Scardino P, Ohori M, Ayala G. Expression of neutral amino acid transporter ASCT2 in human prostate. Anticancer Res. 2003;23:3413–3418. [PubMed] [Google Scholar]
- Li R, Younes M, Wheeler TM, Scardino P, Ohori M, Frolov A, Ayala G. Expression of vascular endothelial growth factor receptor-3 (VEGFR-3) in human prostate. Prostate. 2004b;58:193–199. doi: 10.1002/pros.10321. [DOI] [PubMed] [Google Scholar]
- Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- Magistroni V, Mologni L, Sanselicio S, Reid JF, Redaelli S, Piazza R, Viltadi M, Bovo G, Strada G, Grasso M, et al. ERG deregulation induces PIM1 over-expression and aneuploidy in prostate epithelial cells. PLoS One. 2011;6:e28162. doi: 10.1371/journal.pone.0028162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoner P, Kugler KG, Unterberger K, Kuner R, Mueller LA, Falth M, Schafer G, Seifarth C, Ecker S, Verdorfer I, et al. Characterization of transcriptional changes in ERG rearrangement-positive prostate cancer identifies the regulation of metabolic sensors such as neuropeptide Y. PLoS One. 2013;8:e55207. doi: 10.1371/journal.pone.0055207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlhany SJ, Ayala GE, Frolov A, Ressler SJ, Wheeler TM, Watson JE, Collins C, Rowley DR. Decreased stromal expression and increased epithelial expression of WFDC1/ps20 in prostate cancer is associated with reduced recurrence-free survival. Prostate. 2004;61:182–191. doi: 10.1002/pros.20085. [DOI] [PubMed] [Google Scholar]
- Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, Shen R, Montie JE, Chinnaiyan AM, Shah RB. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- Mertz KD, Horcic M, Hailemariam S, D'Antonio A, Dirnhofer S, Hartmann A, Agaimy A, Eppenberger-Castori S, Obermann E, Cathomas G, et al. Heterogeneity of ERG expression in core needle biopsies of patients with early prostate cancer. Human pathology. 2013;44:2727–2735. doi: 10.1016/j.humpath.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, Simon R, Tennstedt P, Muller J, Scholz L, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5878–5888. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- Minner S, Gartner M, Freudenthaler F, Bauer M, Kluth M, Salomon G, Heinzer H, Graefen M, Bokemeyer C, Simon R, et al. Marked heterogeneity of ERG expression in large primary prostate cancers. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:106–116. doi: 10.1038/modpathol.2012.130. [DOI] [PubMed] [Google Scholar]
- Nakka M, Agoulnik IU, Weigel NL. Targeted disruption of the p160 coactivator interface of androgen receptor (AR) selectively inhibits AR activity in both androgen-dependent and castration-resistant AR-expressing prostate cancer cells. The international journal of biochemistry & cell biology. 2013;45:763–772. doi: 10.1016/j.biocel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hurley G, Prencipe M, Lundon D, O'Neill A, Boyce S, O'Grady A, Gallagher WM, Morrissey C, Kay EW, Watson RW. The analysis of serum response factor expression in bone and soft tissue prostate cancer metastases. The Prostate. 2014;74:306–313. doi: 10.1002/pros.22752. [DOI] [PubMed] [Google Scholar]
- Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Wu SP, Creighton CJ, Dai F, Xie X, Cheng CM, Frolov A, Ayala G, Lin X, Feng XH, et al. COUP-TFII inhibits TGF-beta-induced growth barrier to promote prostate tumorigenesis. Nature. 2013;493:236–240. doi: 10.1038/nature11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AH, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D, Clark J, Flohr P, Edwards S, Berney DM, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. British journal of cancer. 2010;102:678–684. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Tekedereli I, Wang J, Yuca E, Tsang S, Sood A, Lopez-Berestein G, Ozpolat B, Ittmann M. Highly specific targeting of the TMPRSS2/ERG fusion gene using liposomal nanovectors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:6648–6657. doi: 10.1158/1078-0432.CCR-12-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer ES, Johnston RB, Gordon RR, Lucas JM, Ussakli CH, Hurtado-Coll A, Srivastava S, Nelson PS, Porter CR. Prognostic value of ERG oncoprotein in prostate cancer recurrence and cause-specific mortality. The Prostate. 2013;73:905–912. doi: 10.1002/pros.22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, Shaheduzzaman S, Tan SH, Vaidyanathan G, Whitman E, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian TV, Tomavo N, Huot L, Flourens A, Bonnelye E, Flajollet S, Hot D, Leroy X, de Launoit Y, Duterque-Coquillaud M. Identification of novel TMPRSS2:ERG mechanisms in prostate cancer metastasis: involvement of MMP9 and PLXNA2. Oncogene. 2013 doi: 10.1038/onc.2013.176. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Science's STKE : signal transduction knowledge environment. 2004;2004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- Vainio P, Lehtinen L, Mirtti T, Hilvo M, Seppanen-Laakso T, Virtanen J, Sankila A, Nordling S, Lundin J, Rannikko A, et al. Phospholipase PLA2G7, associated with aggressive prostate cancer, promotes prostate cancer cell migration and invasion and is inhibited by statins. Oncotarget. 2011;2:1176–1190. doi: 10.18632/oncotarget.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- Wang J, Cai Y, Shao LJ, Siddiqui J, Palanisamy N, Li R, Ren C, Ayala G, Ittmann M. Activation of NF-{kappa}B by TMPRSS2/ERG Fusion Isoforms through Toll-Like Receptor-4. Cancer Res. 2011;71:1325–1333. doi: 10.1158/0008-5472.CAN-10-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–8524. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington MN, Weigel NL. 1{alpha},25-Dihydroxyvitamin D3 inhibits growth of VCaP prostate cancer cells despite inducing the growth-promoting TMPRSS2:ERG gene fusion. Endocrinology. 2010;151:1409–1417. doi: 10.1210/en.2009-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhao JC, Kim J, Jin HJ, Wang CY, Yu J. ERG is a critical regulator of Wnt/LEF1 signaling in prostate cancer. Cancer research. 2013;73:6068–6079. doi: 10.1158/0008-5472.CAN-13-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Ayala G, De Marzo A, Tian W, Frolov A, Wheeler TM, Thompson TC, Harper JW. Elevated Skp2 protein expression in human prostate cancer: association with loss of the cyclin-dependent kinase inhibitor p27 and PTEN and with reduced recurrence-free survival. Clin Cancer Res. 2002;8:3419–3426. [PubMed] [Google Scholar]
- Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Feng S, Dakhova O, Creighton CJ, Cai Y, Wang J, Li R, Frolov A, Ayala G, Ittmann M. FGFR-4 Arg(3)(8)(8) enhances prostate cancer progression via extracellular signal-related kinase and serum response factor signaling. Clin Cancer Res. 2011;17:4355–4366. doi: 10.1158/1078-0432.CCR-10-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammarchi F, Boutsalis G, Cartegni L. 5′ UTR control of native ERG and of Tmprss2:ERG variants activity in prostate cancer. PLoS One. 2013;8:e49721. doi: 10.1371/journal.pone.0049721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HJ, Yan J, Luo W, Ayala G, Lin SH, Erdem H, Ittmann M, Tsai SY, Tsai MJ. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65:7976–7983. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]