Background: UreE and UreG play important roles in urease activation.
Results: Nickel binding to UreG and its interactions with UreE as well as their roles in Ni2+ transfer were studied.
Conclusion: UreG is a specific nickel-dependent GTPase; formation of (UreE)2-(UreG)2complex is a prerequisite for Ni2+ transfer from UreE to UreG.
Significance: This study provides novel information on Ni2+ transfer among metallochaperones UreE, UreG, and HypA.
Keywords: Helicobacter pylori, Metal Ion-Protein Interaction, Metallochaperone, Metalloprotein, Nickel, Protein-Protein Interaction, Translocation, UreE, UreG, Urease, Urease Maturation
Abstract
The pathogenicity of Helicobacter pylori relies heavily on urease, which converts urea to ammonia to neutralize the stomach acid. Incorporation of Ni2+ into the active site of urease requires a battery of chaperones. Both metallochaperones UreE and UreG play important roles in the urease activation. In this study, we demonstrate that, in the presence of GTP and Mg2+, UreG binds Ni2+ with an affinity (Kd) of ∼0.36 μm. The GTPase activity of Ni2+-UreG is stimulated by both K+ (or NH4+) and HCO3− to a biologically relevant level, suggesting that K+/NH4+ and HCO3− might serve as GTPase elements of UreG. We show that complexation of UreE and UreG results in two protein complexes, i.e. 2E-2G and 2E-G, with the former being formed only in the presence of both GTP and Mg2+. Mutagenesis studies reveal that Arg-101 on UreE and Cys-66 on UreG are critical for stabilization of 2E-2G complex. Combined biophysical and bioassay studies show that the formation of 2E-2G complex not only facilitates nickel transfer from UreE to UreG, but also enhances the binding of GTP. This suggests that UreE might also serve as a structural scaffold for recruitment of GTP to UreG. Importantly, we demonstrate for the first time that UreE serves as a bridge to grasp Ni2+ from HypA, subsequently donating it to UreG. The study expands our horizons on the molecular details of nickel translocation among metallochaperones UreE, UreG, and HypA, which further extends our knowledge on the urease maturation process.
Introduction
Helicobacter pylori is a pathogenic bacterial species that was discovered in human stomach and duodenal mucous membrane (1). This pathogen is responsible for gastritis, peptic ulcer, and even stomach cancer (2). To survive in the acidic environment of the human stomach, H. pylori produces large amounts of urease to turn urea to carbon dioxide and ammonia, which neutralizes the stomach acid (3). The physiological activities for survival in an extreme acidic environment consume huge amounts of energy, and thus H. pylori produces [Ni,Fe]-hydrogenase to oxidize molecular hydrogen to obtain sufficient energy (4). Thus, the survival and successful colonization of H. pylori in human stomach largely rely on the proper function of two enzymes, i.e. urease (5) and [Ni,Fe]-hydrogenase (6). Maturation of both urease and [Ni,Fe]-hydrogenase refers to the assembly of nickel-containing active centers (7, 8). The utilization of nickel must be tightly controlled in H. pylori as a plethora of nickel causes serious cell damage to H. pylori (9). Therefore, H. pylori has developed an elaborate system to tightly regulate the cellular nickel homeostasis from uptake, storage, and delivery to efflux through biosynthesis of a series of metalloproteins and chaperones (10), such as nickel storage protein Hpn and Hpnl (11–14), which are only produced by this pathogen, and HspA (15, 16), which has a distinct His-Cys-rich C terminus with nickel binding ability.
The proper assembly of the metal-containing active site of [Ni,Fe]-hydrogenase depends on a group of accessory proteins (HypA, -B, -C, -D, -E, -F) (8, 17, 18). HpHypA consists of zinc binding and nickel binding domains (22, 23), and binds nickel with an affinity at a micromolar level (20). HpHypB is classified into the P-loop GTPase family with its enzymatic activity being activated by potassium (24). It is commonly believed that HypA and HypB are involved in the nickel insertion into the large subunit of hydrogenase (19–21) via the formation of HypA-HypB complex (20, 25). In addition, SlyD, a protein that binds Ni2+ at its C terminus (28), might also participate in hydrogenase maturation through interaction with HypB (29, 30). The IF (insert-in-flap) domain of SlyD is responsible for the SlyD-HypB interaction, facilitating nickel transfer from the C terminus of SlyD to HypB in H. pylori (31), but stimulates nickel release from HypB in Escherichia coli (29). Interestingly, both HypA and HypB are also involved in urease maturation (19, 26, 27), indicating a cross-talk between urease and hydrogenase maturation processes.
Maturation of urease involves the proper insertion of two nickel ions to its active site and requires at least four accessory proteins (UreE, -F, -G, -H) (9). In addition, accessory proteins for [Ni,Fe]-hydrogenase were also reported to be involved in the process (19, 27). UreE can form a ternary complex with HypA, consisting of one UreE dimer and one HypA monomer. Such an interaction facilitates nickel translocation from HypA to UreE and enhances urease activity (32).
Maturation of urease in H. pylori is not well understood. The formation of apo-urease-(UreG/UreF/UreH) was thought to be a crucial step for nickel insertion to urease (33, 34). It was also demonstrated that UreE-UreG complexation plays an important role in the maturation of apo-urease; however, the molecular mechanism of nickel transfer between the chaperones UreE and UreG is not fully understood. Previously, UreG from H. pylori was demonstrated to bind Zn2+, instead of Ni2+ (35). Recently, the structure of HpUreG in an UreG/F/H complex revealed the metal binding site, consisting of two cysteines and two histidines, which are located at the interface between two UreG monomers. The nickel binding induces GTP-dependent dimerization of UreG (36). A previous isothermal titration calorimetry study showed that H. pylori UreE and UreG form a (UreE)2-(UreG)2 complex (i.e. one UreE dimer with one UreG dimer, or as 2E-2G) and the Zn2+ but not Ni2+ binding stabilized the complex (37), which is quite peculiar given the role of UreE and UreG in nickel delivery. Moreover, a previous study showed that in Klebsiella aerogenes, a (UreE)2-UreG (i.e. 2E-G, one UreE dimer with one UreG monomer) complex was observed (38). It is not clear how nickel transfer is achieved through the interaction of UreE and UreG.
In this study, we have overexpressed and purified UreG and UreE from H. pylori 26695. By using biochemical, biophysical, and molecular biology approaches, we characterize Ni2+ binding properties as well as GTPase activity of UreG. We also carried out detailed studies on the molecular mechanism of the interaction between UreE and UreG. We demonstrate that UreE and UreG form a 2E-2G complex, which preactivates GTPase of UreG through enhancement of GTP binding to facilitate Ni2+ transfer. The roles of UreE, UreG, and HypA in nickel delivery are examined and discussed.
EXPERIMENTAL PROCEDURES
Materials
Restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs. Primers were synthesized by Life Technologies and are listed in Table 1. Phusion High-Fidelity DNA polymerase was obtained from Finnzymes. KOD Hot Start DNA polymerase was obtained from Toyobo Life Science. All chromatography columns and FPLC system were from GE Healthcare. All chemical reagents were purchased from Sigma, unless stated otherwise. All solutions were prepared with Milli-Q water (18.2 megaohms).
TABLE 1.
Primers used for PCR reactions (restriction sites or mutated sites are bolded)
The abbreviations used are: for, forward; rev, reverse.
| For pET28a-ureG | |
| UreG(NdeI)-for | GGAATTCCATATGGTAAAAATTGGAGTTTGTG |
| UreG(EcoRI)-rev | CGGAATTCCTAATCTTCCAATAAAGCGTTGC |
| For pET28a-ureG(C66A) | |
| UreG(C66A)-for | CAGGAGGCGCTCCGCACACGGCTATTAG |
| UreG(C66A)-rev | TTTCTACGCCAATGATCCTCTCTCGTGG |
| For pET28a-ureG(H68A) | |
| UreG(H68A)-for | CAGGAGGCTGTCCGGCCACGGCTATTAG |
| UreG(C66A)-rev | |
| For pET28a-ureE(N100A) | |
| UreE(N100A)-for | CTATGAAATAGGAGCGCGCCATGCG |
| UreE(N100A)-rev | CATATTTTCGCTACTTCTGCCACGC |
| For pET28a-ureE(R101A) | |
| UreE(R101A)-for | CTATGAAATAGGAAACGCCCATGCGG |
| UreE(N100A)-rev | |
| For pET28a-ureE(H102A) | |
| UreE(H102A)-for | CTATGAAATAGGAAACCGCGCTGCGG |
| UreE(N100A)-rev | |
| For pET28a-ureE(H152A) | |
| UreE(H152A)-for | AACCGTGAGCATGCCCGCGAGTGAGC |
| UreE(H152A)-rev | AAGCGTTCTTTGGAATCCAATTTTGAAC |
| For pET-UreA2H | |
| UreA(NdeI)-for | GGAATTCCATATGAAACTCACCCCAAAAGAGTTAG |
| UreH(EcoRI)-rev | CGGAATTCTCAAACCTTTTGCGTGGTGGTTTGC |
| For pET-UreA2HΔG | |
| ΔUreG-for | TGATGAACACTTACGCTCAAGAATCCAAG |
| ΔUreG-rev | TCAAGACATATAAAGGCGCGAGTATAAAC |
Construction of Expression Vectors
The pET28a-ureE and pET28a-ureEΔ158–170 expression plasmids were generated as described previously (32). The ureG gene was amplified by PCR from genomic DNA of H. pylori 26695 and inserted into pET28a after NdeI and EcoRI digest to generate the expression plasmid pET28a-ureG. pET28a-ureE(N100A), pET28a-ureE(R101A), pET28a-ureE(H102A), pET28a-ureE(152A), pET28a-ureG(C66A), and pET28a-ureG(H68A) were generated using Phusion High-Fidelity DNA polymerase. The ligation mixture was subsequently transformed into XL-1 Blue E. coli. All the plasmids were sequenced (BGI) to verify the proper gene sequences. The pET-UreA2HΔG used in urease activity was constructed by insertion of H. pylori urease operon (ureABIEFGH) into the pET-32a vector using the NdeI-EcoRI restriction sites and introduction of the deletion of ureG gene (ΔG) using Phusion High-Fidelity DNA polymerase.
Protein Expression and Purification
UreE and its variants were overexpressed and purified similarly as described previously (32). To express UreG, a single clone of BL21 (DE3) E. coli harboring the pET28a-ureG expression vector was cultured in Luria Broth (LB) medium supplied with 50 μg/ml kanamycin at 37 °C for overnight. The bacteria cultured overnight were diluted 1:100 into 1 liter of LB medium containing a proper antibiotic for subculture at 37 °C for ∼2 h. Until it was grown to an optical density of 0.6–0.8 at 600 nm, expression of His6-UreG fusion protein was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.2 mm. The cells were further cultured at 25 °C overnight (∼16 h).
The following steps were carried out at 4 °C unless stated otherwise. Cells were harvested by centrifugation (4,000 × g, 30 min) and resuspended in buffer A (20 mm Hepes containing 500 mm NaCl, pH 7.5) with 1 mm PMSF as an enzyme inhibitor. After cell lysis by sonication, supernatant was separated from pellets by centrifugation (16,000 × g, 30 min). The supernatant was further filtered through Millex-HA filter (0.45 μm, 33 mm, Millipore) and loaded onto a 5-ml HisTrap column (GE Healthcare) preloaded with nickel ion and pre-equilibrated by buffer A containing 50 mm imidazole. Five column volumes of buffer A with 50 mm imidazole were used to wash away the unspecific binding impurities. The His6-UreG protein was eluted by buffer A supplemented with 300 mm imidazole. The eluted protein fraction was collected and loaded into a 5-ml HiTrap desalting column (GE Healthcare) pre-equilibrated by buffer B (20 mm Hepes, 100 mm NaCl, pH 7.5) to remove excess imidazole for thrombin cleavage. Fusion protein was incubated with ∼50 NIH units of thrombin at 25 °C for 3 h with gentle shaking to cleave His6 tag from free UreG. Trace amounts of uncleaved fusion protein were removed by loading the digested protein sample to the 5-ml HisTrap column again. Fraction of flow-through was collected and incubated with 20 mm EDTA and 1 mm DTT at 4 °C overnight to obtain the apo-form UreG protein, which was further purified by gel filtration using a HiLoad 16/60 Superdex 75 column (GE Healthcare) equilibrated with buffer C (20 mm Hepes, 300 mm NaCl, pH 7.2) containing 500 μm tris(2-carboxyethyl) phosphine (TCEP)2 as a reducing agent. Elution fractions of purified protein were collected and concentrated to ∼2 ml. UreG variants were purified similarly. Protein concentration was determined by the BCA protein assay kit (Novagen), and metal contents of the purified proteins were determined by inductively coupled plasma-mass spectrometry (Agilent 7500a).
UV-visible Spectroscopy
Nickel binding to UreG was monitored by UV-visible spectroscopy. All UV-visible spectra were recorded on a Cary 50 UV-visible spectrometer using a 1-cm quartz cuvette at ambient temperature (∼25 °C). UV-visible spectra were scanned with a rate of 360 nm/min from 600 to 240 nm. Apo-UreG (20 μm) was freshly prepared in 20 mm Hepes, 100 mm NaCl, 500 μm TCEP, pH 7.2, with or without supplementation of 100 μm GTP/GDP and 1 mm Mg2+. Spectrum of the same buffer was used for baseline correction. Aliquots of nickel chloride stock solution (10 mm) were titrated into UreG sample with gentle mixing, and UV-visible spectra were recorded after a 10-min incubation at room temperature. For competition between the Ni2+ and Zn2+ experiments, UreG (20 μm) was freshly prepared in 20 mm Hepes, 100 mm NaCl, 500 μm TCEP, pH 7.2, in the presence of 40 μm Ni2+ or Zn2+, 100 μm GTP, and 1 mm Mg2+, and subsequently, Zn2+ or Ni2+ stock solutions were titrated into the above protein solution. The UV titration curves were fitted to the Ryan-Weber nonlinear equation (39)
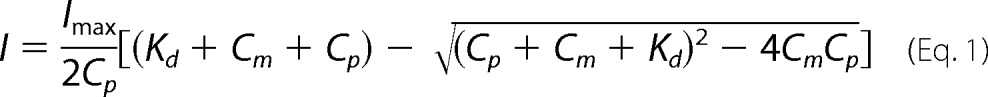 |
where I stands for UV absorbance intensity; Imax stands for maximal UV absorbance; Cp and Cm refer to the total concentrations of proteins and ligands respectively; and Kd is the dissociation constant.
To examine the nickel transfer between UreE and UreG, Ni-UreE (or its variants Ni-CUreE and Ni-UreE-R101A) was titrated stepwise into UreG (25 μm) in the presence of GTP (50 μm) and Mg2+ (1 mm). CUreE is the C-terminal domain of UreE and houses the metal binding site. Both CUreE and UreE-R101A preserve nickel binding ability similarly to the WT-UreE (data not shown). Similarly, Ni-UreE was titrated into UreG in the presence of 50 μm GTP but not Mg2+. To examine the nickel transfer from HypA to UreG via UreE, Ni-Zn-HypA and UreE or Ni-Zn-HypA alone was titrated stepwise into UreG (20 μm) in the presence of GTP (50 μm) and Mg2+ (1 mm).
Analytic Gel Filtration Chromatography and Light Scattering
Analytical gel filtration chromatography was performed on an ÄKTA FPLC system using a Superdex 75 or 200 30/100 GL column (GE Healthcare) precalibrated with the low molecular weight gel filtration calibration kit (GE Healthcare) at 4 °C with a flow rate of 0.5 ml/min. Typically, 40 μm apo-UreG was preincubated with 1.5 molar eq of Ni2+/Zn2+, GTP/GDP, Mg2+, or their combinations for 15 min and subjected to analysis. The column was pre-equilibrated with Hepes buffer (20 mm Hepes, 100 mm NaCl, 200 μm TCEP, pH 7.2). The molecular mass of the protein complex in the elution peaks was further measured by a multiple-angle laser light scattering detector (miniDAWN light scattering detector, Wyatt Technology) or a by DynaPro plate reader (Wyatt Technologies), and the data were analyzed by Astra version 5.3.4.18 (Wyatt Technology) or by the Dynamics software (Wyatt Technologies).
To investigate the interaction among HypA, UreE, and UreG, 50 μm apo-UreG was preincubated with 50 μm apo-UreE and 300 μm Zn-HypA in the absence or presence of 75 μm GTP and MgSO4 (HypA protein was used in excess due to low absorption at 280 nm). The fractions of elution peaks were collected and subjected to 15% SDS-PAGE assay to determine the components of peaks.
GTPase Activity Assay
GTPase activity of UreG was determined by the Malachite Green phosphate assay kit (Abcam). To compare the effect of K+, HCO3−, and KHCO3 on the GTPase activity of Ni-UreG, GTPase assay was carried out with the supplementation of KCl/NH4Cl, NaHCO3, or KHCO3/NH4HCO3 at concentrations of 0, 0.5, 1, 5, 10, 20, 30, 40, 50, 100 mm, and the pH of the buffer was gently adjusted to be 7.5. GTP hydrolysis was triggered by the addition of Ni-UreG (10 μm) into KCl/NaHCO3/KHCO3/NH4HCO3 buffer with incubation at 37 °C for 20 min. To compare the GTPase activities of apo-UreG, Zn-UreG, and Ni-UreG at different pH, the GTPase assay was carried out in a series of buffers with different pH values supplemented with 10 mm KCl/NaHCO3/KHCO3. After incubation at 37 °C for 40 min, the free phosphate from hydrolysis of GTP determined by the kit was used to calculate the percentage of GTP turnover. Similarly, to compare the effect of HypA and UreE on GTPase activity of UreG, apo-UreG was prepared and supplemented with Ni-HypA, Ni-UreE, or Ni-HypA-UreE (10 μm for each protein) in GTPase assay buffer (20 mm Hepes, 100 mm NaCl, 1 mm MgCl2, 100 μm GTP, 1% glycerol, pH 7.5). Hydrolysis of GTP was triggered by the addition of 10 mm KHCO3. After incubation at 37 °C for 40 min, the free phosphate from hydrolysis of GTP was determined. For the time course reaction, a series of 400 μl of 10 μm UreG protein samples was prepared in GTPase assay buffer and was incubated with 10 mm KHCO3at 37 °C. For each time point, 50 μl of reaction mixtures was taken out to allow the free phosphate from hydrolysis of GTP to be determined. Those solutions containing 100 μm GTP but without UreG were also incubated as blank controls to eliminate the self-hydrolysis of GTP. To determine the enzyme kinetic parameters (Km, kcat) of UreG, similar experiments were carried out but with the substrate concentration varied. A series of 50 μl of 10 μm UreG protein samples containing various GTP concentrations ranging from 10 to 500 μm was prepared in GTPase assay buffer with 10 mm KHCO3. After incubation at 37 °C for 40 min, the amounts of free phosphate in the reaction mixtures were determined. The Michaelis-Menten equation was used to obtain kinetic parameters by nonlinear fitting.
Urease Activity Assay
pET-UreA2HΔG was transformed into E. coli strain KMl603 (BL21(DE3) ΔslyD::kan (a gift from Prof. A. R. Davidson, University of Toronto)). The bacterial cells were cultured in Luria Broth with supplementation of 100 μg/ml ampicillin at 37 °C and induced at 25 °C overnight with 0.2 mm isopropyl β-d-1-thiogalactopyranoside when A600 reached 0.8. Cells were collected and washed with 50 mm Hepes buffer, pH 7.5, lysed by sonication, and centrifuged to obtain supernatant. About 50 μl of lysate was mixed with 10 μm Ni-UreG/apo-UreG or 5 μm Ni2+. After supplementation of 250 μl of urea buffer (50 mm Hepes, 25 mm urea, pH 7.5) with or without 10 mm KHCO3, the mixtures were incubated at 37 °C for 1 h to activate urease. Urease activity was measured by the amounts of ammonia released using the phenol-hypochlorite assay. Total protein concentration of lysate was determined by the BCA protein assay kit (Novagen), and the unit of urease activity was defined as nmol of ammonia produced per min/mg of total protein.
RESULTS
Ni2+ Binding Property of HpUreG
UreG has been previously believed to be a specific Zn2+ chaperone with no Ni2+ binding capacity (27), whereas the report studying the UreG/UreF/UreH complex clearly showed that UreG binds Ni2+ (36). To further investigate the nickel binding property of HpUreG, the recombinant H. pylori apo-UreG was titrated with Ni2+ and monitored by UV-visible spectroscopy. The addition of Ni2+ to freshly prepared apo-UreG (20 μm) in 20 mm Hepes containing 1 mm Mg2+ leads to no observable new absorption bands (data not shown), indicative of no binding of Ni2+ to the protein under such a condition. However, upon supplementation of 100 μm GTP (Fig. 1A) but not GDP (Fig. 1B), the stepwise titration of Ni2+ induces the appearance and gradual increases of a peak centered at ∼337 nm, assignable to the π(S)(Cys)→Ni(II) ligand-to-metal charge transfer transition. Also, the intensities of this band are leveled off at a molar ratio of [Ni2+] to [UreG] of over 0.5, indicating that each monomer of UreG binds 0.5 molar eq of Ni2+, in good agreement with a previous report that each UreG dimer binds one Ni2+ (36). By nonlinearly fitting the plot of absorption at 337 nm versus Ni2+ concentrations to the Ryan-Weber equation, the dissociation constant (Kd(Ni-UreG))is determined to be 0.36 ± 0.05 μm. A similar titration experiment was carried out except in the absence of Mg2+, which shows that UreG still binds Ni2+, but with a dissociation constant of ∼10-fold larger (3.50 μm), and complete saturation of UreG by Ni2+ is achieved at ∼2 molar eq of Ni2+ to the protein under this condition (Fig. 1C).
FIGURE 1.
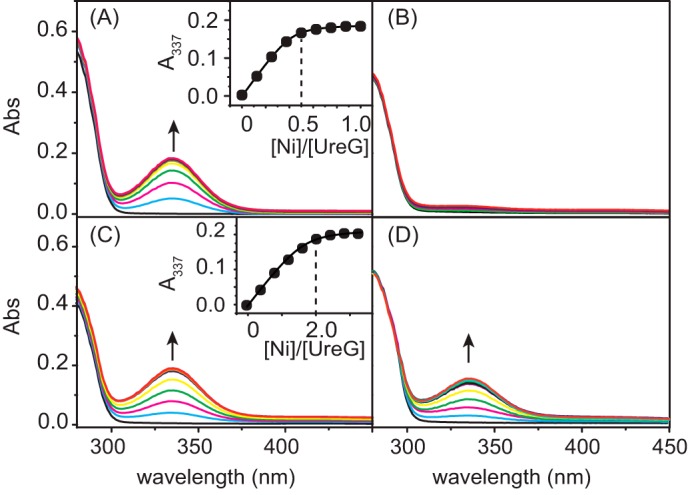
Ni2+ binding to UreG monitored by UV-visible spectroscopy. A–D, Ni2+ was titrated into 20 μm apo-UreG in 20 mm Hepes, 100 mm NaCl, 500 μm TCEP, pH 7.2, with supplementation of 100 μm GTP, 1 mm Mg2+ (A), 100 μm GDP, 1 mm Mg2+ (B), only 100 μm GTP without Mg2+ (C), or 20 μm Zn2+, 100 μm GTP, and 1 mm Mg2+ (D). The titration curve plotted at 337 nm is shown in the inset. Abs, absorbance.
A previous x-ray absorption spectroscopy study showed that the coordination sphere for zinc is formed by residues Cys-66 and His-68 from each of the monomers of HpUreG dimer (40). To investigate whether Cys-66 and His-68 are also involved in nickel binding, we constructed, overexpressed, and purified two UreG variants, UreG-C66A and UreG-H68A, and similarly carried out Ni2+ titration experiments. Mutation of either Cys-66 or His-68 to alanine abolishes Ni2+ binding as judged from the disappearance of the absorption peak at 337 nm (data not shown), suggesting that both Cys-66 and His-68 are essential for nickel binding.
Competitive binding of Zn2+ and Ni2+ to UreG in the presence of GTP was further studied. The addition of Zn2+ to Ni2+-UreG has little effect on the absorption peak at 337 nm (data not shown), indicating that Zn2+ cannot replace Ni2+ from UreG in the presence of GTP. In contrast, titration of Ni2+ into Zn2+-UreG solution results in the appearance of the absorption peak at 337 nm, a characteristic band for nickel binding to sulfur (Fig. 1D), suggesting the displacement of Zn2+ by Ni2+ from the protein. These results demonstrate that HpUreG has a higher binding affinity toward Ni2+ than Zn2+ in the presence of GTP and Mg2+.
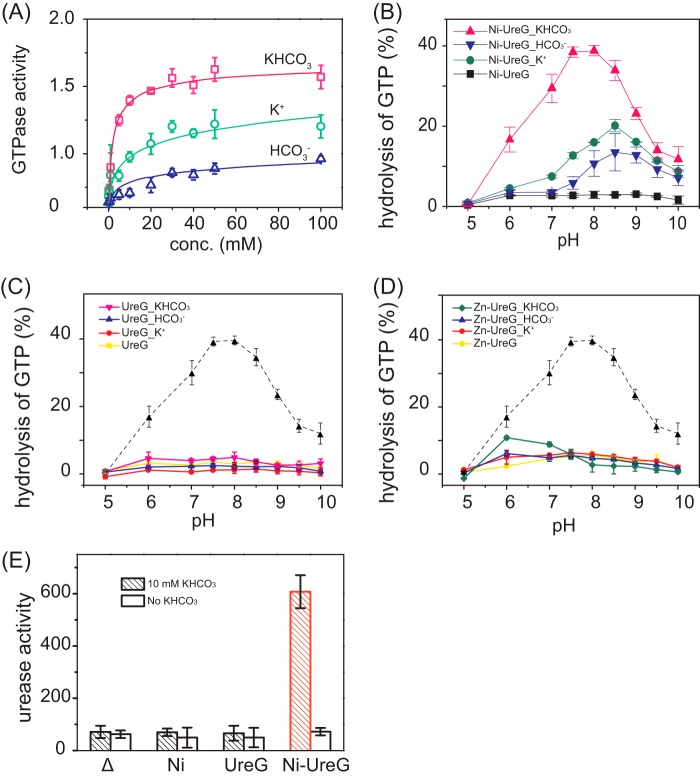
Potassium Bicarbonate Boosts GTPase Activity of Ni2+-HpUreG
It was demonstrated that GTPase of Ni-HpUreG dimer is stimulated by bicarbonate (HCO3−) (36), whereas the HpUreG counterpart, HpHypB, shows a higher GTPase activity in solution containing potassium (K+) (24), implying the potential roles of K+ and HCO3− in GTP hydrolysis. To verify this hypothesis, GTPase assay was carried out in a buffer containing 1 mm MgSO4 supplemented with KCl, NaHCO3, KHCO3, or NH4HCO3 at concentrations of 0, 0.5, 1, 5, 10, 20, 30, 40, 50, 100 mm, and the pH of the buffer was adjusted carefully to be 7.5. Consistent with a previous study (35), Ni2+-UreG does not exhibit GTPase activity in the absence of HCO3− (data not shown), whereas the activity of Ni2+-UreG rises slightly with increasing amounts of HCO3− and is saturated at only ∼0.7 nmol/min/mg of protein (Fig. 2A). Similarly, supplementation of gradient amounts of either K+ (Fig. 2A) or ammonia (NH4+) (data not shown) results in elevated GTPase activities, which saturate at ∼1.1 nmol/min/mg of protein. Interestingly, the GTPase activity of Ni2+-UreG is boosted upon the addition of potassium bicarbonate (KHCO3> 10 mm) or ammonium bicarbonate (data not shown) in a similar concentration gradient to a relatively high level with a rate of ∼1.6 nmol/min/mg of protein. These results indicate that HpUreG may employ potassium bicarbonate or ammonium bicarbonate as GTPase elements to achieve full GTPase activity. Upon GTP hydrolysis, Ni2+ was gradually released from Ni-UreG as judged from gradual losses of the ligand-to-metal charge transfer transition at 337 nm (data not shown).
FIGURE 2.
GTPase activity of UreG and its effect on urease activation. A, GTPase activity of Ni-UreG in the presence of K+ (KCl), HCO3− (NaHCO3), or KHCO3 with concentrations ranging from 0 to 100 mm. (GTPase activity is defined as nmol/min/mg of protein.) B, GTPase activity of Ni-UreG at different pH values in the presence of 10 mm K+, HCO3−, or KHCO3. C, GTPase activity of apo-UreG at different pH values. D, GTPase activity of Zn-UreG at different pH values. The GTPase activity of Ni-UreG is shown as a dotted line for comparison. All GTPase assay buffer contains 1 mm MgSO4. E, effect of Ni-UreG on urease activation. Note that significant enhanced activity of urease was found only in the present of both Ni-UreG and KHCO3. (The unit of urease activity is defined as nmol of ammonia produced per min/mg of total protein.) Error bars indicate means ± S.E.
The effect of pH on the GTP hydrolysis by HpUreG was also investigated in the absence or presence of 10 mm KHCO3/K+/HCO3−. Both apo-UreG and Zn-UreG show negligible GTPase activity at the pH range studied (Fig. 2, C and D). Upon supplementation of either K+ (10 mm) or HCO3− (10 mm) to the buffers, low levels of activity were detected for Ni-UreG and increased with pH from 5.0 to 8.5, with only ∼10–20% of GTP turnover for Ni-UreG. Further increases in pH values lead to decreases in GTPase activity (Fig. 2B). However, in the presence of KHCO3, the catalytic activity of Ni-UreG is enhanced dramatically with ∼40% of GTP turnover, with the highest activity attained at pH 7.5–8 (Fig. 2B).
We further explored the role of GTPase activity of UreG in urease maturation. Urease activation assay was performed by transforming the plasmid pET-UreA2HΔG containing the full set of urease genes ureABIEFH (except ureG) into E. coli. UreG protein or Ni2+ was added into cell lysate in the urease activation assay. In the absence of KHCO3, only weak urease activity (<100 units) was detected in all cases, whereas in the presence of KHCO3, ∼5-fold increases in urease activity were detected for Ni-UreG but not for Ni2+ or apo-UreG (Fig. 2E), confirming that GTP hydrolysis by UreG is essential for urease activation.
Formation of (UreE)2-UreG or (UreE)2-(UreG)2 Complexes
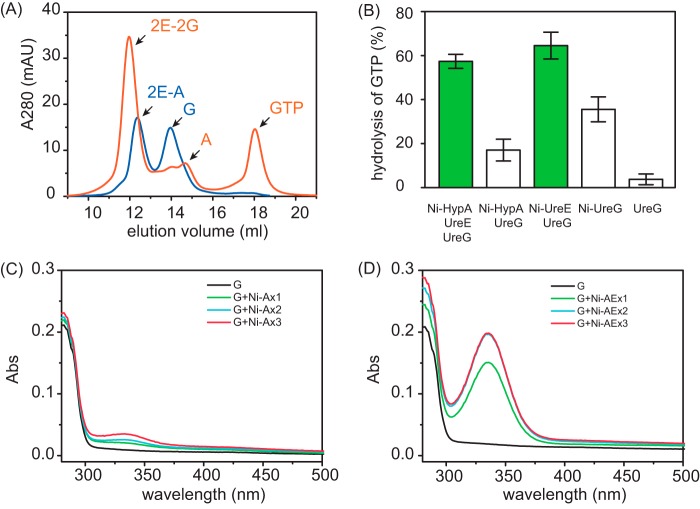
Complexation of UreE with UreG under near physiological conditions was examined by gel filtration chromatography at pH 7.2. As shown in Fig. 3A, apo-UreE and apo-UreG are eluted at ∼13.5 and 13.9 ml, respectively (Fig. 3A, lines 1 and 2), corresponding to molecule masses of ∼40 and 23 kDa, i.e. an UreE dimer and an UreG monomer, respectively. Mixing of equimolar amounts of apo-UreE and apo-UreG resulted in the disappearance of UreE peak and the appearance of a new peak at ∼12.5 ml with a molecular mass of 62 ± 1.6 kDa as determined by light scattering, which can be assigned to (UreE)2-UreG complex (2E-G), consisting of a UreE dimer and a UreG monomer (Fig. 3A, line 3). Similarly, we also investigated the effect of Ni2+, Zn2+, GTP, GDP, or their combinations on the complexation of UreE and UreG. Unexpectedly, neither Ni2+ nor Zn2+ perturbs the formation of UreE-UreG complex with 2E-G complex being observed in in all cases (Fig. 3A). We further carried out similar experiments but with the supplementation of Mg2+ (60 μm) to the reaction buffer. Surprisingly, supplementation of GTP and Mg2+ (60 μm for each) leads to the emergence of a single peak at ∼11.8 ml with a molecular mass of 83 ± 1.4 kDa by light scattering, suggesting the formation of a complex of a UreE dimer and a UreG dimer, i.e. 2E-2G, (Fig. 3B, lines 5, 7, and 9). In conclusion, we show that neither Ni2+ nor Zn2+ perturbs the formation of 2E-G or 2E-2G complexes. Importantly, both GTP and Mg2+ are indispensable for the formation of 2E-2G complex, whereas a 2E-G complex was observed under all conditions without both GTP and Mg2+.
FIGURE 3.
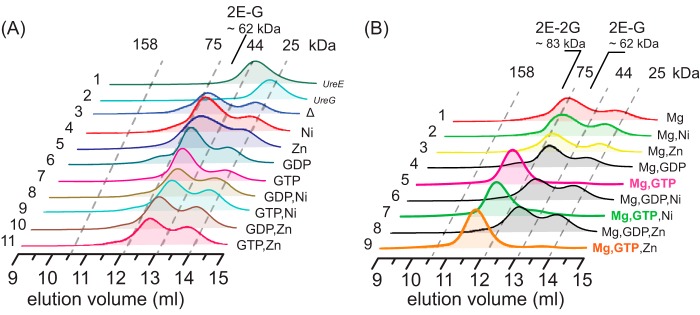
Formation of UreE-UreG complex. UreE and UreG (40 μm for each) samples were incubated with 1.5 molar eq of metal ions (Ni2+/Zn2+/Mg2+) with or without supplementation of guanine nucleotides (GDP or GTP) and loaded onto an analytic column. A, the formation of UreE-UreG complex in the absence of Mg2+. Profiles of free UreE and UreG are shown for comparison (lines 1 and 2). B, the formation of UreE-UreG complex in the presence of Mg2+. Note that 2E-2G complex is formed only in the presence of both GTP and Mg2+ (lines 5, 7, and 9).
Arg-101 of UreE and Cys-66 of UreG Stabilize 2E-2G Complex
Crystal structures of UreE from H. pylori and other species (41–45) reveal a similar homodimer architecture for apo-UreE, consisting of an N-terminal (NUreE) and a C-terminal domain (CUreE) with the latter housing the metal binding site and being responsible for specific HypA-UreE interaction (32). To examine which domain of UreE is responsible for UreE-UreG interaction, both NUreE and CUreE proteins (40 μm) were overexpressed and purified as described previously (32) and then incubated with 1 molar eq of UreG, and the potential protein complexes were monitored by analytic gel filtration chromatography. Surprisingly, upon mixing of either NUreE or CUreE with UreG, no new peaks corresponding to the complexes such as UreG-NUreE or UreG-CUreE were observed, even in the presence of GTP and Mg2+ (data not shown), implying that the scaffold formed between NUreE and CUreE may be essential for the UreG-UreE interaction.
To identify the residues of UreE participating in UreE-UreG interaction, we constructed a series of UreE variants according to two criteria. First, the residues involved in or around the metal binding site were considered in view of their potential roles in UreE-UreG interaction for nickel delivery to urease. Second, highly conserved residues in the C-domain were identified by the alignment of UreE sequences from several species via Clustal W (46). On the basis of these criteria combined with examination of the crystal structure of HpUreE (41, 42), residues for mutagenesis were selected as follows: Asn-100, Arg-101, His-102, His-152, and residues 158–70. Both Asn-100 and Arg-101 are conserved residues and close to the metal binding site. Both His-102 and His-152 are involved in nickel binding. Residues 158–170 are close to the metal binding site upon nickel binding to His-152 and are likely responsible for HypA-UreE interaction (32).
UreE variants were prepared and incubated with wild-type UreG prior to gel filtration analysis. As shown in Fig. 4 (black curves), all mutants of UreE show negligible effect on the formation of 2E-G complex, whereas the formation of 2E-2G complex is not affected by mutations of Asn-100, His-102, His-152, and Δ158–170 (Fig. 4, lines 1, 3, 4, and 5) in the presence of GTP and Mg2+. However, mutation of Arg-101 of UreE to alanine leads to the majority of its mixture with UreG being eluted as 2E-G complex even in the presence of GTP and Mg2+, indicating that residue Arg-101 may play an important role in the stabilization of 2E-2G complex (Fig. 4, line 2, green curve). Interestingly, the addition of excess Ni2+ (120 μm) in the buffer can prompt the mixture of UreE-R101A with UreG to be eluted out as 2E-2G complex (Fig. 4, line 2, broken line).
FIGURE 4.
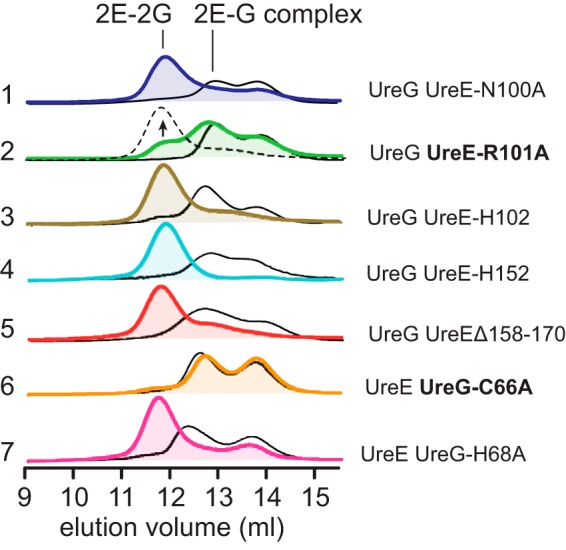
Examination of UreE and UreG mutants on the protein-protein interaction by chromatography. UreE and UreG or their variants (40 μm for each) were incubated in a buffer containing 60 μm Mg2+ in the absence (black curves) or presence (colored curves) of 60 μm GTP. Note that the single mutation R101A on UreE (line 2) and C66A on UreG (line 6) partially or completely disrupts the formation of 2E-2G complex, whereas the excess Ni2+ restores the formation of 2E-2G complex for the UreE-R101A-UreG mixture (line 2, broken line).
Two UreG mutations, UreG-C66A and UreG-H68A, were also prepared to examine their roles on the interaction between UreE and UreG. H68A from UreG shows little effect on the formation of both 2E-G and 2E-2G in the absence or presence of GTP and Mg2+, respectively (Fig. 4, line 7), similar to the wild-type UreG. However, for C66A mutant, mixing UreE and UreG-C66A apo-proteins results in the formation of 2E-G complex both in the absence or in the presence of GTP and Mg2+, but not 2E-2G complex (Fig. 4, line 6), implying that Cys-66 plays a crucial role on the stabilization of 2E-2G complex. Unlike UreE-R101A-UreG, even in the presence of excess Ni2+, no elution peak corresponding to 2E-2G complex can be observed for the UreE-UreG-C66A mixture (data not shown).
Formation of 2E-2G Complex Is a Prerequisite for Ni2+ Transfer from UreE to UreG
To monitor nickel translocation between UreE and UreG, Ni-UreE was titrated stepwise into UreG (25 μm) in the presence of GTP (50 μm) and Mg2+ (1 mm). A typical absorption at 337 nm appeared and increased its intensity with increasing amounts of Ni-UreE, suggesting that Ni2+ is transferred from UreE to UreG (Fig. 5A) as Ni-UreE itself exhibits no UV absorption in this region (data not shown). In contrast, when apo-UreE was stepwise added into Ni-UreG under the same condition, no perturbation on the peak corresponding to nickel binding to UreG was observed (Fig. 5B), indicating that nickel transfer between UreE and UreG is unidirectional.
FIGURE 5.
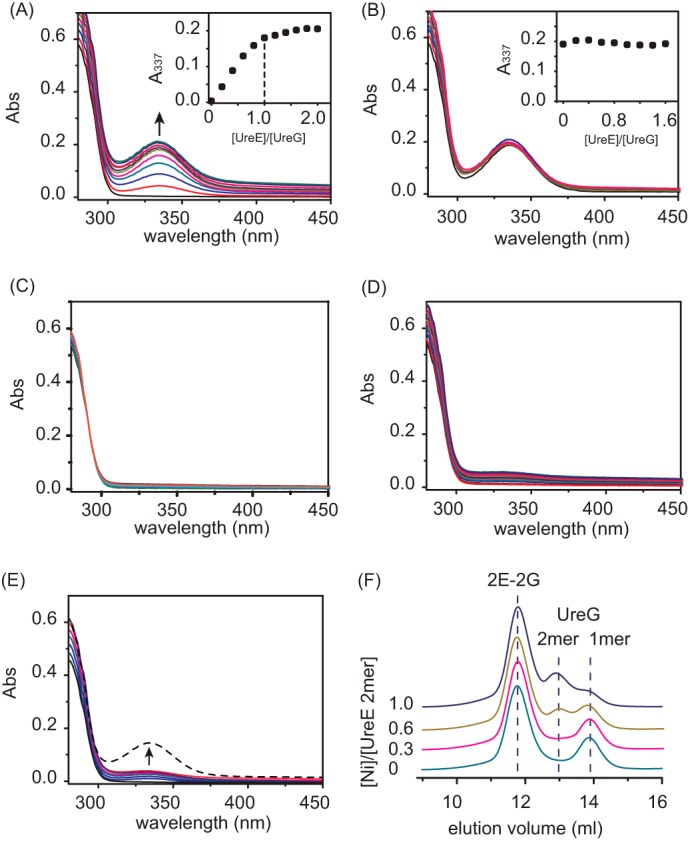
Ni2+ translocation between UreE and UreG monitored by UV-visible spectroscopy and chromatography. A and B, UV-visible spectra of 25 μm UreG (A) and Ni-UreG (B) titrated stepwise with Ni2+-UreE, apo-UreE in Hepes buffer, pH 7.2, containing 50 μm GTP and 1 mm Mg2+. The appearance of peaks at 337 nm in A indicates that Ni2+ is transferred from UreE to UreG. Abs, absorbance. C, UreG (25 μm) was titrated with Ni2+-CUreE in the presence of GTP (50 μm) and Mg2+ (1 mm). D, UreG (25 μm) was titrated with Ni2+-UreE-R101A in the presence of GTP (50 μm) and Mg2+ (1 mm). E, UreG (25 μm) was titrated with Ni2+-UreE in the presence of GTP (50 μm) and absence of Mg2+. The weak peak at 337 nm (D and E) indicates the inefficient Ni2+ transfer via 2E-G complex. The addition of Mg2+ (100 μm) raises the peak intensities at 337 nm (E, broken line), implying that Ni2+ transfer from UreE to UreG is restored upon the formation of 2E-2G complex. F, gel filtration profiles of UreE-UreG mixtures. UreG was incubated with a series of UreE proteins loaded with different molar equivalents of Ni2+ (0, 0.3, 0.6, and 1.0) prior to the injection. The binding of Ni2+ induces dimerization of UreG.
To examine whether Ni2+ transfer is achievable upon UreE-UreG interaction, we carried out similar titration experiments under the same condition except using Ni-CUreE. The stepwise addition of Ni-CUreE does not induce the peaks at 337 nm (Fig. 5C), indicative of no Ni2+ transfer between CUreE and UreG. To examine whether Ni2+ transfer is achievable upon the formation of 2E-G complex, we carried out similar experiments using Ni-UreE-R101A. The addition of Ni-UreE-R101A to UreG in the presence of both GTP and Mg2+ also led to poor nickel transfer from UreE to UreG (Fig. 5D). In parallel, the stepwise addition of Ni-UreE to UreG in the buffer without Mg2+ in which 2E-G complex is formed induces only very weak peaks at 337 nm (Fig. 5E), indicative of an inefficient Ni2+ transfer between UreE and UreG upon the formation of 2E-G complex. However, the further addition of Mg2+ (100 μm) into the reaction buffer enhanced the peak intensities at 337 nm dramatically (Fig. 5E, broken line), implying that the Ni2+ transfer between UreE and UreG has been restored once 2E-2G complex is formed. Taken together, we demonstrate that the formation of 2E-2G complex is crucial for nickel translocation from UreE to UreG.
We further examined the nickel transfer between UreE and UreG by analytical gel filtration chromatography. A series of 40 μm UreE samples loaded with different molar ratios of nickel was incubated with 80 μm apo-UreG in a buffer containing 100 μm GTP and 1 mm MgSO4 for 1 h to allow the potential nickel translocation and then subjected to gel filtration chromatography (Fig. 5F). With an increase of nickel loaded to UreE samples, the intensities of monomer peaks of UreG (∼13.9 ml) decrease accompanied by the increases in the intensities of UreG dimers (∼13 ml) in the presence of both GTP and Mg2+, confirming that Ni2+ has been transferred from UreE to UreG because UreG presents as a monomer in the absence of Ni2+, but presents as a dimer in the presence of higher molar ratios of Ni2+ (36).
UreE Enhances GTPase Activity of UreG
To investigate the biological significance of UreE-UreG interaction, GTPase activity of UreG in the presence of UreE was examined. Similar to Ni2+-UreG (Fig. 6A, black curve), the amounts of GTP turnover increase with the addition of equimolar amounts of UreE loaded with increasing amounts of Ni2+ (0, 0.3, 0.6, and 1.0 molar eq), suggesting that nickel is transferred from UreE to UreG, which enhances the enzyme activity (Fig. 6A). Surprisingly, the addition of 1 molar eq of Ni-UreE to UreG results in much more efficient activity of UreG (with ∼65% of GTP turnover) than that of the same amounts of Ni2+-UreG in the absence of UreE (with ∼30% of GTP turnover) (Fig. 6A, black curve). Moreover, incubation of increasing amounts of apo-UreE (0, 0.3, 0.6, and 1.0 molar eq) with Ni2+-UreG (10 μm) also enhances its activity (Fig. 6B), indicating that there may be an additional channel to elevate the GTPase activity of UreG by UreE besides nickel donation.
FIGURE 6.
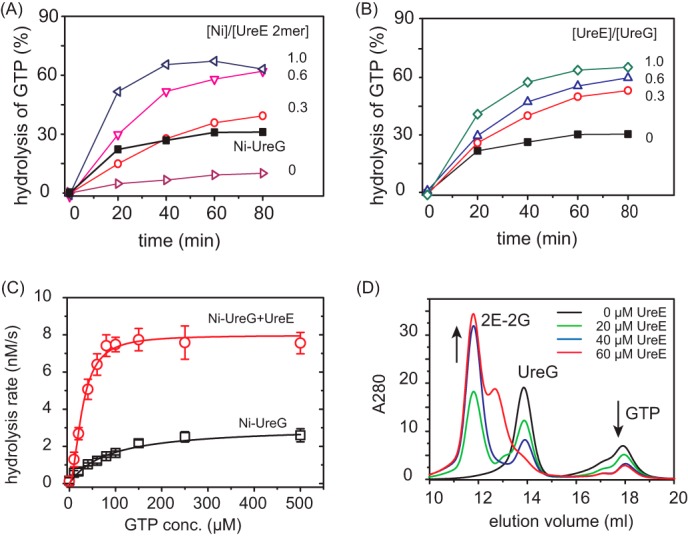
Enhancement of GTPase activity of UreG by UreE. A, time dependence of GTPase activities of apo-UreG in the presence of 1 molar eq of UreE loaded with different amounts of Ni2+ (0-, 0.3-, 0.6-, and 1.0-fold). The black curve represents the GTPase activity of Ni-UreG. B, time dependence of GTPase activities of Ni-UreG in the presence of different amounts of apo-UreE (0-, 0.3-, 0.6-, and 1.0-fold). C, saturation curves of Ni-UreG in the absence and presence of equimolar amounts of apo-UreE. Km values are obtained by fitting the saturation curves to the Michaelis-Menten equation to be ∼82.7 × 10−6 m for Ni-UreG alone and 27.9 × 10−6 m for Ni-UreG and UreE mixture. GTPase assays were carried out in the presence of 10 mm KHCO3. GTP conc., GTP concentration. Error bars indicate means ± S.E. D, gel filtration chromatography profiles of UreG (∼40 μm) preincubated with different amounts of UreE (0, 20, 40, 60 μm) supplemented with ∼40 μm GTP and 1 mm MgSO4. The addition of UreE into UreG leads to the formation of 2E-2G complex, facilitating GTP binding.
The GTPase activity of Ni-UreG upon supplementation of apo-UreE was further determined by fitting the saturation curve to the Michaelis-Menten equation (Fig. 6C) to give rise to kcat of 1.6 ± 0.4 × 10−6 s−1, Km of 27.9 ± 1.8 × 10−6 m, and kcat/Km of ∼57.3 × 10−3 m−1 s−1. In comparison, the activity of the same amounts of Ni-UreG alone was also determined with a kcat of 0.61 ± 0.1 × 10−6 s−1, Km of 82.7 ± 17.3 × 10−6 m, and a 8-fold lower enzyme efficiency (kcat/Km = ∼7.4 × 10−3 m−1 s−1). The lower Km for Ni-UreG and UreE mixture indicates that UreG has a higher affinity for GTP in the protein complex than UreG alone.
The effect of UreE on GTP binding to UreG was further investigated by analytical gel filtration chromatography. Different molar equivalents of UreE (0, 0.5, 1.0, and 1.5 molar eq) were incubated with UreG (40 μm) in the presence of 40 μm GTP and then subjected to gel filtration analysis. As shown in Fig. 6D, increases in UreE concentrations lead to the peak intensities of 2E-2G complex increasing, accompanied by decreases in the intensity of the GTP peak, indicating that UreE enhances GTP binding to UreG due to the formation of 2E-2G complex. Taken together, UreE promotes the enzyme efficiency of UreG not only by donation of nickel but also by serving as a structural scaffold for UreG to recruit GTP.
UreG Extracts Ni2+ from HypA via UreE
We have shown previously that nickel is translocated from HypA to UreE via a direct protein-protein interaction (32). Here, we demonstrate that nickel is transferred from UreE to UreG through the formation of 2E-2G complex (Fig. 5). It is not well understood how nickel is delivered among these three proteins. HypA, UreE, and UreG were mixed and examined by analytic gel filtration, and the elution fractions were collected and further examined by 15% SDS-PAGE. As shown in Fig. 7A, in the absence of GTP and Mg2+, the protein mixture is eluted out as two major peaks at ∼12.2 and ∼14.0 ml (Fig. 7A, blue curve), with molecular weights corresponding to 2E-A complex, i.e. dimer of UreE complexed with monomer of HypA and UreG protein, respectively, implying that UreE dimer tends to bind HypA monomer rather than UreG under this condition. By contrast, in the presence of GTP and Mg2+, almost all UreG protein is eluted out with UreE as the 2E-2G complex, as evidenced by the observation of a peak at ∼11.8 ml corresponding to a molecular mass of ∼80 kDa. Another peak that eluted at ∼14.8 ml corresponds to free HypA protein (Fig. 7A, orange curve), indicating that UreE dimer prefers to bind two UreG monomers instead of HypA in the presence of GTP and Mg2+. Apparently, no stable peaks with molecular weights corresponding to either HypA-UreE-UreG or HypA-UreG complexes were observed under both conditions (data not shown). The nickel translocation among HypA, UreE, and UreG was further examined by GTPase assay and UV-visible spectroscopy. As shown in Fig. 7B, in the GTPase assay, apo-UreG displayed a poor GTPase activity with only ∼4% of GTP turnover, whereas mixing of Ni-HypA with apo-UreG leads to the activity of UreG increased slightly with ∼19% of GTP turnover, which may be attributable to UreG binding to the nickel ions dissociated from HypA. On the other hand, Ni-UreE enhances UreG activity dramatically with ∼65% of GTP turnover. As expected, the addition of apo-UreE into the mixture of Ni-HypA and apo-UreG boosts the GTPase activity of UreG to a level as high as that of the Ni-UreE and UreG mixture, further verifying that UreG can acquire Ni2+ from HypA through UreE. This was further confirmed by subsequent UV titration experiments. When Ni-Zn-HypA was stepwise added into apo-UreG, peaks corresponding to nickel binding to UreG were observed with nearly negligible intensities (Fig. 7C), indicating that nickel transfer between HypA and UreG is inefficient, whereas stepwise titration of a mixture of Ni-Zn-HypA and UreE into apo-UreG led to significant increases in the intensities of the absorption peaks at 337 nm (Fig. 7D), suggesting that nickel was efficiently transferred from HypA to UreG. However, when UreE was present at very low concentrations in the above experiments, Ni2+ transfer was also inefficient, suggesting that UreE serves as a stoichiometric mediator for Ni2+ transfer from HypA to UreG (data not shown).
FIGURE 7.
Complexation of UreE, UreG, and HypA and Ni2+ translocation. A, analytic gel filtration profiles of HypA, UreE, and UreG in the absence (blue curve) or presence (orange curve) of GTP and Mg2+. mAU, milliabsorbance units. B, effect of Ni-HypA on the GTPase activity of UreG. Note that in the presence of UreE, Ni-HypA enhances the activity of UreG to a level similar to that of the Ni-UreE and UreG mixture. GTPase assays were carried out in the presence of 10 mm KHCO3. Error bars indicate means ± S.E. C, UreG (20 μm) was titrated with Ni-Zn-HypA in the presence of GTP (50 μm) and Mg2+ (1 mm). Abs, absorbance. D, UreG (20 μm) was titrated with a mixture of Ni-Zn-HypA and UreE in the presence of GTP (50 μm) and Mg2+ (1 mm).
DISCUSSION
The maturation of urease in H. pylori involves nickel delivery to the metallo-active center and is highly dependent on the cooperation of at least four urease accessory proteins, i.e. UreE, UreG, UreF, and UreH. In addition, hydrogenase accessory proteins HypA and HypB were also found to be crucial for activation of urease, with the former donating nickel ions to UreE by a specific protein-protein interaction (32). Among these chaperones, the nickel binding properties of UreE, UreF, HypA, and HypB have been well studied. Both HpHypA and HpHypB bind Ni2+ with dissociation constants of micromolar levels (20, 22, 24). HpUreF dimer binds two Ni2+ via His-229 and Cys-231 with a dissociation constant of 6.4 ± 0.4 μm (47). HpUreE binds to Ni2+ via at least two His-102 residues from each monomer, with an additional residue (His-152) possibly also participating in the binding, and only one Ni2+ ion binds per UreE dimer with a dissociation constant of 0.15 μm (37). In contrast, there has been a lack of characterization of the nickel binding property of HpUreG. A previous study by isothermal titration microcalorimetry showed that HpUreG binds to Zn2+ specifically, but not to Ni2+, with a dissociation constant of 0.33 μm (35). However, a nickel-bound dimer of UreG has been observed in the study of a complex structure of UreG/F/H (36).
In this study, we demonstrate by UV-visible spectroscopy that HpUreG binds Ni2+ only in the presence of GTP and Mg2+ with a stoichiometry of ∼0.5 Ni2+ ions bound per UreG monomer, and a dissociation constant of ∼0.36 μm. In the absence of Mg2+, UreG exhibits a 10-fold lower binding affinity to Ni2+ (Kd ∼3.6 μm) and requires excess Ni2+ to achieve full saturation (Imax ∼0.2), suggesting a potential communication between the nickel binding site and GTP binding site of UreG, which belongs to the family of magnesium-dependent enzymes (24).
It has been shown that both Ni2+ and Zn2+ bind UreG through coordination of Cys-66 and His-68 (40). Although the binding affinity of UreG to Ni2+ determined in the present study is similar to that of Zn2+ to HpUreG (Kd = 0.33 μm) reported previously by isothermal titration calorimetry (35), our Ni2+-Zn2+ competition experiments reveal that in the presence of GTP and Mg2+, UreG exhibits a higher affinity toward Ni2+ than Zn2+, implying that UreG is a specific nickel binding chaperone. The discrepancy in the binding affinity obtained might be due to the different conditions and methods used.
The UreE-UreG interaction and its role in nickel transfer have been investigated in this study. Our combined gel filtration chromatography and light scattering data show that apo-UreE and apo-UreG proteins form a 2E-2G complex only when both GTP and Mg2+ are present; otherwise a 2E-G complex is formed. Neither Ni2+ nor Zn2+ perturbs the interaction between UreE and UreG (Fig. 3), which is in contrast with a previous study indicating that complexation of UreE with UreG gives rise to 2E-2G in the absence of GTP and Mg2+ and that such a complex is specifically stabilized by excess Zn2+ but not Ni2+ (37). The reason for the discrepancy is not clear; however, considering that UreG is a GTPase, investigation of UreE and UreG interaction in the presence of GTP/Mg2+ should provide more physiologically relevant information. Indeed, UreG from H. pylori does not associate with Ni2+ in the absence of GTP (Fig. 1).
Further biophysical and molecular biology studies show that UreE serves as a structural scaffold to recruit UreG; deletion of either N-domains or C-domains of UreE abolishes the binding. Moreover, replacement of residue Arg-101 of UreE or Cys-66 of UreG to Ala results in disruption of the formation of 2E-2G complex (Fig. 4), implying their critical roles in the stabilization of 2E-2G complex. Interestingly, excess Ni2+ can restore 2E-2G complex for UreE-R101A-UreG but not for UreE-UreG-C66A. This suggests that Ni2+ might also play a certain, although not crucial, role in the stabilization of 2E-2G complex possibly by stabilizing the UreG dimer, which binds to GTP. Although the crystal structure of the UreE-UreG complex is not available, our mutagenesis studies imply that the protein-protein interfaces may be located around the metal binding sites of both UreE and UreG. A previous structural model of 2E-2G complex also revealed that metal binding sites of UreE and UreG “grazed” each other (37); such a structural arrangement might allow Ni2+ translocation readily. Similarly, the metal binding site of UreG is buried in the known UreG/UreF/UreH complex (36), implying that UreE and UreF/H complex may compete with each other for UreG in the process of urease maturation.
Our combined UV-visible spectroscopy and gel filtration chromatography data reveal that Ni2+ is transferred from UreE to UreG but not vice versa and that such a process occurs only when UreE and UreG form the 2E-2G but not 2E-G complex (Fig. 5), because mixing of UreG and a Ni2+-UreE variant with Arg-101 substituted by Ala abolishes the nickel transfer from UreE and UreG. Moreover, when 2E-G complex is formed in the absence of Mg2+, no efficient Ni2+ transfer was observed between UreE and UreG (Fig. 5), indicative of the requirement of 2E-2G but not 2E-G complex for nickel delivery.
Previously, we demonstrated that nickel is translocated via a specific HypA-UreE interaction (32). Therefore, we investigated the roles of these three nickel chaperones in urease maturation process. We show that nickel is transferred from HypA to UreG via UreE, which acts as a stoichiometric instead of a catalytic mediator (Fig. 7) because inefficient Ni2+ transfer was observed when UreE was present at low concentration. Although a transient ternary complex, e.g. HypA-UreE-UreG, still cannot be ruled out, there appears to be no stable ternary protein complex formed although both HypA and UreG bind to UreE at different sites. The residues 158–170 of UreE have been shown to be crucial for HypA recognition (32) but not for UreG (Fig. 4); instead, residue Arg-101 of UreE plays a role in stabilization of 2E-2G complex (Fig. 4). A previous study demonstrated that both HypA and UreG compete with each other for UreE (48). We show here that preference of UreE toward HypA and UreG is elaborately tuned in biological systems and that UreE has a tendency to bind HypA in the absence of GTP and Mg2+, whereas it binds UreG in the presence of GTP and Mg2+.
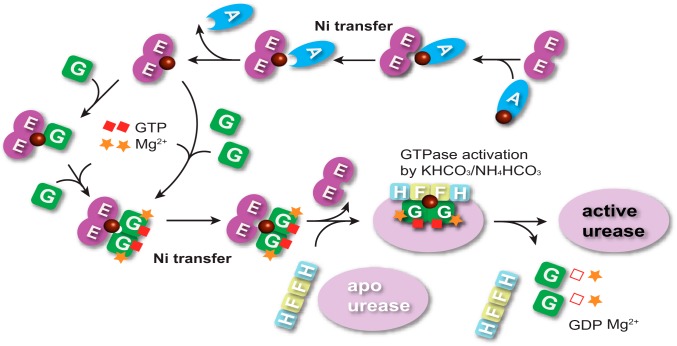
Previously, the GTP hydrolysis by UreG was found to be very slow or even undetectable for UreG from H. pylori and K. aerogenes (35, 49–54). Such a weak GTPase activity of UreG is not consistent with its biological role in urease maturation, which provoked speculation that UreG may need an additional cofactor to stimulate the GTP hydrolysis. A recent study on HypB from H. pylori, an analogue of UreG, demonstrated that GTPase activity is enhanced by K+ by an order of magnitude (24). Similarly, HpUreG activity was also found to be stimulated by bicarbonate (HCO3−) (36). Here, we clearly show that the combination of K+/NH4+ and HCO3− results in an enhanced GTPase activity of HpUreG and further activates urease (Fig. 2), implying potential cooperation between K+/NH4+ and HCO3− in GTP hydrolysis by UreG during urease activation. Interestingly, both NH4+ and HCO3− are the products of urease, whose activity is dependent on the GTPase of UreG. Whether there is a regulation cycle between urease and NH4HCO3-sensitive GTPase activity of UreG warrants further study. Besides K+/NH4+ and HCO3−, GTP hydrolysis by UreG has also been demonstrated to be Ni2+-dependent (Fig. 2), confirming the role of UreG as a nickel chaperone and GTPase enzyme in the process of urease maturation. Moreover, dramatic enhancement of GTPase activity of UreG by Ni2+ donated from UreE (Fig. 6) further confirms the biological significance of UreE-UreG interaction in the process of urease activation. Nevertheless, the significantly high GTPase activity of UreG in 2E-2G complex may not be observable in vivo, as upon receiving Ni2+ from UreE, UreG will bind UreF-H sequentially to form a supercomplex as apo-urease/UreF-H-G, in which the GTP hydrolysis by UreG is catalyzed to finish the final step of nickel insertion into apo-urease (36, 55), whereas besides Ni2+, UreE itself also enhances the efficiency (kcat/Km) of GTP hydrolysis of UreG by facilitating GTP binding to UreG in the GTPase assay in vitro, consistent with our hypothesis that UreE works not only as a stoichiometric nickel mediator for UreG, but also as a structural scaffold for the dimerization and GTP binding of UreG to preactivate GTPase, subsequently facilitating Ni2+ transfer to downstream receptors such as apo-urease (Fig. 8) (36). An overall mechanism of Ni2+ transfer through protein-protein interactions among UreE, UreG, and HypA is therefore proposed and summarized in Fig. 8.
FIGURE 8.
Proposed mechanism of Ni2+ translocation among HypA, UreE, and UreG during urease maturation. Apo-UreE acquires Ni2+ from HypA via specific HypA-UreE interaction. Sequentially, Ni-UreE dimer binds one apo-UreG monomer and then further captures the second UreG monomer, facilitating GTP binding to UreG in the presence of Mg2+, or Ni-UreE dimer binds two apo-UreG monomers in the presence of GTP and Mg2+ to form 2E-2G complex, which triggers nickel translocation from UreE to UreG. Subsequently, UreF-H competes with UreE for Ni-UreG to form the supercomplex as an apo-urease/UreF-H-G (36), in which the GTP hydrolysis by UreG is catalyzed to complete the final step of nickel insertion into the apo-urease in the presence of KHCO3/NH4HCO3.
This work was supported by the Research Grants of Council of Hong Kong (Grants 704909, N_HKU75209, 704612, 703913, and HKU6/11G) and the University of Hong Kong (for an emerging Strategic Research Theme on Integrative Biology).
This article was selected as a Paper of the Week.
- TCEP
- tris(2-carboxyethyl) phosphine.
REFERENCES
- 1. Warren J. R., Marshall B. (1983) Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 321, 1273–1275 [PubMed] [Google Scholar]
- 2. Covacci A., Telford J. L., Del Giudice G., Parsonnet J., Rappuoli R. (1999) Helicobacter pylori virulence and genetic geography. Science 284, 1328–1333 [DOI] [PubMed] [Google Scholar]
- 3. Garner J. A., Cover T. L. (1996) Binding and internalization of the Helicobacter pylori vacuolating cytotoxin by epithelial cells. Infect. Immun. 64, 4197–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maier R. J., Fu C., Gilbert J., Moshiri F., Olson J., Plaut A. G. (1996) Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol. Lett. 141, 71–76 [DOI] [PubMed] [Google Scholar]
- 5. Evans D. J., Jr., Evans D. G., Kirkpatrick S. S., Graham D. Y. (1991) Characterization of the Helicobacter pylori urease and purification of its subunits. Microb. Pathog. 10, 15–26 [DOI] [PubMed] [Google Scholar]
- 6. Li Y., Zamble D. B. (2009) Nickel homeostasis and nickel regulation: an overview. Chem. Rev. 109, 4617–4643 [DOI] [PubMed] [Google Scholar]
- 7. Zambelli B., Musiani F., Benini S., Ciurli S. (2011) Chemistry of Ni2+ in urease: sensing, trafficking, and catalysis. Acc. Chem. Res. 44, 520–530 [DOI] [PubMed] [Google Scholar]
- 8. Leach M. R., Zamble D. B. (2007) Metallocenter assembly of the hydrogenase enzymes. Curr. Opin. Chem. Biol. 11, 159–165 [DOI] [PubMed] [Google Scholar]
- 9. Carter E. L., Flugga N., Boer J. L., Mulrooney S. B., Hausinger R. P. (2009) Interplay of metal ions and urease. Metallomics 1, 207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sydor A. M., Zamble D. B. (2013) Nickel metallomics: general themes guiding nickel homeostasis. Met. Ions Life Sci. 12, 375–416 [DOI] [PubMed] [Google Scholar]
- 11. Zeng Y. B., Yang N., Sun H. (2011) Metal-binding properties of an Hpn-like histidine-rich protein. Chem. Eur. J. 17, 5852–5860 [DOI] [PubMed] [Google Scholar]
- 12. Zeng Y. B., Zhang D. M., Li H., Sun H. (2008) Binding of Ni2+ to a histidine- and glutamine-rich protein, Hpn-like. J. Biol. Inorg. Chem. 13, 1121–1131 [DOI] [PubMed] [Google Scholar]
- 13. Ge R., Zhang Y., Sun X., Watt R. M., He Q. Y., Huang J. D., Wilcox D. E., Sun H. (2006) Thermodynamic and kinetic aspects of metal binding to the histidine-rich protein, Hpn. J. Am. Chem. Soc. 128, 11330–11331 [DOI] [PubMed] [Google Scholar]
- 14. Ge R., Watt R. M., Sun X., Tanner J. A., He Q. Y., Huang J. D., Sun H. (2006) Expression and characterization of a histidine-rich protein, Hpn: potential for Ni2+ storage in Helicobacter pylori. Biochem. J. 393, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cun S., Sun H. (2010) A zinc-binding site by negative selection induces metallodrug susceptibility in an essential chaperonin. Proc. Natl. Acad. Sci. U.S.A. 107, 4943–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cun S., Li H., Ge R., Lin M. C. M., Sun H. (2008) A histidine-rich and cysteine-rich metal-binding domain at the C terminus of heat shock protein A from Helicobacter pylori: implication for nickel homeostasis and bismuth susceptibility. J. Biol. Chem. 283, 15142–15151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Volbeda A., Charon M. H., Piras C., Hatchikian E. C., Frey M., Fontecilla-Camps J. C. (1995) Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373, 580–587 [DOI] [PubMed] [Google Scholar]
- 18. Casalot L., Rousset M. (2001) Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9, 228–237 [DOI] [PubMed] [Google Scholar]
- 19. Olson J. W., Mehta N. S., Maier R. J. (2001) Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39, 176–182 [DOI] [PubMed] [Google Scholar]
- 20. Xia W., Li H., Yang X., Wong K. B., Sun H. (2012) Metallo-GTPase HypB from Helicobacter pylori and its interaction with nickel chaperone protein HypA. J. Biol. Chem. 287, 6753–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Douglas C. D., Ngu T. T., Kaluarachchi H., Zamble D. B. (2013) Metal transfer within the Escherichia coli HypB-HypA complex of hydrogenase accessory proteins. Biochemistry 52, 6030–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xia W., Li H., Sze K. H., Sun H. (2009) Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites. J. Am. Chem. Soc. 131, 10031–10040 [DOI] [PubMed] [Google Scholar]
- 23. Watanabe S., Arai T., Matsumi R., Atomi H., Imanaka T., Miki K. (2009) Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation. J. Mol. Biol. 394, 448–459 [DOI] [PubMed] [Google Scholar]
- 24. Sydor A. M., Lebrette H., Ariyakumaran R., Cavazza C., Zamble D. B. (2014) Relationship between Ni(II) and Zn(II) coordination and nucleotide binding by the Helicobacter pylori [NiFe]-hydrogenase and urease maturation factor HypB. J. Biol. Chem. 289, 3828–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chan K. H., Lee K. M., Wong K. B. (2012) Interaction between hydrogenase maturation factors HypA and HypB is required for [NiFe]-hydrogenase maturation. PLoS One 7, e32592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mehta N., Olson J. W., Maier R. J. (2003) Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J. Bacteriol. 185, 726–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benoit S. L., Zbell A. L., Maier R. J. (2007) Nickel enzyme maturation in Helicobacter hepaticus: roles of accessory proteins in hydrogenase and urease activities. Microbiology 153, 3748–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng T., Li H., Xia W., Sun H. (2012) Multifaceted SlyD from Helicobacter pylori: implication in [NiFe] hydrogenase maturation. J. Biol. Inorg. Chem. 17, 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leach M. R., Zhang J. W., Zamble D. B. (2007) The role of complex formation between the Escherichia coli hydrogenase accessory factors HypB and SlyD. J. Biol. Chem. 282, 16177–16186 [DOI] [PubMed] [Google Scholar]
- 30. Kaluarachchi H., Altenstein M., Sugumar S. R., Balbach J., Zamble D. B., Haupt C. (2012) Nickel binding and[NiFe]-hydrogenase maturation by the metallochaperone SlyD with a single metal-binding site in Escherichia coil. J. Mol. Biol. 417, 28–35 [DOI] [PubMed] [Google Scholar]
- 31. Cheng T., Li H., Yang X., Xia W., Sun H. (2013) Interaction of SlyD with HypB of Helicobacter pylori facilitates nickel trafficking. Metallomics 5, 804–807 [DOI] [PubMed] [Google Scholar]
- 32. Yang X., Li H., Cheng T., Xia W., Lai Y.-T., Sun H. (2014) Nickel translocation between metallochaperones HypA and UreE in Helicobacter pylori. Metallomics 6, 1731–1736 [DOI] [PubMed] [Google Scholar]
- 33. Park I. S., Hausinger R. P. (1996) Metal ion interaction with urease and UreD-urease apoproteins. Biochemistry 35, 5345–5352 [DOI] [PubMed] [Google Scholar]
- 34. Park I. S., Hausinger R. P. (1995) Evidence for the presence of urease apoprotein complexes containing UreD, UreF, and UreG in cells that are competent for in vivo enzyme activation. J. Bacteriol. 177, 1947–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zambelli B., Turano P., Musiani F., Neyroz P., Ciurli S. (2009) Zn2+-linked dimerization of UreG from Helicobacter pylori, a chaperone involved in nickel trafficking and urease activation. Proteins 74, 222–239 [DOI] [PubMed] [Google Scholar]
- 36. Fong Y. H., Wong H. C., Yuen M. H., Lau P. H., Chen Y. W., Wong K. B. (2013) Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease. PLoS Biol. 11, e1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bellucci M., Zambelli B., Musiani F., Turano P., Ciurli S. (2009) Helicobacter pylori UreE, a urease accessory protein: specific Ni2+- and Zn2+-binding properties and interaction with its cognate UreG. Biochem. J. 422, 91–100 [DOI] [PubMed] [Google Scholar]
- 38. Boer J. L., Quiroz-Valenzuela S., Anderson K. L., Hausinger R. P. (2010) Mutagenesis of Klebsiella aerogenes UreG to probe nickel binding and interactions with other urease-related proteins. Biochemistry 49, 5859–5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ryan D. K., Weber J. H. (1982) Fluorescence quenching titration for determination of complexing capacities and stability constants of fulvic acid. Anal. Chem. 54, 986–990 [Google Scholar]
- 40. Martin-Diaconescu V., Bellucci M., Musiani F., Ciurli S., Maroney M. J. (2012) Unraveling the Helicobacter pylori UreG zinc binding site using x-ray absorption spectroscopy (XAS) and structural modeling. J. Biol. Inorg. Chem. 17, 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Banaszak K., Martin-Diaconescu V., Bellucci M., Zambelli B., Rypniewski W., Maroney M. J., Ciurli S. (2012) Crystallographic and x-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking. Biochem. J. 441, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi R., Munger C., Asinas A., Benoit S. L., Miller E., Matte A., Maier R. J., Cygler M. (2010) Crystal structures of apo and metal-bound forms of the UreE protein from Helicobacter pylori: role of multiple metal binding sites. Biochemistry 49, 7080–7088 [DOI] [PubMed] [Google Scholar]
- 43. Stola M., Musiani F., Mangani S., Turano P., Safarov N., Zambelli B., Ciurli S. (2006) The nickel site of Bacillus pasteurii UreE, a urease metallo-chaperone, as revealed by metal-binding studies and x-ray absorption spectroscopy. Biochemistry 45, 6495–6509 [DOI] [PubMed] [Google Scholar]
- 44. Song H. K., Mulrooney S. B., Huber R., Hausinger R. P. (2001) Crystal structure of Klebsiella aerogenes UreE, a nickel-binding metallochaperone for urease activation. J. Biol. Chem. 276, 49359–49364 [DOI] [PubMed] [Google Scholar]
- 45. Remaut H., Safarov N., Ciurli S., Van Beeumen J. (2001) Structural basis for Ni2+ transport and assembly of the urease active site by the metallochaperone UreE from Bacillus pasteurii. J. Biol. Chem. 276, 49365–49370 [DOI] [PubMed] [Google Scholar]
- 46. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 47. Zambelli B., Berardi A., Martin-Diaconescu V., Mazzei L., Musiani F., Maroney M. J., Ciurli S. (2014) Nickel binding properties of Helicobacter pylori UreF, an accessory protein in the nickel-based activation of urease. J. Biol. Inorg. Chem. 19, 319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benoit S. L., McMurry J. L., Hill S. A., Maier R. J. (2012) Helicobacter pylori hydrogenase accessory protein HypA and urease accessory protein UreG compete with each other for UreE recognition. Biochim. Biophys. Acta 1820, 1519–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mehta N., Benoit S., Maier R. J. (2003) Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb. Pathog. 35, 229–234 [DOI] [PubMed] [Google Scholar]
- 50. Moncrief M. B. C., Hausinger R. P. (1997) Characterization of UreG, identification of a UreD-UreF-UreG complex, and evidence suggesting that a nucleotide-binding site in UreG is required for in vivo metallocenter assembly of Klebsiella aerogenes urease. J. Bacteriol. 179, 4081–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zambelli B., Stola M., Musiani F., De Vriendt K., Samyn B., Devreese B., Van Beeumen J., Turano P., Dikiy A., Bryant D. A., Ciurli S. (2005) UreG, a chaperone in the urease assembly process, is an intrinsically unstructured GTPase that specifically binds Zn2+. J. Biol. Chem. 280, 4684–4695 [DOI] [PubMed] [Google Scholar]
- 52. Zambelli B., Musiani F., Savini M., Tucker P., Ciurli S. (2007) Biochemical studies on Mycobacterium tuberculosis UreG and comparative modeling reveal structural and functional conservation among the bacterial UreG family. Biochemistry 46, 3171–3182 [DOI] [PubMed] [Google Scholar]
- 53. Real-Guerra R., Staniscuaski F., Zambelli B., Musiani F., Ciurli S., Carlini C. R. (2012) Biochemical and structural studies on native and recombinant Glycine max UreG: a detailed characterization of a plant urease accessory protein. Plant Mol. Biol. 78, 461–475 [DOI] [PubMed] [Google Scholar]
- 54. Boer J. L., Hausinger R. P. (2012) Klebsiella aerogenes UreF: Identification of the UreG binding site and role in enhancing the fidelity of urease activation. Biochemistry 51, 2298–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Soriano A., Hausinger R. P. (1999) GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]