Background: Voltage-gated K+ channels are generally known to be located in the plasma membrane.
Results: Kv1.3 in the nucleus regulates nuclear membrane potential and activates transcription factors.
Conclusion: Kv1.3 is a functional channel at the nucleus.
Significance: This study provides novel insight about functional nuclear Kv1.3 and expands our knowledge about the function of ion channels within the nucleus.
Keywords: cAMP response element-binding protein (CREB), cell fractionation, ion channel, nucleus, specificity protein 1 (Sp1), Kv channels, UBF1, c-Fos, nuclear membrane potential
Abstract
It is widely known that ion channels are expressed in the plasma membrane. However, a few studies have suggested that several ion channels including voltage-gated K+ (Kv) channels also exist in intracellular organelles where they are involved in the biochemical events associated with cell signaling. In the present study, Western blot analysis using fractionated protein clearly indicates that Kv1.3 channels are expressed in the nuclei of MCF7, A549, and SNU-484 cancer cells and human brain tissues. In addition, Kv1.3 is located in the plasma membrane and the nucleus of Jurkat T cells. Nuclear membrane hyperpolarization after treatment with margatoxin (MgTX), a specific blocker of Kv1.3 channels, provides evidence for functional channels at the nuclear membrane of A549 cells. MgTX-induced hyperpolarization is abolished in the nuclei of Kv1.3 silenced cells, and the effects of MgTX are dependent on the magnitude of the K+ gradient across the nuclear membrane. Selective Kv1.3 blockers induce the phosphorylation of cAMP response element-binding protein (CREB) and c-Fos activation. Moreover, Kv1.3 is shown to form a complex with the upstream binding factor 1 in the nucleus. Chromatin immunoprecipitation assay reveals that Sp1 transcription factor is directly bound to the promoter region of the Kv1.3 gene, and the Sp1 regulates Kv1.3 expression in the nucleus of A549 cells. These results demonstrate that Kv1.3 channels are primarily localized in the nucleus of several types of cancer cells and human brain tissues where they are capable of regulating nuclear membrane potential and activation of transcription factors, such as phosphorylated CREB and c-Fos.
Introduction
Voltage-gated K+ (Kv)3 channels are known to participate in the regulation of membrane potential, neuronal excitability, and signaling (1). Many studies have demonstrated that mutations or abnormal expression of Kv channels occurs in fatal diseases such as episodic ataxia, seizure, and cardiac arrhythmias (2, 3). Kv channels in epithelial cells contribute to multiple cell functions including electrolyte/nutrient transport, cell proliferation, apoptosis, wound healing, O2 sensing, and tumorigenesis (4, 5). Kv1.3 channels in the plasma membrane of T lymphocytes regulate membrane potential, Ca2+ signaling, proliferation, and autoimmune diseases (6, 7). It is noteworthy that Kv1.3 has been identified in the mitochondrial inner membrane of lymphocytes, gerbil hippocampus, and cancer cells (8–10). Szabó et al. (11) demonstrated that the expression of Kv1.3 in mitochondria contributes to apoptotic signaling by binding to pro-apoptotic Bax protein, resulting in the alteration of mitochondrial membrane potential.
Although several ion channels including ATP-sensitive K+ channels (KATP) (12), inwardly rectifying K+ channels (13, 14), Cl− channels (15, 16), voltage-gated Ca2+ channels (17), and Ca2+ activated K+ channels (18) are found to localize in the nuclear region of cells, the functional roles of nuclear ion channels remain unclear. Based on previous studies showing that Kv channels are involved in cell proliferation, apoptosis, and the progression of cancer (19–21), the function of Kv channels could be closely linked to processes that occur inside the nucleus such as gene expression and cell division. We recently reported that Kv1.1 and Kv1.3 blockers suppress A549 proliferation by inhibiting the G1-S cell cycle transition (19, 20).
Although Kv1.3 channels in excitable cells have been extensively characterized using electrophysiological, pharmacological, and molecular biological techniques, their presence in the nucleus has not been previously reported. In the present study, we investigated the localization and the functional roles of Kv1.3 channels in the nuclei of human cancer cells. The results show that functional Kv1.3 channels are expressed in the nucleus and are involved in the activation of specific transcription factors following the inhibition of their transport activity.
EXPERIMENTAL PROCEDURES
Cell Culture
A549 (lung adenocarcinoma cell), SNU-484 (gastric adenocarcinoma cell), and Jurkat (T-cell lymphoblast-like cell) cells were maintained with RPMI 1640 medium (Welgene) containing 10% fetal bovine serum (Welgene) and 1% antibiotic-antimycotic solution (Sigma-Aldrich). MCF-7 (breast adenocarcinoma cell) cells were cultured in DMEM.
Subcellular Fractionation
Cells were fractionated, using a Qproteome cell compartment kit (Qiagen), into cytosol, membrane, and nuclear protein. The cells suspended in extraction buffer CE1 were incubated at 4 °C for 10 min and centrifuged at 1000 × g for 10 min at 4 °C. Supernatant was then transferred into a new microcentrifuge tube (cytosolic fraction). Extraction buffer CE2 was added to the pellets and incubated at 4 °C for 30 min. The extracts were centrifuged at 6000 × g for 10 min at 4 °C, and the supernatant was transferred into a new microcentrifuge tube (membrane fraction). To protect the nuclear fraction, 700 units of nuclease were added to the pellet and incubated for 15 min at room temperature. Extraction buffer CE3 was added to the pellets and incubated at 4 °C for 10 min. The extracts were centrifuged at 6800 × g for 10 min at 4 °C, and the supernatant was transferred into a new microcentrifuge tube (nuclear fraction). Finally, all protein fractions were incubated in ice-cold acetone for 15 min on ice and centrifuged at 12,000 × g for 10 min. The equal protein concentrations of subcellular extracts were resolved in 1× sample buffer to load samples into gels for Western blot analysis.
Nuclear Membrane Purification
The nuclear membrane of A549 cells was isolated by previously reported methods (22, 23). A549 cells were suspended in 4 ml of hypotonic solution containing 10 mm KCl, 1.5 mm MgCl2, 10 mm HEPES-free acid, and 0.5 mm d,l-dithiothreitol (pH 7.9) for 10 min on ice. The swollen cells were spun at 400 × g for 10 min at 4 °C and resuspended in 4 ml of hypotonic solution. The cells were homogenized with 10 strokes of a round glass pestle in a Dounce homogenizer (Wheaton, Millville, NJ). After centrifugation, the deposited nuclei were washed until the supernatant was clear. The nuclei were incubated in nuclear suspension medium containing 250 mm sucrose, 5 mm MgCl2, and 50 mm Tris-Cl (pH 7.4), followed by homogenization with three strokes in a Dounce homogenizer. After adding DNase I and RNase (250 μg/ml), the nuclei were incubated for 1 h at 4 °C. They were spun at 1000 × g and 4 °C for 10 min and resuspended in incubation buffer containing 0.2 mm MgCl2 and 10 mm Tris-Cl (pH 7.4) for 15 min at 4 °C, followed by adding high NaCl buffer containing 2000 mm NaCl, 0.2 mm MgCl2, 10 mm Tris-Cl (pH 7.4), and 2-mercaptoethanol (1%). The nuclear membrane was harvested by centrifugation at 1600 × g and 4 °C for 30 min.
Measurement of Nuclear Membrane Potential by Flow Cytometry
The isolation of the nucleus was performed using the Nuclei Pure Prep nuclei isolation kit (Sigma-Aldrich). Intracellular solution containing 125 mm KCl, 2 mm K2PO4, 40 mm HEPES, 0.1 mm MgCl2, (pH 7.2), 100 nm Ca2+ with 10.2 mm EGTA, and 1.65 mm CaCl2 was added to the isolated nuclei. The nuclei were treated with 1 nm of MgTX (Alomon Labs) or 1 μm of ionomycin (Sigma-Aldrich) for 10 min and then stained with 200 nm of 3,3′-dihexyloxacarbocyanine iodide (DiOC6(3)) (Molecular Probes) at room temperature for 10 min to measure changes of nuclear membrane potential (12, 24, 25). The intensity of DiOC6(3) fluorescence signal was measured using BD FACSCaliburTM (BD Bioscience) and analyzed by BD CellQuestTM Pro software.
To deplete the potassium concentration in the isolated nucleus, the nuclei were incubated in 0 mm [K+] solution containing 125 mm NMDG-Cl, 2 mm K2PO4, 40 mm HEPES, 0.1 mm MgCl2 (pH 7.2), 100 nm Ca2+ with 10.2 mm EGTA, and 1.65 mm CaCl2 for 30 min. The nuclei were changed to extranuclear [K+] solutions (0, 30, 60, or 125 mm) adding 1 nm of MgTX for 5 min and then treated with DiOC6(3) at room temperature for 10 min. For the experiments to examine the effects of blockade of Sp1 expression, A549 cells were treated with 200 nm of mithramycin A to decrease Sp1 proteins for 24 h. The purified nuclei from A549 cells were suspended in intracellular solution and treated with 1 nm of MgTX for 10 min. After the nuclei were stained with DiOC6(3), fluorescence signal was measured using BD ACCURITM C6 (BD Bioscience).
Western Blot Analysis
The isolated nuclei of A549 cells were incubated in intracellular solution supplemented with 1 nm MgTX at room temperature for 10 min to detect the phosphorylation of CREB (pCREB) and CREB. In addition to nuclei, pCREB and c-Fos were measured using A549 cells treated with 10 nm 5-(4-phenoxybutoxy) psoralen (PAP-1) (Sigma-Aldrich) for 10 and 30 min, respectively. Adult normal brain tissue extracts were purchased from Abcam (ab29456, ab29461), GeneTex (GTX29466), and NOVUS Biologicals (NB820–89333, NB820–605700). Tissue extracts were resolved on a 10% polyacrylamide gel and transferred to PVDF membranes. Membranes were probed with Na,K-ATPase (1:1000), ATP5a (1:500), GRP78 BiP (1:1000), lamin A (1:500), Sp1 (1:500), PARP (1:400), Emerin (1:1000) (Abcam), Kv1.1 (1:200, 1:400), Kv1.2 (1:500), Kv1.3 (1:500), Kv2.2 (1:400) (Alomone Labs), CREB (1:200), phospho-CREB (Ser133) (1:200) (Millipore), c-Fos (K-25) (1:200), and β-actin (1:2000) (Santa Cruz Biotechnology) followed by a horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse secondary antibody (1:6000) (Santa Cruz Biotechnology).
Transfection and Establishment of Kv1.3 Knockdown Stable Cell Line
Cells were transfected with sure-silencing Kv1.3 shRNA plasmid DNA (SABioscience) using LipofectamineTM 2000 (Invitrogen). To select transfected cells, cells were grown in medium containing the effective concentration of neomycin (G418; BIOSESANG Inc.).
Co-immunoprecipitation and Mass Spectrometry
Detection of the Kv1.3 protein complex in the nucleus of A549 and MCF-7 cells was performed using the Nuclear Complex Co-IP Kit (Active Motif). Nuclear extracts (400–500 μg) of A549 and MCF-7 cells were incubated with 2 μg of Kv1.3 antibody (Alomone Labs) or guinea pig IgG (Santa Cruz Biotechnology) overnight at 4 °C. Magnetic beads (Invitrogen) were conjugated with antibody by incubating for 1 h at 4 °C. The bounded proteins were resolved on a 10% polyacrylamide gel. Bands detected by silver staining were identified using the Hybrid LC-MS/MS System (Applied Biosystems). MS/MS data were analyzed using a SEQUEST search. Furthermore, a loaded gel was transferred, blocked, and incubated with anti-Kv1.3 (1:500) (Alomone Labs) or anti-UBF1 (1:1000) (Abcam) for Western blot analysis.
Chromatin Immunoprecipitation
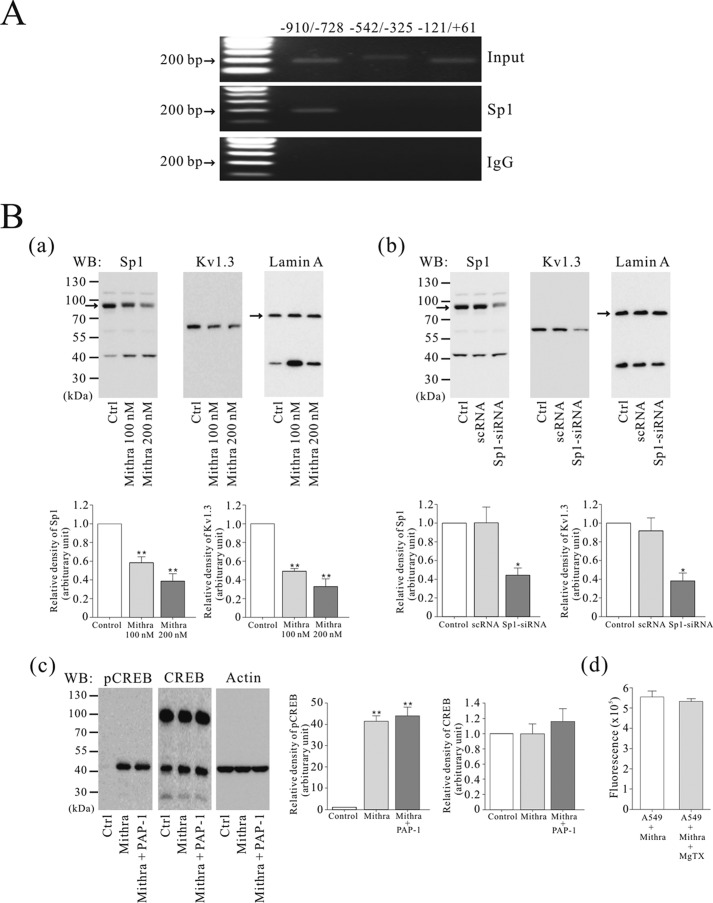
A ChIP-IT® Express Enzymatic kit (Active Motif) was used to perform ChIP using A549 cells. Immunoprecipitation was performed with 2 μg of anti-Sp1 or rabbit IgG (Santa Cruz Biotechnology) at 4 °C for 4 h. The collected DNA was analyzed using RT-PCR with Kv1.3 promoter specific-primers. The sequences of primers were: Kv1.3 promoter (−910/−728) forward, 5′-AAC AAC TAG AGC GCT GCA AA-3′, and reverse, 5′-GCG GGG AAA TAA GAG GAA AA-3′; Kv1.3 promoter (−542/−325) forward, 5′-CCA TCA GCA CCC ATT ACT CC-3′, and reverse, 5′-TCC ACA CCC CTA GGT ACA GC-3′; and Kv1.3 promoter (−121/+61) forward, 5′-ACA ACA TGA GGG CTC TTT CG-3′, and reverse, 5′-CCT CCT CCC TCC TTC TCG-3′.
Transient Transfection of Sp1 siRNA and a Sp1 Inhibitor Mithramycin A
A549 cells were transfected with Sp1 siRNA (Santa Cruz Biotechnology) using LipofectamineTM 2000 reagent (Invitrogen) or treated with mithramycin A (Sigma-Aldrich) for 24 h.
Chemicals
MgTX, ionomycin, mithramycin A, and PAP-1 were dissolved in distilled water or DMSO. The concentration of the stock solution was 10 μm and diluted to the appropriate concentration using PBS or RPMI medium before the experiment.
Statistical Analysis
Statistical significance for the membrane potential experiments was determined using a Wilcoxon signed rank test and unpaired t test. For analysis of Western blot band density, a Mann-Whitney U test was used, and all tests were performed using the SAS program (version 9.1).
RESULTS
Kv1.3 Protein Was Found in the Nucleus of Cancer Cells and Human Brain Tissues
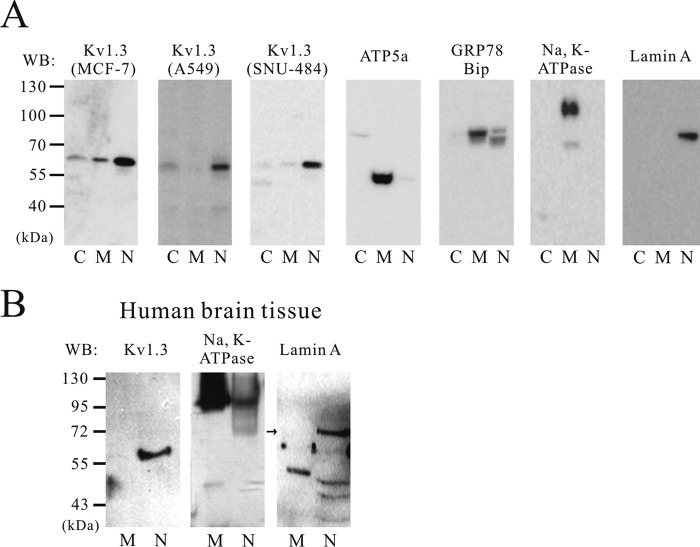
The subcellular localization of Kv1.3 was determined by Western blot analysis using fractionated protein. To confirm the efficiency of the subcellular fractionation of each cell pellet, ATP5a (a mitochondria membrane marker), GRP78 BiP (an endoplasmic reticulum marker), Na,K-ATPase (a plasma membrane marker), and lamin A (a nuclear marker) antibodies were selected. As shown in Fig. 1, localization markers were predominantly expressed in their respective fractions; GRP78 BiP, ATP5a, and Na,K-ATPase were strongly detected in the membrane fraction, and lamin A was only associated with the nuclear fraction. Remarkably, Kv1.3 protein expression was detected in the nuclear fraction of MCF-7, A549, and SNU-484 cells and also in the cytosolic fraction of MCF-7 and A549 cells (Fig. 1A). The specificity of the Kv1.3 antibody was confirmed by using shRNA against Kv1.3 (19). To confirm whether this nuclear localization of Kv1.3 is specific for cancer cell lines, we also tested human brain tissues, which are known to express functional Kv1.3 channels. Kv1.3 protein was observed in the nuclear fraction of human brain tissue. The purity of human brain tissue extracts was validated using lamin A and Na,K-ATPase antibodies (Fig. 1B).
FIGURE 1.
Subcellular localization of Kv1.3 protein in cancer cells and human brain tissues. Western blot (WB) analysis of Kv1.3 shows labeling within the nuclear fraction of three cancer cell lines (MCF7, A549, and SNU-484) (n = 4) (A) and human brain tissue extracts (n = 3) (B). The specificity of the fractionation process was confirmed using specific antibodies including: ATP5a, GRP78 BiP, and Na,K-ATPase, three membrane markers, and lamin A, a marker for the nucleus. Lanes C, cytosol; lanes M, membrane; lanes N, nucleus.
Kv1.3 Protein Was Detected in the Plasma and Mitochondria Membrane in Addition to the Nucleus in Human Jurkat Cells
It has been previously reported that Kv1.3 channels are expressed in the plasma and mitochondria membrane of human Jurkat cells (10, 26). Although Kv1.3 protein was found in the nuclear region in cancer cells such as MCF7, A549, and SNU-484 cells and human brain tissues, Kv1.3 in Jurkat cells was detected in both the plasma membrane and nuclear fractions (Fig. 2). According to our fractionation method, proteins from mitochondria and plasma membrane are also found in the membrane fraction. Therefore, the membrane fraction presumably contains Kv1.3 channels that were localized to the mitochondrial and plasma membranes of Jurkat cells (10, 26). These results suggested the possibility that differential trafficking of Kv1.3 channels to the plasma membrane, mitochondria, and nucleus may be due to the existence of splice variants or chaperone proteins (27, 28). To address the possibility of alternative splicing, we compared the coding sequence of Kv1.3 (GenBankTM accession number L23499.1) between A549 cells and Jurkat cells. We sequenced the coding region of Kv1.3 isolated from both cells but could not detect any splice variants in either cell lines (data not shown). These results indicate that the Kv1.3 coding sequence does not exclusively determine subcellular localization of the channel in A549 and Jurkat cells.
FIGURE 2.
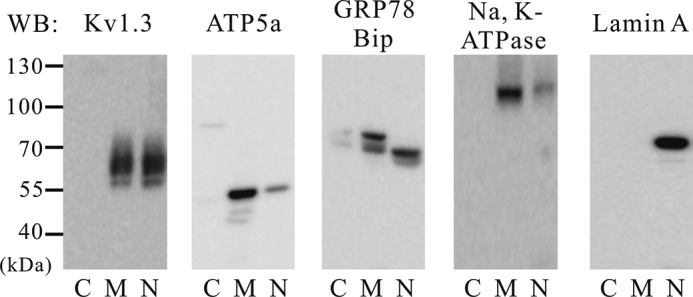
Subcellular localization of Kv1.3 protein in human Jurkat cells. After subcellular fractionation, a band representing Kv1.3 is shown. The expression of Kv1.3 was detected in the plasma and mitochondria membrane and nuclear fractions. The specificity of the fractionation process was confirmed using specific antibodies including: ATP5a, GRP78 BiP, and Na,K-ATPase, three membrane markers, and lamin A, a marker for the nucleus (n = 4). WB, Western blot; lanes C, cytosol; lanes M, membrane; lanes N, nucleus.
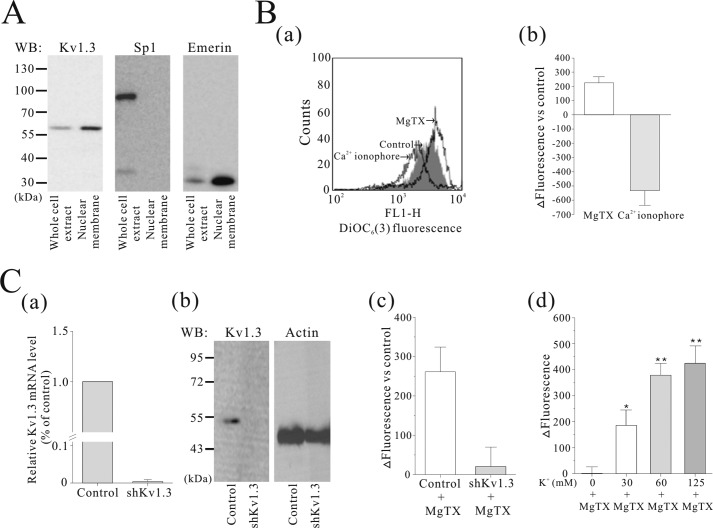
Selective Kv1.3 Inhibition Induces Nuclear Membrane Hyperpolarization
A Western blot assay indicated that Kv1.3 is found in the nuclear membrane of A549 cells purified with the method by Kaufmann et al. (23) (Fig. 3A). To test the hypothesis that Kv1.3 affects the nuclear membrane potential, we treated Kv1.3-selective blocker MgTX in the isolated nuclei from A549 cells and measured changes in nuclear membrane potential using DiOC6(3) fluorescence by flow cytometry. DiOC6(3) is a cationic lipophilic dye that partitions into the nuclear membrane as previously reported by Quesada et al. (12). When the perinuclear space is relatively more negative, DiOC6(3) accumulates in the nuclear envelope, and therefore fluorescence increases. As shown in Fig. 3B (panels a and b), MgTX (1 nm) increased DiOC6(3) fluorescence in isolated nuclei as indicated by a dextral shift of the fluorescence signal, indicating that nuclear membrane was hyperpolarized by a decrease in K+ permeability. The Ca2+ ionophore ionomycin (1 μm) was used as a control, which provoked a decrease in DiOC6(3) fluorescence (left shift), indicating that the nuclear membrane is depolarized. In addition, we confirmed that the MgTX-induced hyperpolarization was due to Kv1.3 channel expression by testing the effects of the toxin in Kv1.3 silenced cells (Fig. 3C, panels a and b). As shown in Fig. 3C (panel c), hyperpolarization induced by MgTX was dramatically diminished following Kv1.3 shRNA transfection into A549 cells. To determine how the K+ gradient affects nuclear membrane hyperpolarization induced by MgTX, we measured changes in membrane potential while varying the K+ concentration gradient across the nuclear membrane. MgTX (1 nm) had little effect on the nuclear membrane potential in 0 mm K+ solution. However, when the K+ concentration outside the nucleus was increased (to 30, 60, and 125 mm), the effect of MgTX was significantly greater (Fig. 3C, panel d). This result demonstrates that the effects of MgTX are dependent on the magnitude of the K+ gradient across the nuclear membrane.
FIGURE 3.
The effect of MgTX on nuclear membrane potential of the isolated nuclei from A549 cells. A, Western blot (WB) analysis indicated that Kv1.3 exists in the nuclear membranes of A549 cells. Nuclear membrane fractionation was confirmed using Sp1 (a nucleoplasmic protein) and Emerin (a nuclear envelope protein) antibodies (n = 3). B, DiOC6(3) was loaded to measure nuclear membrane potential in isolated nuclei. Treatment with 1 nm MgTX induced an increase in DiOC6(3) fluorescence (right shift), whereas 1 μm of ionomycin, a Ca2+ ionophore, caused a decrease in DiOC6(3) fluorescence (left shift) (n = 6) (panels a and b). C, mRNA (n = 5) and protein (n = 2) expression of Kv1.3 were diminished in Kv1.3-silenced A549 cells (panels a and b). MgTX (1 nm) treatment of the isolated nuclei from Kv1.3-silenced A549 cells did not evoke MgTX-induced nuclear membrane hyperpolarization. A549-MgTX refers to A549 cells treated with MgTX (n = 6) (panel c). Isolated nuclei were resuspended in 0 mm [K+] solution for 30 min and then transferred to defined extranuclear [K+] solutions (0, 30, 60, or 125 mm) for 5 min. Effects of MgTX on nuclear membrane potential were measured with different extranuclear [K+] solutions. ΔFluorescence refers to subtraction of the value of when isolated nuclei were suspended in only defined [K+] solutions from in the same [K+] solution with 1 nm MgTX (n = 4) (panel d). The data were normalized as a control (without MgTX) and presented as the means ± S.E. *, p < 0.05; **, p < 0.001.
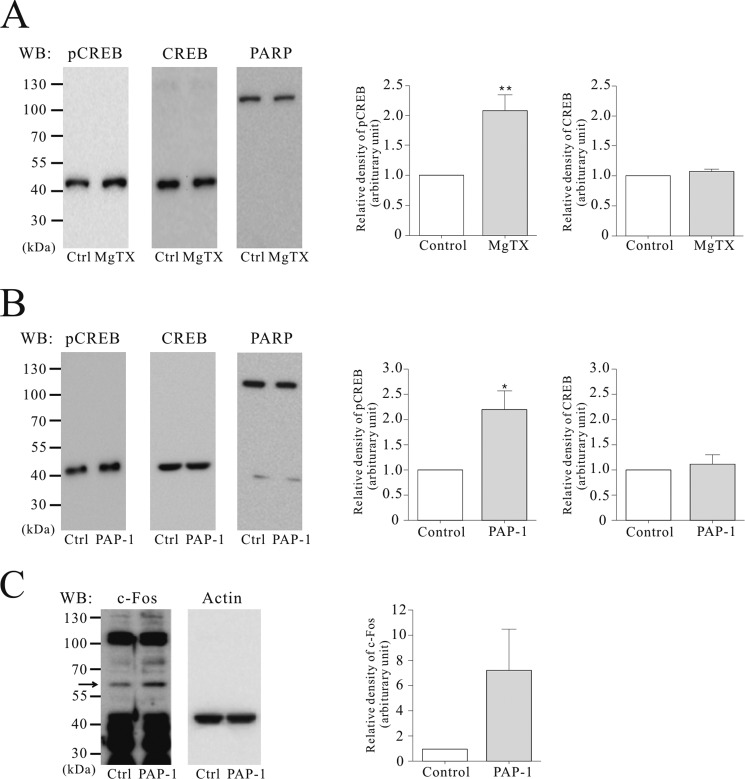
Inhibitors of Nuclear Kv1.3 Channels Lead to Phosphorylation of CREB and c-Fos Activation
To investigate the roles of Kv1.3 in the nucleus, we examined phosphorylation of the transcription factor CREB in isolated nuclei from A549 cells (Fig. 4A). The level of phosphorylated CREB was increased by 2.1-fold following treatment with 1 nm MgTX. In contrast, CREB total protein levels were similar between control and MgTX treatment conditions. In intact A549 cells, the membrane-permeant Kv1.3 blocker, PAP-1 (10 nm) phosphorylated CREB by 2.2-fold (Fig. 4B) and increased c-Fos, an immediate early response transcription factor (Fig. 4C), but there was no significant difference of CREB total protein level between control and PAP-1 treatment conditions.
FIGURE 4.
Up-regulation of phosphorylated CREB and c-Fos induced by Kv1.3 blockers. Representative gel images of Western blots (WB) and the relative expression level of pCREB (n = 4) and CREB (n = 4) induced by 1 nm MgTX in the isolated nuclei of A549 (A), and of pCREB (n = 3), CREB (n = 3), and c-Fos (n = 3) by treatment of 10 nm PAP-1, a membrane-permeable Kv1.3 blocker in intact A549 cells (B and C). The data were normalized relative to the value of controls and presented as the mean ± S.E. (n = 3). *, p < 0.05. Ctrl, control.
Physical Interaction of Nuclear Kv1.3 with Proteins in the Nucleus
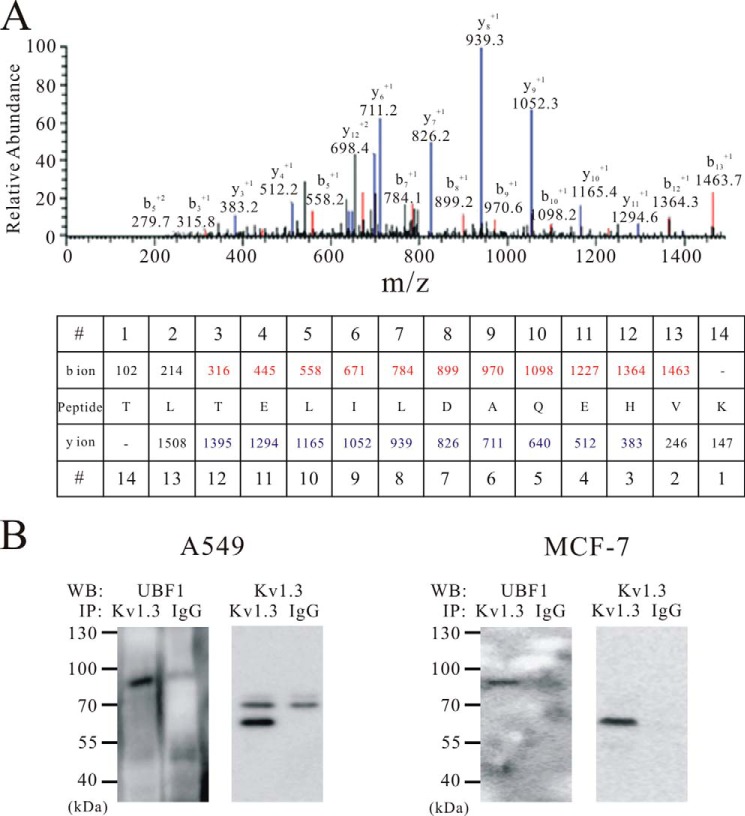
To investigate whether nuclear Kv1.3 interacts with other proteins in the nucleus of A549, we purified Kv1.3 channel complexes using liquid chromatography tandem mass spectrometry (LC-MS/MS) and co-immunoprecipitation in the isolated nuclei. As shown in the MS/MS spectrum from A549 cells (Fig. 5A) and Western blot image using A549 and MCF-7 cells (Fig. 5B), the upstream binding factor 1 (UBF1) was co-purified with Kv1.3 using an antibody that selectively binds Kv1.3 protein.
FIGURE 5.
Co-assembly of UBF1 and nuclear Kv1.3 channel. A, MS/MS spectra of the UBF1-specific peptide obtained by mass spectrometry. The b ions originate from the cleavage of the peptide backbone with N-terminal charge retention, and the y ions indicate peptide fragments with C-terminal charge retention. The b and y ions matched to the UBF1 peptide are indicated in red and blue, respectively. B, co-immunoprecipitation of Kv1.3 with UBF1 from A549 (n = 3) and MCF-7 (n = 3) nuclear extracts. IgG was used as a negative control. WB, Western blot; IP, immunoprecipitation.
Sp1 Transcription Factor Binds to the Kv1.3 Promoter and Controls Kv1.3 Protein Expression in the Nucleus
The Kv1.3 promoter sequence contains TATA-less and GC-rich regions that are required for specificity protein 1 (Sp1) transcription factor binding (29). Therefore, we speculated that Sp1 could bind to the Kv1.3 promoter and regulate the expression of Kv1.3 in the nucleus. Through computational analysis using the JASPAR database and TFSEARCH, several potential Sp1 binding sites were found in the human Kv1.3 promoter (accession number 3180). To determine whether Sp1 is bound to the Kv1.3 promoter, we performed a ChIP assay using A549 cells. PCR analysis using primers divided into the three primers in the Kv1.3 promoter showed that one region (−910/−728) of the Kv1.3 promoter interacts with Sp1 protein (Fig. 6A).
FIGURE 6.
Sp1 transcription factor interaction with the Kv1.3 promoter and down-regulated Kv1.3 protein expression by inhibition of Sp1. A, a ChIP assay showing Sp1 binding to the Kv1.3 promoter in A549 cells (n = 3). Input DNA was used as a control, and rabbit IgG was used as a negative control. B, Western blot (WB) analysis of the nuclear extract of A549 cells treated with 100 and 200 nm mithramycin A for 24 h (Sp1, n = 3; Kv1.3, n = 3) (panel a). A549 cells were transfected with scRNA or Sp1-siRNA. Western blot analysis demonstrating Sp1 (n = 3) and Kv1.3 (n = 3) protein expression levels in nuclear extracts was suppressed by Sp1-siRNA transfection (panel b). Western blot analysis of pCREB (n = 3) and CREB (n = 3) protein levels in the A549 cells treated with 100 nm of mithramycin A for 24 h, followed by 10 nm of PAP-1 for 30 min (panel c). The effect of MgTX on the nuclear membrane potential in isolated nuclei from Sp1-down-regulated A549 cells using DiOC6(3) (n = 5). After the cells were incubated with 200 nm of mithramycin A for 24 h, isolated nuclei from the cells were treated with 1 nm of MgTX for 10 min (panel d). The data were normalized as a value of control and presented as the means ± S.E. *, p < 0.05; **, p < 0.01. Ctrl, control.
To determine whether Sp1 was involved in regulating Kv1.3 expression, mithramycin A, a selective inhibitor of transcription factor binding to the GC-rich promoter, and RNA interference were used to inhibit interaction of Sp1 with the Kv1.3 promoter. As shown in Fig. 6B (panel a), reducing Sp1 protein-promotor interactions using mithramycin A decreased Kv1.3 channel expression compared with control in nuclear extracts. Furthermore, silencing Sp1 mRNA expression by transfection with Sp1-siRNA also reduced Sp1 and Kv1.3 protein expression in nuclear extracts of transfected A549 cells (Fig. 6B, panel b). Given that Sp1 increases the expression of Kv1.3 in the nucleus and that Kv1.3 blockade hyperpolarized the nuclear membrane potential and increased CREB phosphorylation, we examined whether blocking Sp1 altered CREB phosphorylation and the hyperpolarization of the nuclear membrane induced by Kv1.3 blockers. Sp1 blockade significantly increased CREB phosphorylation, and PAP-1 did not increase the phosphorylation of CREB in the A549 cells treated with mithramycin A (Fig. 6B, panel c). Moreover, MgTX treatment did not induce hyperpolarization in purified nuclei from the A549 cells treated with mithramycin A (Fig. 6B, panels d).
Subcellular Localization of other Kv Channels, Kv1.1, Kv1.2, and Kv2.2 in A549, MCF-7, and SNU-484 Cells
To examine whether nuclear localization of Kv1.3 is a general phenomenon or specific to Kv1.3, the subcellular localization of Kv1.1, Kv1.2, and Kv2.2 was determined by Western blot analysis using fractionated protein. We found that Kv1.1 was localized in the nuclear fraction in the A549 and MCF7 cells but appeared to be localized mainly in the membrane in SNU-484 cells. Kv1.2 was localized only in the membrane fraction in the A549 and MCF7 cells but was expressed both in the membrane and in the nuclear fraction in SNU-484 cells. Kv2.2 was localized both in the membrane and nuclear fraction in A549, MCF7, and SNU-484 cells (Fig. 7).
FIGURE 7.
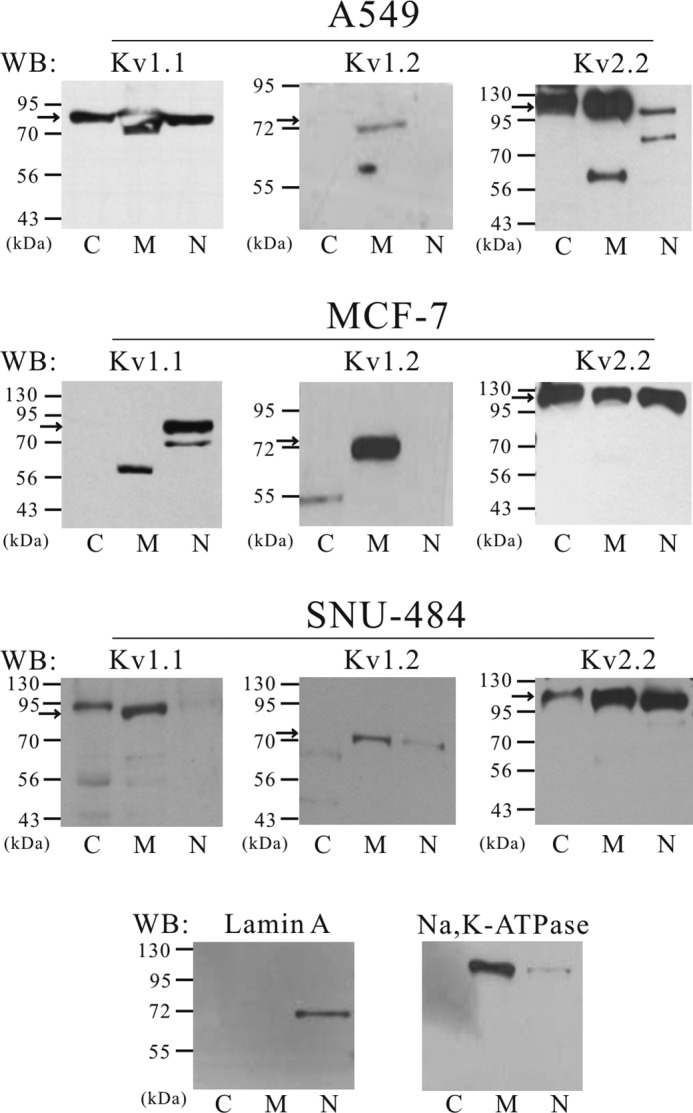
Subcellular localization of Kv1.1, Kv1.2, and Kv2.2 in A549, MCF-7, and SNU-484 cells. Subcellular localization of Kv1.1, Kv1.2, and Kv2.2 was determined by Western blot (WB) analysis using fractionated protein. Kv1.1 was localized in the nuclear fraction in MCF7 cells (n = 2) and A549 cells (n = 2); however, it was mainly localized in the membrane fraction in SNU-484 cells (n = 2). Kv1.2 was only localized in the membrane fraction in MCF7 (n = 2), A549 cells (n = 2) but was expressed both in the membrane and in the nuclear fraction in SNU-484 cells (n = 4). Kv2.2 was localized both in the membrane and nuclear fraction in A549 (n = 2), MCF7 (n = 3) and SNU-484 cells (n = 2). The specificity of the fractionation process was confirmed using Na,K-ATPase, a marker of membrane and lamin A, a marker for the nucleus. Lanes C, cytosol; lanes M, membrane; lanes N, nucleus.
DISCUSSION
K+ selective ion channels have been previously identified in the nucleus of a variety of cell types (18, 30–32). In addition, Ca2+-dependent (18) or independent (31) K+ channel activities were observed in the nucleus of pancreatic acinar cells and rat brain, respectively, and functional KATP channels were previously identified in the nuclear membrane of pancreatic cells (12). Nuclear Kv channel immunoreactivity has also been reported in the paraventricular nucleus projecting into the rostral ventrolateral medulla (33). Recently, eag I channels were found to be localized on the nuclear membrane of Kv10.1-transfected cells, MCF7 cells, and rat cerebellum and hippocampus (34). Interestingly, Mazzanti et al. (35) suggested that nuclear pores may be K+ selective and that K+ channels might form part of the nuclear pore, which provides a conductive pathway between nuclear membranes. In our study, Kv1.3 was localized in the nucleus of human cancer cells. We also confirmed that Jurkat cells express Kv1.3 channels in the plasma and in the mitochondria membrane, in addition to the nucleus region, similar to that reported in a previous study (26).
A previous study (36) showed that Kv1.3 channel was detected by immunoprecipitation in the synaptic plasma membrane from human cerebral cortical gray matter. However, Coleman et al. (36) also described that Kv1.3 is expressed at low levels in the synaptic plasma membrane compared with other Kv channel subunits. Thus, based on the previous observations and the results of the present study, Kv1.3 channels appear to exhibit greater expression in the nucleus of human brain tissues compared with the plasma membrane.
Our search using the PredictNLS algorithm indicated that known nuclear localization signal sequences were not present in Kv1.3 channels. Thus, Kv1.3 channels may contain an nuclear localization signal that has not been previously described. It is worth noting that Kv1.3 channels contain a PDZ domain at the end of the C-terminal sequence (37) and could potentially affect the localization of the channel and allow for interactions with signaling molecules within the nucleus. It has been suggested that alternative splicing or post-translational protein modifications (38) could influence the localization of proteins.
Although potassium channels are known to be located in the nucleus (12–14, 18), little is known about the function of the nuclear channel. Therefore, we examined the roles of nuclear Kv1.3. Blockade of the Kv1.3 channel activity resulted in phosphorylation of CREB, an important transcriptional activator, in the nuclei of A549 cells. Similar to our findings, blockers of the KATP channel in pancreatic beta cells and 4-aminopyridine, a nonselective voltage-gated K+ channel blocker, in vascular smooth muscle myocytes induced phosphorylation of CREB by increasing the Ca2+ concentration in the nucleus (12, 39). CREB phosphorylation is known to be related to cell survival and tumorigenesis in lung cancer and to cell cycle arrest in macrophages (40–42).
Along with CREB, c-Fos was found to be increased by PAP-1 in A549 cells. The c-Fos proto-oncoprotein is a primary component of the activator protein-1 transcription factor complex that is known to be involved in cell growth, differentiation, and the G0/G1 transition (43–45). It has been reported that rat-1 fibroblast and primitive hematopoietic stem cells overexpressing c-Fos inhibit cell cycle progression at the G0/G1 boundary as a result of down-regulated cell cycle-associated proteins (42, 43). This observation corresponds with our previous report showing that silencing Kv1.3 expression or inhibiting channel activity with MgTX induces anti-proliferative effects through inhibition of the G1-S transition (19). Previously, it has been reported that MgTX treatment leads to increased c-Fos mRNA transcription and protein in the rat striatum, which triggers long term neuronal plasticity (46). These results along with data from the present study strongly suggest that ion channels such as Kv1.3 located in the nucleus can influence the regulation of gene expression by modulating the activation of certain transcription factors including CREB and c-Fos.
Kv1.3 channels interact with other proteins through the protein-interaction domain. The PDZ domain in the plasma membrane and mitochondrial Kv1.3 bind to apoptotic protein BAX (9, 11). Therefore, to investigate whether the Kv1.3 channel interacts with nuclear proteins, we used a protein binding assay with nuclear extracts. The results show that nuclear Kv1.3 channel binds to the UBF1 transcription factor, which participates in RNA polymerase I activity, mediating ribosomal RNA synthesis. It was reported that the NR1 subunit of the N-methyl-d-aspartate receptor was found in the nucleoli of hydra cells, which could serve as a regulator for ribosomal subunit production (47). Therefore, nuclear Kv1.3 channels appear to play the role of regulators in ribosome biogenesis, although this possibility needs to be studied further.
As part of the present study, we investigated the regulation of nuclear Kv1.3 channel expression in A549 cells by identifying the transcription factor that mediates the Kv1.3 gene transcription. The Kv1.3 gene has been reported to have GC-rich and TATA-less promoter regions (29). Previous studies have shown that the transcription factor, Sp1, notably binds to GC-rich motifs but also regulates the transcription of other Kv channel genes such as Kv1.5 (48) and Kv4.3 (49). We showed that Sp1 affects Kv1.3 expression through interaction with the Kv1.3 promoter and that silencing Sp1 expression using siRNAs or the Sp1 blocker, mithramycin A, reduces Kv1.3 channel expression in the nucleus. Moreover, treatment with mithramycin A reduced levels of the Sp1 protein and inhibited nuclear hyperpolarization and phosphorylation of CREB induced by Kv1.3 blockers. This is the first report demonstrating that Sp1 plays a crucial role in Kv1.3 gene expression.
The physiological and pathophysiological significance of nuclear Kv channels remains to be determined. One possibility is that nuclear Kv channels may regulate the nuclear membrane potential (35, 50), which may subsequently control the activity of other ion channels within the nuclear membrane (51). Another possible role for nuclear Kv channels is the regulation of cell proliferation and apoptosis of cancer cells. Kv channels are an emerging target for anti-cancer therapy because several Kv channels have been reported as regulators of cancer cell proliferation. However, the mechanism is unclear. As reported previously (52), the K+ current flowing across Kv channels in the nucleus can be closely associated with cancer cell progression through regulatory signaling such as multifunctional CREB or c-Fos transcription factors.
These novel signaling mechanisms linking ion channels and gene expression in the nucleus may not be limited to cancer cells. Nuclear Kv1.3 channels are also observed in human brain tissues where their function may be important in determining neuronal morphology, synaptic plasticity, and neuroprotection, which are also regulated by CREB and c-Fos (53–55).
In addition, we also found that Kv1.1, Kv1.2, and Kv2.2 were localized in nuclear fraction. Therefore, nuclear localization of Kv1.3 is not a specific phenomenon for Kv1.3, and it is possible the Kv channel subunits are localized in different intracellular organelles or in the plasma membrane, depending on the Kv subunits. The localization of Kv channels in specific regions of the cells and their functional consequences need to be further examined.
In conclusion, we suggest that Kv1.3 channels are localized in the nuclei of several types of human cancer cells and human brain tissues. Kv1.3 channels are capable of participating in the regulation of nuclear membrane potential and selective inhibition of nuclear Kv1.3 channel activity, which in turn can trigger gene expression by activating specific transcription factors, including pCREB and c-Fos. Furthermore, nuclear Kv1.3 channels interact with the UBF1 protein, and expression of nuclear Kv1.3 is controlled by Sp1, a transcription factor that directly binds to the Kv1.3 promoter. The results of this study provide biochemical and functional characteristics of nuclear Kv1.3 channels, and these properties could be related to the recently revealed role of Kv channels in proliferation, apoptosis, and differentiation, thus expanding our knowledge about the function of ion channels within the nucleus.
This work was supported by Basic Science Research Program Grant NRF-2010-0011556, the BK21 PLUS Program for Creative Veterinary Science Research, and Priority Research Center Program Grant 2009-0094050 through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology.
- Kv
- voltage-gated potassium channel
- CREB
- cAMP response element-binding protein
- KATP
- ATP-sensitive potassium channel
- MgTX
- margatoxin
- PAP-1
- 5-(4-phenoxybutoxy) psoralen
- pCREB
- phosphorylated cAMP response element-binding protein
- Sp1
- specificity protein 1
- UBF1
- upstream binding factor 1.
REFERENCES
- 1. Hille B. (2001) Ion Channels of Excitable Membranes, 3rd Ed., pp. 131–167, Sinauer Associates, Sunderland, MA [Google Scholar]
- 2. Gutman G. A., Chandy K. G., Grissmer S., Lazdunski M., McKinnon D., Pardo L. A., Robertson G. A., Rudy B., Sanguinetti M. C., Stühmer W., Wang X. (2005) International Union of Pharmacology: LIII. nomenclature and molecular relationships of voltage-gated potassium channels. Pharmacol. Rev. 57, 473–508 [DOI] [PubMed] [Google Scholar]
- 3. Pongs O., Leicher T., Berger M., Roeper J., Bähring R., Wray D., Giese K. P., Silva A. J., Storm J. F. (1999) Functional and molecular aspects of voltage-gated K+ channel beta subunits. Ann. N.Y. Acad. Sci. 868, 344–355 [DOI] [PubMed] [Google Scholar]
- 4. Kunzelmann K. (2005) Ion channels and cancer. J. Membr. Biol. 205, 159–173 [DOI] [PubMed] [Google Scholar]
- 5. O'Grady S. M., Lee S. Y. (2005) Molecular diversity and function of voltage-gated (Kv) potassium channels in epithelial cells. Int. J. Biochem. Cell Biol. 37, 1578–1594 [DOI] [PubMed] [Google Scholar]
- 6. Hu L., Gocke A. R., Knapp E., Rosenzweig J. M., Grishkan I. V., Baxi E. G., Zhang H., Margolick J. B., Whartenby K. A., Calabresi P. A. (2012) Functional blockade of the voltage-gated potassium channel Kv1.3 mediates reversion of T effector to central memory lymphocytes through SMAD3/p21cip1 signaling. J. Biol. Chem. 287, 1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matheu M. P., Beeton C., Garcia A., Chi V., Rangaraju S., Safrina O., Monaghan K., Uemura M. I., Li D., Pal S., de la Maza L. M., Monuki E., Flügel A., Pennington M. W., Parker I., Chandy K. G., Cahalan M. D. (2008) Imaging of effector memory T cells during a delayed-type hypersensitivity reaction and suppression by Kv1.3 channel block. Immunity 29, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bednarczyk P., Kowalczyk J. E., Beresewicz M., Dolowy K., Szewczyk A., Zablocka B. (2010) Identification of a voltage-gated potassium channel in gerbil hippocampal mitochondria. Biochem. Biophys. Res. Commun. 397, 614–620 [DOI] [PubMed] [Google Scholar]
- 9. Leanza L., Henry B., Sassi N., Zoratti M., Chandy K. G., Gulbins E., Szabò I. (2012) Inhibitors of mitochondrial Kv1.3 channels induce Bax/Bak-independent death of cancer cells. EMBO Mol. Med. 4, 577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szabò I., Bock J., Jekle A., Soddemann M., Adams C., Lang F., Zoratti M., Gulbins E. (2005) A novel potassium channel in lymphocyte mitochondria. J. Biol. Chem. 280, 12790–12798 [DOI] [PubMed] [Google Scholar]
- 11. Szabó I., Bock J., Grassmé H., Soddemann M., Wilker B., Lang F., Zoratti M., Gulbins E. (2008) Mitochondrial potassium channel Kv1.3 mediates Bax-induced apoptosis in lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 105, 14861–14866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quesada I., Rovira J. M., Martin F., Roche E., Nadal A., Soria B. (2002) Nuclear KATP channels trigger nuclear Ca2+ transients that modulate nuclear function. Proc. Natl. Acad. Sci. U.S.A. 99, 9544–9549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olsen M. L., Sontheimer H. (2004) Mislocalization of Kir channels in malignant glia. Glia 46, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stonehouse A. H., Grubb B. D., Pringle J. H., Norman R. I., Stanfield P. R., Brammar W. J. (2003) Nuclear immunostaining in rat neuronal cells using two anti-Kir2.2 ion channel polyclonal antibodies. J. Mol. Neurosci. 20, 189–194 [DOI] [PubMed] [Google Scholar]
- 15. Buyse G., Trouet D., Voets T., Missiaen L., Droogmans G., Nilius B., Eggermont J. (1998) Evidence for the intracellular location of chloride channel (ClC)-type proteins: co-localization of ClC-6a and ClC-6c with the sarco/endoplasmic-reticulum Ca2+ pump SERCA2b. Biochem. J. 330, 1015–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Valenzuela S. M., Mazzanti M., Tonini R., Qiu M. R., Warton K., Musgrove E. A., Campbell T. J., Breit S. N. (2000) The nuclear chloride ion channel NCC27 is involved in regulation of the cell cycle. J. Physiol. 529, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gomez-Ospina N., Tsuruta F., Barreto-Chang O., Hu L., Dolmetsch R. (2006) The C terminus of the L-type voltage-gated calcium channel CaV1.2 encodes a transcription factor. Cell 127, 591–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maruyama Y., Shimada H., Taniguchi J. (1995) Ca2+-activated K+-channels in the nuclear envelope isolated from single pancreatic acinar cells. Pflugers Arch. 430, 148–150 [DOI] [PubMed] [Google Scholar]
- 19. Jang S. H., Choi S. Y., Ryu P. D., Lee S. Y. (2011) Anti-proliferative effect of Kv1.3 blockers in A549 human lung adenocarcinoma in vitro and in vivo. Eur. J. Pharmacol. 651, 26–32 [DOI] [PubMed] [Google Scholar]
- 20. Jang S. H., Ryu P. D., Lee S. Y. (2011) Dendrotoxin-κ suppresses tumor growth induced by human lung adenocarcinoma A549 cells in nude mice. J. Vet. Sci. 12, 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang W., Xiao J., Adachi M., Liu Z., Zhou J. (2011) 4-Aminopyridine induces apoptosis of human acute myeloid leukemia cells via increasing [Ca2+]i through P2X7 receptor pathway. Cell Physiol. Biochem. 28, 199–208 [DOI] [PubMed] [Google Scholar]
- 22. Franco-Obregón A., Wang H. W., Clapham D. E. (2000) Distinct ion channel classes are expressed on the outer nuclear envelope of T- and B-lymphocyte cell lines. Biophys. J. 79, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaufmann S. H., Gibson W., Shaper J. H. (1983) Characterization of the major polypeptides of the rat liver nuclear envelope. J. Biol. Chem. 258, 2710–2719 [PubMed] [Google Scholar]
- 24. Koning A. J., Lum P. Y., Williams J. M., Wright R. (1993) DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil. Cytoskeleton 25, 111–128 [DOI] [PubMed] [Google Scholar]
- 25. Loew L. M. (1993) Fluorescent and Luminescent Probes for Biological Activity (Mason W.T., ed) pp. 150–160, Academic Press, San Diego, CA [Google Scholar]
- 26. Panyi G., Bagdány M., Bodnár A., Vámosi G., Szentesi G., Jenei A., Mátyus L., Varga S., Waldmann T. A., Gáspar R., Damjanovich S. (2003) Colocalization and nonrandom distribution of Kv1.3 potassium channels and CD3 molecules in the plasma membrane of human T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 100, 2592–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Napp J., Monje F., Stühmer W., Pardo L. A. (2005) Glycosylation of Eag1 (Kv10.1) potassium channels: intracellular trafficking and functional consequences. J. Biol. Chem. 280, 29506–29512 [DOI] [PubMed] [Google Scholar]
- 28. Solé L., Roura-Ferrer M., Pérez-Verdaguer M., Oliveras A., Calvo M., Fernández-Fernández J. M., Felipe A. (2009) KCNE4 suppresses Kv1.3 currents by modulating trafficking, surface expression and channel gating. J. Cell Sci. 122, 3738–3748 [DOI] [PubMed] [Google Scholar]
- 29. Simon M., Conley E. C., Shelton P. A., Gutman G. A., Chandy K. G. (1997) Transcription of the T-cell potassium channel Kv1.3 is regulated by a GC-rich TATA-less promoter. Cell Physiol. Biochem. 7, 243–250 [Google Scholar]
- 30. Bustamante J. O. (1992) Nuclear ion channels in cardiac myocytes. Pflugers Arch. 421, 473–485 [DOI] [PubMed] [Google Scholar]
- 31. Draguhn A., Börner G., Beckmann R., Buchner K., Heinemann U., Hucho F. (1997) Large-conductance cation channels in the envelope of nuclei from rat cerebral cortex. J. Membr. Biol. 158, 159–166 [DOI] [PubMed] [Google Scholar]
- 32. Mazzanti M., DeFelice L. J., Cohn J., Malter H. (1990) Ion channels in the nuclear envelope. Nature 343, 764–767 [DOI] [PubMed] [Google Scholar]
- 33. Sonner P. M., Stern J. E. (2007) Functional role of A-type potassium currents in rat presympathetic PVN neurones. J. Physiol. 582, 1219–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Y., Sánchez A., Rubio M. E., Kohl T., Pardo L. A., Stühmer W. (2011) Functional Kv10.1 channels localize to the inner nuclear membrane. PLoS One 6, e19257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mazzanti M., DeFelice L. J., Smith E. F. (1991) Ion channels in murine nuclei during early development and in fully differentiated adult cells. J. Membr. Biol. 121, 189–198 [DOI] [PubMed] [Google Scholar]
- 36. Coleman S. K., Newcombe J., Pryke J., Dolly J. O. (1999) Subunit composition of Kv1 channels in human CNS. J. Neurochem. 73, 849–858 [DOI] [PubMed] [Google Scholar]
- 37. Marks D. R., Fadool D. A. (2007) Post-synaptic density perturbs insulin-induced Kv1.3 channel modulation via a clustering mechanism involving the SH3 domain. J. Neurochem. 103, 1608–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schulz D. J., Temporal S., Barry D. M., Garcia M. L. (2008) Mechanisms of voltage-gated ion channel regulation: from gene expression to localization. Cell Mol. Life Sci. 65, 2215–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coussin F., Scott R. H., Nixon G. F. (2003) Sphingosine 1-phosphate induces CREB activation in rat cerebral artery via a protein kinase C-mediated inhibition of voltage-gated K+ channels. Biochem. Pharmacol. 66, 1861–1870 [DOI] [PubMed] [Google Scholar]
- 40. Conkright M. D., Montminy M. (2005) CREB: the unindicted cancer co-conspirator. Trends Cell Biol. 15, 457–459 [DOI] [PubMed] [Google Scholar]
- 41. Linnerth N. M., Baldwin M., Campbell C., Brown M., McGowan H., Moorehead R. A. (2005) IGF-II induces CREB phosphorylation and cell survival in human lung cancer cells. Oncogene 24, 7310–7319 [DOI] [PubMed] [Google Scholar]
- 42. Sawka-Verhelle D., Escoubet-Lozach L., Fong A. L., Hester K. D., Herzig S., Lebrun P., Glass C. K. (2004) PE-1/METS, an antiproliferative Ets repressor factor, is induced by CREB-1/CREM-1 during macrophage differentiation. J. Biol. Chem. 279, 17772–17784 [DOI] [PubMed] [Google Scholar]
- 43. Balsalobre A., Jolicoeur P. (1995) Fos proteins can act as negative regulators of cell growth independently of the fos transforming pathway. Oncogene 11, 455–465 [PubMed] [Google Scholar]
- 44. Okada S., Fukuda T., Inada K., Tokuhisa T. (1999) Prolonged expression of c-fos suppresses cell cycle entry of dormant hematopoietic stem cells. Blood 93, 816–825 [PubMed] [Google Scholar]
- 45. Thomas D. P., Sunters A., Gentry A., Grigoriadis A. E. (2000) Inhibition of chondrocyte differentiation in vitro by constitutive and inducible overexpression of the c-fos proto-oncogene. J. Cell Sci. 113, 439–450 [DOI] [PubMed] [Google Scholar]
- 46. Saria A., Fischer H. S., Humpel C., Pfattner A., Schatz D. S., Schuligoi R. (2000) Margatoxin and iberiotoxin, two selective potassium channel inhibitors, induce c-fos like protein and mRNA in rat organotypic dorsal striatal slices. Amino Acids 19, 23–31 [DOI] [PubMed] [Google Scholar]
- 47. Kass-Simon G., Zompa M. A., Scappaticci A. A., Zackroff R. V., Hufnagel L. A. (2009) Nucleolar binding of an anti-NMDA receptor antibody in hydra: a non-canonical role for an NMDA receptor protein? J. Exp. Zool A Ecol. Genet. Physiol. 311, 763–775 [DOI] [PubMed] [Google Scholar]
- 48. Fountain S. J., Cheong A., Li J., Dondas N. Y., Zeng F., Wood I. C., Beech D. J. (2007) Kv1.5 potassium channel gene regulation by Sp1 transcription factor and oxidative stress. Am. J. Physiol. Heart Circ. Physiol. 293, H2719–H2725 [DOI] [PubMed] [Google Scholar]
- 49. Li Q., Zhang Y., Sheng Y., Huo R., Sun B., Teng X., Li N., Zhu J. X., Yang B. F., Dong D. L. (2012) Large T-antigen up-regulates Kv4.3 K+ channels through Sp1, and Kv4.3 K+ channels contribute to cell apoptosis and necrosis through activation of calcium/calmodulin-dependent protein kinase II. Biochem. J. 441, 859–867 [DOI] [PubMed] [Google Scholar]
- 50. Bustamante J. O. (1993) Restricted ion flow at the nuclear envelope of cardiac myocytes. Biophys. J. 64, 1735–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bkaily G., Avedanian L., Jacques D. (2009) Nuclear membrane receptors and channels as targets for drug development in cardiovascular diseases. Can. J. Physiol. Pharmacol. 87, 108–119 [DOI] [PubMed] [Google Scholar]
- 52. Mazzanti M., Bustamante J. O., Oberleithner H. (2001) Electrical dimension of the nuclear envelope. Physiol. Rev. 81, 1–19 [DOI] [PubMed] [Google Scholar]
- 53. Kaczmarek L., Nikolajew E. (1990) c-fos protooncogene expression and neuronal plasticity. Acta Neurobiol. Exp. (Wars.) 50, 173–179 [PubMed] [Google Scholar]
- 54. Sakamoto K., Karelina K., Obrietan K. (2011) CREB: a multifaceted regulator of neuronal plasticity and protection. J. Neurochem. 116, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang J., Zhang D., McQuade J. S., Behbehani M., Tsien J. Z., Xu M. (2002) c-fos regulates neuronal excitability and survival. Nat. Genet. 30, 416–420 [DOI] [PubMed] [Google Scholar]