Background: IgA responses are important for human gut mucosal barrier function.
Results: Gnotobiotic Rag1−/− mice harboring a human gut bacterial symbiont and a single, naturally primed IgA revealed species/strain/epitope level specificities and biological effects of the antibody.
Conclusion: Bacteroides thetaiotaomicron maintains a highly conserved immunogenic epitope important for its fitness in vivo.
Significance: Our approach can yield diagnostic antibodies and new understanding of microbiota-immune system co-evolution.
Keywords: Immunoglobulin A (IgA), Microbiome, Molecular Evolution, Mucosal Immunology, Symbiosis, Immunoglobulin A
Abstract
The adaptive immune response to the human gut microbiota consists of a complex repertoire of antibodies interacting with a broad range of taxa. Fusing intestinal lamina propria lymphocytes from mice monocolonized with Bacteroides thetaiotaomicron to a myeloma fusion partner allowed us to recover hybridomas that captured naturally primed, antigen-specific antibody responses representing multiple isotypes, including IgA. One of these hybridomas, 260.8, produced a monoclonal antibody that recognizes an epitope specific for B. thetaiotaomicron isolates in a large panel of hospital- and community-acquired Bacteroides. Whole genome transposon mutagenesis revealed a 19-gene locus, involved in LPS O-antigen polysaccharide synthesis and conserved among multiple B. thetaiotaomicron isolates, that is required for 260.8 epitope expression. Mutants in this locus exhibited marked fitness defects in vitro during growth in rich medium and in gnotobiotic mice colonized with defined communities of human gut symbionts. Expression of the 260.8 epitope was sustained during 10 months of daily passage in vitro and during 14 months of monocolonization of gnotobiotic wild-type, Rag1−/−, or Myd88−/− mice. Comparison of gnotobiotic Rag1−/− mice with and without subcutaneous 260.8 hybridomas disclosed that this IgA did not affect B. thetaiotaomicron population density or suppress 260.8 epitope production but did affect bacterial gene expression in ways emblematic of a diminished host innate immune response. Our study illustrates an approach for (i) generating diagnostic antibodies, (ii) characterizing IgA responses along a continuum of specificity/degeneracy that defines the IgA repertoire to gut symbionts, and (iii) identifying immunogenic epitopes that affect competitiveness and help maintain host-microbe mutualism.
Introduction
The human gut microbiota and immune system co-develop following birth with each influencing the properties of the other to maintain a homeostasis that is mutually advantageous (1, 2). The biological barrier that supports this peaceful co-existence is made up of mucus (3, 4), immunoglobulins, and antimicrobial peptides “backed up” by an epithelial monolayer knitted together by tight junction proteins. Studies in animal models have shown that innate signaling pathways in the gut mucosa couple sensing of microbial ligands with secretion of antibacterial peptides that prevent microbes from penetrating the mucus layer and underlying epithelium (5–8). Activation of the adaptive arm of the immune system in the intestinal mucosa induces production of antibodies, notably secretory IgA, directed against gut microbes (9, 10).
IgA responses can develop in a T cell-dependent or -independent manner (11) and may exhibit either broad or limited reactivity (12–15). Although class switching and plasma cell maturation in the gut were classically described in Peyer patches, evidence suggests that these processes may also occur locally in the intestinal lamina propria in response to cues from the microbiota, epithelium, and stromal compartment (16). In this model, a decentralized immune response permits the host to rapidly respond to microbial encroachment by activating the adaptive immune system at the site of insult. To study the kinetics and plasticity of this response, Hapfelmeier et al. (9) used a model in which germ-free mice were reversibly colonized with an auxotrophic strain of Escherichia coli that was unable to replicate within the gut, leaving these animals germ-free after a brief period of colonization. Induction of a specific IgA response in these animals required high doses of bacteria (∼109 colony-forming units (cfu)). Subsequent colonization with a stable microbiota lacking E. coli decreased titers of anti-E. coli antibody, demonstrating the lability of the intestinal IgA response. Together, these observations support the notion that IgA responses develop in both secondary intestinal lymphoid structures and the lamina propria, and where the antibody repertoire is altered continuously in response to local or regional antigenic stimulation (17).
Characterizing the specificity of IgA responses has been a challenge due to the complexity of the response and the microbiota (12, 13, 18–21). Cullender et al. (19) reported that Toll-like receptor 5 drives production of IgA and that this response modulates Toll-like receptor 5 ligand (flagellin) expression. More recently, we have used fluorescence-activated cell sorting (FACS) to identify bacterial targets of gut mucosal IgA responses in fecal samples obtained from children with healthy growth phenotypes or with varying degrees of undernutrition, as well as fecal samples harvested from gnotobiotic mice harboring the microbiota and fed the diets of these human gut community donors. FACS-purified viable IgA-targeted bacterial taxa were also transferred to a second round of germ-free animals to examine their functional properties. The results established that IgA responses can be used as biomarkers of disease in undernourished children, that they mediate a diet-dependent enteropathy characterized by small intestinal and colonic epithelial barrier dysfunction, and that certain IgA-targeted microbes purified from healthy donor microbiota can prevent development of this mucosal barrier disruption (22).
To explore the species, strain, and epitope level specificities of the IgA response and its effects on the biological properties and fitness of targeted components of the microbiota, we created a simplified, defined gnotobiotic mouse model (23). Bacteroides thetaiotaomicron is a prominent member of the adult human gut microbiota. The genome of the type strain, VPI-5482, encodes a larger complement of carbohydrate-active enzymes (glycoside hydrolases, polysaccharide lyases, and carbohydrate esterases) than the number of these enzymes specified by our human genome. This repertoire of carbohydrate-active enzymes is embedded in 88 polysaccharide utilization loci, composed of 866 genes comprising 18% of its genome, that allow B. thetaiotaomicron to sense, acquire, and degrade otherwise indigestible polysaccharides in our diets and to forage on mucus glycans when these polysaccharides are absent from the diet (24, 25). Colonization of adult germ-free C57BL/6J mice with B. thetaiotaomicron VPI-5482 leads to increased levels of serum IgA and IgG (particularly IgG3 and IgG2b) within 14 days. Fusing intestinal lamina propria B cells isolated from these monocolonized gnotobiotic animals with a myeloma fusion partner allowed us to recover hybridoma cell lines that “captured” naturally primed, antigen-specific antibody responses representing multiple isotypes, including IgA. Using a transposon mutant library of B. thetaiotaomicron, we identified the cognate epitope for the IgA product of one of the hybridoma cell lines, 225.4, as the product of one of its eight capsular polysaccharide synthesis (CPS)4 loci, CPS4. This 225.4 hybridoma cell line was then injected subcutaneously into the backs of adult germ-free mice with no T or B cells (C57BL/6J Rag1−/−). After a period had elapsed during which time the hybridomas grew to a substantial size, animals were colonized with B. thetaiotaomicron VPI-5482. Comparisons of monocolonized mice with and without the 225.4 hybridoma “backpack” revealed that the presence of the monoclonal antibody (mAb 225.4) decreased 225.4 epitope expression by the bacterium. When an in vivo competition was performed using isogenic wild-type and a CPS4-null (225.4-negative) mutant strain, we observed that the presence of mAb 225.4 reduced the wild-type to mutant ratio by a factor of 10 (23). Moreover, GeneChip-based whole genome profiling of bacterial and intestinal transcriptional responses disclosed that without the 225.4 IgA B. thetaiotaomicron elicited a more robust innate immune response (e.g. inducible NOS) and reacted to this response by inducing bacterial genes that metabolize host oxidative products (e.g. those involved in nitrate metabolism). Thus, the presence of the IgA reduced intestinal proinflammatory signaling and bacterial epitope expression, thereby balancing suppression of the oxidative burst with the negative impact of the antibody on bacterial fitness.
To determine whether these features were unique to mAb 225.4 or apply more generally to the naturally primed antibody repertoire elicited by colonization, we have now characterized another monoclonal IgA (mAb 260.8) generated from a different lamina propria fusion. Unlike mAb 225.4, which was specific for the VPI-5482 type strain, mAb 260.8 exhibits strong specificity and sensitivity for multiple B. thetaiotaomicron isolates from established clinical microbiology laboratory culture collections as well as new culture collections generated from previously frozen human fecal samples. Genetic mapping revealed a 19-gene locus required for expression of the 260.8 epitope. Mutants in this locus, which appears to be involved in LPS O-antigen polysaccharide synthesis and is conserved among multiple B. thetaiotaomicron isolates, display a profound fitness defect in vitro and in vivo. Unlike the capsular polysaccharide-specific 225.4 IgA, studies in gnotobiotic C57BL/6J Rag1−/− mice with and without 260.8 hybridoma backpacks indicate that expression of the 260.8 epitope is not down-regulated in the presence of its cognate IgA. Moreover, epitope expression was sustained during 300 days of continuous growth in vitro and during 14 months of monocolonization of wild-type C57BL/6J gnotobiotic mice. Together, our results indicate that bacterial surface epitopes may vary widely within a species both in terms of their genetic conservation and expression. Our findings emphasize how a human gut symbiont can maintain highly conserved, species-specific genetic loci that encode immunogenic epitopes that, despite being targeted by host IgA and important for fitness in vitro and in vivo, are not transcriptionally suppressed in the presence of a specific IgA or negatively selected against during adaptation to a mammalian gut. Finally, the approach provides a means for creating new clinical diagnostic reagents.
EXPERIMENTAL PROCEDURES
Colonization of Germ-free Mice
C57BL/6J wild-type, Rag1−/−, and Myd88−/− mice were obtained from The Jackson Laboratory. Mice were rederived as germ-free and maintained in plastic, flexible film gnotobiotic isolators under a strict 12-h light cycle. Animals were fed an autoclaved, polysaccharide-rich, standard rodent chow diet (B&K Universal, East Yorkshire, UK) ad libitum. All experiments involving mice were performed using protocols approved by the Washington University Animal Studies Committee.
B. thetaiotaomicron strain VPI-5482 was harvested from stationary phase cultures that had been grown in TYG medium (1% tryptone, 0.5% yeast extract, 0.2% glucose) supplemented with 100 mm potassium phosphate buffer, pH 7.2, 4.1 mm cysteine, 200 mm histidine, 6.8 mm CaCl2, 140 nm FeSO4, 81 mm MgSO4, 4.8 mm NaHCO3, 1.4 mm NaCl, 1.9 mm hematin, and 5.8 mm vitamin K3. 108 cfu were gavaged (in 100 μl of TYG medium) into 6–8-week-old male germ-free recipient animals.
Generating the 260.8 Hybridoma Cell Line from Lamina Propria Fusions
Wild-type C57BL/6J gnotobiotic mice were killed 2 weeks after gavage with just wild-type B. thetaiotaomicron VPI-5482. The small intestine from a given mouse was removed immediately and flushed with Dulbecco's modified Eagle's medium (DMEM) containing penicillin (100 units/ml) and streptomycin (100 μg/ml). The excised small intestine was opened along its cephalocaudal axis and minced into small (∼2–5-mm) pieces. The pieces were incubated for 15 min at 37 °C in 50 ml of Hanks' buffered salt solution containing 5 mm EDTA to remove epithelial cells and intraepithelial lymphocytes. Tissue fragments were placed in DMEM containing collagenase (100 units/ml; Sigma) and Dispase (0.5 unit/ml; Fisher) and incubated for 2 h at 37 °C with vigorous shaking for 30 s every 30 min over a 2-h period. Released cells were separated from the remaining tissue fragments by sedimentation on the bench top for 1 min, and the resulting cell suspension (supernatant fraction) was passed through a Nytex filter (70-μm pore diameter; BD Biosciences). Cells in the filtrate were then combined with the myeloma fusion partner (P3X63.Ag8). The combined population was washed three times in serum-free DMEM, and cells were fused by adding PEG1500 (50%, w/v; Roche Applied Science; Refs. (26 and 27). Cloned IgA-producing lines were identified using procedures detailed in a previous publication (23).
Purification of mAb 260.8 and mAb 225.4
Hybridoma cell lines were grown in bioreactors, and their monoclonal antibodies were subsequently purified from the bioreactor supernatant through a two-step process that used standard ammonium sulfate precipitations followed by hydroxyapatite column chromatography (Leinco Technologies Inc., St. Louis, MO).
Enzyme-linked Immunosorbent Assay (ELISA)
Cultured bacterial strains, bacterial cell lysates or capsular polysaccharides prepared from the bacterial strains, or extracts of cecal contents harvested from gnotobiotic mice colonized with B. thetaiotaomicron VPI-5482 (for details, see below) were assayed by ELISA in 96-well plates (Nunc Maxisorp, Nalge Nunc, Rochester, NY). Samples were diluted in sodium bicarbonate coating buffer (15 mm sodium carbonate, 35 mm sodium bicarbonate, and 3 mm sodium azide, pH 8.5). All assays were performed either at 4 °C overnight or at room temperature for 2 h. mAb 260.8 or 225.4 was added (10 μg/ml in PBS containing 1% BSA; 50 μl/well) followed by horseradish peroxidase (HRP)-conjugated goat anti-mouse IgA (Southern Biotech; 1:1,000 dilution, 50 μl/well). All ELISAs were developed using 1 mm 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Roche Applied Science) in citrate buffer (100 mm citric acid, 50 mm sodium phosphate, pH 4.2) containing 0.03% H2O2. Reactions were read in an ELISA plate reader (ThermoMax, Molecular Devices) at 405 nm.
Bacterial Lysates
Lysates were generated by suspending 1010 cfu of bacteria in 0.5 ml of sterile PBS and sonicating the mixture in a Misonix XL-2020 sonicator (10 min at a setting of 10). This mixture was then diluted 1:1,000 in bicarbonate coating buffer, and a 50-μl aliquot was added to each well of an ELISA plate. Because these lysates contained capsular antigens, membrane and cell wall antigens, and intracellular antigens, they were used for initial screening of hybridoma fusions.
Cell Surface Glycans
Surface glycans (capsular polysaccharides and LPS) were isolated from B. thetaiotaomicron VPI-5482 cells that had been harvested after they entered stationary phase in TYG medium. Briefly, bacteria were incubated at 65 °C in phenol:water (1:1) with constant stirring. Phases were separated by gravity, and the top phase was dialyzed using 10-kDa-cutoff dialysis tubing for 3 days at 4 °C against deionized water. The dialyzed material was subjected to ultracentrifugation (65,000 × g for 2 h at 4 °C) and then lyophilized. This material was suspended in sterile PBS at a concentration of 1 mg/ml and used in the ELISA after additional dilution in bicarbonate coating buffer.
Extracts of Cecal Contents from Mice Monocolonized with B. thetaiotaomicron
Cecal contents were harvested and frozen at −80 °C immediately after gnotobiotic mice were killed. After thawing, 0.5 ml of a 1:100 dilution of cecal contents (in PBS) was added to 0.5 ml of phenol, and the mixture was incubated at 65 °C for 2 h with vortexing (30 s every 30 min). Following centrifugation at 20,000 × g for 30 min at 4 °C, the upper phase was recovered, serial dilutions (in bicarbonate buffer) were made, and an ELISA was performed.
To estimate the amount of mAb 260.8-reactive epitope present in cecal contents, ELISA results were standardized to a curve constructed from serial dilutions of stationary phase B. thetaiotaomicron (grown in TYG medium) in bicarbonate buffer. This stationary phase culture was arbitrarily designated as having 1 × 109 epitope units/ml, which is equal to the number of cfu under these conditions; i.e. in TYG medium-grown B. thetaiotaomicron, 1 260.8 epitope unit is equivalent to 1 cfu.
Isolation of mAb 260.8-reactive Colonies of B. thetaiotaomicron from Human Fecal Samples
Fecal samples were collected from four monozygotic and two dizygotic twin pairs and their mothers enrolled in the Missouri Adolescent Female Twin Study (28). Procedures for recruitment of participants and protocols for sample collection and deidentification were approved by the Human Research Protection Office of Washington University. Samples were frozen at −20 °C immediately after they were produced and stored at −80 °C.
The presence of B. thetaiotaomicron was detected by PCR of fecal DNA using primers directed at variable region 2 of bacterial 16 S rRNA genes, and the resulting amplicons were subjected to multiplex 454 pyrosequencing (FLX standard chemistry) (28). An aliquot of each selected frozen fecal sample harboring B. thetaiotaomicron 16 S rRNA genes was plated on brain heart infusion (BHI) blood agar supplemented with 200 μg/ml gentamicin. Plates were incubated at 37 °C in an anaerobic chamber (atmosphere, 5% H2, 20% CO2, 75% N2; Coy Laboratory Products, Grass Lake, MI). Plates were screened for the presence of 260.8-positive colonies using a filter-based assay (29). Single 260.8-positive colonies were restreaked on fresh BHI blood agar supplemented with 200 μg/ml gentamicin, and four to eight single colonies from each individual donor microbiota were tested by ELISA for 260.8 reactivity. Colonies confirmed to be positive were inoculated into single wells of a sterile 96-well plate with each well containing 200 μl of TYG medium. After an overnight incubation under anaerobic conditions at 37 °C, 25 μl of each culture was transferred into 25 μl of coating buffer in fresh 96-well plates (Maxisorp) for ELISA. Colonies positive by ELISA were restreaked, and single colonies were picked and tested once again by an ELISA. The colony with the highest signal was then cultured in TYG broth. DNA was isolated from an aliquot of the culture using the Qiagen DNeasy kit (glycerol stocks were prepared from the remaining culture). The 16 S rRNA gene was amplified using standard PCR primers 8F and 1391R. Amplicons were purified with the Qiagen PCR cleanup kit and sequenced using the dideoxy chain termination method and the following three primers: 8F (5′-AGAGTTTGATCCTGGCTCAG), 1391R (5′-GACGGGCGGTGWGTRCA), and 515F (5′-GTGCCAGCMGCCGCGGTAA) (W = A or T; R = A or G; M = A or C).
XplorSeq (Phyloware) was used to assemble contigs and trim sequence files based on a Phred Q40 quality cutoff. Sequences were aligned using ClustalW and trimmed to the shortest common sequence length. Errors in alignment were manually corrected in MEGA. Phylogenetic trees were produced in MEGA (30) using the nearest neighbor algorithm based on “number of differences.”
Whole Genome Transposon Mutagenesis
Mutagenesis of B. thetaiotaomicron VPI-5482 was performed using pEP4351 (λpir-dependent R6K oriV; RP4 oriT; Cmr Tcr (Emr); Tn4351 mutagenesis vector (31, 32)). Mutants were isolated based on their growth on BHI blood agar plates containing erythromycin (10 μg/ml to select for transposon-containing B. thetaiotaomicron strains) and 600 μg/ml gentamicin (to select against any persistent E. coli harboring the transposon mutagenesis vector). Individual colonies were picked into 96-well plates and grown overnight in TYG medium in anaerobic jars (n = 55 plates; 4,800 mutants). Plates were replicated with one replicate frozen in 25% glycerol for later studies. Each isolate in each well was then subjected to ELISA screening with mAb 260.8 and mAb 225.4 (the latter mAb served as a positive control for the presence of B. thetaiotaomicron VPI-5482; see the Introduction).
Sites of transposon insertion in 225.4-positive, 260.8 antibody-negative colonies were identified using arbitrarily primed PCR. The arbitrarily primed PCR protocol consisted of a nested PCR with the following primers: Round 1, S3794 (5′-ATCAGTATGCTTTGTGTGTG) and either AR7 (5′-GGCCACGCGTCGACTAGTACN10GTAAT) or AR8 (5′-GGCCACGCGTCGACTAGTACN10GATGC); Round 2, ISF (5′-TCGGTTATATGTTTGCTCATCTGC) and AR2 (5′-GGCCACGCGTCGACTAGTAC). The PCR cycling conditions were 95 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min (a total of 30 cycles). Round 2 used the same conditions but for 40 cycles. The product of the second round of PCR was purified (QIAquick PCR purification kit, Qiagen) and sequenced from the right arm of the transposon into flanking chromosomal DNA using the primer IS4908S (5′-ATCCATTCAGAGTGAGAGAAAG). The results were compared with the genome sequence of B. thetaiotaomicron VPI-5482 (GenBankTM accession number NC_004663) and Tn4351 (GenBank accession number M17124); sequences aligning to both were considered to be positive hits.
A second library of transposon mutants was independently generated with a modified mariner transposon delivery vector, pSAM, that contains (i) an antibiotic resistance cassette flanked by MmeI-modified mariner inverted repeats, (ii) the himar1C9 transposase, and (iii) machinery for replication in the donor strain and for conjugative transfer (33). The MmeI sites allow cleavage at chromosomal sites located 16–17 bp from the inserted transposon so that the mutants can be mapped (see below). Approximately 4,200 colonies from this second library were picked and grown within 96-well plates containing TYG medium (n = 48 plates; each plate replicated so that frozen glycerol stocks could be archived). The contents of each well were screened by ELISA using both mAb 260.8 and mAb 225.4. DNA from each mutant strain that was mAb 260.8-negative and mAb 225.4-positive was subjected to arbitrary PCR to map the site of insertion of the transposon.
A third library was constructed using the pSAM vector: this library was clonally arrayed into 96-well plates, and the location of each mutant was defined using a bar-coding strategy (33). This mapped, arrayed library was composed of 7,155 members and was used to identify additional mutants in the region defined from libraries 1 and 2 as producing a 260.8-negative/225.4-positive phenotype. The pattern of 260.8 and 225.4 reactivity in these newly identified mutants was subsequently determined using ELISA.
B. thetaiotaomicron CPS Locus Mutants
B. thetaiotaomicron VPI-5482 contains eight distinct capsular polysaccharide synthesis loci, each composed of 15–32 genes. A panel of eight isogenic mutants, generated with the suicide vector pGERM to block expression of genes in each locus (23), was used to test the CPS epitope specificity of mAb 260.8.
Comparative Genomic Hybridization Analyses
Genomic DNA was prepared from a number of B. thetaiotaomicron isolates with a mAb 225.4-positive and a 260.8-positive (or 260.8-negative) phenotype. DNA was sonicated on ice for 10 min using a Misonix XL-2020 sonicator (10 min at a setting of 10). Fragmentation efficiency was confirmed on a 1% agarose gel (300–400-bp fragments). DNA (5 μg) was labeled using the Enzo BioArray terminal labeling kit. Samples were denatured at 95 °C for 5 min and hybridized to custom GeneChips generated from the finished B. thetaiotaomicron VPI-5482 genome sequence (24). Genomic hybridization analysis was performed using the DNA Chip Analyzer software package (dChip).
Sequencing the Genomes of B. thetaiotaomicron Strains DH3731 and BT7330-1
B. thetaiotaomicron strains DH3731 and BT7330-1 were isolated from fecal samples obtained from two unrelated normal healthy volunteers. Using an ABI 3730xl capillary sequencer, we obtained >8× >Q20 coverage of each strain's genome. In addition, >15× coverage was obtained using a 454 pyrosequencer. Hybrid assemblies of 3730xl and 454 reads yielded 205 contigs spanning 7.1 Mb for the BT3731 strain, whereas strain BT7330 was assembled into 478 contigs spanning 6.8 Mb. Predicted ORFs were annotated based on BLASTP queries of the Clusters of Orthologous Groups of proteins (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG), and NCBI non-redundant (NR) databases (genes assigned to best BLAST hit, e-value < 10−5).
Experimental in Vivo and in Vitro Evolution of B. thetaiotaomicron VPI-5482
These experiments were designed to determine whether the 260.8 epitope was expressed by B. thetaiotaomicron VPI-5482 during periods of prolonged growth in defined medium and over the course of the lifespan of monocolonized gnotobiotic mice. The in vitro experimental protocol involved culturing the bacterium anaerobically in minimal medium (100 mm KH2PO4, 15 mm NaCl, 8.5 mm (NH4)2SO4, 28 mm glucose) at 37 °C in glass culture tubes. A 10-μl aliquot of the culture was transferred every 24 h to fresh medium. This procedure was repeated 300 times in eight replicate experiments. Aliquots of the culture were frozen every 50 transfers and stored in 20% glycerol at −80 °C.
For the in vivo experiments, the same starting culture of B. thetaiotaomicron VPI-5482 (derived from a single colony) that was used for the in vitro experiment was inoculated into 8–10-week-old male germ-free C57BL/6J wild-type, Rag1−/−, or Myd88−/− mice. Gnotobiotic mice were maintained as described above. Animals were individually caged (n = 4 wild type, n = 3 Rag1−/−, and n = 2 Myd88−/−). Sterile bedding (Aspen wood shavings, NEPCO) was replaced in each cage every 7 days. Fecal samples were obtained from each animal once a week for the first month of the experiment and every 2 weeks thereafter. Fecal samples were frozen at −80 °C immediately after they had been produced by the animal. The experiment continued throughout the lifespan of each mouse (maximum, ∼26 months).
Samples from the in vivo and in vitro experiments were characterized by ELISA. Fecal samples from days 5 and 433 (representing early and late time points in the experiment) were suspended in 500 μl of PBS. Aliquots were taken for ELISA, assay of total DNA (QuantiFluor dsDNA dye, Promega), and serial dilution plating on BHI blood agar. Individual colonies from these fecal samples and from the in vitro experiments were tested for 260.8 and 225.4 epitope expression by ELISA following growth to stationary phase in TYG medium (positive control, B. thetaiotaomicron VPI-5842; negative control, B. thetaiotaomicron strain 8702).
Insertion Sequencing (INSeq) Analysis of B. thetaiotaomicron VPI-5482 Grown in Vitro and in Gnotobiotic Mice
INSeq is based on the modified mariner transposon delivery vector, pSAM, that as noted above contains an antibiotic resistance cassette flanked by MmeI-modified mariner inverted repeats, the himar1C9 transposase, and machinery for replication in the donor strain and for conjugative transfer (33). To identify transposon insertion sites, genomic regions adjacent to the transposons were amplified by linear PCR with a biotinylated primer and subsequently bound to magnetic beads where they were digested with MmeI and linkers were appended to each restriction fragment. After limited PCR amplification, fragments were sequenced with an Illumina GA II instrument (42-nucleotide unidirectional reads), and each read was used to map the location of each transposon mutant in the bacterial genome (33).
The INSeq library (∼35,000 mutant strains; 72% of predicted ORFs covered by at least one transposon) was grown in TYG medium in a continuously fed batch fermenter (two independent experiments). Aliquots were subjected to INSeq analysis using an experimental and computational pipeline described in our earlier publication (33). For in vivo experiments, the mutant library was introduced alone or in combination with three defined consortia of sequenced members of the normal human gut microbiota. Prior to gavage, community members were assembled in equal proportion under anaerobic conditions (see Ref. 33 for details).
Hybridoma Backpack Experiments in Rag1−/− Mice
260.8 hybridoma cells were grown in DMEM supplemented with penicillin (100 units/ml) and streptomycin (100 μg/ml) until they were 50–75% confluent and subsequently washed three times in pyrogen-free saline. Cells were introduced into the gnotobiotic isolators in a way that preserved sterility (34) and then injected subcutaneously on the dorsum of germ-free 6–8-week-old male C57BL/6J Rag1−/− recipients (2 × 106 cells/mouse). Ten days after implantation, each mouse with a palpable subcutaneous hybridoma was colonized with a single gavage of 108 cfu of B. thetaiotaomicron. Mice were sacrificed 10 days following colonization, and cecal contents were recovered.
GeneChip-based Transcriptional Profiling of B. thetaiotaomicron in Vivo
Cecal contents were flash frozen in liquid nitrogen immediately after their harvest from each mouse in each treatment group (n = 4 mice/group with hybridoma backpacks and controls). An aliquot (∼200 mg) was thawed in 2–3 volumes of RNAProtect (Qiagen) and centrifuged (3,000 × g for 10 min), and 500 μl of 200 mm NaCl, 20 mm EDTA was added to the resulting pellet together with 200 μl of 20% SDS and 500 μl of phenol:chloroform:isoamyl alcohol, pH 4.5 (125:24:1; Ambion). Acid-washed silica beads (Sigma; 212–300-μm diameter; 250 mg) were added to the mixture, and bacteria were lysed (Mini-Beadbeater, Biospec; “high” setting for 5 min at room temperature). Following centrifugation (13,000 × g for 3 min at 4 °C), the sample was re-extracted in phenol:chloroform:isoamyl alcohol, precipitated with 60 μl of 3 m sodium acetate, pH 5.2 plus 600 μl of cold isopropanol. RNA was purified (RNeasy kit, Qiagen). Residual genomic DNA was subsequently removed by treatment with DNAfree (Ambion), and the relative proportion of bacterial versus host RNA in each preparation was defined by quantitative RT-PCR using primers directed at B. thetaiotaomicron 16 S rRNA (forward, 5′-GGTAGTCCACACAGTAAACGATGAA; reverse, 5′-CCCGTCAATTCCTTTGAGTTTC) and mouse 18 S rRNA (forward, 5′-CATTCGAACGTCTGCCCTATC; reverse, 5′-CCTGTGCCTTCCTTGGA). The results disclosed that ≥90% of the recovered rRNA in each RNA preparation was bacterial.
GeneChip targets were generated as described (24). The cDNA product was isolated (QiaQuick spin columns, Qiagen), fragmented by treatment with DNase I (Amersham Biosciences), and biotinylated (BioArray terminal labeling kit). Standard Affymetrix protocols were used for hybridization of the cDNA targets to each B. thetaiotaomicron GeneChip (23, 24). After normalization of the data, model-based expression values were generated (perfect match-mismatch model). Signals from spiked-in control transcripts and oligo-B2 (Affymetrix) were used to assess the quality of target preparation and target hybridization, respectively. Comparisons were performed on specified GeneChip data sets to identify genes up- or down-regulated in the experimental group (E) relative to the baseline group (B). The following criteria were used to identify significantly up-regulated genes: (i) E/B > 1.5; (ii) E − B ≥ 100; (iii) E = B, p < 0.05; and (iv) called “Present” in ≥75% of “E” GeneChips.
Assaying the Effect of the mAbs on Growth and Gene Expression in Vitro
B. thetaiotaomicron VPI-5482 was cultured anaerobically in TYG medium to stationary phase, and an aliquot was diluted to an A600 of 0.01 in 5 ml of fresh TYG medium. Optical density was measured over time in three replicate monocultures using a Spectronic 20D+ spectrophotometer (Thermo).
When monocultures grown as described in the preceding paragraph had reached an A600 of ∼ 0.1, 50 μl of either PBS, mAb 225.4 (1 mg/ml in PBS), mAb 260.8 (1 mg/ml in PBS), or an equal mixture of mAb 225.4 plus mAb 260.8 (each at 1 mg/ml) were added, and tubes were resealed. An additional set of cultures was grown anaerobically in the presence of an isotype monoclonal IgA (anti-CD36; 1 mg/ml; clone JC63.1, Abcam). Prior to its use, this antibody was dialyzed at 4 °C against three washes of PBS using a 10-kDa-cutoff membrane (Slide-A-Lyzer 10K Dialysis Cassettes, Thermo) to remove glycerol and traces of azide.
Replicate cultures were harvested into 10 ml of RNAProtect (Qiagen) after 10.5 h of growth when all cultures had reached an A600 of 0.5–0.7 (note that no differences in growth kinetics were observed between cultures inoculated with antibody compared with control cultures inoculated with PBS). After a 5-min incubation at 24 °C, cells were centrifuged at 3,000 × g for 10 min. The supernatant was removed, and cell pellets were frozen at −80 °C until further use.
RNA was isolated as above with the exception that following isopropanol precipitation pellets were washed with 750 μl of ice-cold 70% ethanol and resuspended in 100 μl of nuclease-free water (Ambion). RNA was then cleaned up twice using MEGAclear (Life Technologies) and DNase-treated with TURBO DNase (Life Technologies) and Baseline-ZERO DNase (Epicentre). RNA integrity was verified by gel electrophoresis, and genomic DNA depletion was confirmed using a PCR assay directed at the 16 S rRNA gene (primers 8F and 1391R). rRNA was depleted using Ribo-Zero (Epicentre) according to the manufacturer's instructions. cDNA was synthesized using the SuperScript II kit (Life Technologies) and then sheared for 10 min using a bath sonicator (Bioruptor, Diagenode) with alternating cycles of 30 s of sonication and a 30-s pause (sonication was carried out at 4 °C). Sonicated DNA was concentrated using MinElute kits (Qiagen). Blunt end synthesis and 5′ nucleotide phosphorylation was performed using T4 polynucleotide kinase and T4 polymerase (New England Biolabs; incubation at 25 °C for 30 min). A-tails were generated using Taq polymerase (Invitrogen; incubation at 75 °C for 20 min). Illumina adapters were ligated to the DNA with T4 DNA ligase (New England Biolabs), and cDNAs were size-selected by 2% agarose gel electrophoresis. PCR of the adapter-ligated product was performed using the following cycling conditions: 98 °C for 30 s, 98 °C for 10 s, 65 °C for 30 s, and 72 °C for 45 s (a total of 18 cycles). Samples were pooled in equal proportion and sequenced using an Illumina HiSeq 2500 instrument (unidirectional 50-nucleotide reads). The resulting microbial RNA-Seq data set was analyzed as described previously (35) with the following exceptions. (i) After splitting barcodes, reads were mapped to the B. thetaiotaomicron VPI-5482 reference genome using Bowtie (36), and (ii) analysis of differential expression was performed using the DESeq2 package in R (37). Functional annotation of genes with significantly different expression after false discovery rate correction was performed using the NCBI online genome browser.
RESULTS
Phylogenetic Distribution of 260.8 Epitope
In a previous study, we showed that 24 h after gavage of adult germ-free C57BL/6J mice with 108 cfu of the sequenced B. thetaiotaomicron type strain VPI-5482 bacteria achieve a density in the small intestine and cecum that does not change significantly over the ensuing 6 months and that within 2 weeks of colonization B. thetaiotaomicron-reactive B cells are well represented in the intestine and can be captured by hybridoma fusion (23). This allowed us to immortalize a single naturally primed, bacterial epitope-specific IgA response (see the Introduction).
One of the cloned IgA-producing hybridoma cell lines, designated 260.8, produces a mAb that recognized polysaccharides extracted with hot water/phenol from wild-type B. thetaiotaomicron VPI-5482 grown in rich TYG medium and from the same strain isolated from the ceca of gnotobiotic mice 180 days after monocolonization (data not shown). To define the phylogenetic distribution of the epitope recognized by the 260.8 mAb, we surveyed two existing collections of Bacteroides isolates. The “Salyers collection” was provided by Abigail Salyers (University of Illinois, Urbana/Champaign) and contained 22 isolates recovered by the microbiology laboratories of the Virginia Polytechnic Institute (Blacksburg, VA), the Wadsworth Veterans Affairs Hospital (Los Angeles, CA), the Loyola Veterans Affairs Hospital (Maywood, IL), the Carle Hospital (Urbana-Champaign, IL), and the Marine Biology Laboratory (Woods Hole, MA) (Table 1). The “NEMC Collection” from the Special Studies Laboratory at New England Medical Center contained 78 strains isolated by clinical microbiology laboratories within the United States in 2006 (38) (Table 2). These isolates had been characterized by using a commonly used diagnostic method (API RapID ANA II System) and/or by other standard biochemical techniques. Using 16 S rRNA gene sequencing, we found that these methods had misidentified 41% of the isolates, a rate higher than reported by others (39). In addition to these pre-existing strain panels, we tested the ability of mAb 260.8 to identify B. thetaiotaomicron from individual human fecal microbiota samples. Aliquots of frozen fecal samples obtained from six healthy adult female twin pairs and their mothers were cultured on BHI blood agar plates under anaerobic conditions, and the resulting colonies were blotted onto nitrocellulose filters. These filters were subsequently incubated with mAb 260.8. Using this approach, we identified mAb 260.8-reactive isolates in 15 of the 18 donors surveyed (Table 3). Single colonies that reacted with the mAb were then cultured in TYG medium and characterized by ELISA using both 225.4 and 260.8 mAbs (see “Experimental Procedures”). DNA was extracted from these isolates; near full-length amplicons were subsequently generated from their 16 S rRNA genes by PCR and sequenced using the dideoxy chain termination method.
TABLE 1.
Type strains and Salyers culture collection
ELISA reactivity and 16 S rRNA sequence comparisons for type strains and Bacteroides isolates are shown. ID, nucleotide sequence identity; RDP, Ribosomal Database Project, and (T) indicates type strain.
| Isolate name | Percentage of VPI-5482 260.8 ELISA | Percent ID to VPI-5482 16 S rRNA | Best match in RDP database of 16 S rRNA sequences from type strains | Percent ID to best hit in RDP |
|---|---|---|---|---|
| Bacteroides stercoris 2030 | 0 | 93.1 | B. stercoris (T); ATCC 43183; X83953 | 99 |
| Bacteroides caccae 43185T | 25 | 94.5 | B. caccae (T); ATCC 43185T; X83951 | 98 |
| Bacteroides coprocola | 0 | 90.9 | B. coprocola (T); M16; AB200224 | 100 |
| Bacteroides dorei | 0 | 90.7 | B. dorei (T); JCM 13471; 175; AB242142 | 100 |
| Bacteroides finegoldii | 1 | 96.5 | B. finegoldii (T); JCM 13345; 199T; AB222699 | 100 |
| B. fragilis 9343 | 2 | 94.5 | B. fragilis (T); NCTC 9343; CR626927 | 100 |
| Bacteroides intestinalis | 2 | 93.9 | B. intestinalis (T); JCM 13265; 341; AB214328 | 100 |
| Bacteroides ovatus | 1 | 96.4 | B. ovatus (T); JCM 5824T; AB050108 | 99 |
| Bacteroides plebeius M35 | 0 | 89.6 | B. plebeius (T); M12; AB200217 | 98 |
| Bacteroides uniformis | 0 | 93.9 | B. uniformis (T); JCM 5828T; AB050110 | 98 |
| Bacteroides vulgatus | 11 | 90.8 | B. vulgatus ATCC 8482; CP000139 | 99 |
| BT0633-1 | 99 | 99.8 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| BT3164 | 90 | 99.6 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| BT3443 | 101 | 99.2 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| BT5951 | 98 | 99.8 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| BT7853 | 113 | 99.8 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| BT8669 | 105 | 99.5 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| BT8702 | 1 | 97.8 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 97 |
| BT8713 | 86 | 99.8 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| BT8736 | 104 | 97.6 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 |
| J19-343 | 103 | 99.6 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| DH3730 | 120 | 99.4 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| DH3731 | 136 | 99.7 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| DH3941 | 2 | 96.6 | B. ovatus (T); JCM 5824T; AB050108 | 98 |
| DH4072 | 3 | 97.0 | Bacteroides xylanisolvens (T); XB1A; AM230650 | 99 |
| DH4087 | 105 | 99.2 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 |
| DH4108 | 96 | 98.8 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 |
| DH4111 | 102 | 99.0 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
| WH2 | 0 | 94.9 | Bacteroides cellulosilyticus (T); CRE21; | 98 |
| WH302 | 2 | 96.9 | B. xylanisolvens (T); XB1A; AM230650 | 99 |
| WH305 | 1 | 96.8 | B. xylanisolvens (T); XB1A; AM230650 | 99 |
| WH406 | 90.8 | Bacteroides massiliensis (T); B84634; AY126616 | 99 | |
| BT-DOT2 | 103 | 99.7 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 |
TABLE 2.
NEMC culture collection
ELISA reactivity, 16 S rRNA sequence comparisons, and biochemical assignments for Bacteroides isolates in the NEMC collection are shown. ID, nucleotide sequence identity; RDP, Ribosomal Database Project, and (T) indicates type strain.
| Isolate name | Percent ID to VPI-5482 16 S rRNA | Percentage of VPI-5482 260.8 ELISA | Best match in RDP database 16 S rRNA sequences from type strains | Percent ID to best hit in RDP | Biochemical assignment |
|---|---|---|---|---|---|
| 22361 | 93.9 | 0 | B. caccae (T); ATCC 43185T; X83951 | 98 | B. caccae |
| 22547 | 94.9 | 0 | B. finegoldii (T); JCM 13345; 199T; AB222699 | 97 | B. caccae |
| 22566 | 93.7 | 4 | B. caccae (T); ATCC 43185T; X83951 | 97 | B. caccae |
| 22599 | 94.0 | 0 | B. caccae (T); ATCC 43185T; X83951 | 97 | B. caccae |
| 22866 | 99.6 | 108 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 22911 | 99.3 | 0 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 22913 | 98.7 | 93 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 22946 | 99.3 | 123 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 22953 | 98.5 | 107 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 22959 | 99.6 | 92 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 22967 | 98.9 | 77 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 22987 | 99.8 | 104 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 22998 | 93.5 | 8 | B. fragilis (T); NCTC 9343; CR626927 | 98 | B. thetaiotaomicron |
| 23041 | 98.4 | 83 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 23113 | 99.3 | 7 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23114 | 99.4 | 103 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 23149 | 99.8 | 112 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23150 | 98.4 | 110 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 23156 | 99.7 | 92 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23161 | 98.8 | 103 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 23188 | 99.3 | 99 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23190 | 99.4 | 53 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23205 | 94.5 | 3 | B. caccae (T); ATCC 43185T; X83951 | 98 | B. thetaiotaomicron |
| 23212 | 93.8 | 7 | B. fragilis (T); NCTC 9343; CR626927 | 99 | B. thetaiotaomicron |
| 23213 | 99.8 | 36 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23218 | 98.6 | 90 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 23243 | 99.6 | 99 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23251 | 97.8 | 113 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 97 | B. thetaiotaomicron |
| 23257 | 90.4 | 0 | B. dorei (T); JCM 13471; 175; AB242142 | 99 | B. thetaiotaomicron |
| 23262 | 92.7 | 1 | B. caccae (T); ATCC 43185T; X83951 | 96 | B. caccae |
| 23269 | 98.5 | 3 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 23272 | 99.8 | 79 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23367 | 99.4 | 128 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23368 | 99.5 | 110 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23369 | 96.4 | 1 | Bacteroides salyersiae (T); WAL 10018; AY608696 | 99 | B. thetaiotaomicron |
| 23411 | 94.5 | 0 | B. caccae (T); ATCC 43185T; X83951 | 98 | B. caccae |
| 23412 | 94.1 | 0 | B. caccae (T); ATCC 43185T; X83951 | 98 | B. caccae |
| 23418 | 93.8 | 0 | B. caccae (T); ATCC 43185T; X83951 | 97 | B. caccae |
| 23447 | 99.8 | 112 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23500 | 94.0 | 1 | B. caccae (T); ATCC 43185T; X83951 | 98 | B. caccae |
| 23559 | 94.0 | 2 | B. uniformis (T); JCM 5828T; AB050110 | 99 | B. uniformis |
| 23575 | 96.3 | 1 | B. ovatus (T); JCM 5824T; AB050108 | 99 | B. thetaiotaomicron |
| 23576 | 94.6 | 2 | B. fragilis (T); NCTC 9343; CR626927 | 99 | B. uniformis |
| 23580 | 99.8 | 90 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23584 | 99.8 | 96 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23596 | 98.5 | 107 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 23601 | 93.7 | 1 | B. cellulosilyticus (T); CRE21; | 98 | B. thetaiotaomicron |
| 23618 | 94.9 | 6 | B. fragilis (T); NCTC 9343; CR626927 | 99 | B. thetaiotaomicron |
| 23621 | 96.3 | 4 | B. ovatus (T); JCM 5824T; AB050108 | 99 | B. uniformis |
| 23655 | 94.5 | 3 | B. fragilis (T); NCTC 9343; CR626927 | 99 | B. thetaiotaomicron |
| 23656 | 94.9 | 3 | B. fragilis (T); NCTC 9343; CR626927 | 99 | B. ovatus |
| 23657 | 94.8 | 3 | B. fragilis (T); NCTC 9343; CR626927 | 99 | B. fragilis |
| 23658 | 94.9 | 5 | B. fragilis (T); NCTC 9343; CR626927 | 99 | B. fragilis |
| 23661 | 94.8 | 3 | B. fragilis (T); NCTC 9343; CR626927 | 99 | B. fragilis |
| 23673 | 90.4 | 3 | B. dorei (T); JCM 13471; 175; AB242142 | 96 | B. fragilis |
| 23677 | 99.0 | 2 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. fragilis |
| 23680 | 97.6 | 3 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. fragilis |
| 23681 | 95.7 | 2 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 96 | B. vulgatus |
| 23682 | 90.6 | 4 | B. dorei (T); JCM 13471; 175; AB242142 | 99 | B. vulgatus |
| 23685 | 99.4 | 89 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23688 | 97.6 | 97 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 23689 | 96.3 | 2 | B. ovatus (T); JCM 5824T; AB050108 | 99 | B. thetaiotaomicron |
| 23692 | 99.8 | 98 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. vulgatus |
| 23693 | 91.0 | 2 | B. dorei (T); JCM 13471; 175; AB242142 | 97 | B. thetaiotaomicron |
| 23698 | 91.0 | 3 | B. dorei (T); JCM 13471; 175; AB242142 | 97 | B. thetaiotaomicron |
| 23699 | 99.5 | 3 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. thetaiotaomicron |
| 23708 | 96.0 | 3 | B. ovatus (T); JCM 5824T; AB050108 | 97 | B. thetaiotaomicron |
| 23711 | 94.7 | 6 | B. cellulosilyticus (T); CRE21; | 98 | B. vulgatus |
| 23722 | 99.3 | 107 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 99 | B. vulgatus |
| 23723 | 96.3 | 2 | B. ovatus (T); JCM 5824T; AB050108 | 99 | B. thetaiotaomicron |
| 23724 | 94.0 | 3 | B. uniformis (T); JCM 5828T; AB050110 | 99 | B. thetaiotaomicron |
| 23728 | 93.5 | 3 | B. intestinalis (T); JCM 13265; 341; AB214328 | 99 | B. ovatus |
| 23730 | 98.1 | 106 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. uniformis |
| 23734 | 96.3 | 2 | B. ovatus (T); JCM 5824T; AB050108 | 99 | B. thetaiotaomicron |
| 23735 | 96.0 | 2 | B. ovatus (T); JCM 5824T; AB050108 | 99 | B. ovatus |
| 23742 | 99.0 | 92 | B. thetaiotaomicron (T); VPI-5482; AE015928 | 98 | B. thetaiotaomicron |
| 23748 | 96.4 | 1 | B. ovatus (T); JCM 5824T; AB050108 | 98 | B. thetaiotaomicron |
| 23836 | 92.2 | 4 | B. uniformis (T); JCM 5828T; AB050110 | 97 | B. thetaiotaomicron |
TABLE 3.
Twin isolates
ELISA reactivity and 16 S rRNA sequence comparisons for mAb 260.8-reactive isolates from twin pairs and mothers enrolled in the Missouri Adolescent Female Twin Study are shown. ID, nucleotide sequence identity; RDP, Ribosomal Database Project; MZ, monozygotic; DZ, dizygotic.
| Family | Family ID-relationship | Isolate ID | Percent ID to VPI-5482 16 S rRNA | Percentage of VPI-5482 260.8 ELISA | Percentage of VPI-5482 225.4 ELISA | Percent ID to best hit in RDP |
|---|---|---|---|---|---|---|
| 1 | TS19-MZ twin | APG1.1 | 98.2 | 52 | 0 | 98 |
| 1 | TS19-MZ twin | APG1.2 | 98.0 | 59 | 1 | 98 |
| 1 | TS19-MZ twin | APG4.10 | 97.8 | 78 | 0 | 98 |
| 1 | TS19-MZ twin | APG4.11 | 97.9 | 88 | 0 | 98 |
| 1 | TS19-MZ twin | APG4.12 | 97.5 | 71 | 0 | 97 |
| 1 | TS19-MZ twin | APG4.13 | 98.1 | 91 | 1 | 98 |
| 1 | TS19-MZ twin | APG4.14 | 97.4 | 57 | 3 | 97 |
| 1 | TS19-MZ twin | APG4.3 | 97.8 | 50 | 3 | 98 |
| 1 | TS19-MZ twin | APG4.5 | 97.8 | 64 | 0 | 97 |
| 1 | TS19-MZ twin | APG4.6 | 98.0 | 62 | 1 | 97 |
| 1 | TS19-MZ twin | APG4.7 | 97.8 | 79 | 0 | 97 |
| 1 | TS19-MZ twin | APG4.9 | 98.1 | 79 | 0 | 98 |
| 1 | TS20-MZ twin | APG2.1 | 99.1 | 95 | 0 | 98 |
| 1 | TS20-MZ twin | APG2.10 | 99.1 | 92 | 0 | 99 |
| 1 | TS20-MZ twin | APG2.11 | 99.3 | 91 | 0 | 99 |
| 1 | TS20-MZ twin | APG2.13 | 97.8 | 105 | 0 | 97 |
| 1 | TS20-MZ twin | APG2.2 | 99.8 | 75 | 0 | 99 |
| 1 | TS20-MZ twin | APG2.3 | 98.9 | 91 | 0 | 98 |
| 1 | TS20-MZ twin | APG2.4 | 99.2 | 91 | 0 | 98 |
| 1 | TS20-MZ twin | APG2.5 | 99.8 | 99 | 0 | 99 |
| 1 | TS20-MZ twin | APG2.6 | 98.6 | 99 | 1 | 97 |
| 1 | TS20-MZ twin | APG2.7 | 99.8 | 88 | 0 | 99 |
| 1 | TS20-MZ twin | APG2.8 | 99.7 | 86 | 0 | 99 |
| 1 | TS20-MZ twin | APG2.9 | 98.8 | 93 | 0 | 97 |
| 1 | TS21-mother | APG2.2 | 98.2 | 106 | 0 | 97 |
| 1 | TS21-mother | APG3.3 | 98.6 | 112 | 0 | 98 |
| 1 | TS21-mother | APG5.6 | 98.6 | 110 | 0 | 98 |
| 1 | TS21-mother | APG5.7 | 98.3 | 106 | 0 | 97 |
| 1 | TS21-mother | APG5.8 | 98.3 | 91 | 0 | 97 |
| 1 | TS21-mother | APG5.9 | 98.6 | 106 | 0 | 98 |
| 1 | TS21-mother | APG6.10 | 96.9 | 86 | 0 | 96 |
| 2 | TS130-MZ twin | APG1.2 | 99.6 | 50 | 0 | 99 |
| 2 | TS130-MZ twin | APG1.3 | 99.6 | 67 | 0 | 99 |
| 2 | TS130-MZ twin | APG1.4 | 99.6 | 53 | 0 | 99 |
| 2 | TS130-MZ twin | APG1.6 | 99.6 | 40 | 0 | 99 |
| 2 | TS131-MZ twin | APG1.1 | 99.0 | 81 | 0 | 98 |
| 2 | TS131-MZ twin | APG1.2 | 98.8 | 85 | 0 | 98 |
| 2 | TS131-MZ twin | APG1.3 | 98.9 | 78 | 0 | 98 |
| 2 | TS131-MZ twin | APG1.4 | 99.0 | 34 | 0 | 98 |
| 2 | TS131-MZ twin | APG1.5 | 99.2 | 105 | 0 | 99 |
| 2 | TS131-MZ twin | APG1.6 | 98.9 | 36 | 0 | 98 |
| 2 | TS131-MZ twin | APG1.7 | 99.0 | 38 | 0 | 98 |
| 2 | TS131-MZ twin | APG1.8 | 99.0 | 63 | 0 | 98 |
| 2 | TS131-MZ twin | APG1.9 | 99.1 | 38 | 0 | 99 |
| 2 | TS132-mother | APG1.1 | 99.0 | 103 | 0 | 98 |
| 2 | TS132-mother | APG1.10 | 99.1 | 79 | 0 | 99 |
| 2 | TS132-mother | APG1.2 | 99.1 | 106 | 0 | 98 |
| 2 | TS132-mother | APG1.3 | 99.3 | 117 | 0 | 99 |
| 2 | TS132-mother | APG1.4 | 99.1 | 118 | 0 | 98 |
| 2 | TS132-mother | APG1.5 | 99.1 | 111 | 0 | 99 |
| 2 | TS132-mother | APG1.6 | 99.0 | 93 | 0 | 99 |
| 2 | TS132-mother | APG1.7 | 99.1 | 117 | 1 | 99 |
| 2 | TS132-mother | APG1.8 | 99.2 | 108 | 1 | 99 |
| 2 | TS132-mother | APG1.9 | 98.9 | 114 | 0 | 98 |
| 3 | TS118-DZ twin | APG1.1 | 99.7 | 127 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.10 | 99.5 | 116 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.11 | 99.9 | 111 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.12 | 99.9 | 66 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.14 | 99.7 | 99 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.15 | 99.8 | 91 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.16 | 99.7 | 117 | 1 | 99 |
| 3 | TS118-DZ twin | APG1.2 | 99.7 | 84 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.3 | 99.8 | 80 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.5 | 99.9 | 91 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.6 | 99.7 | 125 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.7 | 99.9 | 99 | 1 | 99 |
| 3 | TS118-DZ twin | APG1.8 | 99.7 | 110 | 0 | 99 |
| 3 | TS118-DZ twin | APG1.9 | 99.5 | 112 | 1 | 99 |
| 3 | TS119-DZ twin | APG1.1 | 99.1 | 71 | 4 | 98 |
| 3 | TS119-DZ twin | APG1.3 | 99.7 | 60 | 0 | 99 |
| 3 | TS120-mother | APG1.1 | 99.4 | 85 | 1 | 99 |
| 3 | TS120-mother | APG1.2 | 97.3 | 88 | 2 | 96 |
| 4 | TS1-MZ twin | APG1.01 | 99.7 | 86 | 99 | |
| 4 | TS1-MZ twin | APG1.02 | 99.6 | 83 | 1 | 99 |
| 4 | TS1-MZ twin | APG1.03 | 99.7 | 92 | 0 | 99 |
| 4 | TS1-MZ twin | APG1.04 | 99.8 | 75 | 0 | 99 |
| 4 | TS1-MZ twin | APG1.05 | 99.7 | 89 | 0 | 99 |
| 4 | TS1-MZ twin | APG1.06 | 99.6 | 105 | 0 | 99 |
| 4 | TS3-mother | APG1.03 | 99.8 | 102 | 0 | 99 |
| 4 | TS3-mother | APG1.05 | 99.8 | 16 | 0 | 99 |
| 4 | TS3-mother | APG1.06 | 99.8 | 96 | 0 | 99 |
| 4 | TS3-mother | APG1.07 | 99.8 | 34 | 0 | 99 |
| 5 | TS61-DZ twin | APG1.1 | 99.6 | 62 | 21 | 99 |
| 5 | TS61-DZ twin | APG1.10 | 99.6 | 123 | 74 | 99 |
| 5 | TS61-DZ twin | APG1.4 | 99.6 | 95 | 14 | 99 |
| 5 | TS61-DZ twin | APG1.5 | 99.6 | 122 | 178 | 99 |
| 5 | TS61-DZ twin | APG1.6 | 99.6 | 98 | 17 | 99 |
| 5 | TS61-DZ twin | APG1.7 | 99.4 | 119 | 125 | 99 |
| 5 | TS61-DZ twin | APG1.8 | 99.6 | 115 | 15 | 99 |
| 5 | TS61-DZ twin | APG1.9 | 99.6 | 99 | 101 | 99 |
| 5 | TS62-mother | APG1.1 | 99.8 | 41 | 0 | 99 |
| 5 | TS62-mother | APG1.2 | 99.8 | 71 | 0 | 99 |
| 6 | TS4-MZ twin | D6G4 | 94.5 | 29 | 0 | 99 |
| 6 | TS6-mother | A7 | 99.7 | 88 | 0 | 98 |
Combining the results of the ELISA and 16 S rRNA gene sequencing revealed that the 260.8 mAb is an excellent diagnostic reagent with strong discriminatory power for B. thetaiotaomicron. Receiver operating characteristic curves generated by plotting the fraction of true positives among all positives (true positive rate) versus the fraction of false positives of the negatives (false positive rate) demonstrated an area under the curve of 0.934 for distinguishing isolates whose 16 S rRNA genes had >97% nucleotide sequence identity with the 16 S rRNA gene in the finished genome of the VPI-5482 type strain from isolates whose 16 S rRNA genes had <97% nucleotide sequence identity (Fig. 1, A and B).
FIGURE 1.
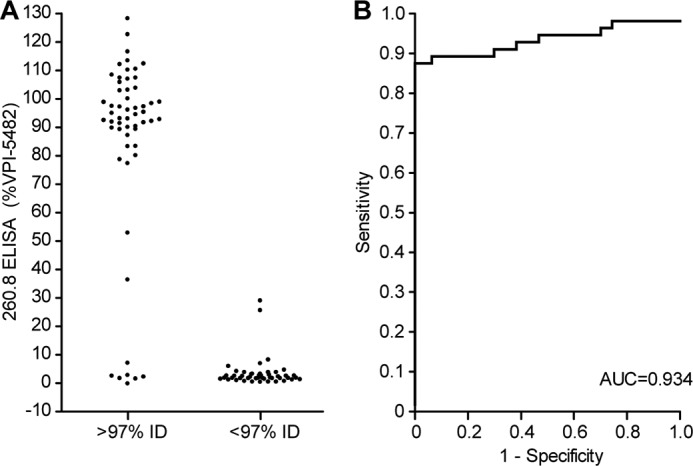
Sensitivity and specificity of mAb 260.8 for correct identification of B. thetaiotaomicron. A, ELISA reactivity of B. thetaiotaomicron isolates. A threshold of ≥97% nucleotide sequence identity (% ID) to the 16 S rRNA gene in the type strain VPI-5482 was used to define species membership and to characterize the species specificity of the mAb 260.8 ELISA. B, receiver-operator characteristics of the 260.8 ELISA signal (absorbance measured at 405 nm). AUC, area under the curve.
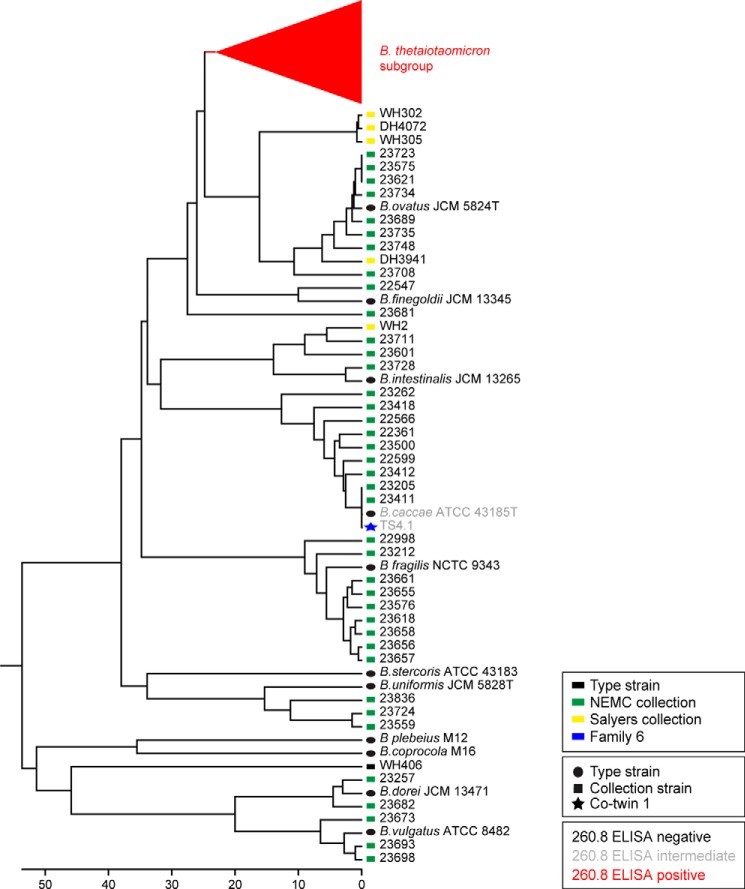
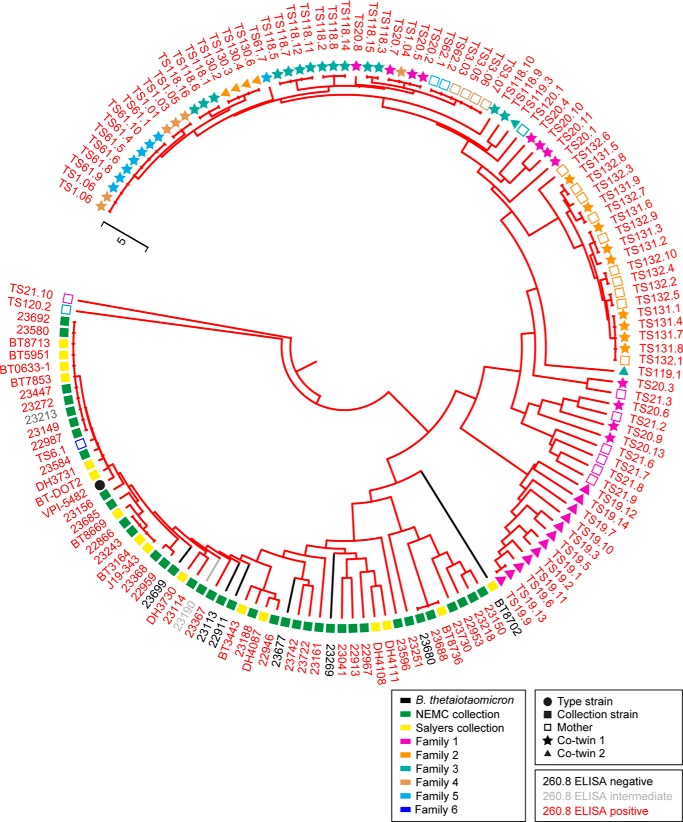
Isolates from the readily cultured component of fecal microbiota samples that were initially identified as positive in the 260.8-based dot blot assay and confirmed by ELISA had a similar distribution of 16 S rRNA sequence identity to strain VPI-5482 as isolates in the previously established Salyers and NEMC collections (see Tables 1 and 2, and Figs. 2 and 3 for 16 S rRNA-based phylogenetic trees of the isolates). Each of the isolates identified by dot blot assay as being positive (260.8-reactive) after three rounds of streaking for clonality was antigen-positive in the ELISA, and 93 of 94 were confirmed to be B. thetaiotaomicron by 16 S rRNA sequencing (Table 3). We subsequently screened this collection of 260.8-positive isolates for mAb 225.4 reactivity and found that all strains from all but one of the donors (TS61) were 225.4-negative (Table 3).
FIGURE 2.
16 S rRNA-based phylogenetic tree of human gut Bacteroides isolates and their reactivity with mAb 260.8 as measured by ELISA. Icons accompanying the isolate name are color-coded according to the source (i.e. type strain, culture collection, or the human family with twins from which the isolate was derived). Branch and text color reflect the ELISA reactivity.
FIGURE 3.
16 S rRNA-based phylogenetic tree of human gut B. thetaiotaomicron isolates and their reactivity with mAb 260.8 as measured by ELISA. Shown is an expansion of the B. thetaiotaomicron subgroup depicted in Fig. 2. Icons accompanying the isolate name are color-coded according to the source (i.e. type strain, culture collection, or the human family with twins from which the isolate was derived). Branch and isolate name color reflect the ELISA reactivity.
Identification of a Conserved Locus in the B. thetaiotaomicron Genome Required for Production of the 260.8 Epitope
To determine whether the 260.8 mAb, like the previously characterized 225.4 mAb, recognizes a capsular polysaccharide epitope, cultures of wild-type B. thetaiotaomicron VPI-5482 and eight isogenic mutant strains, each harboring a polar insertion of a suicide vector in the first gene of each of the organism's eight capsular polysaccharide synthesis loci, were extracted with hot phenol/water (which retains capsular polysaccharides and LPS), and the extracts were examined by ELISA. Levels of this epitope were not significantly different in the wild-type strain grown to stationary phase in TYG medium compared with any member of the panel of isogenic mutant strains (each strain assayed in triplicate; data not shown).
After eliminating these CPS loci as a source of the 260.8 epitope, we screened three libraries of isogenic transposon mutants of B. thetaiotaomicron VPI-5482 to identify genes required for its expression (see “Experimental Procedures” for details of how these libraries were generated and characterized). The results allowed us to attribute 260.8 epitope production to a 19-gene region spanning BT3362–BT3380 (Fig. 4). The genes in this locus are predicted to encode glycosyltransferases belonging to carbohydrate-active enzyme glycosyltransferase families 2, 4, and 14. Mutants in BT3362–BT3364 were not observed in any of the three transposon mutant libraries, suggesting that these genes may be essential for in vitro growth and survival under the conditions used to create the transposon library. We have provisionally included them in the 260.8 locus on the basis of RNA-Seq data indicating that BT3362–BT3364 are co-transcribed with the downstream genes that we observed were required for epitope expression (35). BT3362 and BT3363 are homologs of WaaC/YibC (heptosyltransferase) and LpsA involved in 3-deoxy-d-manno-octulosonic acid-lipid A modification, suggesting that at least some of the downstream glycosyltransferases are involved in LPS O-antigen polysaccharide synthesis and modification in B. thetaiotaomicron. These findings are consistent with the observation that mAb 260.8 reacts with hot water/phenol-extracted material that contains LPS. None of the transposon mutants had rough colony morphology, indicating that there is likely redundancy in the capacity for LPS O-antigen biosynthesis in the B. thetaiotaomicron genome.
FIGURE 4.

The 260.8 epitope is produced by a locus spanning BT3362–BT3380 of the B. thetaiotaomicron VPI-5482 genome. Annotation and predicted functions of genes in the locus are shown. Sites of insertion of transposons (Tn) from the three mutant libraries that are associated with loss of reactivity to mAb 260.8 in the ELISA are indicated by color-coded vertical lines.
To further define which genes in the BT3362–BT3380 locus are conserved and required for producing the epitope recognized by mAb 260.8, we characterized 17 B. thetaiotaomicron strains using a combination of comparative genomic hybridization with custom GeneChips that contained probe sets for >98% of the 4,779 known or predicted genes in the type strain and by generating deep draft assemblies of the genomes of two 260.8-positive human gut-derived isolates from the Salyers collection (strains DH3731 and BT7330-1; N50 contig lengths = 97,716 and 217,107 bp, respectively (Fig. 5A)). Comparison with ELISA data from the same isolates allowed us to determine that several genes in the locus are not required for expression of the 260.8 epitope. For example, several strains that lacked BT3369–BT3371, a region that encodes three putative glycosyltransferases, retained ELISA reactivity. Comparative genomic hybridization also established that the one 260.8-negative strain (8702) was missing all but seven of the conserved genes from the BT3362–BT3380 locus. In contrast, the CPS4 locus responsible for production of the 225.4 epitope was only conserved in the type strain, consistent with the observed pattern of 225.4 ELISA reactivity (Fig. 5B).
FIGURE 5.
Comparative genomic hybridization and analysis of draft genome assemblies of B. thetaiotaomicron isolates reveal that the BT3362–BT3380 locus is well conserved. Genomic DNA from the indicated isolates was hybridized to B. thetaiotaomicron GeneChips. A, isolate strain names colored red are 260.8-positive in the ELISA, whereas those colored black are 260.8-negative. Genes called “present” are shown in gray, whereas those called “absent” are white. The locus required for production of the 260.8 epitope is compared between the VPI-5482 type strain and two 260.8-positive B. thetaiotaomicron isolates (DH3731 and BT7330-1). Percent nucleotide sequence identities (% ID) for ORFs in the draft genomes are expressed relative to the type strain. Data obtained from other Bacteroides species are shown for reference. B, comparable comparative genomic hybridization data for the CPS4 locus that is the source of the epitope recognized by mAb 225.4.
Role of the 260.8 locus in bacterial fitness
Examining transposon mutants under different growth conditions allowed us to define the impact of the genes that are essential for 260.8 epitope formation on fitness in vitro and in vivo. A library of 34,554 mutants of B. thetaiotaomicron generated with a modified mariner transposon that contains MmeI sites at its flanking inverted repeat sites had allowed us to use a method known as INSeq to quantify the proportional representation of mutants in the library before and after a series of selections were applied (33). These selections included (i) growth during early and late log phase in batch fermenters containing TYG medium; (ii) introduction into wild-type, Rag1−/−, or Myd88−/− C57BL/6J mice without other gut bacterial species for 14 days (n = 4–5 mice/group); and (iii) introduction into wild-type mice together with one of three types of defined communities of sequenced members of the human gut microbiota. These communities were assembled from strains representing three bacterial phyla that numerically dominate the human distal gut microbiota and included a community of six other Bacteroidetes, a community composed of eight members of the Firmicutes and Actinobacteria, and a third community consisting of all 14 of these representatives (see Table S7 of Ref. 33 for the species composition of these communities; note that all communities also contained the B. thetaiotaomicron INSeq mutant library).
Analysis of these previously generated INSeq data sets (33) now enabled us to identify BT3362–BT3368 as well as BT3378–BT3380 in the BT3362–BT3380 260.8 locus as required for fitness during growth in TYG medium in a continuously fed batch fermenter and under all in vivo conditions tested. Mutants in BT3373–BT3376 (encoding a putative spermidine N1-acetyltransferase, carbamoyl-phosphate synthase, aspartate aminotransferase, and a nucleotide-sugar transaminase; Fig. 4) transcribed from the opposite strand as the genes required for 260.8 expression exhibited fitness defects in gnotobiotic mice that were robust to wild-type, Rag1−/−, and Myd88−/− host genotypes but sensitive to microbial community context in wild-type C57BL/6J hosts (mutants were proportionally underrepresented in the Firmicutes/Actinobacteria and full communities compared with the Bacteroidetes-only community) (Fig. 6).
FIGURE 6.
INSeq analysis of the importance of genes spanning BT3362–BT3380 to the fitness of B. thetaiotaomicron VPI-5482 under different environmental conditions. The frequency (relative abundances) of mutants in the input INSeq library (left column) is compared with their frequency in output communities recovered (i) after 15 or 45 h of growth in TYG medium in a continuously fed batch fermenter; (ii) 10 days after monocolonization of C57BL/6J wild-type, Rag1−/−, or Myd88−/− mice; or (iii) 10 days after colonization of mice with a six-member community of human gut Bacteroidetes, an eight-member community of human gut Firmicutes and Actinobacteria, or a 14-member community composed of the Bacteroidetes, Firmicutes, and Actinobacteria. Note that data for BT3381–BT3384, which are not part of the locus required for 260.8 epitope production, are included as reference controls.
To validate our findings from the INSeq data set and to further assess the impact of the 260.8 epitope (genes) on fitness in vivo, we performed a competition experiment in wild-type C57BL/6J mice. A 1:1 mixture of wild-type VPI-5482 and an isogenic mutant strain (3.1.G12) containing a transposon disruption of BT3366 (yibD glycosyltransferase) were introduced into germ-free mice by gavage. After 10 days, we observed a 40-fold lower level of the mutant strain in the cecal contents of these animals (p < 0.0001, unpaired t test; n = 8 mice per group; Fig. 7).
FIGURE 7.
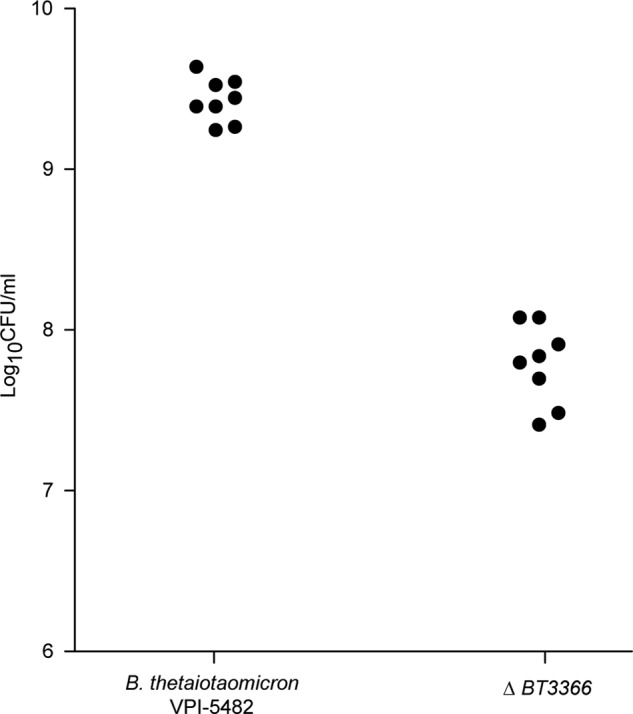
In vivo competition assay between wild-type B. thetaiotaomicron and an isogenic strain lacking BT3366. Wild-type germ-free adult male C57BL/6J mice (n = 8) received a 1:1 mixture of wild-type VPI-5482 and an isogenic mutant strain containing a transposon disruption of BT3366. Animals were sacrificed 10 days later. Wild-type and mutant stains were quantified by replicate plating on selective medium (BHI blood agar with or without erythromycin). Each filled circle represents the results from a given animal.
Expression of the 260.8 and 225.4 Epitopes during Growth in Vivo and in Vitro
To determine whether expression of the 260.8 and 225.4 epitopes was stable during prolonged periods of growth in vitro and in vivo, two experiments were performed. In the first experiment, wild-type B. thetaiotaomicron VPI-5482 was inoculated into wild-type, Myd88−/−, and Rag1−/− mice. Animals were caged individually (n = 2 animals/genotype/isolator), and fecal samples were collected regularly throughout the duration of their natural lives with no further intervention. In the second experiment, a separate aliquot of the gavage sample was used to inoculate eight individual broth cultures that were subsequently passaged daily over the course of 300 days of in vitro growth.
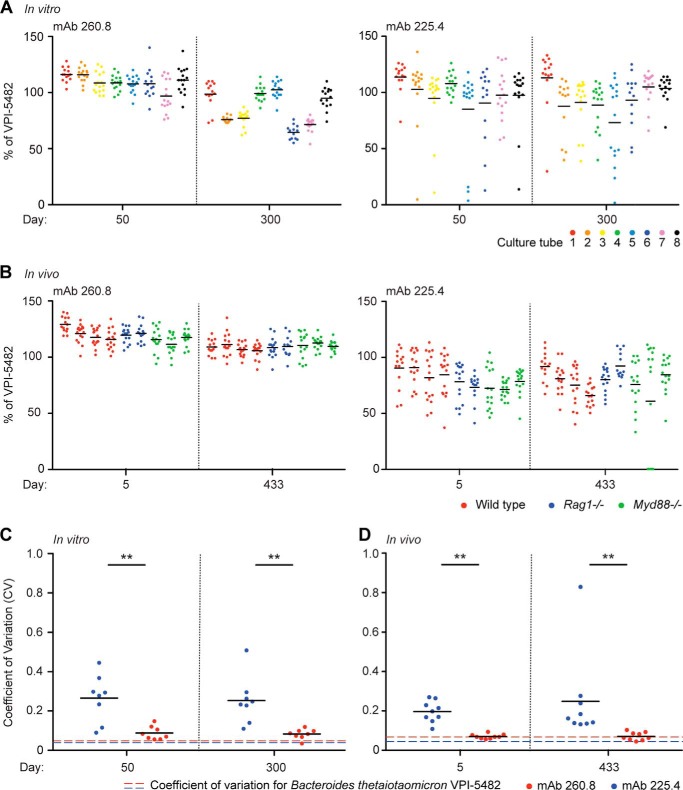
ELISA of the intact fecal bacterial communities sampled at early and late time points from both experiments revealed no change in overall levels of 225.4 or 260.8 epitope expression (data not shown). Because phase variable expression of the capsular polysaccharide has been demonstrated in a related Bacteroides spp. (Bacteroides fragilis; Ref. 40) and hypothesized for several others (41), we examined the extent to which expression of these epitopes varied among members of the population during prolonged growth in vivo and in vitro. Thus, for each sample, 10 or more colonies were isolated from early and late time point samples (in vivo, days 5 and 433; in vitro, days 50 and 300). Colonies were picked into TYG medium, grown to stationary phase, and evaluated by ELISA. These experiments allowed us to conclude that the 260.8 epitope is expressed at a relatively uniform level between isolates from a given sample both in vitro and in vivo (Fig. 8). In contrast, individual isolates varied widely with respect to expression of the 225.4 epitope at all time points for nearly all in vitro and in vivo samples (Fig. 8). These patterns of ELISA reactivity support a model in which the expression of the 260.8 epitope is relatively stable across isolates in a given sample, whereas expression of the 225.4 epitope is under phase variable control. Consistent with this model, homologs of UpxY and UpxZ, which help mediate phase variation of capsular polysaccharides in B. fragilis (42), are present in the 225.4 locus but not in the 260.8 locus.
FIGURE 8.
Expression of the 260.8 and 225.4 epitopes in monocolonized gnotobiotic mice and continuously passaged cultures. A, samples taken at the indicated time points from B. thetaiotaomicron VPI-5482 monocultures that were passaged daily for 300 days in eight separate culture tubes containing minimal medium. B, fecal samples collected from wild-type, Rag1−/−, or Myd88−/− gnotobiotic mice 5 and 433 days after monocolonization with B. thetaiotaomicron were streaked onto TYG agar. Individual colonies were then picked into TYG broth, grown to late stationary phase, and analyzed by ELISA. Epitope expression was normalized to a late stationary phase culture of the type strain. Each colored circle represents results from a single colony. Horizontal lines represent means for each sample type at the time point surveyed. C and D, coefficients of variation (CV) for the group of samples obtained at the indicated time points during the in vitro and in vivo experiments. **, p < 0.01 for the comparison between mAb 225.4 and mAb 260.8 (Wilcoxon rank sum test).
The Effects of the 260.8 IgA on B. thetaiotaomicron in Vivo
To examine the impact of the 260.8 IgA on B. thetaiotaomicron in vivo, adult male germ-free Rag1−/− mice were injected subcutaneously with 260.8 hybridoma cells (n = 11 animals). Germ-free Rag1−/− and wild-type C57BL/6J controls (n = 5/group) did not receive hybridoma cells. After 10 days, all groups of animals were colonized with a single gavage of wild-type B. thetaiotaomicron VPI-5482 and then sacrificed 10 days later. At the time of sacrifice, IgA was detected in the serum and cecal contents of animals with the hybridoma backpack and their wild-type counterparts (cecal concentrations, 124 ± 18 and 372 ± 65 μg/ml, respectively; Fig. 9A) but not in Rag1−/− (no backpack) controls. No statistically significant differences in bacterial density were observed among the treatment groups as measured by quantifying cfu obtained after culturing cecal contents on BHI blood agar plates (data not shown). ELISA of hot phenol/water-extracted cecal contents indicated no significant change in 260.8 epitope levels (or the control 225.4 epitope in the absence of a circulating IgA that recognizes it) in Rag1−/− (mAb 260.8) backpack mice compared with Rag1−/− (no backpack) or wild-type controls (Fig. 9B).
FIGURE 9.

In vivo analysis of the effects of mAb260.8 production on expression of its epitope in gnotobiotic mice monocolonized with B. thetaiotaomicron VPI-5482. A, Rag1−/− mice were injected with 260.8 hybridoma cells. Ten days later they were gavaged with B. thetaiotaomicron. At the time of sacrifice, 10 days after gavage, IgA was measured in serum and in cecal contents. B, 260.8 epitope expression in cecal contents of monocolonized Rag1−/− mice injected subcutaneously with 260.8 hybridoma cells (hybridoma backpack) and wild-type and Rag1−/− C57BL/6 mice without hybridoma cells. Cecal contents were extracted with hot phenol/water, and epitope levels were measured by ELISA using the 260.8 IgA or 225.4 IgA mAb. BT, B. thetaiotaomicron.
Whole genome transcriptional profiling using B. thetaiotaomicron GeneChips and RNA prepared from cecal contents harvested at the time of sacrifice revealed a total of 63 bacterial genes whose levels of expression satisfied our criteria (see “Experimental Procedures”) as being significantly different in Rag1−/− (mAb 260.8) backpack and wild-type C57BL/6J mice versus Rag1−/− (no backpack) controls (Fig. 10). Unlike the 225.4 epitope associated with CPS4 (BT1338–BT1358), there was not a significant difference in expression of genes in the BT3362–BT3380 locus that specifies the 260.8 epitope. Like previously reported Rag1−/− mice with 225.4 hybridoma backpacks, we observed significantly lower levels of expression of bacterial oxidative stress genes (Fig. 10) as well as genes encoding nitrite reductase (BT1415 and BT1416). In this earlier study, we showed that bacterial nitrite reductase expression directly correlates with host inducible NOS expression (23). Although we did not directly measure inducible NOS levels in the current study, our results are consistent with the notion that the 260.8 mAb-bacterium interaction leads to a more muted innate immune response. Finally, unsupervised clustering of B. thetaiotaomicron GeneChip data sets showed that the monocolonized Rag1−/− (260.8) backpack mice cluster with monocolonized wild-type animals, indicating that the effects of the monospecific IgA immune system on the bacterium shares similarities with the effects of a far less restricted polyclonal immune response to colonization in wild-type gnotobiotic animals.
FIGURE 10.
GeneChip profiling of the effects of mAb 260.8 on B. thetaiotaomicron gene expression. Hierarchical clustering of GeneChip data sets generated from B. thetaiotaomicron VPI-5482 harvested from the ceca of Rag1−/− mice with and without 260.8 hybridoma backpacks (BP) and from wild-type C57BL/6J controls is shown. Genes whose expression was defined as significantly different in either direction (increased versus decreased; see “Experimental Procedures”) between Rag1−/− (mAb 260.8) backpack and Rag1−/− (no backpack) and between Rag1−/− and wild-type C57BL/6J mice are shown. Duplicated gene names (i.e. Bt-tRNA5 and BT2157) reflect the presence of these probe sets at multiple sites on the GeneChip where they served as internal controls. Genes involved in nitrite reduction are highlighted in red.
The Effects of the 260.8 and 225.4 IgA on B. thetaiotaomicron in Vitro
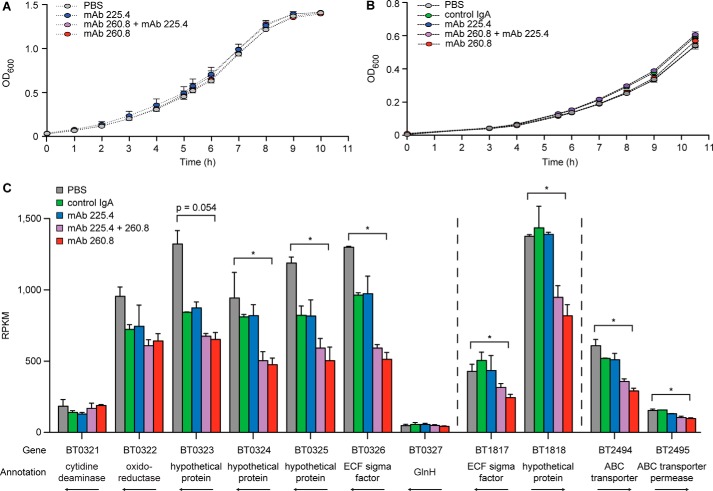
To test whether the changes in gene expression that we observed in vivo were directly mediated by antibody binding or involved host-dependent pathways, we cultured B. thetaiotaomicron VPI-5482 in the presence of (i) mAb 260.8 alone, (ii) mAb 225.4 alone, (iii) an equal mixture of the two antibodies, or (iv) a mouse isotype control IgA that does not bind B. thetaiotaomicron. There were no differences in the growth characteristics of B. thetaiotaomicron in any of the four treatment conditions compared with control cultures inoculated with PBS alone (Fig. 11, A and B). We subsequently performed microbial RNA-Seq of midlog phase cultures. The results disclosed that the changes in gene expression we observed in vivo were not recapitulated by antibody binding in vitro. In vitro binding of the 260.8 or the 225.4 mAb produced no change in expression of the loci responsible for production of their epitopes. No genes were up-regulated by the addition of IgA. Modest down-regulation in expression of three loci (BT0323–BT0326, BT1817-BT1818, and BT2494-BT2495) was observed in cultures grown in the presence of mAb 260.8. BT0323–BT0326 and BT1817-BT1818 each specify an extracytoplasmic function-type σ factor, a group of transcription factors involved in transduction of signals from the extracellular environment (43). INSeq analysis revealed that genes encoding these two extracytoplasmic function σ factors (BT1817 and BT0326) and a hypothetical protein (BT0323) were not present above the limit of detection used to identify membership in our input INSeq library (indicating that they are presumably essential for growth under the in vitro conditions used to generate the library), whereas two hypothetical proteins (BT0324 and BT1818) were significant fitness determinants in hosts with or without deficient innate or adaptive immune systems.
FIGURE 11.
Assaying the effects of mAb 260.8 and mAb 225.4 on in vitro growth of B. thetaiotaomicron and its pattern of gene expression. A, B. thetaiotaomicron VPI-5482 was grown in TYG medium in the presence of mAb 225.4, mAb 260.8, an equal amount of both mAbs, or the antibody buffer (PBS) alone. B, cultures grown under the same conditions as those used in A that were used for microbial RNA-Seq (note that an additional isotype control has been included). Mean values ±S.D. (error bars) are shown in A and B. C, microbial RNA-Seq results. Results are expressed as reads per kilobase per million (RPKM) for three loci that were differentially expressed in the presence of mAb 260.8 (mean values ±S.E. (error bars) are plotted). *, p < 0.05 (Wald test). ABC, ATP-binding cassette; ECF, extracytoplasmic function.
DISCUSSION
The adaptive immune response to the gut microbiota consists of a complex repertoire of potential antibody specificities interacting with a broad range of microbial species and strains. We have developed an approach that examines the specificity of individual antibodies from a naturally primed immune response to a prominent human gut mutualist. The 260.8 mAb, like our previously characterized 225.4 mAb, is specific to B. thetaiotaomicron; mAb 225.4, which recognizes a product of one of the organism's capsular polysaccharide synthesis loci, exhibits strain specificity. The 260.8 mAb, which recognizes the product of a conserved B. thetaiotaomicron locus apparently involved in LPS O-antigen polysaccharide synthesis, has species specificity that encompasses the vast majority of strains we tested from many different human hosts. As such, the process by which we produced and immortalized the naturally primed 260.8 antibody illustrates a method for generating diagnostic antibodies of value and perhaps antibodies that may have therapeutic utility.
Undoubtedly, this approach will yield antibodies along a continuum of specificity and degeneracy that define the aggregate repertoire. It remains to be seen precisely how this repertoire shapes the microbiota. We can conceptualize the immune response as a malleable tool that can be forged to “fit” any microbial community, responding to both common and unique epitopes. We show that whereas the presence of the CPS4-specific 225.4 IgA leads to down-regulation of epitope expression in vivo, we did not see that adaptability in the case of the 260.8 epitope. It is interesting that the epitope that was more responsive in expression (225.4) is specified by a non-conserved locus in the B. thetaiotaomicron pan-genome, whereas expression of the conserved 260.8-associated locus was not changed significantly by the presence of the antibody in vivo. We speculate that it is beneficial for a bacterial species/strain to have immunogenic epitopes that impact its competitiveness. If it were impervious to IgA effector functions, then it could expand or penetrate the functional barrier created by IgA and in this sense behave as a pathogen. A successful evolutionary strategy may be balanced immunogenicity as well as susceptibility to the immune system to preserve a place for an organism in the gut community while preventing overgrowth of that species.
Both 260.8 and 225.4 IgAs could represent the innate/natural antibody/B1 B cell repertoire as they both recognize bacterial polysaccharides and were developed early in the immune response to colonization (i.e. within 2 weeks). However, the level of specificity that we have observed in both hybridomas does not fit that concept. Nonetheless, recent studies support the concept of bacterium-specific natural antibodies and memory formation independent of T cells (44, 45). The antibodies we have characterized thus far are not responsive to a universal bacterial epitope such as phosphocholine but rather to structures that are likely restricted to B. thetaiotaomicron. Because these epitopes are polysaccharides and repetitive, we postulate that they can be both low affinity and high specificity. As such, it is questionable how important T cell help is in the generation of this B cell response. As future experiments examine the immune response to the gut microbiota, there is a need to address further this general question of the specificity and degeneracy of antibody responses directed against individual strains within a (defined) microbial community. Additionally, further experiments will be needed to delineate how binding of intestinal IgA affects the fitness and behavior of microbes along the length of the gut and whether IgA binding leads to changes in the localization of these organisms within the mucus layer or their spatial distribution between the mucosa and luminal contents.
Acknowledgments
We thank the late Abigail Salyers for generously supplying us with B. thetaiotaomicron strains, David Snydman for providing the NEMC collection strains, and David O'Donnell and Maria Karlsson for assistance with gnotobiotic mouse husbandry.
This work was supported, in whole or in part, by National Institutes of Health Grants DK30292, DK078669, K08-AI07660902, and GM007200, a Washington University Medical Scientist Training Program grant (to J. P.). This work was also supported by National Science Foundation Grant EF-0333284 and the Crohn's and Colitis Association of America. J. I. G is co-founder of Matatu, Inc., a company characterizing the role of diet-by-microbiota interactions in animal health.
Draft genome sequences for B. thetaiotaomicron strains BT7330-1 and DH3731 and 16 S rRNA gene sequences have been deposited in the European Nucleotide Archive (ENA study accession number PRJEB8679). GeneChip and RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO accession number GSE65456).
- CPS
- capsular polysaccharide synthesis
- BHI
- brain heart infusion
- INSeq
- insertion sequencing
- RNA-Seq
- RNA sequencing
- NEMC
- New England Medical Center.
REFERENCES
- 1. Hooper L. V., Littman D. R., Macpherson A. J. (2012) Interactions between the microbiota and the immune system. Science 336, 1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blumberg R., Powrie F. (2012) Microbiota, disease, and back to health: a metastable journey. Sci. Transl. Med. 4, 137rv7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sommer F., Adam N., Johansson M. E., Xia L., Hansson G. C., Bäckhed F. (2014) Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS One 9, e85254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pelaseyed T., Bergström J. H., Gustafsson J. K., Ermund A., Birchenough G. M., Schütte A., van der Post S., Svensson F., Rodríguez-Piñeiro A. M., Nyström E. E., Wising C., Johansson M. E., Hansson G. C. (2014) The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 260, 8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cash H. L., Whitham C. V., Behrendt C. L., Hooper L. V. (2006) Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vaishnava S., Behrendt C. L., Ismail A. S., Eckmann L., Hooper L. V. (2008) Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U.S.A. 105, 20858–20863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vaishnava S., Yamamoto M., Severson K. M., Ruhn K. A., Yu X., Koren O., Ley R., Wakeland E. K., Hooper L. V. (2011) The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gallo R. L., Hooper L. V. (2012) Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12, 503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hapfelmeier S., Lawson M. A., Slack E., Kirundi J. K., Stoel M., Heikenwalder M., Cahenzli J., Velykoredko Y., Balmer M. L., Endt K., Geuking M. B., Curtiss R., 3rd, McCoy K. D., Macpherson A. J. (2010) Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fagarasan S., Kawamoto S., Kanagawa O., Suzuki K. (2010) Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol. 28, 243–273 [DOI] [PubMed] [Google Scholar]
- 11. He B., Xu W., Santini P.A., Polydorides A. D., Chiu A., Estrella J., Shan M., Chadburn A., Villanacci V., Plebani A., Knowles D. M., Rescigno M., Cerutti A. (2007) Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity 26, 812–826 [DOI] [PubMed] [Google Scholar]
- 12. Shroff K. E., Meslin K., Cebra J. J. (1995) Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 63, 3904–3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shimoda M., Inoue Y., Azuma N., Kanno C. (1999) Natural polyreactive immunoglobulin A antibodies produced in mouse Peyer's patches. Immunology 97, 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stoel M., Jiang H.-Q., van Diemen C. C., Bun J. C., Dammers P. M., Thurnheer M. C., Kroese F. G., Cebra J. J., Bos N. A. (2005) Restricted IgA repertoire in both B-1 and B-2 cell-derived gut plasmablasts. J. Immunol. 174, 1046–1054 [DOI] [PubMed] [Google Scholar]
- 15. Benckert J., Schmolka N., Kreschel C., Zoller M. J., Sturm A., Wiedenmann B., Wardemann H. (2011) The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J. Clin. Investig. 121, 1946–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suzuki K., Fagarasan S. (2008) How host-bacterial interactions lead to IgA synthesis in the gut. Trends Immunol. 29, 523–531 [DOI] [PubMed] [Google Scholar]
- 17. Cerutti A., Chen K., Chorny A. (2011) Immunoglobulin responses at the mucosal interface. Annu. Rev. Immunol. 29, 273–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Macpherson A. J., Gatto D., Sainsbury E., Harriman G. R., Hengartner H., Zinkernagel R. M. (2000) A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 [DOI] [PubMed] [Google Scholar]
- 19. Cullender T. C., Chassaing B., Janzon A., Kumar K., Muller C. E., Werner J. J., Angenent L. T., Bell M. E., Hay A. G., Peterson D. A., Walter J., Vijay-Kumar M., Gewirtz A. T., Ley R. E. (2013) Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14, 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Palm N. W., de Zoete M. R., Cullen T. W., Barry N. A., Stefanowski J., Hao L., Degnan P. H., Hu J., Peter I., Zhang W., Ruggiero E., Cho J. H., Goodman A. L., Flavell R. A. (2014) Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mirpuri J., Raetz M., Sturge C. R., Wilhelm C. L., Benson A., Savani R. C., Hooper L. V., Yarovinsky F. (2014) Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 5, 28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kau A. L., Planer J. D., Liu J., Rao S., Yatsunenko T., Trehan I., Manary M. J., Liu T.-C., Stappenbeck T. S., Maleta K. M., Ashorn P., Dewey K. G., Houpt E. R., Hsieh C.-S., Gordon J. I. (2015) Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl. Med. 7, 276ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peterson D. A., McNulty N. P., Guruge J. L., Gordon J. I. (2007) IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 [DOI] [PubMed] [Google Scholar]
- 24. Sonnenburg J. L. (2005) Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 [DOI] [PubMed] [Google Scholar]
- 25. Martens E. C., Lowe E. C., Chiang H., Pudlo N. A., Wu M., McNulty N. P., Abbott D. W., Henrissat B., Gilbert H. J., Bolam D. N., Gordon J. I. (2011) Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9, e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Köhler G., Milstein C. (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 [DOI] [PubMed] [Google Scholar]
- 27. Galfrè G., Milstein C., Wright B. (1979) Rat × rat hybrid myelomas and a monoclonal anti-Fd portion of mouse IgG. Nature 277, 131–133 [DOI] [PubMed] [Google Scholar]
- 28. Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., Sogin M. L., Jones W. J., Roe B. A., Affourtit J. P., Egholm M., Henrissat B., Heath A. C., Knight R., Gordon J. I. (2009) A core gut microbiome in obese and lean twins. Nature 457, 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Faith J. J., Guruge J. L., Charbonneau M., Subramanian S., Seedorf H., Goodman A. L., Clemente J. C., Knight R., Heath A. C., Leibel R. L., Rosenbaum M., Gordon J. I. (2013) The long-term stability of the human gut microbiota. Science 341, 1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shoemaker N. B., Getty C., Gardner J. F., Salyers A. A. (1986) Tn4351 transposes in Bacteroides spp., and mediates the integration of plasmid R751 into the Bacteroides chromosome. J. Bacteriol. 165, 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salyers A. A., Bonheyo G., Shoemaker N. B. (2000) Starting a new genetic system: lessons from Bacteroides. Methods 20, 35–46 [DOI] [PubMed] [Google Scholar]
- 33. Goodman A. L., McNulty N. P., Zhao Y., Leip D., Mitra R. D., Lozupone C. A., Knight R., Gordon J. I. (2009) Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crawford P. A., Gordon J. I. (2005) Microbial regulation of intestinal radiosensitivity. Proc. Natl. Acad. Sci. U.S.A. 102, 13254–13259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rey F. E., Faith J. J., Bain J., Muehlbauer M. J., Stevens R. D., Newgard C. B., Gordon J. I. (2010) Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. 285, 22082–22090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Love M. I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Snydman D. R., Jacobus N. V., McDermott L. A., Ruthazer R., Golan Y., Goldstein E. J., Finegold S. M., Harrell L. J., Hecht D. W., Jenkins S. G., Pierson C., Venezia R., Yu V., Rihs J., Gorbach S. L. (2007) National survey on the susceptibility of Bacteroides fragilis group: report and analysis of trends in the United States from 1997 to 2004. Antimicrob. Agents Chemother. 51, 1649–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simmon K. E., Mirrett S., Reller L. B., Petti C. A. (2008) Genotypic diversity of anaerobic isolates from bloodstream infections. J. Clin. Microbiol. 46, 1596–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krinos C. M., Coyne M. J., Weinacht K. G., Tzianabos A. O., Kasper D. L., Comstock L. E. (2001) Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414, 555–558 [DOI] [PubMed] [Google Scholar]
- 41. Xu J., Mahowald M. A., Ley R. E., Lozupone C. A., Hamady M., Martens E. C., Henrissat B., Coutinho P. M., Minx P., Latreille P., Cordum H., Van Brunt A., Kim K., Fulton R. S., Fulton L. A., Clifton S. W., Wilson R. K., Knight R. D., Gordon J. I. (2007) Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chatzidaki-Livanis M., Weinacht K. G., Comstock L. E. (2010) Trans locus inhibitors limit concomitant polysaccharide synthesis in the human gut symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. U.S.A. 107, 11976–11980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martens E. C., Roth R., Heuser J. E., Gordon J. I. (2009) Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J. Biol. Chem. 284, 18445–18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang Y., Ghosn E. E., Cole L. E., Obukhanych T. V., Sadate-Ngatchou P., Vogel S. N., Herzenberg L. A., Herzenberg L. A. (2012) Antigen-specific memory in B-1a and its relationship to natural immunity. Proc. Natl. Acad. Sci. U.S.A. 109, 5388–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang Y., Ghosn E. E., Cole L. E., Obukhanych T. V., Sadate-Ngatchou P., Vogel S. N., Herzenberg L. A., Herzenberg L. A. (2012) Antigen-specific antibody responses in B-1a and their relationship to natural immunity. Proc. Natl. Acad. Sci. U.S.A. 109, 5382–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]