Background: The functional activities of membrane-bound cytochrome P450s (P450s) are affected by lipids.
Results: Stronger interactions between P450 2B4 (CYP2B4) and cytochrome b5 (cytb5) are observed in bicelles as compared with lipid-free solution or micelles.
Conclusion: Lipid bilayers enhance the interaction between CYP2B4 and cytb5.
Significance: The findings provide insights into the important role of membrane in P450-cytb5 complex formation.
Keywords: cytochrome P450, lipid bilayer, membrane bilayer, membrane protein, nuclear magnetic resonance (NMR), cytochrome b5
Abstract
Mammalian cytochrome P450 (P450) is a membrane-bound monooxygenase whose catalytic activities require two electrons to be sequentially delivered from its redox partners: cytochrome b5 (cytb5) and cytochrome P450 reductase, both of which are membrane proteins. Although P450 functional activities are known to be affected by lipids, experimental evidence to reveal the effect of membrane on P450-cytb5 interactions is still lacking. Here, we present evidence for the influence of phospholipid bilayers on complex formation between rabbit P450 2B4 (CYP2B4) and rabbit cytb5 at the atomic level, utilizing NMR techniques. General line broadening and modest chemical shift perturbations of cytb5 resonances characterize CYP2B4-cytb5 interactions on the intermediate time scale. More significant intensity attenuation and a more specific protein-protein binding interface are observed in bicelles as compared with lipid-free solution, highlighting the importance of the lipid bilayer in stabilizing stronger and more specific interactions between CYP2B4 and cytb5, which may lead to a more efficient electron transfer. Similar results observed for the interactions between CYP2B4 lacking the transmembrane domain (tr-CYP2B4) and cytb5 imply interactions between tr-CYP2B4 and the membrane surface, which might assist in CYP2B4-cytb5 complex formation by orienting tr-CYP2B4 for efficient contact with cytb5. Furthermore, the observation of weak and nonspecific interactions between CYP2B4 and cytb5 in micelles suggests that lipid bilayer structures and low curvature membrane surface are preferable for CYP2B4-cytb5 complex formation. Results presented in this study provide structural insights into the mechanism behind the important role that the lipid bilayer plays in the interactions between P450s and their redox partners.
Introduction
Cytochrome P450 (P450)2 monooxygenases are a ubiquitous superfamily of enzymes found in all living kingdoms, including plants, animals, bacteria, and fungi (1). Eukaryotic P450s are membrane-bound proteins, usually containing a large soluble domain and a single α-helical transmembrane (TM) domain (2). A total of 57 human P450s have been discovered (3, 4) and are responsible for the metabolism of a wide range of endogenous and exogenous substrates, including sterols, vitamins, fatty acids, environmental pollutants, and over 50% of marketed drugs (1, 3, 5). One of the most studied functions of P450s is the insertion of a single hydroxyl group into hydrophobic compounds, rendering them more hydrophilic for easier excretion from the kidneys (6). Completion of the hydroxylation reaction requires two electrons to be sequentially delivered to P450, with the first one coming from cytochrome P450 reductase (CPR) and the second one from either CPR or cytochrome b5 (cytb5) (7–9).
Mammalian P450s and their redox partners (CPR and cytb5) are membrane-bound proteins primarily located on the cytoplasmic side of the endoplasmic reticulum (ER) of hepatic cells (10). The structure of most mammalian P450s is composed of a large soluble domain and a single α-helical TM domain; cytb5 contains a soluble domain, a single α-helical TM domain, and a linker connecting the aforementioned two domains (1, 11). It is well documented that the environment provided by the cell membrane, including phospholipids in the ER membrane, is closely tied to the functional activities of many membrane proteins (12). Therefore, it is not surprising that phospholipids comprising the ER membrane have been demonstrated to be essential for optimal P450 activities (13–16). It is widely believed that the TM domain of mammalian P450s is not the sole membrane binding segment, but a secondary binding site on the P450 lacking the N-terminal TM domain (tr-P450) exists. A number of different P450s have been reported to bind the membrane in the absence of the TM domain (17–22). It has been proposed that several loop regions in the tr-P450s interact with the membrane, allowing for a significant portion of the protein surface to be buried inside the membrane (23, 24); this interaction could further serve in assisting access of hydrophobic substrates to the catalytic active site and holding P450 in an orientation that allows optimal contact with its redox partners for efficient electron transfer (23, 25). Furthermore, it is reported that substrate turnover could be stimulated if phospholipids are present (16, 26). Extensive studies on the interaction and electron transfer between P450 and CPR in the presence and absence of phospholipids have been carried out and revealed stronger interactions between the two proteins and faster electron transfer from CPR to P450 in the presence of lipids or membrane mimetics (27–30). However, studies demonstrating the effect of phospholipid membrane on P450-cytb5 interactions are still lacking in the literature.
Although cytb5 is only capable of donating the second electron due to its high redox potential as compared with ferric P450, it plays a key role in the P450 enzyme system in the catalysis of a variety of compounds and significantly regulates the functional activities of P450s (31, 32). It is reported that cytb5 could stimulate, inhibit, or have no effect on P450 activities, depending on the P450 isozyme studied, the substrate involved, or the particular experimental conditions employed (33–36). The mechanism underlying the differential effects of cytb5 on P450 activities is not fully understood. A generally well accepted explanation is that cytb5 and CPR possess an overlapping but non-identical binding surface on P450, resulting in competitive binding between the two proteins (37, 38). When P450 predominantly binds cytb5 due to a high cytb5 concentration, the first electron to be delivered from CPR is inhibited (1, 38). To obtain an insight into the influence of cytb5 on P450 activities, an in-depth understanding of P450-cytb5 interaction is necessary. A recent study on the interaction between the truncated cytochrome P450 17A1 and the soluble domain of human cytb5 in a lipid-free environment revealed a binding interface located on the upper cleft of cytb5 (37). Because both proteins are naturally membrane-bound, our study aims to characterize the interactions of P450 and cytb5 in a native-like membrane environment in order to obtain a more physiologically relevant view. Although the microsomes resemble the ER membrane most, it is a very complex system containing a variety of different types of lipids, cholesterol, carbohydrates, proteins, etc. (39, 40) For mechanistic studies on the P450 system, a model membrane is needed that can both mimic the native membrane environment and be easily characterized and controlled. Among the most suitable membrane mimetics for NMR studies, detergent micelles and phospholipid/detergent isotropic bicelles have been most frequently and successfully applied to the investigation of the structure and function of a number of different membrane proteins during the past few decades, as reviewed in Refs. 41 and 42.
In this study, we report an investigation of the interaction between full-length rabbit cytochrome P450 2B4 (CYP2B4) and full-length rabbit cytb5 in different membrane mimetic environments, including lipid-free environment and isotropic bicelles and micelles, utilizing NMR techniques. Our study provides the first structural evidence on the importance of the phospholipid bilayer in governing the interaction between these two proteins. The mechanism by which the membrane affects CYP2B4-cytb5 interaction is also explored. By comparing the effects of different membrane environments in assisting complex formation, we propose that phospholipid bilayers enhance both the affinity and specificity in the interaction between CYP2B4 and cytb5.
EXPERIMENTAL PROCEDURES
Materials
1,2-Dilauroyl-sn-glycero-3-phosphocholine (DLPC), 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC), and n-dodecylphosphocholine (DPC) were purchased from Avanti Polar Lipids Inc. Potassium phosphate monobasic and dibasic, benzphetamine glycerol, and sodium dithionite were purchased from Sigma-Aldrich. Deuterium oxide was purchased from Cambridge Isotope Laboratories, Inc. The 5-mm symmetrical D2O-matched Shigemi NMR microtubes were purchased from Shigemi, Inc.
Protein Expression and Purification
Full-length wild-type rabbit CYP2B4 (wt-CYP2B4) and 15N-labeled full-length wild-type rabbit cytb5 were expressed and purified individually as described previously (11, 38, 43, 44). Briefly, the pLW01 plasmid containing the gene for the wt-CYP2B4 was transformed to Escherichia coli. C41 cells and then plated on a Luria-Bertani (LB) agar plate with 0.24 mm carbenicillin overnight at 37 °C. Three colonies were selected from the plate and then incubated in 50–140 ml of LB medium/carbenicillin medium for 16 h at 30 °C with shaking at 200 rpm. The overnight culture was transferred to TB medium (100-fold dilution) supplemented with final concentrations of 500 μm δ-aminolevulinic acid and 0.24 mm carbenicillin. The cultures were incubated at 22–23 °C with shaking at 120 rpm. CYP2B4 expression was induced by isopropyl 1-thio-β-d-galactopyranoside when the cell density reached A600 = 4–6. After induction, the cultures were grown at 22–23 °C with shaking at 120 rpm. The P450 and P420 content of the cultures was monitored. The cells were harvested between 76 and 110 h after isopropyl 1-thio-β-d-galactopyranoside induction. The cells were lysed and solubilized by detergents and then applied to a DE52 column, Reactive Red-agarose column, octyl-Sepharose column, and hydroxyl apatite column sequentially for purification of the protein (44). The full-length rabbit 15N-cytb5 was expressed and purified similarly as wild-type CYP2B4 except that cells were grown in 15N Celtone complete medium with additional supplements as detailed previously (43). The harvested cells were lysed and broken by sonication and were solubilized by detergents. The proteins went through two DEAE columns and one size exclusion column for purification.
Truncated rabbit CYP2B4 (tr-CYP2B4) was expressed and purified similarly with wild-type CYP2B4, as described (45). Amino acids at the N terminus from 3 to 21 were truncated. In order to increase the solubility of tr-CYP2B4, which in turn increases the expression level, the mutations E2A, G22K, H23K, P24T, K25S, A26S, H27K, and R29K were introduced, which made it possible to release the proteins from the membrane under high-salt conditions (45). These mutations do not affect the binding affinity between tr-CYP2B4 and cytb5, as indicated by the identical Kd values measured for wild-type tr-CYP2B4-cytb5 and mutant tr-CYP2B4-cytb5 (data not shown). No affinity tags were used for the expression and purification of any of the three proteins.
Determination of the Dissociation Constant between cytb5 and wt-CYP2B4
Measurements of the equilibrium dissociation constant of the wt-CYP2B4-cytb5 complex were carried out as described previously (38). Briefly, wt-CYP2B4 (0.3 μm) was titrated by cytb5 at concentrations of 0, 0.01, 0.05, 0.1, 0.3, 0.5, 0.8, and 1.2 μm in the presence of 0.3 μm benzphetamine in 100 mm potassium phosphate buffer containing 5%(w/v) glycerol at pH 7.4. The titrations were performed in the presence and absence of 30 μm DLPC, respectively. The UV-visible spectra were collected on a Cary 4000 spectrophotometer at 25 °C. The absorbance at 420 and 385 nm was recorded. The total change in absorbance (ΔA = ΔA420 + ΔA385) was plotted against the concentration of cytb5 and fitted using an equation reported previously (38) to obtain the Kd values for the wt-CYP2B4-cytb5 complex.
Preparation of Membrane Mimetics
A DLPC/DHPC isotropic bicelle (q = [DLPC]/[DHPC] = 0.25) was prepared by mixing the appropriate amount of DLPC and DHPC in chloroform. The mixture was vortexed and dried under nitrogen to make a thin film, which was further dried under vacuum overnight. The film was then hydrated in 100 mm potassium phosphate buffer, containing 5% (w/v) glycerol, pH 7.4 (referred to as NMR buffer). A DPC micelle solution was prepared by dissolving DPC powder into NMR buffer followed by vortexing.
Carbon Monoxide (CO) Assay
A solution containing 1 μm CYP2B4 in NMR buffer was added to a cuvette with a 1-cm path length. The protein was then reduced by an excess of sodium dithionite, after which CO gas was allowed to flow over the solution for 1 min. UV-visible absorption spectra were recorded from 400 to 600 nm on a Cary 4000 spectrophotometer before and after treatment with CO gas. This assay was performed in the presence of 10% (w/v) DLPC/DHPC isotropic bicelles and 2 mm DPC micelles, respectively, to check the activity of CYP2B4.
Circular Dichroism (CD)
CD experiments were performed on a Jasco J-715 spectropolarimeter fitted with a 150-watt xenon lamp at 25 °C using a 1-mm cuvette. Spectra were recorded in the far UV region with 16 scans accumulated and averaged. DLPC/DHPC isotropic bicelles (q = 0.25) or DPC micelles were titrated into a solution containing 1 μm CYP2B4 in NMR buffer. The following final concentrations were achieved throughout the titrations: 0, 1, 2, 20, and 40 mm for bicelles and 0, 0.5, 0.8, 1.1, and 2 mm for DPC micelles. The CMC of DPC is 1.1 mm. Background (with everything present except CYP2B4) was subtracted for all experiments. Quantitative data analysis was performed by CDPro software package using the Continll program (46, 47).
NMR Titration Experiments and Data Analysis
NMR titration experiments were performed at 298 K on a Bruker Avance II 600-MHz spectrometer equipped with a cryoprobe. For the titration experiments, spectra were first recorded with 0.2 mm free 15N-cytb5 either in NMR buffer or incorporated into 10% (w/v) membrane mimetics in NMR buffer, followed by a titration of either wt-CYP2B4 or tr-CYP2B4 at molar ratios (cytb5/CYP2B4) of 1:0, 1:0.25, 1:0.5, 1:0.75, and 1:1. The CYP2B4 stock solutions used for titration were at low concentration (75 μm) in order to avoid any loss of CYP2B4 sample at high concentrations due to protein precipitation. After the spectrum of free 15N-cytb5 was acquired, a 0.25 m equivalent of CYP2B4 stock solution was added to 15N-cytb5. Subsequently, the mixture was concentrated to 300 μl so that the final 15N-cytb5 concentration remained constant. The rest of the titration experiments were carried out identically. For the control experiments, 15N/1H TROSY-HSQC spectra were collected from 15N cytb5 in complex with wt-CYP2B4 in DLPC/DHPC isotropic bicelles (q = 0.25) at the following cytb5 concentrations: 50, 80, and 100 μm. All of the three experiments were performed at a cytb5/wt-CYP2B4 molar ratio of 1:1.5 in NMR buffer.
All NMR experiments were recorded using two-dimensional 15N/1H TROSY-HSQC spectra with 64 scans and 256 t1 increments. Data were processed using TopSpin version 2.0 (Bruker) and analyzed using Sparky (48). The backbone chemical shift assignments of the rabbit cytb5 have been reported previously (11). The weighted amide chemical shift perturbation (Δδavg) was calculated using the equation,
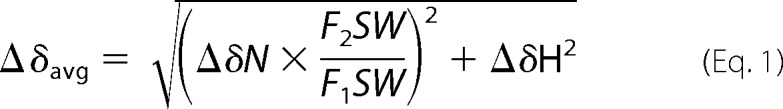 |
where ΔδN and ΔδH are the changes in the amide's nitrogen and hydrogen chemical shifts, respectively, and F1SW and F2SW represent the spectral width in the first and second dimension, respectively; all parameters are in ppm (49, 50).
RESULTS
Characterization of the Effect of Lipids on the Binding Affinity between CYP2B4 and cytb5
The Kd values between wt-CYP2B4 and cytb5 were determined, based on the Type I spectral change (Fig. 1A) of CYP2B4 induced by cytb5, to be 0.008 ± 0.015 and 0.14 ± 0.02 μm in the presence (Fig. 1B) and absence (Fig. 1C) of DLPC lipids, respectively. This implies the ability of lipids to facilitate tighter binding between wt-CYP2B4 and cytb5. Our results are in agreement with the previous findings that phospholipids enhance the interactions between P450s and the other redox partner, CPR (27–30).
FIGURE 1.
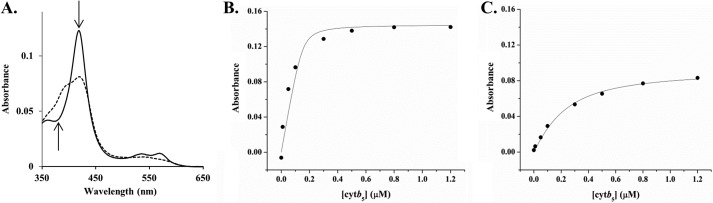
Determination of the dissociation constant Kd between cytb5 and wt-CYP2B4. A. Illustration of Type I spectral shift of CYP2B4 induced by cytb5: decrease at 420 nm and increase at 385 nm. Solid line, CYP2B4; dashed line, CYP2B4 bound with cytb5 (the absolute cytb5 spectrum is subtracted). B and C, titration of cytb5 into 0.3 μm wt-CYP2B4 in the presence (B) and absence (C) of 30 μm DLPC lipid bilayers. Both titrations were performed in 100 mm potassium phosphate buffer, containing 5% (w/v) glycerol, 0.3 μm benzphetamine, pH 7.4. The Kd values for wt-CYP2B4-cytb5 were determined to be 0.008 ± 0.015 μm in DLPC lipid bilayers (B) and 0.14 ± 0.02 μm in the absence of DLPC lipid bilayers (C).
Interaction between cytb5 and wt-CYP2B4
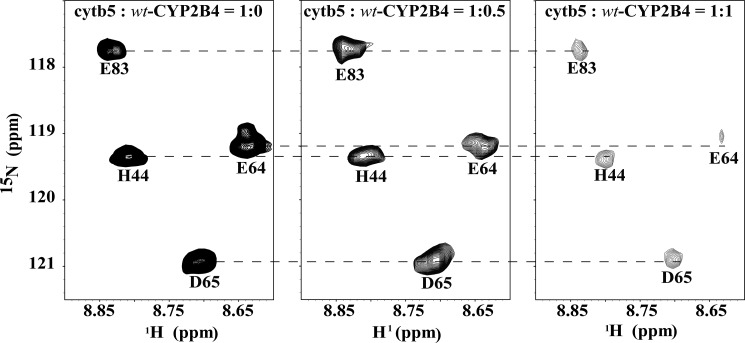
In order to investigate the influence of different membrane mimetic environments on CYP2B4-cytb5 interaction, two-dimensional 15N/1H TROSY-HSQC NMR spectra were recorded to monitor the interaction between 15N-labeled cytb5 and unlabeled wt-CYP2B4 in lipid-free solution, DLPC/DHPC isotropic bicelles, and DPC micelles. In all three experiments, broadening of resonances and moderate chemical shift perturbations (CSPs) (Δδavg ≤ 0.1 ppm) for amide nuclei of cytb5 were observed upon interaction with wt-CYP2B4. A representative region of the two-dimensional spectra of free cytb5 and those in complex with wt-CYP2B4 in DLPC/DHPC isotropic bicelles are shown in Fig. 2. In order to assess the effect of nonspecific oligomerization/aggregation of CYP2B4 on the resonance intensities of cytb5 upon titration, experiments were performed on a wt-CYP2B4-cytb5 complex at a cytb5/CYP2B4 molar ratio of 1:1.5 in DLPC/DHPC bicelles at different protein concentrations. The average relative resonance intensities are 44.84, 46.87, and 46.98% at a cytb5 concentration of 50, 80, and 100 μm, respectively. The small S.D. value (1.20%) between the three sets of average relative intensities reveals no significant difference between the average relative intensities of cytb5 at different protein concentrations, suggesting no (or negligible) interference of CYP2B4 oligomerization/aggregation with the interpretation of the observed resonance intensities of cytb5 (Table 1).
FIGURE 2.
Representative two-dimensional 15N/1H TROSY-HSQC spectra for titration of unlabeled wt-CYP2B4 into 15N-labeled cytb5. The titration was performed in NMR buffer (100 mm potassium phosphate buffer, 5% glycerol, pH 7.4) containing 10% (w/v) DLPC/DHPC isotropic bicelles. Interaction between wt-CYP2B4 and cytb5 in isotropic bicelles causes a combination of moderate chemical shift perturbation and significant line broadening of cytb5 resonances, which is indicative of wt-CYP2B4-cytb5 interaction on an intermediate exchange time scale.
TABLE 1.
A comparison of the average relative intensities of the cytb5 resonances in complex with wt-CYP2B4 at different protein concentrations
| [cytb5] | Average relative intensity of the cytb5 resonances |
|---|---|
| μm | % |
| 50 | 44.84 |
| 80 | 46.87 |
| 100 | 46.98 |
| S.D. | 1.20 |
The average chemical shift changes observed for lipid-free solution, bicelles, and micelles are 0.009, 0.023, and 0.017 ppm, respectively. The small and widespread chemical shift perturbations could be occurring for two reasons: first, fast-to-intermediate chemical exchange between free- and bound-cytb5, which would also explain the overall broadening of cytb5 resonances (11, 37); second, the formation of an ensemble of dynamic “encounter complexes,” which causes the “averaging out” of the chemical shift changes, thus leading to small CSP values (51–55). Due to the small magnitude of the CSP values and the dispersive perturbation pattern of encounter complexes, residue-specific CSPs only provide rough estimations of the binding interface and interaction strength between CYP2B4 and cytb5.
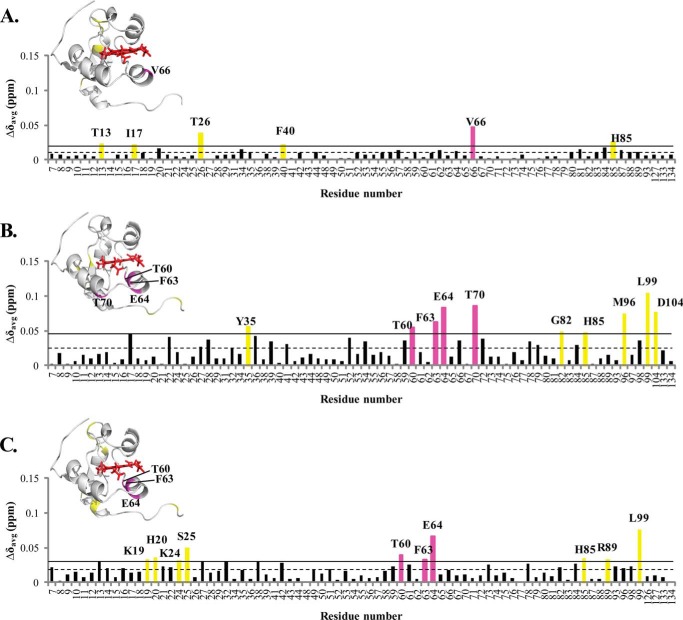
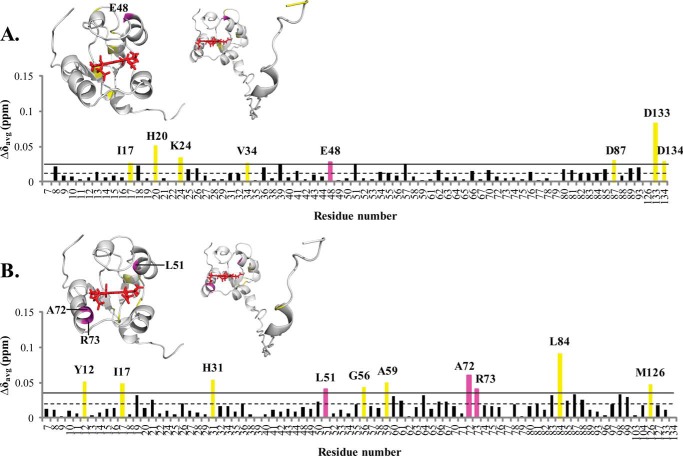
Histograms showing the weighted chemical shift perturbations (Δδavg) observed for the amide resonances of cytb5 upon interaction with wt-CYP2B4 at 1:1 molar ratio in different membrane mimetic environments are presented in Fig. 3. Residues of cytb5 with Δδavg larger than the average value plus one S.D. are considered to be most perturbed among all residues and are therefore most likely to be involved in the interaction with CYP2B4.
FIGURE 3.
Chemical shift perturbation of cytb5 resonances upon complex formation with wt-CYP2B4. CSPs are characterized by the small magnitude and wide dispersion, rendering accurate binding interface mapping impossible but allowing rough estimations of epitopes involved in wt-CYP2B4-cytb5 interactions. Weighted averages of chemical shift differences (Δδavg) for backbone amides of cytb5 upon interaction with a molar equivalent of wt-CYP2B4 in a lipid-free solution (A), solution containing DLPC/DHPC isotropic bicelles (B), and solution containing DPC micelles (C) are plotted against the cytb5 residue number. The dashed line represents the mean chemical shift change among all residues, and the solid line represents the mean chemical shift change plus one S.D. Residues with Δδavg above the solid line are considered to be the most affected among all of the cytb5 residues upon the addition of wt-CYP2B4. The residues most affected are highlighted in magenta for those located on the front face of cytb5 around the heme edge and yellow for those not in this area, both on the histogram and in the three-dimensional structures of cytb5 above each corresponding histogram. Because no residue in the TM domain of cytb5 is found to be perturbed, only the structure of the soluble domain of cytb5 is shown. The structure of cytb5 has been determined previously by our group (11).
Spectra obtained in lipid-free solution reveal only the six most perturbed residues, and they are spread over an extensive area of cytb5 (Fig. 3A), whereas the spectra acquired in bicelles indicate 10 most perturbed residues, which are mostly localized on the front face of cytb5 around the heme edge and in the linker region (Fig. 3B). These areas largely overlap with our previously published CSP mapping (11): around Tyr35 and region Asp88–Asp104. The perturbations in the active site of cytb5 (Thr60–Thr70) are more pronounced in the current work than in our previous work. The discrepancies could be attributable to the fact that the bicelles used in our previous work were DMPC/DHPC bicelles, whereas the bicelles used in the current work are DLPC/DHPC bicelles. The different membrane compositions (varying the hydrophobic thickness of the lipid bilayer) might induce different membrane topologies of CYP2B4 in terms of its depth of insertion into the lipid membrane and its soluble domain orientation on the membrane surface, which could lead to different binding epitopes on cytb5, whereas the major areas still roughly overlap. Results obtained from DPC micelles also indicate the 10 most perturbed residues located on the front face of cytb5 around the heme edge, the flexible loop region on the back of cytb5, and the linker region (Fig. 3C). Among all three different sample conditions, bicelles exhibited the largest average CSP value, implying the ability of bicelles to facilitate stronger interactions between wt-CYP2B4 and cytb5.
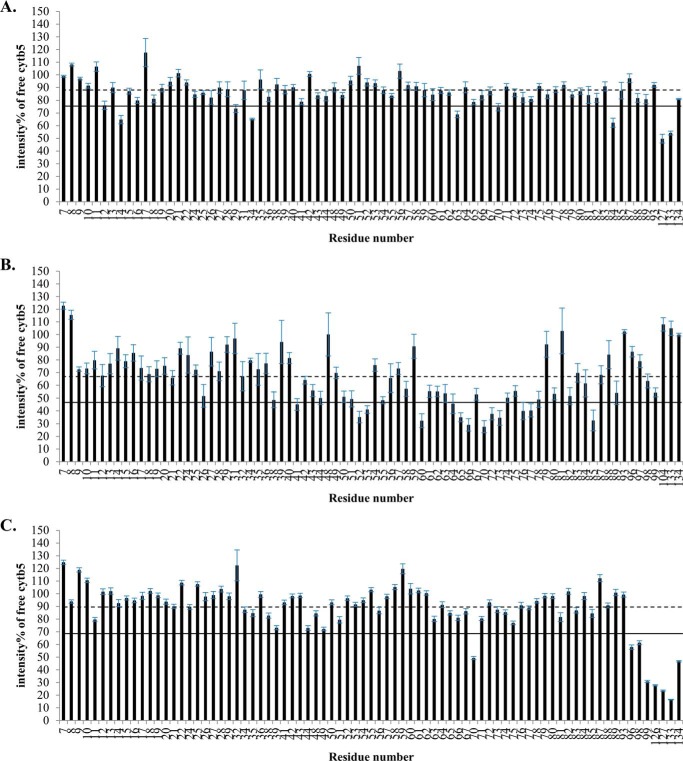
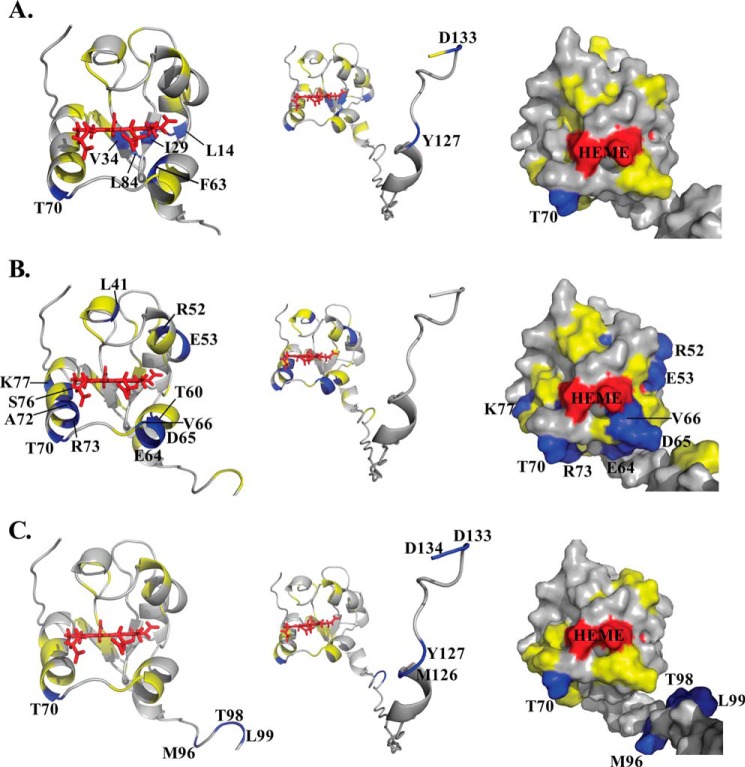
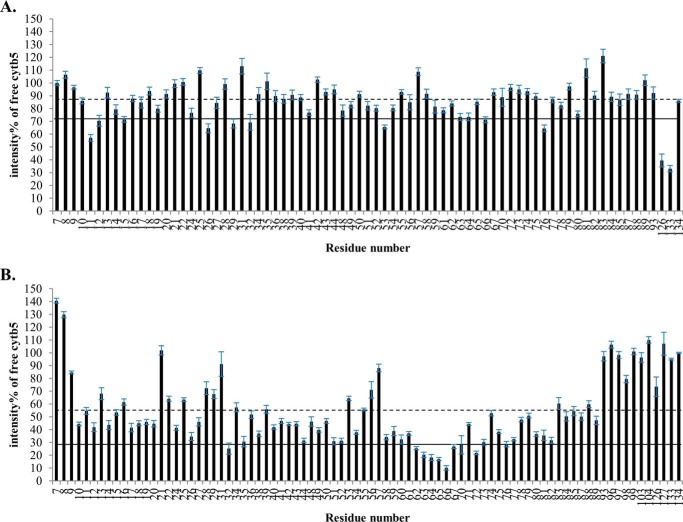
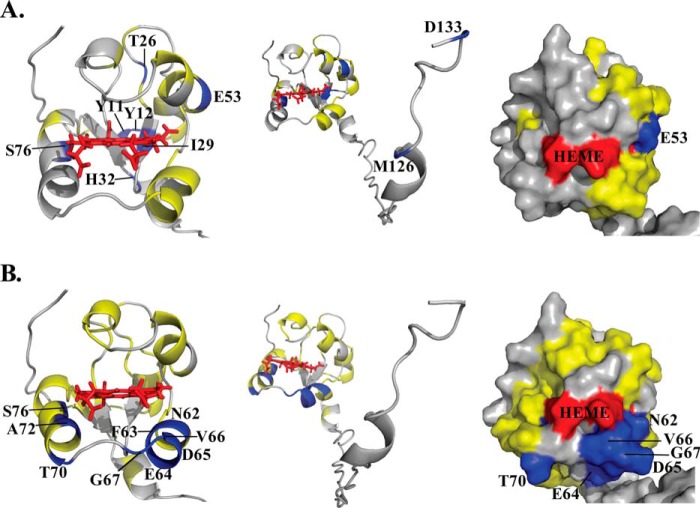
In addition to the modest chemical shift perturbations, interaction between cytb5 and wt-CYP2B4 also causes a global decrease in signal intensities. In Fig. 4, the relative resonance peak heights measured from the 1:1 cytb5-wt-CYP2B4 complex, represented as a percentage of the corresponding resonance peak heights in free cytb5 are plotted as a function of cytb5 residue number for lipid-free solution (Fig. 4A), bicelles (Fig. 4B), and micelles (Fig. 4C). A significant loss in signal intensity is defined as more than one S.D. below the average relative peak height of cytb5 upon complex formation with wt-CYP2B4 at a 1:1 molar ratio. For spectra obtained from lipid-free solution, only a slight reduction in signal intensities is observed, with an average relative cytb5 peak height of 87% at a 1:1 molar ratio with wt-CYP2B4. Residues considered significantly affected upon interaction with wt-CYP2B4 are spread over an extensive area on the soluble domain of cytb5 (Fig. 5A, left and right); therefore, no specific region could be highlighted as the interaction interface between cytb5 and wt-CYP2B4. Additionally, residues in the C terminus of the TM domain of cytb5 are also significantly affected upon the addition of wt-CYP2B4 (Fig. 5A, middle), which is probably due to non-native interactions between cytb5 TM domain and the hydrophobic patches on wt-CYP2B4 due to the absence of a membrane environment. On the other hand, in bicelles, the average relative cytb5 peak height drops to as low as 67%, and a closer inspection of Fig. 4B reveals differential line broadening of cytb5 resonances upon interaction with wt-CYP2B4. The majority of the significantly affected residues lie on the front face of cytb5 around the solvent-exposed edge of the heme, among which Leu41, Arg52, and Glu53 lying on the upper cleft and Thr60, Glu64, Asp65, Val66, Thr70, Ala72, Arg73, Ser76, and Lys77 lying on the lower cleft of cytb5 exhibit the most significant line broadening upon interaction with wt-CYP2B4 (Fig. 5B, left and right). This is consistent with our previous study of cytb5-wt-CYP2B4 complex formation in DMPC/DHPC isotropic bicelle solution (11). However, the binding interface mapped out in the current study differs from a previous study (37) on the interaction between a truncated cytb5 and truncated CYP17A1 in a lipid-free environment, in which residues Glu47–Val50 were shown to form the major complex interface. This could be attributed to the following reasons: first, the membrane environment regulates the interactions between cytb5 and P450, resulting in a different binding interface; second, rabbit full-length cytb5-CYP2B4 complex could intrinsically possess a binding interface different from that of the human truncated cytb5-CYP17A1 complex. As we have shown in this study, the presence of membrane enhances the binding affinity between the two proteins. In addition, the absence of membrane does not render a specific binding interface between the proteins. Therefore, it is more likely that the presence of membrane is the dominant factor contributing to the discrepancy between our results and the published study. Residue Leu99 in the linker domain is also observed to be affected, which can be attributed to the restriction of motions in the linker upon complex formation (56). No significant perturbation is observed in the C-terminal TM domain (Fig. 5B, middle), in contrast to the observation in lipid-free solution. This could be attributable to the primary interaction between the cytb5 TM domain and the bicelles, which aids in proper anchoring of the protein and diminishes non-native interactions. In DPC micelles, the average relative cytb5 peak height is 89%, similar to that observed in lipid-free solution, and is significantly higher than that observed in bicelles. Differential line broadening is also observed in micelles, where the subset of residues showing significant intensity loss is not localized around the functionally active heme edge but centered at the end of C-terminal TM domain and the flexible linker region (Fig. 5C, left and right). CO assay of CYP2B4 suggests marked inactivation of the protein upon the addition of DPC (Fig. 6). Moreover, CD titration experiments reveal partial unfolding of CYP2B4 helices induced by the presence of DPC (Fig. 7). It is likely that the disruption in both the secondary structure and the conformation of the CYP2B4 active site accounts for why cytb5 and wt-CYP2B4 cannot properly interact with each other in the presence of DPC micelles.
FIGURE 4.
Differential line broadening of cytb5 resonances upon complex formation with wt-CYP2B4. Relative intensities for backbone amides of cytb5 at a 1:1 molar ratio with wt-CYP2B4 shown as a percentage of free cytb5 resonance intensities, demonstrate differential line broadening for cytb5 upon interaction with wt-CYP2B4 in a lipid-free solution (A), a solution containing DLPC/DHPC isotropic bicelles (B), and a solution containing DPC micelles (C). The dashed line represents the average relative intensity, and the solid line represents the mean value minus one S.D. Error bars are calculated based on the signal/noise ratio extracted from Sparky (48). Residues of which the relative intensities are below the solid line are mapped onto the three-dimensional structure of cytb5 and colored blue in Fig. 3. The structure of cytb5 in solution has been determined previously by our group (11).
FIGURE 5.
Mapping of differential line broadening of cytb5 residues upon interaction with wt-CYP2B4. Differential line broadening of cytb5 residues is mapped onto the cytb5 structure upon interaction with wt-CYP2B4 at a molar ratio of 1:1 in a lipid-free solution (A), a solution containing DLPC/DHPC isotropic bicelles (B), and a solution containing DPC micelles (C). Residues are categorized and color-coded according to their relative intensities: blue for significantly perturbed residues upon interaction with wt-CYP2B4 with relative intensities more than one S.D. below the average, yellow for moderately perturbed residues with relative intensities in the range from average value to one S.D. below the average, and gray for residues with negligible to no perturbation, of which the relative intensities are above the average value. Heme is colored in red. Significantly perturbed residues are also labeled by amino acid name and the sequence number. Ribbon representations of the soluble domain of cytb5 are presented in the left panel. The active site of cytb5 (front face around the heme edge) is presented in surface representations in the right panel. The full-length structure of cytb5 is shown by ribbon representations in the middle panel to demonstrate the perturbations in the TM domain if present.
FIGURE 6.
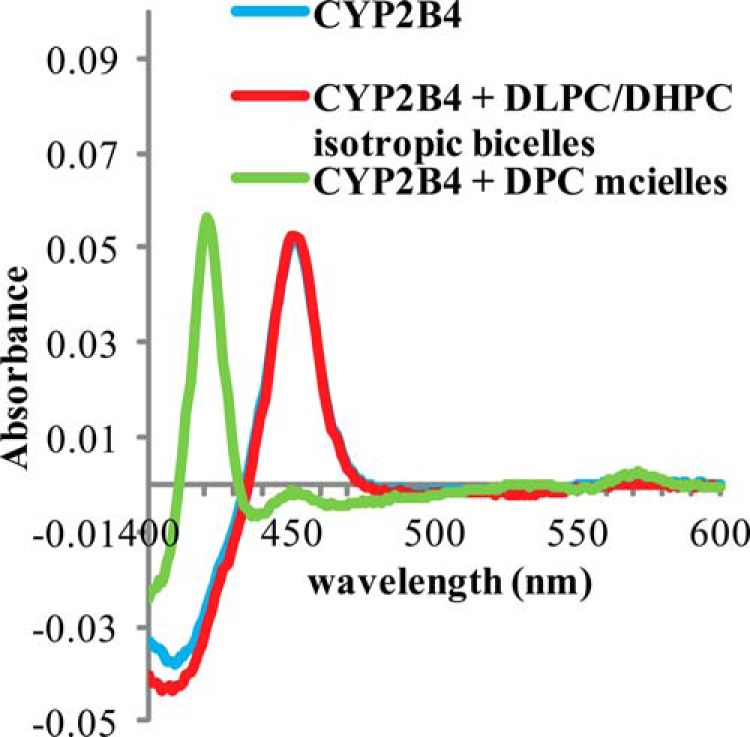
Carbon Monoxide assay of wt-CYP2B4 in different membrane environments. CO assays were performed on 1 μm CYP2B4 at 25 °C in 100 mm potassium phosphate buffer with 5% (w/v) glycerol, pH 7.4, containing no lipid (blue curve) as control, 10% (w/v) DLPC/DHPC isotropic bicelles (red curve), and 2 mm DPC micelles (green curve). An absorption maximum at 450 nm is observed in DLPC/DHPC isotropic bicelle solution, indicative of CYP2B4 in a functionally active P450 form. In DPC micelle solution, CYP2B4 turns into an inactive cytochrome P420 form as shown by a peak at 420 nm in the green curve.
FIGURE 7.
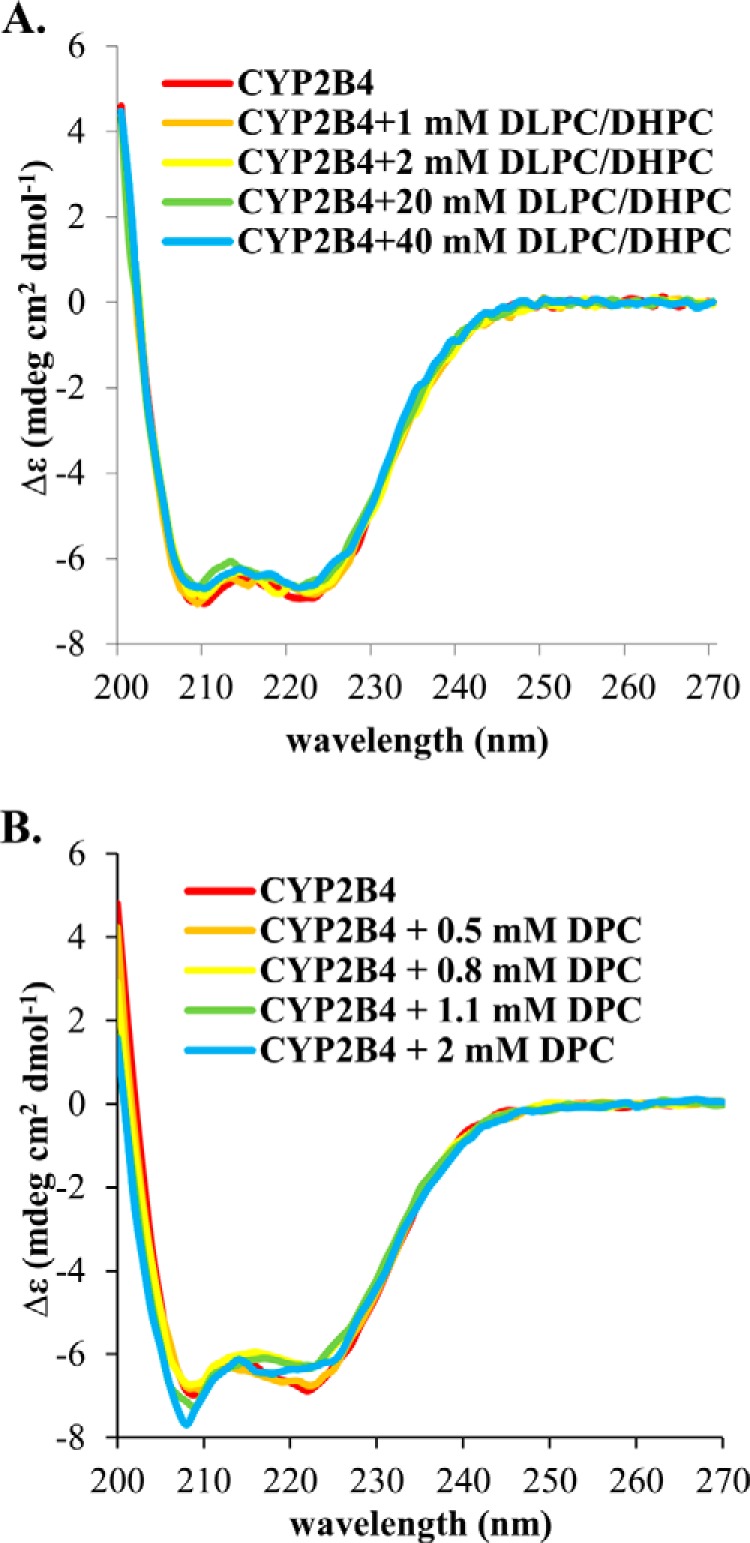
Circular Dichroism titration experiments of wt-CYP2B4. CD titrations of DLPC/DHPC isotropic bicelles (A) and DPC micelles (B) into 1 μm CYP2B4 were performed at 25 °C in 100 mm potassium phosphate buffer with 5% (w/v) glycerol, pH 7.4. Loss in α-helix content is observed in DPC titration (from 49.1 ± 2.1% at the beginning of the titration to 40.1 ± 2.2% at the end of the titration). While in DLPC/DHPC titration, the CYP2B4 secondary structure stays roughly unperturbed throughout the experiment (with α-helix content changing from 48.5 ± 2.0 to 46.5 ± 2.2%).
Interaction between cytb5 and tr-CYP2B4
In order to investigate the role of the membrane in the interaction between cytb5 and the tr-CYP2B4, a series of 15N/1H TROSY-HSQC NMR spectra were recorded to monitor 15N-labeled cytb5-unlabeled-tr-CYP2B4 interaction in lipid-free solution and DLPC/DHPC isotropic bicelles. The addition of tr-CYP2B4 to a solution of cytb5 leads to a general increase in line width and moderate chemical shift perturbation (Δδavg ≤ 0.1 ppm) for amide resonances of cytb5 in aqueous solution and bicelles. For bicelles, the average chemical shift change is 0.019 ppm, and in lipid-free solution, the value is even smaller (0.012 ppm).
Perturbations of chemical shifts at a 1:1 molar ratio of the two proteins are depicted in the histograms presented in Fig. 8. The residues of cytb5 most affected upon interaction with tr-CYP2B4 are observed across all regions of cytb5 under both sample conditions. However, among the most affected residues, three residues are found to be located on the front face of cytb5 surrounding the heme edge in bicelles (Fig. 8B), whereas only one is found in this area in lipid-free solution (Fig. 8A), although the affected areas observed in bicelles are not as specific as those observed for the cytb5-wt-CYP2B4 complex under the same conditions.
FIGURE 8.
Chemical shift perturbation of cytb5 resonances upon complex formation with tr-CYP2B4. As with wt-CYP2B4-cytb5 interactions, CSPs from tr-CYP2B4-cytb5 are also characterized by the small magnitude and wide dispersion, rendering accurate binding interface mapping impossible but allowing rough estimations of epitopes involved in tr-CYP2B4-cytb5 interactions. Weighted averages of chemical shift differences (Δδavg) for backbone amides of cytb5 upon interaction with tr-CYP2B4 at a 1:1 molar ratio in a lipid-free solution (A) and solution containing DLPC/DHPC isotropic bicelles (B) are plotted against cytb5 residue number. The dashed line represents the mean chemical shift change among all residues, and the solid line represents the mean chemical shift change plus one S.D. Residues with Δδavg above the solid line are considered to be the most affected among all of the cytb5 residues upon the addition of tr-CYP2B4. The residues most affected are highlighted in magenta for those located on the front face of cytb5 around the heme edge and yellow for those not in this area, both on the histogram and in the three-dimensional structures of cytb5 above each corresponding histogram. The structure of the soluble domain of cytb5 is shown on the left, and that of the full-length cytb5 is shown on the right.
Line broadening of cytb5 resonances caused by interaction with tr-CYP2B4 is presented in Fig. 9, in which the relative cytb5 resonance peak heights are plotted against cytb5 residue number at a 1:1 molar ratio with tr-CYP2B4. Residues with relative peak heights of more than one S.D. below the average value are considered to be significantly affected upon interaction with tr-CYP2B4. Similar to cytb5-wt-CYP2B4 interaction in lipid-free solution, no significant line broadening is observed for cytb5 upon the addition of tr-CYP2B4 under the same condition. The average relative peak height of cytb5 at a 1:1 molar ratio of the two proteins is 86%. Little perturbations could be found around the heme edge of cytb5, and the residues significantly affected are located on the back of cytb5 and in the C terminus of the TM domain (Fig. 10A). This is consistent with the case of cytb5-wt-CYP2B4 complex in a lipid-free environment, which is probably due to non-native interactions between the cytb5 TM domain and the hydrophobic patches on tr-CYP2B4, probably the F-G loop region. In contrast to the results from lipid-free solution, cytb5 resonances exhibit extensive reduction in signal intensity when interacting with tr-CYP2B4 in bicelles, with an average relative peak height of only 55%. This result reveals that even in the absence of the TM domain, CYP2B4 still interacts with cytb5 in a membrane mimetic environment created by the bicelles, in a similar manner to cytb5-wt-CYP2B4 interaction. The majority of the residues involved in the interaction between cytb5 and tr-CYP2B4 are localized around the solvent exposed heme edge (Fig. 10B), overlapping with the cytb5-wt-CYP2B4 interface under the same conditions. The area highlighted by the most significantly affected residues, including Asn62, Phe63, Glu64, Asp65, Val66, Gly67, Thr70, Ala72, and Ser73, are located mostly on the α4 helix, the loop between the α4 and α5 helices, and the beginning of the α5 helix (Fig. 10B, left and right), which is slightly different from what is observed for the cytb5-wt-CYP2B4 complex (Fig. 5B).
FIGURE 9.
Differential line broadening of cytb5 resonances upon complex formation with wt-CYP2B4. Relative intensities of backbone amides of cytb5 at a 1:1 molar ratio with tr-CYP2B4 shown as a percentage of free cytb5 resonance intensities, demonstrate differential line broadening for cytb5 upon interaction with tr-CYP2B4 in a lipid-free solution (A) and a solution containing DLPC/DHPC isotropic bicelles (B). The dashed line represents the average resonance intensity, and the solid line represents the mean value minus one S.D. Error bars are calculated based on the signal/noise ratio extracted from Sparky (48). Residues of which the relative intensities are below the solid line are mapped onto the three-dimensional structure of cytb5 and colored blue in Fig. 7.
FIGURE 10.
Mapping of differential line broadening of cytb5 residues upon interaction with tr-CYP2B4. Differential line broadening of cytb5 residues is mapped onto the cytb5 structure upon interaction with tr-CYP2B4 at a molar ratio of 1:1 in a lipid-free solution (A) and a solution containing DLPC/DHPC isotropic bicelles (B). Residues are color-coded according to the same category in Fig. 3. Heme is colored in red. Significantly perturbed residues are also labeled by amino acid name and the sequence number. Ribbon representations of the soluble domain of cytb5 are presented on the left. The active site of cytb5 (front face around the heme edge) is presented in a surface representations on the right. The full-length structure of cytb5 is shown by ribbon representations in the middle panel to demonstrate the perturbations in the TM domain if present.
DISCUSSION
Effect of DLPC/DHPC Isotropic Bicelles on CYP2B4-cytb5 Interaction
The importance of phospholipids in the cytochrome P450 reconstituted system has been known since the first successful reconstitution of cytochrome P450-dependent activities (13–16, 57, 58). It has been reported that the addition of phospholipids into the cytochrome P450 reconstituted system has a remarkable stimulating effect, as shown by an increase in P450 catalytic activity (16, 26, 30, 59, 60). Although evidence has shown that the interaction between cytochrome P450 and its reductase is affected by the presence of phospholipids (27–30, 61), especially phosphatidylcholine-containing lipids, the mechanism of how membrane affects P450-cytb5 interactions at the atomic level still remains unknown.
In order to provide insights into the role that membrane plays in the interaction between P450 and cytb5, we have investigated the interaction between wt-CYP2B4 and full-length rabbit cytb5 in the presence and absence of DLPC/DHPC isotropic bicelles by NMR. A titration of unlabeled wt-CYP2B4 into 15N-labeled cytb5 in DLPC/DHPC isotropic bicelles results in extensive line broadening and modest chemical shift perturbation, which is characteristic of binding on an intermediate exchange time scale (nanoseconds to microseconds). A subset of residues exhibiting the most significant reductions in signal intensity is found to be localized around the catalytically active heme edge on the front face of cytb5, suggesting a predominant site for interaction with wt-CYP2B4. Chemical shift perturbations, although small in magnitude, also show the largest changes at the same site, with an additional site in the flexible linker region probably due to conformational change upon interaction with wt-CYP2B4. In contrast, when wt-CYP2B4 is added to cytb5 in a lipid-free solution, no significant line broadening or CSPs are observed for cytb5 resonances, suggesting very weak interactions between the two proteins in the absence of a membrane environment. The average CSP is only 0.009 ppm, much smaller than that observed in bicelles (0.023 ppm). The average relative cytb5 resonance peak height, when mixed with a molar equivalent of wt-CYP2B4, is 87% of the corresponding free cytb5 resonances, which is significantly higher than the 67% observed in DLPC/DHPC isotropic bicelles. Furthermore, in lipid-free solution, the potential residues involved in the interactions are quite dispersed with almost no area specified for a predominant binding site, judged by differential line broadening, whereas the interaction gives a specific binding interface in bicelles as mentioned above. The largest signal intensity reductions in lipid-free solution are found for residues at the C terminus of the TM domain (Fig. 4A), which is most likely due to non-native interactions when the TM domain is not anchored in the membrane mimetic, implying inefficient interactions between the two proteins, whereas such interaction is not observed in bicelles. Our results indicate that in the presence of bicelles, the interaction between wt-CYP2B4 and cytb5 is greatly enhanced, and the binding site is much more specific, which would allow more efficient electron transfer between the two redox partners, in comparison with that in lipid-free solution. This may be one of the reasons why P450 catalytic activities are promoted by the presence of phospholipids.
The possible reasons for the capability of isotropic bicelles to enhance cytb5-CYP2B4 interactions could be that 1) both proteins are anchored into the membrane through their TM domains so that they are in close proximity to each other, or 2) the membrane directly interacts with the non-TM part of CYP2B4, the tr-CYP2B4, which assists in orienting the protein for optimal contact with cytb5. Although a large body of evidence has been reported to support the hypothesis that interactions between the membrane surface and tr-P450s (lacking the TM domain) exist (17–22, 62–66), it remains vague what role this potential interaction plays in the complex formation between cytb5 and P450. Therefore, it is of great interest to explore the interactions between cytb5 and the tr-P450 under the influence of a membrane environment.
In the present study, we have investigated the interaction between tr-CYP2B4 and cytb5 in the presence and absence of the DLPC/DHPC isotropic bicelle environment by NMR spectroscopy. Similar to wt-CYP2B4, titration of tr-CYP2B4 into cytb5 in bicelles leads to extensive line broadening and modest chemical shift perturbations. Again, this is characteristic of an intermediate exchange process on the NMR time scale. The predominant interaction interface on cytb5 highlighted by differential line broadening is on the front face of cytb5 surrounding the heme edge. The significant decrease in the signal intensities and the specific interaction interface identified are indicative of strong interactions between cytb5 and tr-CYP2B4. The tr-CYP2B4-cytb5 interactions reveal overlapping but non-identical binding interface compared with wt-CYP2B4-cytb5; in both cases, the interfaces are located surrounding the heme edge; however, Asn62, Phe63, and Gly67 are only observed on the tr-CYP2B4-cytb5 interface (Fig. 10B), whereas Leu41, Arg52, Glu53, Thr60, Arg73, and Lys77 can only be found on the wt-CYP2B4-cytb5 interface (Fig. 5B). The difference between the interfaces most likely results from the presence/absence of the TM domain of CYP2B4. The TM domain of wt-CYP2B4 serves as an anchor of the protein to the membrane, which leads to tighter binding between the wt-CYP2B4 and the membrane than the tr-CYP2B4 alone and/or aids in better orientation of CYP2B4 for interacting with cytb5 and transferring electrons (66). A solid-state NMR study on the TM domain of CYP2B4 revealed a 17° tilting angle between the TM helix and the membrane normal, suggesting a characteristic orientation of the TM domain of CYP2B4 when anchored to the membrane, which probably restricts the soluble domain to certain orientations on the membrane (67, 68). The absence of the TM domain could therefore lead to different membrane topologies of the tr-CYP2B4 in terms of its depth of insertion into the lipid bilayer and its orientation on the surface of the bicelle, which may result in different surface areas of CYP2B4 being exposed and available for interacting with cytb5 and hence divergent binding epitopes on cytb5. The dissimilar interaction interfaces between wt-CYP2B4 and cytb5 and between tr-CYP2B4 and cytb5 are probably the dominating factors contributing to the distinct average relative intensities observed for the two cases (Figs. 4B and 9B). In lipid-free solution, only a slight decrease in signal intensities could be observed, with widely dispersed perturbations found on cytb5. This indicates weak interactions between cytb5 and tr-CYP2B4 in lipid-free solution compared with what is observed in bicelles. Our results show that a bicelle environment could enhance not only cytb5-wt-CYP2B4 but also cytb5-tr-CYP2B4 interactions. The possible explanation for this observation could be that parts of the tr-CYP2B4, most likely the F-G and B-C loops (23, 24), directly interact with the lipid bilayer of bicelles. This hypothesis is well supported by studies reported in the literature. Both tr-CYP2B4 and a number of other P450 isoforms lacking the NH2-terminal membrane anchor region expressed in bacteria (17–20), yeast (21) and mammalian cells (22) have been shown to retain much of their membrane localizations, suggesting that the TM domain is not the sole determinant for membrane binding and that direct interaction exists between the tr-P450s and the membrane. Both an atomic force microscopy study (64) and a Langmuir-Blodgett monolayer study (69) have suggested that a segment of the tr-P450 is buried in the membrane; the observed slow rotation rate for membrane-embedded cytochrome P450s could be well explained by a secondary binding interaction between the tr-P450 and the membrane (62, 63). A recent simulation study on CYP3A4 also supports partial insertion of the tr-P450 into a membrane mimetic (25). We propose that this potential interaction between tr-CYP2B4 and the membrane could lead to some specific orientations of the tr-CYP2B4 on the membrane, which facilitates a more efficient interaction with cytb5 and probably further stimulates electron transfer between the two redox partners, as compared with what would be achieved if the tr-P450 simply tumbles isotropically in the solution flexibly coupled to the TM domain through a linker, like that in cytb5. A model depicting P450-cytb5 interaction facilitated by lipid bilayers is proposed in Fig. 11.
FIGURE 11.
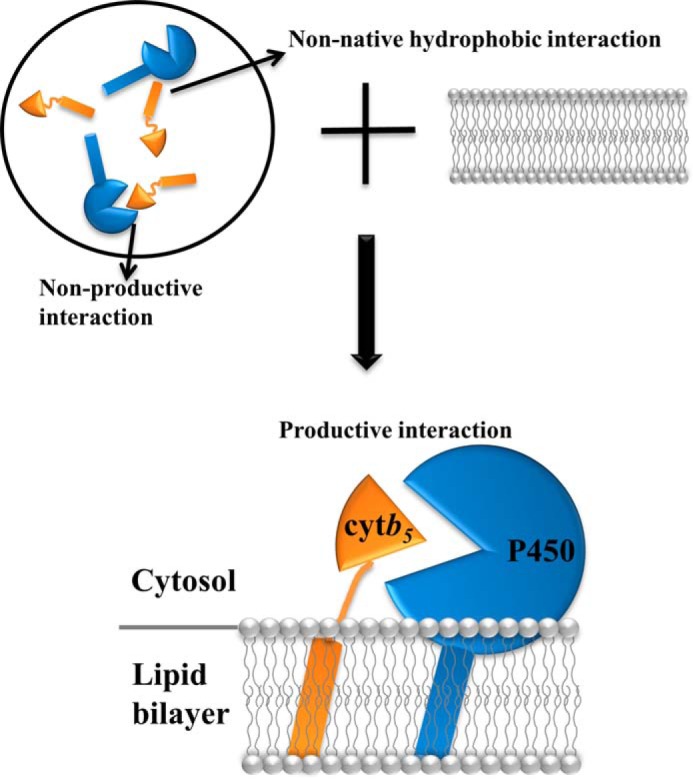
A model of the role of lipid bilayer in the interaction between P450 and cytb5. P450 and cytb5 are randomly oriented in a membrane-free environment (top left), giving rise to non-native hydrophobic interactions and non-productive interactions. With the assistance of lipid bilayers (top right and bottom), both proteins are anchored to the membrane, rendering them in close proximity to each other. Additionally, the interaction between the soluble domain of P450 and the lipid bilayer poses the protein in certain orientations that favor the interaction with cytb5, leading to more productive complex formation between the two redox partners.
Comparison between the Influence of DLPC/DHPC Isotropic Bicelles and That of DPC Micelles on wt-CYP2B4-cytb5 Interaction
Experiments carried out to study wt-CYP2B4-cytb5 interactions in DPC micelles generally revealed less significant line broadening of cytb5 resonances and smaller CSPs as compared with what was observed in bicelles, implying much weaker interactions between the two proteins in DPC micelles as compared with bicelles. Interestingly, in DPC micelles, resonances in both the flexible linker region and the C terminus of the TM domain demonstrate exceptionally large line broadening effects (relative intensity around 20%) as compared with the rest of the cytb5 residues (average relative intensity 89%). The perturbation observed in the linker region could be due to restriction of motions upon interactions between the soluble domains of the two proteins (11, 56). On the other hand, this observation could also be due to direct interaction with CYP2B4. Judging from the fact that the residues perturbed in this region mostly contain hydrophobic side chains (Met96 and Leu99), it is likely that the perturbation in this region, together with the perturbations found at the C terminus of the TM domain, mostly comes from direct non-native hydrophobic interactions with CYP2B4. An additional region exhibiting chemical shift perturbations is observed in the loop region on the back of the cytb5 soluble domain. Residues involved mostly contain positively charged side chains (Lys19, His20, and Lys24), which probably participate in the nonspecific, long range electrostatic interactions with the negatively charged area of the surface of CYP2B4. Overall, unlike the strong and specific wt-CYP2B4-cytb5 interactions observed in bicelles, the interaction observed in DPC micelles is weak and involves more nonspecific/artificial interactions, which could lead to the formation of nonproductive complexes. This observation could be attributed to the fact that both the catalytically active site and the secondary structure of CYP2B4 are disturbed upon interaction with DPC micelles (Figs. 6 and 7), which probably further affects its interaction with cytb5. Other membrane proteins have also been reported to suffer a loss in activity when embedded in micelles (70, 71). The monolayer lipid packing in micelles and the large curvature at the micelle surface render them unsuitable for mimicking the natural membrane environment, which might be the major reason leading to both structural and functional disruption of CYP2B4 (65, 72).
In summary, a phospholipid bilayer containing membrane mimetic environment regulates strong and specific interactions between wt-/tr-CYP2B4 and cytb5, as revealed by two-dimensional 15N/1H TROSY-HSQC NMR titration experiments. Proper interactions between tr-CYP2B4 and the lipid bilayer probably pose the protein into optimal orientations that favor interactions with cytb5, which could lead to more efficient productive complex formation between the two proteins. Unlike in bicelles, the interaction between CYP2B4 and cytb5 is less evident both in complex affinity and interface specificity in DPC micelles. The loss of activity and partial structure unfolding of CYP2B4 upon interaction with DPC micelles might be the cause of unfavorable complex formation between CYP2B4 and cytb5 in this membrane condition.
Acknowledgment
We thank Dr. Patrick Walsh for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM084018 and GM095640 (to A. R.) and GM35533 (to L. W.). This work was also supported by a VA Merit grant (to L. W.).
- P450 and P420
- cytochrome P450 and P420, respectively
- tr-P450
- truncated cytochrome P450 (cytochrome P450 lacking the N-terminal transmembrane domain)
- cytb5
- cytochrome b5
- CYP2B4
- cytochrome P450 2B4
- TM
- transmembrane
- CPR
- cytochrome P450 reductase
- wt- and tr-CYP2B4
- full-length wild-type and truncated cytochrome P450 2B4, respectively
- ER
- endoplasmic reticulum
- DLPC
- 1,2-dilauroyl-sn-glycero-3-phosphocholine
- DHPC
- 1,2-dihexanoyl-sn-glycero-3-phosphocholine
- DPC
- n-dodecylphosphocholine
- TROSY-HSQC
- transverse relaxation optimized spectroscopy-heteronuclear single quantum correlation
- CSP
- chemical shift perturbation.
REFERENCES
- 1. Dürr U. H. N., Waskell L., Ramamoorthy A. (2007) The cytochromes P450 and b5 and their reductases: promising targets for structural studies by advanced solid-state NMR spectroscopy. Biochim. Biophys. Acta 1768, 3235–3259 [DOI] [PubMed] [Google Scholar]
- 2. Danielson P. B. (2002) The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr. Drug. Metab. 3, 561–597 [DOI] [PubMed] [Google Scholar]
- 3. Guengerich F. P., Wu Z.-L., Bartleson C. J. (2005) Function of human cytochrome P450s: characterization of the orphans. Biochem. Biophys. Res. Commun. 338, 465–469 [DOI] [PubMed] [Google Scholar]
- 4. Nelson D. R. (2003) Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution. Arch. Biochem. Biophys. 409, 18–24 [DOI] [PubMed] [Google Scholar]
- 5. Nebert D. W., Russell D. W. (2002) Clinical importance of the cytochromes P450. Lancet 360, 1155–1162 [DOI] [PubMed] [Google Scholar]
- 6. Ortiz de Montellano P. R. (2010) Hydrocarbon hydroxylation by cytochrome P450 enzymes. Chem. Rev. 110, 932–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang H., Hamdane D., Im S.-C., Waskell L. (2008) Cytochrome b5 inhibits electron transfer from NADPH-cytochrome P450 reductase to ferric cytochrome P450 2B4. J. Biol. Chem. 283, 5217–5225 [DOI] [PubMed] [Google Scholar]
- 8. Guengerich F. P. (2006) Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J. 8, E101–E111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gruenke L. D., Konopka K., Cadieu M., Waskell L. (1995) The stoichiometry of the cytochrome P-450-catalyzed metabolism of methoxyflurane and benzphetamine in the presence and absence of cytochrome b5. J. Biol. Chem. 270, 24707–24718 [DOI] [PubMed] [Google Scholar]
- 10. Guengerich F. P. (2003) Cytochromes P450, drugs, and diseases. Mol. Interv. 3, 194–204 [DOI] [PubMed] [Google Scholar]
- 11. Ahuja S., Jahr N., Im S.-C., Vivekanandan S., Popovych N., Le Clair S. V., Huang R., Soong R., Xu J., Yamamoto K., Nanga R. P., Bridges A., Waskell L., Ramamoorthy A. (2013) A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data. J. Biol. Chem. 288, 22080–22095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coleman R. (1973) Membrane-bound enzymes and membrane ultrastructure. Biochim. Biophys. Acta 300, 1–30 [DOI] [PubMed] [Google Scholar]
- 13. Lu A. Y. H., Strobel H. W., Coon M. J. (1969) Hydroxylation of benzphetamine and other drugs by a solubilized form of cytochrome P-450 from liver microsomes: lipid requirement for drug demethylation. Biochem. Biophys. Res. Commun. 36, 545–551 [DOI] [PubMed] [Google Scholar]
- 14. Lu A. Y. H., Junk K. W., Coon M. J. (1969) Resolution of the cytochrome P-450-containing ohgr-hydroxylation system of liver microsomes into three components. J. Biol. Chem. 244, 3714–3721 [PubMed] [Google Scholar]
- 15. Lu A. Y. H., Strobel H. W., Coon M. J. (1970) Properties of a solubilized form of the cytochrome P-450-containing mixed-function oxidase of liver microsomes. Mol. Pharmacol. 6, 213–220 [PubMed] [Google Scholar]
- 16. Strobel H. W., Lu A. Y. H., Heidema J., Coon M. J. (1970) Phosphatidylcholine requirement in the enzymatic reduction of hemoprotein P-450 and in fatty acid, hydrocarbon, and drug hydroxylation. J. Biol. Chem. 245, 4851–4854 [PubMed] [Google Scholar]
- 17. Pernecky S. J., Larson J. R., Philpot R. M., Coon M. J. (1993) Expression of truncated forms of liver microsomal P450 cytochromes 2B4 and 2E1 in Escherichia coli: influence of NH2-terminal region on localization in cytosol and membranes. Proc. Natl. Acad. Sci. U.S.A. 90, 2651–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gillam E. M., Baba T., Kim B. R., Ohmori S., Guengerich F. P. (1993) Expression of modified human cytochrome P450 3A4 in Escherichia coli and purification and reconstitution of the enzyme. Arch. Biochem. Biophys. 305, 123–131 [DOI] [PubMed] [Google Scholar]
- 19. Sagara Y., Barnes H. J., Waterman M. R. (1993) Expression in Escherichia coli of functional cytochrome P450c17 lacking its hydrophobic amino-terminal signal anchor. Arch. Biochem. Biophys. 304, 272–278 [DOI] [PubMed] [Google Scholar]
- 20. Larson J. R., Coon M. J., Porter T. D. (1991) Alcohol-inducible cytochrome P-450IIE1 lacking the hydrophobic NH2-terminal segment retains catalytic activity and is membrane-bound when expressed in Escherichia coli. J. Biol. Chem. 266, 7321–7324 [PubMed] [Google Scholar]
- 21. Cullin C. (1992) Two distinct sequences control the targeting and anchoring of the mouse P450 1A1 into the yeast endoplasmic reticulum membrane. Biochem. Biophys. Res. Commun. 184, 1490–1495 [DOI] [PubMed] [Google Scholar]
- 22. Clark B. J., Waterman M. R. (1991) The hydrophobic amino-terminal sequence of bovine 17α-hydroxylase is required for the expression of a functional hemoprotein in COS 1 cells. J. Biol. Chem. 266, 5898–5904 [PubMed] [Google Scholar]
- 23. Williams P. A., Cosme J., Sridhar V., Johnson E. F., McRee D. E. (2000) Mammalian microsomal cytochrome P450 monooxygenase. Mol. Cell 5, 121–131 [DOI] [PubMed] [Google Scholar]
- 24. Zhao Y., White M. A., Muralidhara B. K., Sun L., Halpert J. R., Stout C. D. (2006) Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J. Biol. Chem. 281, 5973–5981 [DOI] [PubMed] [Google Scholar]
- 25. Baylon J. L., Lenov I. L., Sligar S. G., Tajkhorshid E. (2013) Characterizing the membrane-bound state of cytochrome P450 3A4: structure, depth of insertion, and orientation. J. Am. Chem. Soc. 135, 8542–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Hoeven T. A., Coon M. J. (1974) Preparation and properties of partially purified cytochrome P-450 and reduced nicotinamide adenine dinucleotide phosphate-cytochrome P-450 reductase from rabbit liver microsomes. J. Biol. Chem. 249, 6302–6310 [PubMed] [Google Scholar]
- 27. Müller-Enoch D., Churchill P., Fleischer S., Guengerich F. P. (1984) Interaction of liver microsomal cytochrome P-450 and NADPH-cytochrome P-450 reductase in the presence and absence of lipid. J. Biol. Chem. 259, 8174–8182 [PubMed] [Google Scholar]
- 28. Miwa G. T., Lu A. Y. H. (1981) Studies on the stimulation of cytochrome P-450-dependent monooxygenase activity by dilauroylphosphatidylcholine. Arch. Biochem. Biophys. 211, 454–458 [DOI] [PubMed] [Google Scholar]
- 29. Coon M. J., Haugen D. A., Guengerich F. P., Vermilion J. L., Dean W. L. (1976) in The Structural Basis of Membrane Function (Hatefi Y., Djavadi-Ohaniance L., eds) pp. 409–427, Academic Press, Inc., New York [Google Scholar]
- 30. Imaoka S., Imai Y., Shimada T., Funae Y. (1992) Role of phospholipids in reconstituted cytochrome P450 3A form and mechanism of their activation of catalytic activity. Biochemistry 31, 6063–6069 [DOI] [PubMed] [Google Scholar]
- 31. Kominami S., Ogawa N., Morimune R., D-Ying H., Takemori S. (1992) The role of cytochrome b5 in adrenal microsomal steroidogenesis. J. Steroid Biochem. Mol. Biol. 42, 57–64 [DOI] [PubMed] [Google Scholar]
- 32. Finn R. D., McLaughlin L. A., Ronseaux S., Rosewell I., Houston J. B., Henderson C. J., Wolf C. R. (2008) Defining the in vivo role for cytochrome b5 in cytochrome P450 function through the conditional hepatic deletion of microsomal cytochrome b5. J. Biol. Chem. 283, 31385–31393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morgan E. T., Coon M. J. (1984) Effects of cytochrome b5 on cytochrome P-450-catalyzed reactions: studies with manganese-substituted cytochrome b5. Drug. Metab. Dispos. 12, 358–364 [PubMed] [Google Scholar]
- 34. Canova-Davis E., Chiang J. Y. L., Waskell L. (1985) Obligatory role of cytochrome b5 in the microsomal metabolism of methoxyflurane. Biochem. Pharmacol. 34, 1907–1912 [DOI] [PubMed] [Google Scholar]
- 35. Shimada T., Mernaugh R. L., Guengerich F. P. (2005) Interactions of mammalian cytochrome P450, NADPH-cytochrome P450 reductase, and cytochrome b5 enzymes. Arch. Biochem. Biophys. 435, 207–216 [DOI] [PubMed] [Google Scholar]
- 36. Im S.-C., Waskell L. (2011) The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b5. Arch. Biochem. Biophys. 507, 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Estrada D. F., Laurence J. S., Scott E. E. (2013) Substrate-modulated cytochrome P450 17A1 and cytochrome b5 interactions revealed by NMR. J. Biol. Chem. 288, 17008–17018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bridges A., Gruenke L., Chang Y. T., Vakser I. A., Loew G., Waskell L. (1998) Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J. Biol. Chem. 273, 17036–17049 [DOI] [PubMed] [Google Scholar]
- 39. DePierre J. W., Ernster L. (1977) Enzyme topology of intracellular membranes. Annu. Rev. Biochem. 46, 201–262 [DOI] [PubMed] [Google Scholar]
- 40. Depierre J. W., Dallner G. (1975) Structural aspects of the membrane of the endoplasmic reticulum. Biochim. Biophys. Acta 415, 411–472 [DOI] [PubMed] [Google Scholar]
- 41. Dürr U. H. N., Gildenberg M., Ramamoorthy A. (2012) The magic of bicelles lights up membrane protein structure. Chem. Rev. 112, 6054–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arora A., Tamm L. K. (2001) Biophysical approaches to membrane protein structure determination. Curr. Opin. Struct. Biol. 11, 540–547 [DOI] [PubMed] [Google Scholar]
- 43. Dürr U. H. N., Yamamoto K., Im S.-C., Waskell L., Ramamoorthy A. (2007) Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b5. J. Am. Chem. Soc. 129, 6670–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saribas A. S., Gruenke L., Waskell L. (2001) Overexpression and purification of the membrane-bound cytochrome P450 2B4. Protein Expr. Purif. 21, 303–309 [DOI] [PubMed] [Google Scholar]
- 45. Scott E. E., Spatzenegger M., Halpert J. R. (2001) A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch. Biochem. Biophys. 395, 57–68 [DOI] [PubMed] [Google Scholar]
- 46. van Stokkum I. H. M., Spoelder H. J. W., Bloemendal M., van Grondelle R., Groen F. C. A. (1990) Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem. 191, 110–118 [DOI] [PubMed] [Google Scholar]
- 47. Provencher S. W., Glöckner J. (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 48. Kneller D. G., Kuntz I. D. (1993) UCSF Sparky-an NMR display, annotation, and assignment tool. J. Cell Biochem. 53, 254–254 [Google Scholar]
- 49. Williamson R. A., Carr M. D., Frenkiel T. A., Feeney J., Freedman R. B. (1997) Mapping the binding site for matrix metalloproteinase on the N-terminal domain of the tissue inhibitor of metalloproteinases-2 by NMR chemical shift perturbation. Biochemistry 36, 13882–13889 [DOI] [PubMed] [Google Scholar]
- 50. Williamson M. P. (2013) Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 [DOI] [PubMed] [Google Scholar]
- 51. Prudêncio M., Ubbink M. (2004) Transient complexes of redox proteins: structural and dynamic details from NMR studies. J. Mol. Recognit. 17, 524–539 [DOI] [PubMed] [Google Scholar]
- 52. Volkov A. N., Ferrari D., Worrall J. A. R., Bonvin A. M. J. J., Ubbink M. (2005) The orientations of cytochrome c in the highly dynamic complex with cytochrome b5 visualized by NMR and docking using HADDOCK. Protein Sci. 14, 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tang C., Iwahara J., Clore G. M. (2006) Visualization of transient encounter complexes in protein-protein association. Nature 444, 383–386 [DOI] [PubMed] [Google Scholar]
- 54. Volkov A. N., Ubbink M., van Nuland N. A. J. (2010) Mapping the encounter state of a transient protein complex by PRE NMR spectroscopy. J. Biomol. NMR 48, 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suh J.-Y., Tang C., Clore G. M. (2007) Role of electrostatic interactions in transient encounter complexes in protein-protein association investigated by paramagnetic relaxation enhancement. J. Am. Chem. Soc. 129, 12954–12955 [DOI] [PubMed] [Google Scholar]
- 56. Koberova M., Jecmen T., Sulc M., Cerna V., Kizek R., Hudecek J., Stiborova M., Hodek P. (2013) Photo-cytochrome b5: a new tool to study the cytochrome P450 electron-transport chain. Int. J. Electrochem. Sci. 8, 125–134 [Google Scholar]
- 57. Lu A. Y. H., Coon M. J. (1968) Role of hemoprotein P-450 in fatty acid ohgr-hydroxylation in a soluble enzyme system from liver microsomes. J. Biol. Chem. 243, 1331–1332 [PubMed] [Google Scholar]
- 58. French J. S., Guengerich F. P., Coon M. J. (1980) Interactions of cytochrome P-450, NADPH-cytochrome P450 reductase, phospholipid, and substrate in the reconstituted liver microsomal enzyme system. J. Biol. Chem. 255, 4112–4119 [PubMed] [Google Scholar]
- 59. Yang C. S. (1977) Interactions between solubilized cytochrome P-450 and hepatic microsomes. J. Biol. Chem. 252, 293–298 [PubMed] [Google Scholar]
- 60. Ingelman-Sundberg M., Glaumann H. (1977) Reconstitution of the liver microsomal hydroxylase system into liposomes. FEBS Lett. 78, 72–76 [DOI] [PubMed] [Google Scholar]
- 61. Blanck J., Smettan G., Ristau O., Ingelman-Sundberg M., Ruckpaul K. (1984) Mechanism of rate control of the NADPH-dependent reduction of cytochrome P-450 by lipids in reconstituted phospholipid vesicles. Eur. J. Biochem. 144, 509–513 [DOI] [PubMed] [Google Scholar]
- 62. Kawato S., Gut J., Cherry R. J., Winterhalter K. H., Richter C. (1982) Rotation of cytochrome P-450: I. Investigations of protein-protein interactions of cytochrome P-450 in phospholipid vesicles and liver microsomes. J. Biol. Chem. 257, 7023–7029 [PubMed] [Google Scholar]
- 63. Ohta Y., Sakaki T., Yabusaki Y., Ohkawa H., Kawato S. (1994) Rotation and membrane topology of genetically expressed methylcholanthrene-inducible cytochrome P-450IA1 lacking the N-terminal hydrophobic segment in yeast microsomes. J. Biol. Chem. 269, 15597–15600 [PubMed] [Google Scholar]
- 64. Bayburt T. H., Sligar S. G. (2002) Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc. Natl. Acad. Sci. U.S.A. 99, 6725–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Matthews E. E., Zoonens M., Engelman D. M. (2006) Dynamic helix interactions in transmembrane signaling. Cell 127, 447–450 [DOI] [PubMed] [Google Scholar]
- 66. Pernecky S. J., Olken N. M., Bestervelt L. L., Coon M. J. (1995) Subcellular localization, aggregation state, and catalytic activity of microsomal P450 cytochromes modified in the NH2-terminal region and expressed in Escherichia coli. Arch. Biochem. Biophys. 318, 446–456 [DOI] [PubMed] [Google Scholar]
- 67. Yamamoto K., Dürr U. H. N., Xu J., Im S.-C., Waskell L., Ramamoorthy A. (2013) Dynamic interaction between membrane-bound full-length cytochrome P450 and cytochrome b5 observed by solid-state NMR spectroscopy. Sci. Rep. 3, 2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yamamoto K., Gildenberg M., Ahuja S., Im S.-C., Pearcy P., Waskell L., Ramamoorthy A. (2013) Probing the transmembrane structure and topology of microsomal cytochrome-p450 by solid-state NMR on temperature-resistant bicelles. Sci. Rep. 3, 2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shank-Retzlaff M. L., Raner G. M., Coon M. J., Sligar S. G. (1998) Membrane topology of cytochrome P450 2B4 in Langmuir-Blodgett monolayers. Arch. Biochem. Biophys. 359, 82–88 [DOI] [PubMed] [Google Scholar]
- 70. Sanders C. R., 2nd, Landis G. C. (1995) Reconstitution of membrane proteins into lipid-rich bilayered mixed micelles for NMR studies. Biochemistry 34, 4030–4040 [DOI] [PubMed] [Google Scholar]
- 71. Garavito R. M., Ferguson-Miller S. (2001) Detergents as tools in membrane biochemistry. J. Biol. Chem. 276, 32403–32406 [DOI] [PubMed] [Google Scholar]
- 72. Lee D., Walter K. F. A., Brückner A.-K., Hilty C., Becker S., Griesinger C. (2008) Bilayer in small bicelles revealed by lipid-protein interactions using NMR spectroscopy. J. Am. Chem. Soc. 130, 13822–13823 [DOI] [PubMed] [Google Scholar]