Background: Signaling pathways regulating NAD+ homeostasis and their complex interplay with cell longevity remain unclear.
Results: Attenuated SPS (Ssy1-Ptr3-Ssy5) signaling extends replicative life span, which requires increased nicotinamide riboside salvage and functional NADH shuttle.
Conclusion: Enhanced NAD+ homeostasis contributes to SPS-induced cell longevity.
Significance: Studying SPS signaling as a novel longevity factor helps elucidate the complex regulation of NAD+ homeostasis.
Keywords: cell metabolism, metabolic regulation, NAD biosynthesis, yeast genetics, yeast metabolism, nicotinamide riboside salvage
Abstract
Attenuated nutrient signaling extends the life span in yeast and higher eukaryotes; however, the mechanisms are not completely understood. Here we identify the Ssy1-Ptr3-Ssy5 (SPS) amino acid sensing pathway as a novel longevity factor. A null mutation of SSY5 (ssy5Δ) increases replicative life span (RLS) by ∼50%. Our results demonstrate that several NAD+ homeostasis factors play key roles in this life span extension. First, expression of the putative malate-pyruvate NADH shuttle increases in ssy5Δ cells, and deleting components of this shuttle, MAE1 and OAC1, largely abolishes RLS extension. Next, we show that Stp1, a transcription factor of the SPS pathway, directly binds to the promoter of MAE1 and OAC1 to regulate their expression. Additionally, deletion of SSY5 increases nicotinamide riboside (NR) levels and phosphate-responsive (PHO) signaling activity, suggesting that ssy5Δ increases NR salvaging. This increase contributes to NAD+ homeostasis, partially ameliorating the NAD+ deficiency and rescuing the short life span of the npt1Δ mutant. Moreover, we observed that vacuolar phosphatase, Pho8, is partially required for ssy5Δ-mediated NR increase and RLS extension. Together, our studies present evidence that supports SPS signaling is a novel NAD+ homeostasis factor and ssy5Δ-mediated life span extension is likely due to concomitantly increased mitochondrial and vacuolar function. Our findings may contribute to understanding the molecular basis of NAD+ metabolism, cellular life span, and diseases associated with NAD+ deficiency and aging.
Introduction
Reduced nutrient signaling activity has been shown to extend life span in a variety of species, but the molecular mechanisms are not completely understood. Because of its short life span and well established molecular genetic techniques, the budding yeast, Saccharomyces cerevisiae, is an efficient model, enabling identification and study of novel longevity factors at the molecular level. In budding yeast, several conserved nutrient-sensing pathways such as the cAMP-PKA (cAMP-dependent protein kinase A), TOR (target of rapamycin), and Sch9/AKT (ortholog of mammalian S6 kinase) pathways, regulate longevity, and were shown to also play a role in low glucose-mediated calorie restriction (CR)2 (1–3). Metabolic factors regulating the downstream processes of these pathways are likely to play pivotal roles in longevity signaling. Due to the complexity of these pathways, determining certain molecular aspects of the underlying mechanisms of longevity is difficult. Unraveling the cross-regulation of metabolic longevity factors, nutrient signaling, and their downstream targets will help elucidate the complex interplay of these factors.
NAD+ (nicotinamide adenine dinucleotide) metabolism has emerged as a metabolic regulator of longevity. NAD+ homeostasis is important for many cellular processes; NAD+, and its reduced form, NADH, mediate essential redox reactions in cellular metabolism. In addition, NAD+ and its derivatives function as substrates and signaling molecules in key cellular processes such as regulation of Ca2+ signaling, chromatin structure, DNA repair, circadian rhythm, metabolic responses, and life span (4, 5). In S. cerevisiae, NAD+ is synthesized from two key intermediates, nicotinic acid mononucleotide and nicotinamide mononucleotide. Nicotinic acid mononucleotide is mainly produced via the de novo and nicotinic acid/nicotinamide salvaging pathways. Nicotinic acid mononucleotide is produced by transferring the phosphoribose moiety of phosphoribosyl pyrophosphate to salvaged (or imported) nicotinic acid or to de novo tryptophan-derived quinolinic acid, which is catalyzed by phosphoribosyltransferase Npt1 and Bna6, respectively (4). More recently, nicotinamide riboside (NR) was identified as an efficient NAD+ precursor that is converted to nicotinamide mononucleotide by Nrk1 kinase in a branch of NR salvaging (6, 7). Functional NR and nicotinic acid/nicotinamide salvaging pathways are essential for NAD+ homeostasis and life span (1, 8, 9). To date, signaling pathways regulating NAD+ homeostasis remain unclear because of the dynamic nature of these factors. Although the phosphate-responsive signaling (PHO) pathway has been connected to regulation of NR salvaging (10), detailed mechanisms underlying the cross-regulation of NAD+ homeostasis, PHO and other nutrient sensing pathways are still unclear.
In this study we characterized a long-lived ssy5Δ mutant and showed that NR/NAD+ homeostasis plays an important role in ssy5Δ-mediated life span extension. SSY5 is part of the SPS (Ssy1-Ptr3-Ssy5) amino acid nutrient-sensing pathway (11, 12), which is inactive without extracellular amino acids (Fig. 1A, left panel). SPS pathway is activated when Ssy1 senses extracellular amino acids and sends a signal via conformational changes that trigger subsequent phosphorylations of Ptr3 and Ssy5 by the kinases Yck1 and Yck2 (Fig. 1A, right panel) (11, 13). Once phosphorylated, the prodomain and catalytic domain of Ssy5 disassociate. The Ssy5 catalytic domain then cleaves the N-terminal cytoplasmic retention domain of both transcription factors, Stp1 and Stp2 (11, 14, 15). Without the cytoplasmic retention domain, Stp1 and Stp2 can bypass the surveillance of inner nuclear membrane ubiquitin ligase Asi complex components, Asi1, Asi2, and Asi3, entering the nucleus to modulate (activate or inhibit) SPS pathway downstream gene expression (15–17). Here we show that SPS signaling is a novel longevity factor and provide evidence linking SPS signaling to NAD+ homeostasis. Our studies may advance the understanding of the interconnection and cross-regulation of NAD+ homeostasis, nutrient-sensing, and longevity pathways.
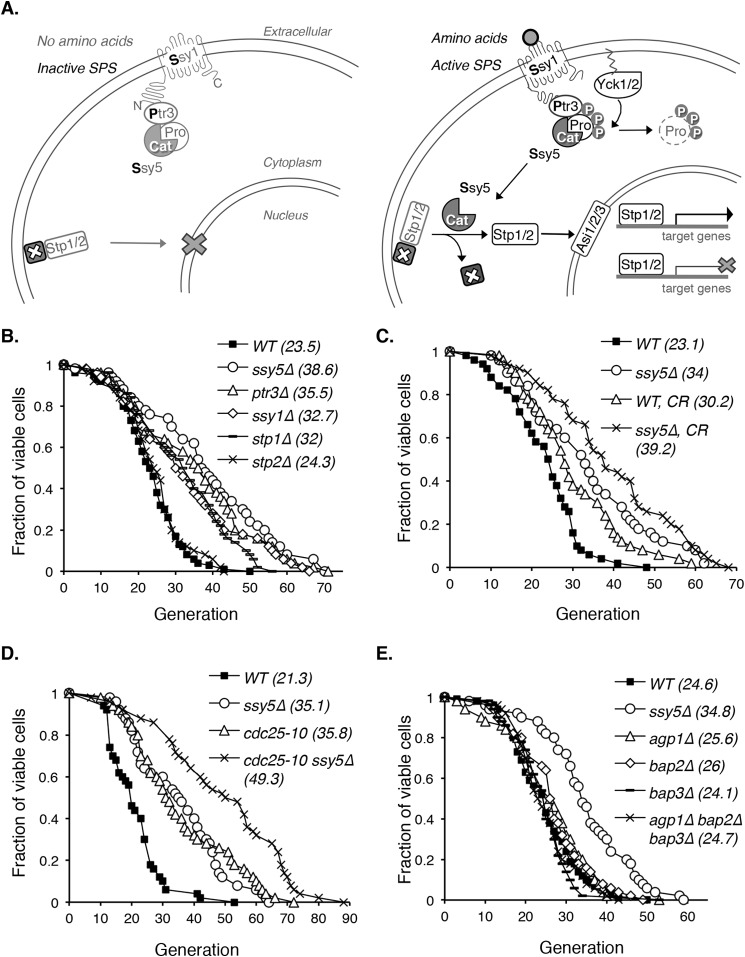
FIGURE 1.
Yeast mutants with reduced SPS signaling show increased replicative life span. A, a simple model of the SPS nutrient-sensing pathway in S. cerevisiae. The left panel shows inactive SPS without amino acid stimulation. Upon activation by extracellular amino acids, SPS activates transcription factors Stp1 and Stp2 (right panel) to modulate (activate or inhibit) downstream gene expression. Pro, Ssy5 prodomain; Cat, Ssy5 catalytic domain; P, phosphorylation. B, deletions of various SPS pathway components extend RLS. The RLS of all mutants (except for stp2Δ) are significantly increased (p < 0.005) when compared with the WT. Δ: gene deletions. C, moderate CR further extends RLS in the SPS mutant ssy5Δ background (ssy5Δ versus ssy5Δ, CR; p < 0.05). CR, 0.5% glucose (versus 2% glucose in standard growth media). D, cdc25-10, a CR genetic mimic, further extends life span in the ssy5Δ background (ssy5Δ versus cdc25-10 ssy5Δ; p < 0.005). E, reduced amino acid uptake is not the main cause of RLS extension in ssy5Δ cells. Unlike ssy5Δ, deletion of SPS downstream amino acid permeases AGP1, BAP2, and BAP3 does not extend RLS. Data shown are representative of multiple independent experiments. Statistical analysis of RLS is determined by the Wilcoxon rank sum test.
EXPERIMENTAL PROCEDURES
Yeast Strains, Growth Media, and Plasmids
Yeast strain BY4742 MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 acquired from Open Biosystems (18) was used for this study. Rich media (YPD) and synthetic media were made as described (19). All gene deletions were generated by replacing wild type genes with reusable loxP-Kanr-loxP cassettes as described (20). Multiple deletions were carried out by removing the Kanr marker using a galactose-inducible Cre recombinase (20). The original ssy5 mutant was identified as a temperature sensitive (ts) mutant (ssy5ts, Ser-75 to Pro) in a screen for longevity genes carried out as described (21). In brief, we employed the cdc25-10ts mutant in an accelerated cell death system to look for genes that (when mutated or overexpressed) can extend the survival of this mutant. CDC25 encodes a GTP-GDP exchange factor that activates Ras in the cAMP/PKA pathway in response to glucose (22). When shifted to non-permissive temperature at 38 °C, the cdc25-10ts mutant exhibits phenotypes similar to G0 stage cells and survives only ∼3 days (21). We have previously identified Bmh1 as a longevity factor using similar screening conditions (21). In a pilot study for optimizing the screening condition, a colony carrying a library plasmid with a LEU2 marker survived >3 days. However, this extended survival phenotype was not due to the plasmid but instead was due to an unknown ts mutation. The identity of this ts mutant was revealed by introducing WT SSY5 using the genomic DNA library, which complemented the ts phenotype of ssy5ts. Defective SPS signaling impairs leucine uptake; therefore, deleting components of SPS signaling is lethal in leucine auxotrophic backgrounds (23–25) such as BY4742. To make viable SPS mutants, a pPP81 plasmid carrying the LEU2 gene was introduced before SPS gene deletion. As controls, wild type and other non-SPS mutants used in this study carry pPP81 (26).
Measurement(s) of NAD+, NADH, and NR
Total intracellular levels of NAD+ and NADH were determined using enzymatic cycling reactions as described (26, 27). Relative NR levels were determined by a liquid-based cross-feeding bioassay (8). To prepare cell extracts for intracellular NR determination, ∼2.5 × 109 (∼250 A600 unit) cells (donors of interest) grown to late log-phase (∼16 h growth from an A600 of 0.1) were lysed by bead-beating (Biospec Products) in 800 μl of ice-cold 50 mm ammonium acetate solution. After filter sterilization, 16 μl of clear extract was used to supplement 8-ml cultures of recipient cells (the NR dependent npt1Δbna6Δpho5Δ mutant) (10) with a starting A600 of 0.05 in YPD. To determine NR release levels, the supernatant of the donor cell culture was collected and filter-sterilized, and then 1 ml was added to 7 ml of recipient cell culture with a total starting A600 of 0.05 in YPD. A control culture of recipient cells in YPD without supplementation was included in all experiments. After incubation at 30 °C for 24 h, growth of the recipient cells (A600) was measured and normalized to the cell number of each donor strain. A600 readings were then converted to NR concentrations using the NR standard curve as previously described (8)
Replicative Life Span
All replicative life span (RLS) analyses were carried out on YPD plates with 50 cells per strain for each experiment (27) using a micromanipulator. Statistical analysis was carried out using the JMP statistics software (SAS), and the Wilcoxon rank-sum test p values were calculated for each pair of life spans.
Quantitative PCR (qPCR) Analysis of Gene Expression Levels
Cells were grown to log-phase or late log-phase in YPD (∼6 or ∼16 h of growth from A600 of 0.1). Total RNA was isolated using the RNeasy Mini kit, and cDNA was synthesized using Quantitect Reverse Transcription kit (Qiagen) according to the manufacturer's instructions. For each qPCR reaction, 50 ng of cDNA and 500 nm concentrations of each primer were used. qPCR reaction was run on Roche LightCycler 480 using LightCycler 480 SYBR Green I Master Mix (Roche Applied Science) as previously described (28). Average size of the amplicon for each gene was ∼150 bp. The target mRNA transcript levels were normalized to ACT1 transcript levels.
Pho8-dependent Alkaline Phosphatase Activity Assay
The cell extract-based alkaline phosphatase activity assay was carried out as previously described (29) with modifications. ∼2–3 A600 units of cells grown in YPD for 16 h were collected and washed in 0.85% NaCl with PMSF as described (29). The resultant cell pellet was then resuspended in 600 μl of lysis buffer (20 mm PIPES, 0.5%, Triton X-100, 50 mm KCl, 100 mm potassium acetate, 10 mm MgSO4, 10 μm ZnSO4, 1 mm PMSF) followed by bead-beating. Cell lysates were then centrifuged at 13,200 rpm for 5 min at 4 °C to collect supernatant. 200 μl of supernatant was added to 300 μl of reaction buffer (333 mm Tris-HCl, pH 8.5, 0.53% Triton X-100, 133 mm MgSO4, 13.3 μm ZnSO4, with/without 1.66 mm para-nitrophenyl phosphate) and then incubated at 37 °C for 20 min. Reactions were stopped by adding 500 μl of stop buffer (1 m glycine/KOH, pH 11.0). Supernatants were then collected by centrifugation. The alkaline phosphatase activities were determined by normalizing A400 readings of colorimetric phosphatase product para-nitrophenol to cell number and expressed as 10−6 nmol of para-nitrophenyl produced/min/cell (29).
Chromatin Immunoprecipitation (ChIP) Assay
Log phase yeast cultures (400 ml) in YPD were induced with phenylalanine at 5 mm for 30 min (30). Induced cells were cross-linked with 1% formaldehyde for 30 min at room temperature and stopped by adding glycine to a final concentration of 125 mm. Cells were pelleted by centrifugation and washed two times with cold Tris-buffered saline. Cells were then suspended in 1 ml of FA-140 lysis buffer (50 mm HEPES, 140 mm NaCl, 1% Triton X-100, 1 mm EDTA, 0.1% sodium deoxycholate, 0.1 mm PMSF, 1× protease inhibitor mixture (Pierce)) (31) and lysed by bead-beating. The cell lysate was drawn off the beads and centrifuged at a maximum speed (13,200 rpm) for 30 min at 4 °C. The chromatin pellet was resuspended in 1 ml of FA-140 lysis buffer and sonicated on ice 8 times with 20-s pulses using a Branson 450 Sonifer (output control set at 1.5 and duty cycle held at constant) to shear chromatin to an average length of ∼500 bp. Sonicated chromatin solution was centrifuged twice at 10,000 rpm for 10 min at 4 °C. The supernatant was then aliquoted into three tubes (labeled “input”, “IP,” and “no-Ab”). The IP sample was incubated overnight at 4 °C with anti-HA monoclonal antibody (ab1424, Abcam) at a dilution of 1:150. Both IP and no-Ab samples were incubated with 60 μl of ChIP-grade protein G beads (Cell Signaling Technology) for 2 h at 4 °C and then washed as described (31). DNA was then eluted from the beads 2 times with 125 μl of elution buffer (5× Tris-EDTA plus 1% SDS). The combined DNA solution and input samples were incubated at 65 °C overnight to reverse the cross-linking. The purified DNA samples were analyzed by qPCR, and results were compared with a standard curve prepared from input DNA. The amount of immunoprecipitated DNA from the experimental promoters (OAC1, MAE1, and PHO8), the positive control promoter (AGP1) (30), and the negative control (ACT1) (32) was determined relative to no-Ab DNA. The oligonucleotide sequences used are provided in Fig. 5A. S.D. were calculated from the results for three independent biological replicates.
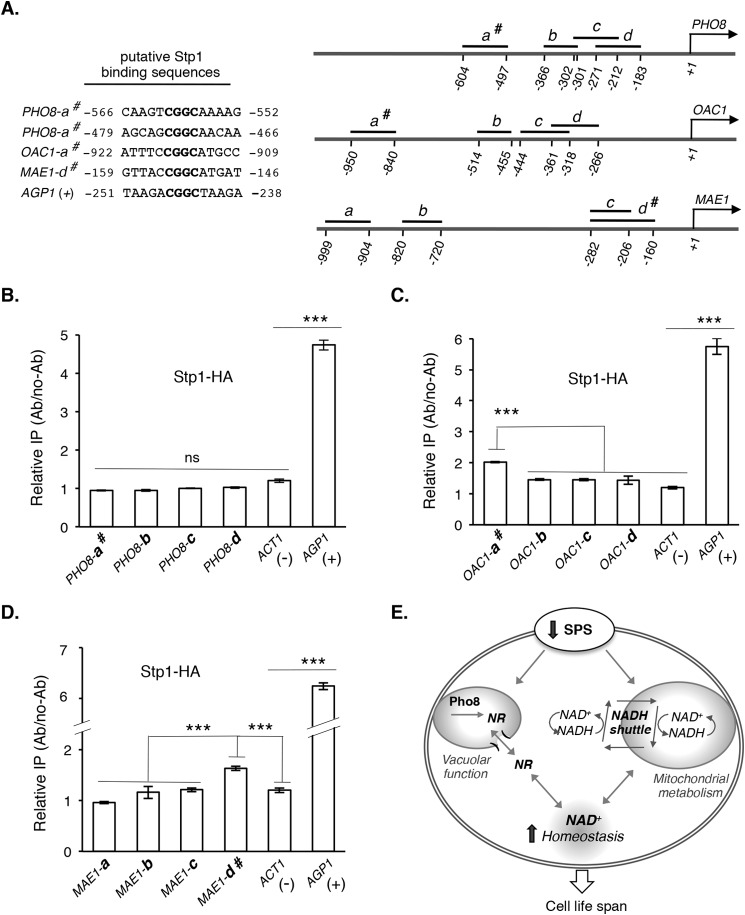
FIGURE 5.
Stp1, a transcription factor of SPS signaling pathway, may directly bind to the promoter regions of NADH shuttle components. A, putative binding sites of Stp1 in the promoter regions of PHO8, OAC1, and MAE1 are shown in the left panel. DNA fragments used for ChIP analysis are shown in the right panel. # denotes fragments containing a putative Stp1 binding site. B, HA-tagged Stp1 (Stp1-HA) does not significantly bind to various promoter regions of PHO8 compared with AGP1 as positive control (+) and ACT1 as negative control (−), determined by qPCR. C, Stp1-HA significantly binds to OAC1-a# but not other promoter regions of OAC1. D, Stp1-HA significantly binds to MAE1-d# but not other promoter regions of MAE1. E, proposed model of how reduced SPS signaling leads to increased NAD+ homeostasis (via NR salvage and NADH shuttle), which works in concert with mitochondrial and vacuolar metabolism to maintain cell function and extend life span. For clarity, additional factors and interactions are not shown. Data shown are representative of multiple independent experiments. Error bars denote S.D. derived from triplicate samples. The p values were calculated using Student's t test (ns, not significant; ***, p < 0.005).
RESULTS
Yeast Mutants with Reduced SPS Signaling Exhibit Increased Replicative Life Span
A temperature-sensitive ssy5 loss-of-function mutant was identified in a screen for long-lived mutants (see “Experimental Procedures”). SSY5 is part of the SPS nutrient-sensing pathway in S. cerevisiae (Fig. 1A) (11, 12, 33). We first examined whether deleting SSY5 indeed extended life span. As shown in Fig. 1B, cells lacking SSY5 exhibited an ∼50% extension in RLS (cell division potential) compared with wild type (WT). To determine whether the observed life span extension was due to reduced SPS signaling activity, we analyzed mutants lacking specific SPS components. As shown in Fig. 1B, all SPS mutants examined, except for stp2Δ, showed significantly increased RLS (p < 0.005) compared with WT. STP1 and STP2 are homologous SPS downstream transcription factors that share overlapping function (30, 34). These results support that the RLS extension observed in the ssy5Δ mutant was due to overall reduction in SPS activity. Henceforth ssy5Δ was used as a positive control for RLS extension representative of reduced SPS pathway.
Lowered SPS activity may mimic nutrient restriction; thus, we tested whether ssy5Δ functions in previously characterized CR pathways to extend life span. As shown in Fig. 1, C and D, both moderate CR (0.5% glucose versus 2% glucose as the normal condition) (1, 27) (Fig. 1C) and a genetic mimic of CR, cdc25-10 (1) (Fig. 1D), further extended the long life span of ssy5Δ cells. Therefore, reduced SPS activity and CR appeared to modulate life span by different mechanisms.
Reduced Amino Acid Uptake Does Not Mimic the Long Life Span of ssy5Δ
As an amino acid sensor, major downstream targets of SPS include several amino acid permeases (35), some of which have been shown to modulate life span (36). Lowered SPS signaling reduced the expression of these permeases (35), raising the possibility that the long life span of ssy5Δ was due to reduced amino acid uptake. To address this, we determined the RLS of deletions of amino acid permeases AGP1, BAP2, and BAP3, which are downstream targets of SPS. Agp1 is a low affinity broad range amino acid permease, which transports asparagine, glutamine, and other amino acids (37, 38). The paralogs Bap2 and Bap3 are branched-chain amino acid permeases that transport leucine, isoleucine, and valine (39, 40). Unlike ssy5Δ, both the single deletions (agp1Δ, bap2Δ, or bap3Δ) and triple deletion (agp1Δbap2Δbap3Δ) of these permeases did not significantly extend RLS (Fig. 1E). Therefore, reduced amino acid uptake mediated by these permeases was not responsible for the life span extension observed in ssy5Δ cells.
Components of NADH Shuttle System Are Required for SPS Signaling-mediated Life Span Extension
To understand how ssy5Δ extends RLS, we examined additional potential downstream targets of SPS. Several components of the putative malate-pyruvate NADH shuttle system (41) (Fig. 2A) were suggested to be transcriptionally regulated by SPS (35, 42).
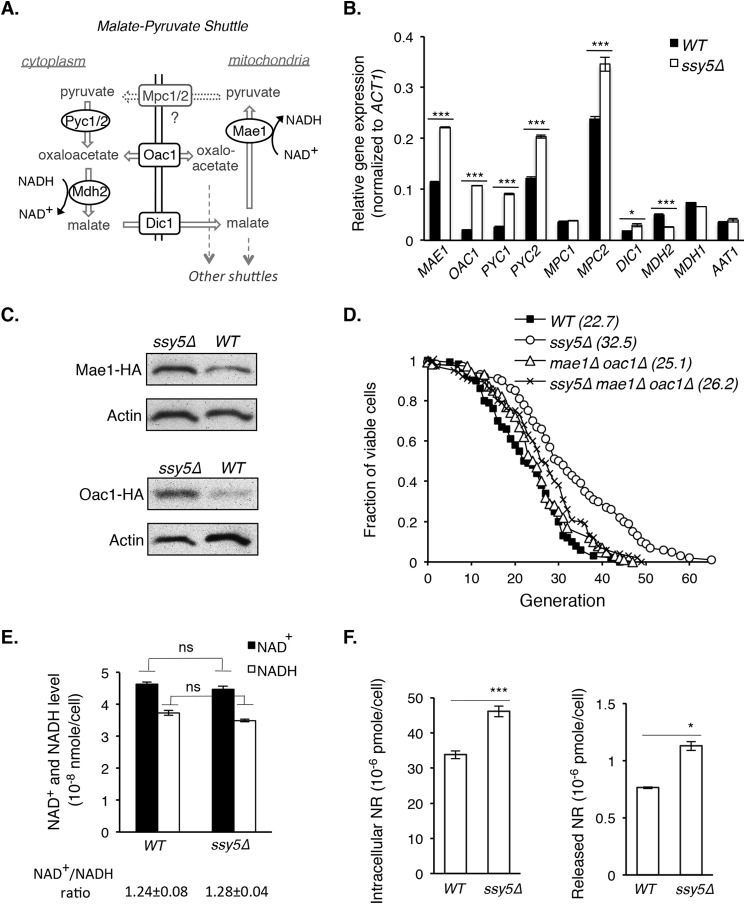
FIGURE 2.
SPS signaling-mediated life span extension requires components of NADH shuttle systems. A, a simplified model of the putative malate-pyruvate NADH shuttle. Cellular NAD+/NADH ratio is regulated by groups of cytoplasmic (e.g. Pyc1/Pyc2 and Mdh2) and mitochondrial (e.g. Mae1) enzymes, which produce permeable or small metabolites that can be transported (via carrier proteins Dic1, Oac1, and Mpc1/Mpc2) across the inner membrane. As a result, the NAD+/NADH ratio and metabolites are balanced between the mitochondrial and cytoplasmic pools. B, gene expression of the malate-pyruvate shuttle components increases in the ssy5Δ mutant. Results show gene expression comparisons between WT and ssy5Δ cells determined by qPCR. MDH1 and AAT1 are components of another NADH shuttle, shown as a control. C, Mae1 and Oac1 protein levels increase in the ssy5Δ mutant. Results show Western blot analysis of HA-tagged Oac1 and Mae1 in both WT and ssy5Δ cells. D, deletions of both MAE1 and OAC1 largely abolish the life span extension by ssy5Δ (ssy5Δ versus ssy5Δmae1Δoac1Δ; p < 0.005). E, the levels and ratio of NAD+ and NADH are not changed by ssy5Δ. F, the level of nicotinamide riboside (NR, a NAD+ precursor) increases in ssy5Δ cells. Data shown are representative of multiple independent experiments. Error bars denote S.D. derived from triplicate samples. The p values were calculated using Student's t test (ns, not significant; *, p < 0.05; ***, p < 0.005) except for D (Wilcoxon rank sum test).
The malate-pyruvate NADH shuttle system consists of mitochondrial enzymes (Mae1; Fig. 2A) and cytoplasmic enzymes (Pyc1/Pyc2 and Mdh2; Fig. 2A) (41). These enzymes produce permeable or small metabolites that can be transported via carrier proteins (Dic1, Oac1, and Mpc1/Mpc2; Fig. 2A) across the mitochondrial inner membrane (41, 43). Because some of these enzymes require NAD+ or NADH for redox reactions, the NAD+/NADH ratio is concomitantly balanced between the mitochondrial and cytoplasmic pools. Thus, this group of enzymes is described as a NADH shuttle although they do not shuttle NADH directly (41).
As shown in Fig. 2B, expression of these shuttle components indeed significantly increased in ssy5Δ cells. Expression of the negative controls, MDH1 and AAT1, components of another shuttle (malate-aspartate shuttle), was not increased by ssy5Δ. Increased expression of MAE1 and OAC1 was further validated by Western blot analysis using HA-tagged Mae1 and Oac1 (Fig. 2C). Moreover, deleting both MAE1 and OAC1 largely abolished the life span extension in ssy5Δ cells (ssy5Δ versus ssy5Δmae1Δoac1Δ; p < 0.005) (Fig. 2D), suggesting the malate-pyruvate NADH shuttle contributes to ssy5Δ-mediated RLS.
To discern the mechanism, we first examined the NAD+/NADH ratio in ssy5Δ cells. As shown in Fig. 2E, the NAD+/NADH ratio was not significantly changed by ssy5Δ, suggesting that the increased malate-pyruvate shuttle expression (Fig. 2B) did not alter the ratio. Nevertheless, the malate-pyruvate shuttle is entwined with the TCA cycle, which produces various biosynthetic intermediates including amino acid precursors (44). Due to reduced amino acid uptake (23–25), ssy5Δ may increase the malate-pyruvate shuttle expression (Fig. 2B) to produce amino acid precursors via the TCA cycle. This predicts increased demand for NAD+ and NADH in ssy5Δ cells, prompting us to study whether SPS signaling modulates NAD+ homeostasis.
NR Production Contributes to NAD+ Homeostasis in ssy5Δ Cells
Because NAD+ and NADH levels did not change in ssy5Δ cells (Fig. 2E), we determined the level of NR, a NAD+ precursor shown to contribute to NAD+ homeostasis and life span (7, 8). Interestingly, both intracellular and released NR levels were elevated in ssy5Δ cells (Fig. 2F), suggesting NR salvaging activity was increased, which may contribute to NAD+ biosynthesis. To identify the source of elevated NR in ssy5Δ cells (Fig. 2F), we examined the gene expression of pathways known to produce and salvage NAD+ and NR (8, 10, 45–47). Expression levels of the genes in NAD+ synthetic pathways generally remained similar between WT and ssy5Δ cells as we expected (Fig. 3A), as no change in NAD+ and NADH levels was observed between these strains (Fig. 2E). Therefore, the increased NR in ssy5Δ cells (Fig. 2F) was not likely from canonical NAD+ biosynthetic pathways. Instead, elevated NR in ssy5Δ cells is more likely due to increased expression of the phosphate-sensing PHO pathway (35, 48, 49) components (Fig. 3B), some of which were previously shown to produce NR, and therefore contribute to NAD+ homeostasis (4, 10).
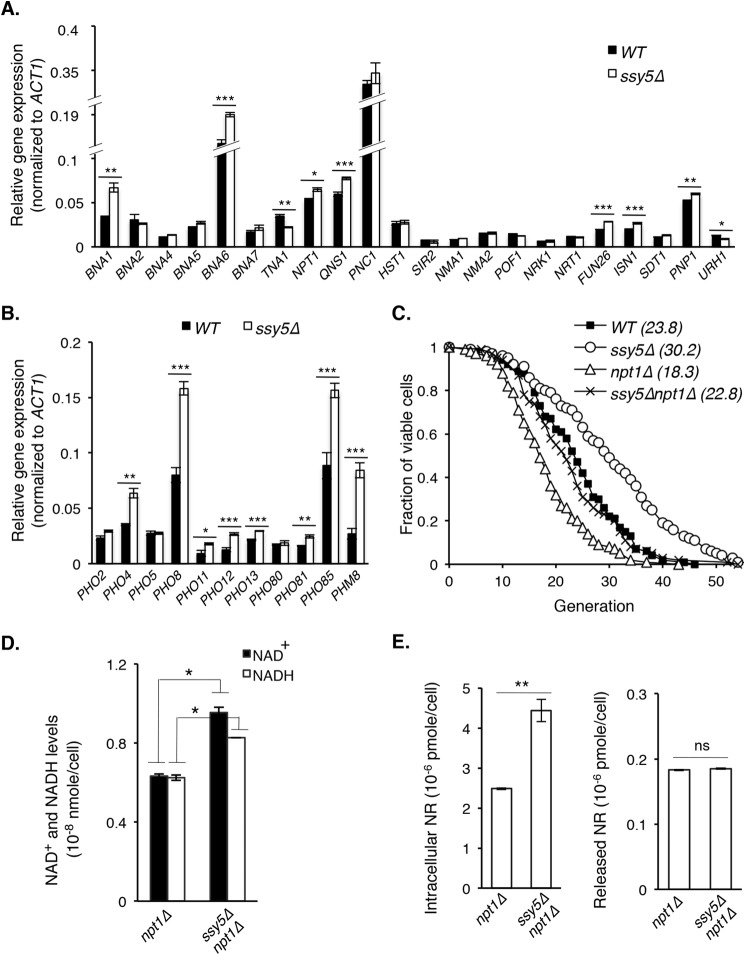
FIGURE 3.
NR production contributes to enhanced NAD+ homeostasis in ssy5Δ cells. A, overall gene expression of the NAD+ biosynthetic pathway remains largely similar in ssy5Δ cells compared with WT. Results show relative gene expression (normalized to ACT1) determined by qPCR. B, deletion of SSY5 increases gene expression of the phosphate sensing (PHO) pathway components. Results show gene expression comparisons between WT and ssy5Δ cells. C, deletion of SSY5 rescues the short RLS of the NAD+ biosynthesis-deficient npt1Δ mutant (npt1Δ versus ssy5Δnpt1Δ; p < 0.005). D, deletion of SSY5 increases both NAD+ and NADH levels in npt1Δ cells. E, deletion of SSY5 increases intracellular NR levels in npt1Δ cells. Data shown are representative of multiple independent experiments. Error bars denote S.D. derived from triplicate samples. The p values were calculated using Student's t test (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.005) except for C (Wilcoxon rank sum test).
Previously, NR supplementation was shown to rescue the short life span and reduced NAD+ level of the npt1Δ mutant (7). Because we did not observe increases in NAD+ and NADH levels by deleting SSY5 in WT background, we examined whether increased NR in ssy5Δ cells would improve RLS and NAD+ homeostasis in the npt1Δ mutant. First, we tested if ssy5Δ could rescue the short RLS of the npt1Δ mutant. As shown in Fig. 3C, deletion of SSY5 rescued the RLS of npt1Δ mutant to WT level (npt1Δ versus ssy5Δnpt1Δ; p < 0.005). This was accompanied by increases in NAD+ and NADH levels (Fig. 3D) as well as intracellular NR levels (Fig. 3E) compared with the npt1Δ mutant. These results support that elevated NR in ssy5Δ cells contributes to NAD+ homeostasis and plays a role in ssy5Δ-mediated RLS extension.
Reduced SPS Signaling Increases the Expression of Specific PHO Signaling and NADH Shuttle Components Independent of PHO2 and PHO4
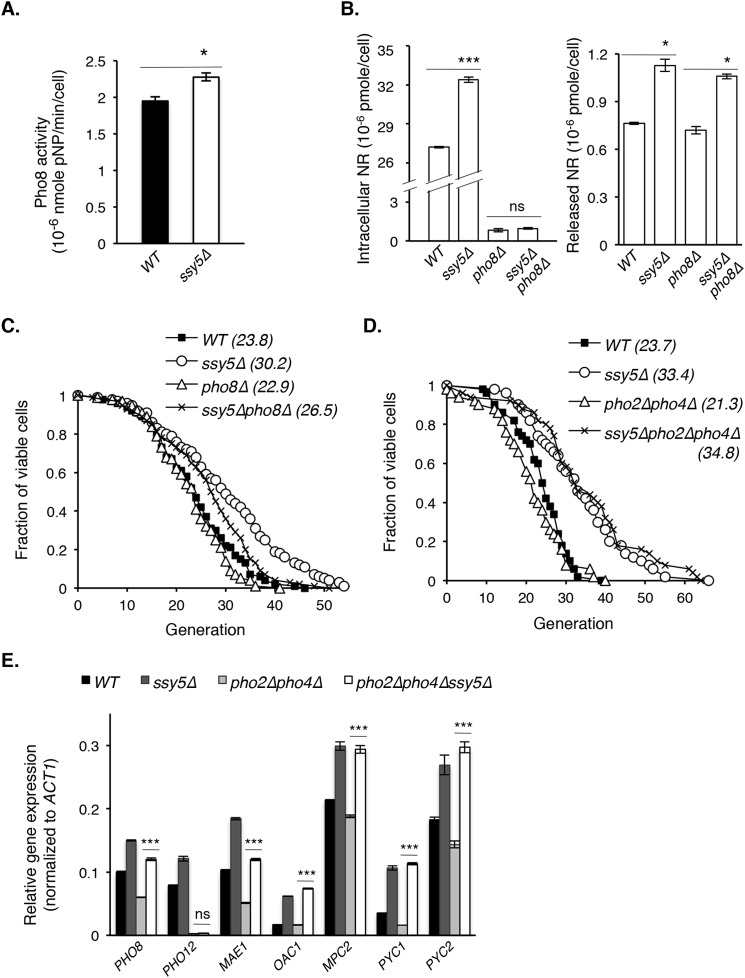
Next, we sought to identify factors responsible for ssy5Δ-induced NR increase. A likely candidate was Pho8, whose expression was increased by ssy5Δ (Fig. 3B). Pho8 is a vacuolar phosphatase (50) shown to produce NR from nicotinamide mononucleotide (10). We examined Pho8 activity using cell lysates of WT and ssy5Δ cells as previously described (29). Indeed, ssy5Δ cells showed a slight increase in Pho8 activity (Fig. 4A). In addition, deleting PHO8 abolished the intracellular (Fig. 4B, left panel) but not the released (Fig. 4B, right panel), NR increase in ssy5Δ cells and correspondingly mitigated the life span extension in ssy5Δ cells (Fig. 4C). Interestingly, deletion of both PHO2 and PHO4, the transcription factors that regulate PHO pathway (49), did not block life span extension in ssy5Δ cells (Fig. 4D), suggesting that ssy5Δ regulates certain PHO downstream components independent of PHO2 and PHO4. Indeed, as shown in Fig. 4E, SSY5 deletion still increased PHO8 expression in the pho2Δpho4Δ background. In contrast, PHO12 transcript (negative control) is under strict PHO2/PHO4 regulation (51) and was undetectable in pho2Δpho4Δ background (Fig. 4E).
FIGURE 4.
Reduced SPS signaling increases the expression of specific PHO signaling and NADH shuttle components independent of PHO2 and PHO4. A, vacuolar Pho8 phosphatase activity is slightly increased in ssy5Δ cells. Results show Pho8 phosphatase activity using para-nitrophenyl phosphate as a substrate for cell lysate derived from WT and ssy5Δ cells. Pho8 activity is reflected by production rate of colorimetric para-nitrophenol (pNP), determined at A400. B, deletion of PHO8 abolishes intracellular NR increase in ssy5Δ cells (left panel) but not released NR, which mostly originates from cytosolic phosphatases (right panel). C, deletion of PHO8 partially abolishes the life span extension in ssy5Δ cells (ssy5Δ versus ssy5Δpho8Δ; p < 0.05). D, deletions of both PHO2 and PHO4, transcription factors that regulate PHO pathway, do not alter the life span extension in ssy5Δ cells. E, ssy5Δ increases gene expression of PHO8 and the malate-pyruvate shuttle components independent of PHO2 and PHO4. Results show relative gene expression (normalized to ACT1) of WT, ssy5Δ, pho2Δpho4Δ, and ssy5Δpho2Δpho4Δ cells determined by qPCR. Data shown are representative of multiple independent experiments. Error bars denote S.D. derived from triplicate samples. The p values were calculated using Student's t test (ns, not significant; *, p < 0.05; ***, p < 0.005) except for C and D (Wilcoxon rank sum test).
In addition to PHO8, components of the malate-pyruvate shuttle were required to extend RLS in ssy5Δ (Fig. 2D). Although potential Pho2/Pho4 binding sequences (CACGTG) (52) were found in the promoter regions of OAC1 and PYC1, deletion of both PHO2 and PHO4 did not block ssy5Δ-induced increases in gene expression of the malate-pyruvate shuttle components examined (Fig. 4E). However, the basal level expression of MAE1 and PHO8 decreased in pho2Δpho4Δ background (Fig. 4E), suggesting some control by Pho2/Pho4. Overall, these observations are in line with the result that ssy5Δ extends RLS independent of Pho2 and Pho4 (Fig. 4D) and supports a role for NR and NAD+ homeostasis in ssy5Δ-mediated RLS extension.
Stp1, a Transcription Factor of the SPS Signaling Pathway, May Directly Bind to the Promoter Regions of Shuttle System Components
In this study we show PHO8, MAE1, and OAC1 are required for ssy5Δ-induced RLS (Figs. 2D and 4C) and that the expression of these three genes is controlled by SPS signaling (Figs. 2B and 3B). When SPS-activated Stp1 binds to target promoters, it may activate or inhibit gene expression (Fig. 1A). That expression of PHO8, MAE1, and OAC1 increased in SPS-defective ssy5Δ cells (Figs. 2B and 3B) suggests that SPS-activated Stp1 binding inhibits expression of these promoters. Thus, in SPS-defective ssy5Δ cells, Stp1 is retained in the cytoplasm, resulting in increased expression of these genes. However, it remains unclear whether these genes are direct targets of Stp1. Therefore, we searched for and found putative Stp1 binding sequences (53–55) on the promoter of PHO8, MAE1, and OAC1 (Fig. 5A, left panel). Next, we carried out ChIP analysis of the promoter fragments of these genes using HA-tagged Stp1 (Stp1-HA) to examine whether Stp1 directly binds to these promoters. The AGP1 promoter fragment AGP1 (+) was used as a positive control for Stp1 binding as previously described (30). Surprisingly, Stp1 did not appear to bind to the DNA fragment containing two Stp1 binding sites (PHO8-a#) (Fig. 5A) under the condition examined (Fig. 5B). Thus, PHO8 is less likely to be a direct target of Stp1. On the other hand, Stp1-HA showed significantly increased binding to the DNA fragments containing putative Stp1 binding sites (OAC1-a# and MAE1-d#) (Fig. 5A) for both OAC1 (Fig. 5C) and MAE1 (Fig. 5D). Together, these results suggest that both MAE1 and OAC1 are direct targets of Stp1 and that the regulation of PHO8 expression by SPS signaling likely requires an additional transcription factor(s).
DISCUSSION
In this study we characterized a low SPS activity mutant, ssy5Δ and showed that NAD+ homeostasis plays an important role in ssy5Δ-mediated life span extension (Fig. 5E). Expression of malate-pyruvate NADH shuttle components was increased in ssy5Δ cells (Fig. 2B). Of these components, deleting MAE1 and OAC1 significantly reduced ssy5Δ-induced life span extension (Fig. 2D). We also showed that Stp1, a transcription factor of the SPS pathway, directly binds to the promoter of MAE1 and OAC1 to regulate their expression (Fig. 5, C and D). In addition to the shuttle components, ssy5Δ enhances NAD+ homeostasis by increasing NR salvaging. Deletion of SSY5 increases NR level (Fig. 2F), which partially restores the NAD+ pool (Fig. 3D) and life span (Fig. 3C) of the short-lived and NAD+-depleted npt1Δ mutant. A main source of increased NR in ssy5Δ cells is the vacuolar phosphatase Pho8 (Fig. 4B), which is also required for full life span extension by ssy5Δ (Fig. 4C). Interestingly, ssy5Δ-induced PHO8 expression is independent of the canonical PHO pathway (Fig. 4E). Moreover, SPS-mediated regulation of Pho8 likely involves additional yet-to-be identified transcription factors as Stp1 does not appear to bind to PHO8 promoter (Fig. 5B) under the conditions tested. Together, our studies have unraveled SPS signaling as a regulator of NAD+ homeostasis. Enhanced NAD+ homeostasis due to reduced SPS signaling may concomitantly increase mitochondrial and vacuolar function, all contributing to life span extension (Fig. 5E).
Although ssy5Δ synergizes with glucose restriction to extend life span (Fig. 1, C and D), suggesting they function in parallel pathways, they also modulated different components in similar pathways. For example, ssy5Δ and CR require different NADH shuttles for life span extension. The malate-pyruvate shuttle plays an important role in the ssy5Δ pathway (Fig. 2D), whereas the malate-aspartate shuttle is important for CR (26). Interestingly, increased NAD+/NADH ratio was observed in cells under CR (26) but not in ssy5Δ cells (Fig. 2E). Perhaps increased shuttle activity in ssy5Δ cells does not alter the ratio but instead maintains balance between the mitochondrial and cytoplasmic NAD+(H) pools. Another possibility is that increased expression of NR salvaging factors by ssy5Δ (Fig. 3, A and B) leads to NR production (Fig. 2F and 3E) and replenishes the NAD+ pool (Fig. 3D), thereby compensating depletion of NAD+ or NADH. Notably, CR-induced increase in the NAD+/NADH ratio is due to decreased NADH level (56); however, the mechanisms underlying this decrease remain unclear.
One amino acid permease target of SPS signaling, AGP1, has been linked to TOR signaling and shown to extend chronological life span (cell survival at a non-dividing G0 state) when deleted (36). In ssy5Δ cells, low SPS activity results in decreased AGP1 expression, and therefore, longer chronological life span is anticipated. Interestingly, ssy5Δ only extends RLS (Fig. 1B) but not chronological life span (data not shown). In addition, deleting AGP1 and additional SPS downstream amino acid permeases, BAP2 and BAP3, is not sufficient to mimic ssy5Δ and extend RLS (Fig. 1E). These studies support that reduced amino acid uptake mediated by these specific amino acid permeases is not the major cause of ssy5Δ-induced RLS extension, although it plays important roles in chronological life span. Instead, our studies suggest that enhanced mitochondrial metabolism plays a key role in ssy5Δ-induced life span. The malate-pyruvate NADH shuttle is entwined with the TCA cycle, which supports mitochondrial energy metabolism and produces intermediates for various biosynthetic pathways (44). Because lowered SPS signaling reduces amino acid uptake, ssy5Δ cells may compensate by up-regulating expression of malate-pyruvate shuttle components (Fig. 2B) to increase amino acid biosynthesis via the TCA cycle. This is in line with ssy5Δ cells requiring functional malate-pyruvate shuttle for life span extension (Fig. 2D). Overall, this evidence supports a key role for enhanced mitochondrial metabolism in the extended life span of ssy5Δ cells. In mammals, impaired mitochondrial metabolism and NADH shuttles have also been implicated in age-associated diseases such as diabetes (57, 58).
Our studies identified Pho8 as an important player in ssy5Δ-increased NR salvaging. Pho8 produces NR from nicotinamide mononucleotide as a major source of intracellular NR and contributes to NAD+ homeostasis (10). Because deletion of Pho8 only partially abrogates ssy5Δ-induced life span extension (Fig. 4C), additional NR homeostasis factors likely play a role. In fact, deleting Pho8 abolishes only intracellular, not released, NR increases in ssy5Δ cells (Fig. 4B). We previously showed that intracellular NR level largely reflects the NR stored in the vacuole (produced mainly via Pho8) (10). On the other hand, released NR level reflects the NR produced in the cytoplasm (mainly via nucleotidases, Isn1 and Sdt1) (9) because cytoplasmic NR dynamically exchanges with the extracellular environment (4, 10, 28). It would be interesting to determine whether other NR homeostasis factors, such as Isn1 and Sdt1, also play a role in ssy5Δ-mediated NR salvaging. Increased Pho8 activity in ssy5Δ cells not only increases NR and NAD+ homeostasis but is also likely to contribute to vacuolar function. The vacuolar H+-ATPase (v-ATPase) was previously shown to play essential roles in preserving mitochondrial function in yeast (59). Overexpression of VPH1, VMA1 (v-ATPase subunits), VPH2 (required for v-ATPase function and assembly), and AVT1 (a H+-dependent vacuolar amino acid importer) preserved vacuolar acidity and amino acid import and extended RLS (59). These factors are suggested to extend RLS by preserving vacuolar function, allowing proper transport of amino acids and metabolites between the cytoplasm and vacuole. Compromised vacuolar function led to mitochondrial fragmentation and malfunction perhaps due to abnormal accumulation of metabolites and reduced metabolic flexibility (59). Interestingly, AVT1, VMA2, and VMA4 have been suggested to be downstream targets of SPS signaling pathway (60–62). Thus, enhanced overall vacuolar function is tightly connected to proper mitochondrial metabolism, and both play important roles in ssy5Δ-mediated life span extension.
The vacuole also plays an important role in NR and NAD+ homeostasis. We previously showed the vacuole is a major source of NR production (via Pho8) and storage (10). Our studies suggest NR produced by Pho8 plays an important role in ssy5Δ cells, as deleting PHO8 largely abolished intracellular NR increase (Fig. 4B) and mitigated life span extension induced by ssy5Δ (Fig. 4C). Because NAD+ biosynthesis takes place in the cytoplasm and nucleus, how is vacuolar NR converted to NAD+? We have previously identified Fun26 as a putative NR transporter on the vacuolar membrane (10). Fun26 is a homolog of the human ENT (equilibrative nucleoside transporter) protein family that facilitates transport of various purine and pyrimidine nucleosides (63). Deleting FUN26 causes a significant increase in NR release and accumulation (10). Interestingly, expression of FUN26 is slightly increased in ssy5Δ cells (Fig. 3A), suggesting enhanced NR exchange between the cytoplasmic and vacuolar pools. Overall, these studies support a role for vacuolar NR mediating NAD+ homeostasis in ssy5Δ-mediated life span extension.
Maintaining NAD+ homeostasis is essential for cellular function: aberrant NAD+ metabolism has been implicated in numerous metabolic and age-associated diseases (5). Due to its complex nature, factors regulating NAD+ metabolism and homeostasis are not completely understood, although recent studies have identified several regulators of NAD+ homeostasis. For example, the mitochondria and vacuole were shown to play important roles in the biosynthesis and metabolism of pyridine nucleotides such as NAD+(H) and NADP+(H) (4, 41, 64, 65). Our studies similarly support that compartmentalization of pyridine metabolites is pivotal for regulating NAD+ homeostasis. Recently, NAD+ metabolism was linked to the PHO pathway in yeast. Activation of the PHO pathway is associated with increased NR production and mobilization (10). Cross-regulation of PHO and multiple CR-related nutrient-sensing pathways (cAMP-PKA, TOR, Sch9/AKT) have been reported (4); however, detailed mechanisms remain unclear. Our current studies link SPS signaling to PHO signaling in which reduced SPS signaling activates PHO gene expression (Fig. 3B). However, SPS signaling also regulates certain PHO downstream targets (such as Pho8) independent of the canonical PHO transcription factors Pho2/4 (Fig. 4E), highlighting the complex regulation of these pathways. Most likely, the interplay between longevity signaling pathways is essential for conferring metabolic flexibility in different growth conditions. Future studies to identify and characterize additional signaling factors that regulate cross-talk between NAD+ homeostasis and longevity regulating signaling pathways are highly anticipated. Overall, our studies contribute to comprehending how nutrient signaling pathways regulate NAD+ homeostasis and may also provide insight into the underlying mechanisms of diseases related to defects in NAD+ metabolism.
Acknowledgments
We thank Dr. C. Skinner and T. Croft for critical reading of this manuscript and discussion, Dr. D. Meyer for assistance with ChIP assay, and A. Rymer, E. Su, K. Mehta, and S. Yang for assistance with yeast strain construction. We also thank Dr. T. Powers, Dr. B. Liu, and Dr. K. Shiozaki for suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM102297 (NIGMS).
- CR
- calorie restriction
- NR
- nicotinamide riboside
- SPS
- Ssy1-Ptr3-Ssy5
- YPD
- yeast extract/peptone/dextrose
- RLS
- replicative life span
- qPCR
- quantitative PCR.
REFERENCES
- 1. Lin S. J., Defossez P. A., Guarente L. (2000) Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128 [DOI] [PubMed] [Google Scholar]
- 2. Fabrizio P., Pozza F., Pletcher S. D., Gendron C. M., Longo V. D. (2001) Regulation of longevity and stress resistance by Sch9 in yeast. Science 292, 288–290 [DOI] [PubMed] [Google Scholar]
- 3. Kaeberlein M., Powers R. W., 3rd, Steffen K. K., Westman E. A., Hu D., Dang N., Kerr E. O., Kirkland K. T., Fields S., Kennedy B. K. (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196 [DOI] [PubMed] [Google Scholar]
- 4. Kato M., Lin S. J. (2014) Regulation of NAD+ metabolism, signaling and compartmentalization in the yeast Saccharomyces cerevisiae. DNA Repair 23, 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Imai S., Guarente L. (2014) NAD+ and sirtuins in aging and disease. Trends Cell Biol. 24, 464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bieganowski P., Brenner C. (2004) Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117, 495–502 [DOI] [PubMed] [Google Scholar]
- 7. Belenky P., Racette F. G., Bogan K. L., McClure J. M., Smith J. S., Brenner C. (2007) Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129, 473–484 [DOI] [PubMed] [Google Scholar]
- 8. Lu S. P., Kato M., Lin S. J. (2009) Assimilation of endogenous nicotinamide riboside is essential for calorie restriction-mediated life span extension in Saccharomyces cerevisiae. J. Biol. Chem. 284, 17110–17119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bogan K. L., Evans C., Belenky P., Song P., Burant C. F., Kennedy R., Brenner C. (2009) Identification of Isn1 and Sdt1 as glucose- and vitamin-regulated nicotinamide mononucleotide and nicotinic acid mononucleotide [corrected] 5′-nucleotidases responsible for production of nicotinamide riboside and nicotinic acid riboside. J. Biol. Chem. 284, 34861–34869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu S. P., Lin S. J. (2011) Phosphate-responsive signaling pathway is a novel component of NAD+ metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 286, 14271–14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Omnus D. J., Pfirrmann T., Andréasson C., Ljungdahl P. O. (2011) A phosphodegron controls nutrient-induced proteasomal activation of the signaling protease Ssy5. Mol. Biol. Cell 22, 2754–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abdel-Sater F., Jean C., Merhi A., Vissers S., André B. (2011) Amino acid signaling in yeast: activation of Ssy5 protease is associated with its phosphorylation-induced ubiquitylation. J. Biol. Chem. 286, 12006–12015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Z., Thornton J., Spírek M., Butow R. A. (2008) Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol. Cell. Biol. 28, 551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andréasson C., Heessen S., Ljungdahl P. O. (2006) Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 20, 1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Omnus D. J., Ljungdahl P. O. (2014) Latency of transcription factor Stp1 depends on a modular regulatory motif that functions as cytoplasmic retention determinant and nuclear degron. Mol. Biol. Cell 25, 3823–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zargari A., Boban M., Heessen S., Andréasson C., Thyberg J., Ljungdahl P. O. (2007) Inner nuclear membrane proteins Asi1, Asi2, and Asi3 function in concert to maintain the latent properties of transcription factors Stp1 and Stp2. J. Biol. Chem. 282, 594–605 [DOI] [PubMed] [Google Scholar]
- 17. Foresti O., Rodriguez-Vaello V., Funaya C., Carvalho P. (2014) Quality control of inner nuclear membrane proteins by the Asi complex. Science 346, 751–755 [DOI] [PubMed] [Google Scholar]
- 18. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 19. Burke D., Dawson D., Sterns T. (2000) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, pp. 171–174, Cold Spring Harbor, NY [Google Scholar]
- 20. Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang C., Skinner C., Easlon E., Lin S. J. (2009) Deleting the 14-3-3 protein Bmh1 extends life span in Saccharomyces cerevisiae by increasing stress response. Genetics 183, 1373–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Broach J. R. (2012) Nutritional control of growth and development in yeast. Genetics 192, 73–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forsberg H., Hammar M., Andréasson C., Molinér A., Ljungdahl P. O. (2001) Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 158, 973–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klasson H., Fink G. R., Ljungdahl P. O. (1999) Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19, 5405–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 26. Easlon E., Tsang F., Skinner C., Wang C., Lin S. J. (2008) The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 22, 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Easlon E., Tsang F., Dilova I., Wang C., Lu S. P., Skinner C., Lin S. J. (2007) The dihydrolipoamide acetyltransferase is a novel metabolic longevity factor and is required for calorie restriction-mediated life span extension. J. Biol. Chem. 282, 6161–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kato M., Lin S. J. (2014) YCL047C/POF1 Is a Novel Nicotinamide mononucleotide adenylyltransferase (NMNAT) in Saccharomyces cerevisiae. J. Biol. Chem. 289, 15577–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noda T., Klionsky D. J. (2008) The quantitative Pho8Δ60 assay of nonspecific autophagy. Methods Enzymol. 451, 33–42 [DOI] [PubMed] [Google Scholar]
- 30. Wielemans K., Jean C., Vissers S., André B. (2010) Amino acid signaling in yeast: post-genome duplication divergence of the Stp1 and Stp2 transcription factors. J. Biol. Chem. 285, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li M., Petteys B. J., McClure J. M., Valsakumar V., Bekiranov S., Frank E. L., Smith J. S. (2010) Thiamine biosynthesis in Saccharomyces cerevisiae is regulated by the NAD+-dependent histone deacetylase Hst1. Mol. Cell. Biol. 30, 3329–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lodhi N., Tulin A. V. (2011) PARP1 genomics: chromatin immunoprecipitation approach using anti-PARP1 antibody (ChIP and ChIP-seq). Methods Mol. Biol. 780, 191–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ljungdahl P. O. (2009) Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 37, 242–247 [DOI] [PubMed] [Google Scholar]
- 34. Tumusiime S., Zhang C., Overstreet M. S., Liu Z. (2011) Differential regulation of transcription factors Stp1 and Stp2 in the Ssy1-Ptr3-Ssy5 amino acid sensing pathway. J. Biol. Chem. 286, 4620–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eckert-Boulet N., Nielsen P. S., Friis C., dos Santos M. M., Nielsen J., Kielland-Brandt M. C., Regenberg B. (2004) Transcriptional profiling of extracellular amino acid sensing in Saccharomyces cerevisiae and the role of Stp1p and Stp2p. Yeast 21, 635–648 [DOI] [PubMed] [Google Scholar]
- 36. Powers R. W., 3rd, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S. (2006) Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20, 174–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schreve J. L., Sin J. K., Garrett J. M. (1998) The Saccharomyces cerevisiae YCC5 (YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagine and glutamine. J. Bacteriol. 180, 2556–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Forsberg H., Gilstring C. F., Zargari A., Martínez P., Ljungdahl P. O. (2001) The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol. Microbiol. 42, 215–228 [DOI] [PubMed] [Google Scholar]
- 39. Grauslund M., Didion T., Kielland-Brandt M. C., Andersen H. A. (1995) BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1269, 275–280 [DOI] [PubMed] [Google Scholar]
- 40. Regenberg B., Düring-Olsen L., Kielland-Brandt M. C., Holmberg S. (1999) Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36, 317–328 [DOI] [PubMed] [Google Scholar]
- 41. Bakker B. M., Overkamp K. M., van Maris A. J., Kötter P., Luttik M. A., van Dijken J. P., Pronk J. T. (2001) Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25, 15–37 [DOI] [PubMed] [Google Scholar]
- 42. Kodama Y., Omura F., Takahashi K., Shirahige K., Ashikari T. (2002) Genome-wide expression analysis of genes affected by amino acid sensor Ssy1p in Saccharomyces cerevisiae. Curr. Genet. 41, 63–72 [DOI] [PubMed] [Google Scholar]
- 43. Bricker D. K., Taylor E. B., Schell J. C., Orsak T., Boutron A., Chen Y. C., Cox J. E., Cardon C. M., Van Vranken J. G., Dephoure N., Redin C., Boudina S., Gygi S. P., Brivet M., Thummel C. S., Rutter J. (2012) A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337, 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ljungdahl P. O., Daignan-Fornier B. (2012) Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190, 885–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bogan K. L., Brenner C. (2008) Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28, 115–130 [DOI] [PubMed] [Google Scholar]
- 46. Tempel W., Rabeh W. M., Bogan K. L., Belenky P., Wojcik M., Seidle H. F., Nedyalkova L., Yang T., Sauve A. A., Park H. W., Brenner C. (2007) Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol. 5, e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohashi K., Kawai S., Murata K. (2013) Secretion of quinolinic acid, an intermediate in the kynurenine pathway, for utilization in NAD+ biosynthesis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 12, 648–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lenburg M. E., O'Shea E. K. (1996) Signaling phosphate starvation. Trends Biochem. Sci. 21, 383–387 [PubMed] [Google Scholar]
- 49. Wykoff D. D., O'Shea E. K. (2001) Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159, 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaneko Y., Tamai Y., Toh-e A., Oshima Y. (1985) Transcriptional and post-transcriptional control of PHO8 expression by PHO regulatory genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5, 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Serrano R., Ruiz A., Bernal D., Chambers J. R., Ariño J. (2002) The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46, 1319–1333 [DOI] [PubMed] [Google Scholar]
- 52. Pinson B., Vaur S., Sagot I., Coulpier F., Lemoine S., Daignan-Fornier B. (2009) Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes Dev. 23, 1399–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de Boer M., Nielsen P. S., Bebelman J. P., Heerikhuizen H., Andersen H. A., Planta R. J. (2000) Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 28, 974–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Abdel-Sater F., Iraqui I., Urrestarazu A., André B. (2004) The external amino acid signaling pathway promotes activation of Stp1 and Uga35/Dal81 transcription factors for induction of the AGP1 gene in Saccharomyces cerevisiae. Genetics 166, 1727–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gordân R., Murphy K. F., McCord R. P., Zhu C., Vedenko A., Bulyk M. L. (2011) Curated collection of yeast transcription factor DNA binding specificity data reveals novel structural and gene regulatory insights. Genome Biol. 12, R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin S. J., Ford E., Haigis M., Liszt G., Guarente L. (2004) Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 18, 12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eto K., Tsubamoto Y., Terauchi Y., Sugiyama T., Kishimoto T., Takahashi N., Yamauchi N., Kubota N., Murayama S., Aizawa T., Akanuma Y., Aizawa S., Kasai H., Yazaki Y., Kadowaki T. (1999) Role of NADH shuttle system in glucose-induced activation of mitochondrial metabolism and insulin secretion. Science 283, 981–985 [DOI] [PubMed] [Google Scholar]
- 58. Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 59. Hughes A. L., Gottschling D. E. (2012) An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Forsberg H., Ljungdahl P. O. (2001) Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21, 814–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brown K. M., Landry C. R., Hartl D. L., Cavalieri D. (2008) Cascading transcriptional effects of a naturally occurring frameshift mutation in Saccharomyces cerevisiae. Mol. Ecol. 17, 2985–2997 [DOI] [PubMed] [Google Scholar]
- 62. Gertz J., Riles L., Turnbaugh P., Ho S. W., Cohen B. A. (2005) Discovery, validation, and genetic dissection of transcription factor binding sites by comparative and functional genomics. Genome Res. 15, 1145–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Young J. D., Yao S. Y., Sun L., Cass C. E., Baldwin S. A. (2008) Human equilibrative nucleoside transporter (ENT) family of nucleoside and nucleobase transporter proteins. Xenobiotica 38, 995–1021 [DOI] [PubMed] [Google Scholar]
- 64. Sies H. (1982) Metabolic Compartmentation, pp. 205–231, Academic Press, Orlando, FL [Google Scholar]
- 65. Lewis C. A., Parker S. J., Fiske B. P., McCloskey D., Gui D. Y., Green C. R., Vokes N. I., Feist A. M., Vander Heiden M. G., Metallo C. M. (2014) Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263 [DOI] [PMC free article] [PubMed] [Google Scholar]