Background: GEFs can serve as targets in Ras family GTPase activity signaling.
Results: A virtual screening coupled with the experimental screening platform is established targeting a Ras GEF enzyme.
Conclusion: The multiple tier screening platform is useful for lead discovery targeting GEFs of Ras-like small GTPases.
Significance: This study provides a unique screening approach for inhibitors of Ras and Ras-like GTPase activities.
Keywords: chemical biology, drug discovery, guanine nucleotide exchange factor (GEF), signal transduction, small GTPase
Abstract
The Ras family small GTPases regulate multiple cellular processes, including cell growth, survival, movement, and gene expression, and are intimately involved in cancer pathogenesis. Activation of these small GTPases is catalyzed by a special class of enzymes, termed guanine nucleotide exchange factors (GEFs). Herein, we developed a small molecule screening platform for identifying lead hits targeting a Ras GEF enzyme, SOS1. We employed an ensemble structure-based virtual screening approach in combination with a multiple tier high throughput experimental screen utilizing two complementary fluorescent guanine nucleotide exchange assays to identify small molecule inhibitors of GEF catalytic activity toward Ras. From a library of 350,000 compounds, we selected a set of 418 candidate compounds predicted to disrupt the GEF-Ras interaction, of which dual wavelength GDP dissociation and GTP-loading experimental screening identified two chemically distinct small molecule inhibitors. Subsequent biochemical validations indicate that they are capable of dose-dependently inhibiting GEF catalytic activity, binding to SOS1 with micromolar affinity, and disrupting GEF-Ras interaction. Mutagenesis studies in conjunction with structure-activity relationship studies mapped both compounds to different sites in the catalytic pocket, and both inhibited Ras signaling in cells. The unique screening platform established here for targeting Ras GEF enzymes could be broadly useful for identifying lead inhibitors for a variety of small GTPase-activating GEF reactions.
Introduction
The Ras family of small guanosine triphosphatases (GTPases) is a large family of proteins consisting of over 20 members (1). This class of proteins regulates numerous cellular processes, such as cell growth, survival, movement, cytoskeletal organization, polarity, and gene expression (1, 2). Most Ras GTPases are regulated by two major classes of modulators, which control the cycling of these proteins between their inactive GDP-bound form and active GTP-bound form. The guanine nucleotide exchange factors (GEFs)2 catalyze the exchange of GDP for GTP upon small GTPases leading to their active GTP-bound form, thus allowing the GTPases to actively signal to downstream effectors in the cell leading to regulation of cellular processes (2). Conversely, the GTPase-activating proteins catalyze the hydrolysis of the GTP-bound form of active GTPases to GDP, leading to the inactive GDP-bound form and termination of signaling in the cell (2).
The critical involvement of Ras GTPases in the regulation of numerous cellular activities readily implicates them in many human diseases and disorders. In particular, H-, N-, and K-Ras are well documented to frequently have gain of function mutations in a myriad of human cancers (e.g. lung, breast, pancreatic, ovarian, colon, and prostate) (3). Interestingly, the activating GEF enzymes have been found overexpressed and/or contain mutations in developmental disorders and human cancers (e.g. Noonan syndrome, acute myeloid leukemia, prostate cancer, and breast cancer) (4–10). These suggest that the activity of the small GTPases and their activating GEF enzymes may serve as drug targets in diseases in which the small GTPase signaling is aberrant.
Although there has been much effort in targeting various proteins involved in Ras signaling,(1, 11), until recently there has been little effort in targeting the GEF enzyme activators. A flurry of studies came out recently targeting the GEF and effector binding pockets on Ras GTPase itself through various approaches, including fragment-based screening, high throughput screening, and rational design of peptidomimetics (12–16). Recently, we have identified chemical inhibitors that could specifically suppress a Ras GEF (17). Here, we report an affordable academic drug screening platform for targeting potentially more druggable GEF enzymes, using the Ras GEF, SOS1, as a model study. We employed an ensemble structure-based docking approach in combination with a multiple tier high throughput experimental screening scheme utilizing two distinct, complementary fluorescent guanine nucleotide exchange assays, i.e. fluorescein-based GDP-dissociation assay and Texas Red-based GTP-loading assay. Applying this unique screening platform, we identified two chemically distinct small molecule inhibitors of SOS1 catalytic activity. Our study establishes an academically feasible drug screening platform that can easily be used to identify lead inhibitors of a variety of small GTPase GEFs.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
Guanosine 5′-triphosphate sodium salt solution, Tween 20, sodium deoxycholate, sodium chloride (NaCl), and Trizma® hydrochloride were purchased from Sigma. Magnesium chloride (MgCl2) was purchased from Acros Organics (Gael, Belgium). Guanosine diphosphate (GDP), dimethyl sulfoxide (DMSO), and Nonidet P-40 alternative were purchased from Calbiochem®-EMD Millipore (Billerica, MA). Human epidermal growth factor (EGF) was purchased from Roche Applied Science. BODIPY® fluorescein (FL)-GDP, BODIPY® Texas Red (TR)-GTP, One Shot® BL21(DE3) chemically competent Escherichia coli, and DTT were purchased from Life Technologies, Inc.
Residues 1–166 of human H-Ras were subcloned from the full-length human H-Ras/pcDNA3.1+ mammalian expression vector (Missouri S&T cDNA Resource Center, Rolla, MO) into the bacterial pET15b bacteria vector using the XhoI and HindIII restriction enzyme sites. Residues 1–182 of human Cdc42 were subcloned from a full-length human Cdc42/pET vector into the pET15b vector using the XhoI and HindIII restriction enzyme sites. The Ras exchanger motif and Cdc25 catalytic domains (residues 564–1049) of human SOS1 (SOS1cat) were subcloned from the full-length human SOS1 MGC clone (accession number BC140215) (Life Technologies, Inc.) into the bacterial expression vector pET15b (Novagen®-EMD Millipore) using the XhoI and HindIII restriction enzyme sites. The DH and PH catalytic domains (residues 1227–1571) of human Intersectin 1 (ITSN1DH-PH) were subcloned from a pGEX bacterial expression vector backbone into the bacterial expression vector pET15b (Novagen®-EMD Millipore) using the XhoI and HindIII restriction enzyme sites. The single and double SOS1 alanine mutants (L687A, W809A, T810A, K814A, L822A, M824A, I825A, R826A, T828A, T829A, N869A, N879A, E902A, H911A, Y912A, F929A, F930A, L934A, T935A, L938A, K939A, I825A/W809A, I825A/K814A, I825A/N869A, I825A/T935A, and I825A/F929A) were inserted into the SOS1cat/pET15b bacterial expression vector using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA).
Computational Screening and SAR Analysis
To identify candidate small molecules that could disrupt the interaction between SOS1 and Ras by binding to the Ras interaction interface(s) in SOS1, in silico screening was carried out. Using multiple SOS1 crystal structures, docking simulations using rigid body docking for virtual screening were performed using AutoDock version 4.2 (18) in conjunction with the Center for Protein Informatics' computational pipelines on a Linux cluster with upwards of 512 CPU at the Cincinnati Children's Hospital Medical Center. The crystal structure of SOS1 in complex with Ras (bound at both the catalytic and allosteric sites, PDB code 1XD2) (19), as well as the unbound SOS1 structure (PDB code 1XD4) (19) were used to build alternative models of the SOS1 enzyme for docking simulations. Probis and CASTp were used to analyze structurally equivalent sites and overall similarity between the catalytic and allosteric Ras-binding interfaces within SOS1.
The University of Cincinnati-Drug Discovery Center's collection of 350,000 compounds was used for virtual screening. Gesteiger partial charges were used for both the SOS1 enzyme and chemical compounds. A multiple step virtual screening approach was implemented, using increasingly stringent parameters and gradually more extensive sampling by increasing the number of energy evaluations (from 1.0 × 105 to 1.0 × 107), Genetic Algorithm runs (10 to 33) and population size (from 75 to 150), as discussed in detail previously (20). Upon completion of initial fast screening, 30,000 top candidate compounds with high estimates of binding affinities were retained, followed by re-scoring using improved sampling in the refinement stage. The top 3000 candidate compounds were again re-scored, using extensive sampling, and then clustered to avoid over-representation of some chemical classes.
As a result, two sets of hits were obtained, 315 for the catalytic site and 392 for the allosteric site, which were selected using both their predicted median inhibition constant and entropy of clustering of poses in multiple docking runs (excluding compounds with entropies larger than 0.4 to filter out likely nonspecific binders). An increasing level of overlap between top hits for these two sites (as tested by direct overlap of lists as well as clustering of the superset) was observed with subsequent runs and improved sampling, consistent with the observed structural similarity between the two sites. This was used as indication of the convergence of the sampling. Because of the overlap between the two sets, the set of hits selected for experimental validation consisted of 418 chemical compounds that were sufficient to cover the observed classes and clusters of hits.
Candidate compounds were further assessed from the point of view of their properties. Chemmine was used to cluster and visually analyze candidate molecules. Sppider and Polyview were used to analyze protein-protein complexes and to assess predicted docking poses for candidate inhibitors (the latter task facilitated by Polyview-MM) (21). Additionally, the predicted docking poses of candidate inhibitors were used to rationally guide which amino acid residues to mutate in SOS1 for site of action mutagenesis studies.
Using the BIOVIA Pipeline Pilot (version 8.5.0.200), computational structural similarity searches based upon both UC-773587 and UC-857993 were performed using the University of Cincinnati-Drug Discovery Center's 350,000 compound library collection. As a result of these searches, 1 structurally similar compound to UC-773587 and 10 structurally similar compounds to UC-857993 were selected for experimental testing. In addition, Chemmine was used to perform computational structural similarity searches for UC-773587 on a subset of 118,500 drug-like synthetic compounds from the NCI/DTP Open Chemical Repository (dtp.cancer.gov). As result of this search, seven structurally similar compounds to UC-773587 were selected for experimental testing.
Protein Purifications
Wild-type and mutant recombinant human SOS1cat (residues 564–1049) containing both the REM and Cdc25 domains, recombinant human ITSN1DH-PH (residues 1227–1571) containing both the DH and PH domains, recombinant human C-terminal truncated Cdc42 (residues 1–182), and recombinant human C-terminal truncated H-Ras (residues 1–166) were expressed in BL21(DE3) Escherichia coli cells as the N-terminal His6-tagged fusion proteins using the pET expression system (Novagen). For the wild-type and mutant recombinant human guanine nucleotide exchange factors (SOS1cat and ITSN1DH-PH), E. coli cells were grown to an A600 between 0.4 and 0.8 and induced with 1 mm isopropyl β-d-1-thio-galactopyranoside overnight at room temperature. For the recombinant C-terminal truncated human GTPases (H-Ras and Cdc42), E. coli cells were grown to an A600 between 0.4 and 0.8 and induced with 1 mm isopropyl β-d-1-thio-galactopyranoside for 4 h at 37 °C. Subsequently, the histidine-tagged fusion proteins were purified using Ni2+-agarose affinity chromatography followed by gel filtration with a HiLoad 16/60 Superdex 200 column and FPLC AKTA Purifier UPC 10 system (GE Healthcare).
Experimental High Throughput Guanine Nucleotide Exchange Assays
For both the FL-based dissociation and the TR-based GTP loading, high throughput screening exchange assays, the Plate::ExplorerTM automated uHTS system (PerkinElmer Life Sciences) at the University of Cincinnati-Drug Discovery Center was employed to carry out the experimental screening scheme. For the FL-based dissociation assay, purified His6-H-Ras (residues 1–166) was pre-loaded with BODIPY-FL-GDP for 1 h at room temperature with a stoichiometric ratio of 4:1, respectively. Exchange buffer (EB) (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm MgCl2, 0.01% Nonidet P-40 alternative, 1 mm DTT), 50 nm purified His6-SOS1cat, 50-fold excess GTP, and DMSO or 100 μm chemical compounds were sequentially added to wells of a black 384-well NBSTM plate (mouse embryonic fibroblast 3655) using 384-well automated liquid dispensers in conjunction with the Plate::ExplorerTM automated uHTS system (PerkinElmer Life Sciences). Subsequently, after a brief 30-s shake, the plate was incubated for 10 min at room temperature. Then, the kinetic reaction was initiated by adding 2 μm purified His6-H-Ras (residues 1–166) pre-loaded with BODIPY-FL-GDP nucleotide with a 384-well automated liquid dispenser in conjunction with the Plate::ExplorerTM automated system (PerkinElmer Life Sciences) (30 μl per well, final reaction volume). The validity of this assay for HTS was determined by calculating the statistical coefficient, Z′ factor, (22), using Equation 1.
For the TR-based GTP loading assay, EB, 50 nm purified His6-SOS1cat, and DMSO or 100 μm chemical compounds were sequentially added to wells of a black 384-well NBSTM plate (Corning Glass, catalog no. 3655) using 384-well automated liquid dispensers in conjunction with the Plate::ExplorerTM automated uHTS system (PerkinElmer Life Sciences). Subsequently, after a brief 30-s shake, the plate was incubated for 10 min at room temperature. Then, the kinetic reaction was initiated by adding 2 μm purified His6-H-Ras (residues 1–166) immediately followed by 2.5-fold excess of BODIPY-TR-GTP with 384-well automated liquid dispensers in conjunction with the Plate::ExplorerTM automated system (PerkinElmer Life Sciences) (30 μl per well, final reaction volume). The kinetic reaction for both exchange assays was measured continuously for 15 min at room temperature in an EnVision plate reader (PerkinElmer Life Sciences) (fluorescein channel: excitation, 485 nm; emission, 535 nm; Texas Red channel: excitation, 560 nm; emission, 635 nm).
96-Well Guanine Nucleotide Exchange Assays
Initially, for the fluorescein-based dissociation exchange assay, purified His6-H-Ras (residues 1–166) was pre-loaded with BODIPY-FL-GDP for 1 h at room temperature with a stoichiometric ratio of 4:1, respectively. For both the FL-based dissociation exchange assay, EB (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm MgCl2, 0.01% Nonidet P-40 alternative, 1 mm DTT), 50-fold excess of free GTP, and 50 nm purified His6-SOS1cat were sequentially added to wells of a black 96-well plate (Corning Glass, catalog no. 3915). Subsequently, DMSO and the indicated concentrations of UC-773587 or UC-857993 were added to appropriate wells, followed by shaking the plate for 30 s and 10 min of incubation at room temperature. The reaction was initiated by adding 2 μm of pre-loaded H-Ras to each well (final reaction volume, 100 μl).
For the TR-based GTP loading exchange assay, EB and 100 nm purified His6-SOS1cat were sequentially added to the wells of a black 96-well plate. Subsequently, DMSO and the indicated concentrations of UC-773587 and UC-857993 were added to the appropriate wells, followed by shaking the plate for 30 s and 10 min of incubation at room temperature. The reaction was initiated by adding 2 μm purified His6-H-Ras (residues 1–166) followed by 2.5-fold excess of BODIPY-TR-GTP to each well (final reaction volume, 100 μl). The kinetic reactions for both exchange assays were measured continuously for 15 min at room temperature in an EnVision plate reader (PerkinElmer Life Sciences) (fluorescein channel: excitation, 485 nm; emission, 535 nm; Texas Red channel: excitation, 560 nm; emission, 635 nm).
Microscale Thermophoresis (MST) Binding Assays
Purified wild-type and mutant His6-SOS1cat and C-terminally truncated His6-H-Ras were labeled with the red NT-647 amine-reactive dye using a protein labeling kit (NanoTemper Technologies). The indicated concentrations of UC-773587 or UC-857993 were incubated with NT-647-labeled SOS1cat (100 nm) or H-Ras (100 nm) in binding buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm MgCl2, 0.1% Triton X-100, and 2% DMSO) at room temperature in the dark for 30 min. For the competition of binding experiments, purified His6-SOS1cat was labeled with the red NT-647 cysteine-reactive dye using a protein labeling kit (NanoTemper Technologies). 0.2 to 100 μm unlabeled purified C-terminally truncated His6-H-Ras were incubated with NT-647-labeled SOS1-cat (50 nm) in the presence of the indicated concentrations of UC-773587 or UC-857993 in binding buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm MgCl2, 0.1% Tween 20, and 2% DMSO) at room temperature in the dark for 30 min. Subsequently, samples were loaded into hydrophilic capillaries (NanoTemper Technologies), and binding events were measured with a Monolith NT.115 reader (NanoTemper Technologies). Binding data were analyzed using Thermophoresis or Thermophoresis with Temperature Jump analysis as described previously (23, 24). Data were normalized using fraction-bound binding (23, 24).
Cell Culture
The NIH/3T3 mouse fibroblast cell line was maintained at 37 °C in 5% CO2 in complete Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The production and maintenance of the immortalized parental Cdc42f/f (wild type) and retrovirally transduced MIEG3-H-RasG12V mouse embryonic fibroblasts (MEFs) were described previously (25). The human DU-145 prostate cancer cell line was a kind gift from Dr. Zhongyun Dong (Dept. of Internal Medicine, University of Cincinnati,). The human BxPC-3, MIA-PaCa-2, and Panc-1 pancreatic cancer cell lines were obtained from the ATCC (Manassas, VA). The human DU-145, BxPC-3, and Panc-1 cell lines were maintained at 37 °C in 5% CO2 in complete DMEM containing 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The MIA-PaCa-2 cell line was maintained at 37 °C in 5% CO2 in complete DMEM containing 10% heat-inactivated fetal bovine serum, 2.5% heat-inactivated horse serum, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Western Blot Analysis
NIH/3T3 or DU-145 cells were grown in 10-cm dishes to a cell confluency of 80%. Cells were lysed in lysis buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm DTT, 0.4% sodium deoxycholate, 5 mm NaF) supplemented with the EDTA-free protease inhibitor mixture (Roche Applied Science) and the Simple StopTM 1 phosphatase inhibitor mixture (Gold Biotechnology, St. Louis, MO) and were subsequently clarified. Protein concentrations of the samples were determined and normalized by the Bradford assay using the protein assay dye reagent from Bio-Rad. Lysates were separated in 4–15% Mini-PROTEAN® TGXTM precast polyacrylamide gels by SDS-PAGE and transferred onto PVDF membranes with the Trans-Blot® TurboTM transfer system (Bio-Rad). The membranes were blocked with 5% BSA in TBS-T (20 mm Tris-HCl, pH 7.6, 150 mm NaCl, and 0.1% Tween 20) overnight at 4 °C or for 30–60 min at room temperature and then probed with primary antibodies and subsequently with horseradish peroxidase (HRP)-coupled (GE Healthcare) secondary antibodies for chemiluminescence analysis. The anti-pan-Ras (catalog no. 05-516) antibody was purchased from Millipore (Billerica, MA). The anti-phospho-ERK1/2 (catalog no. 4370), anti-ERK1/2 (catalog no. 9102), and anti-GAPDH (catalog no. 5174) antibodies were purchased from Cell Signaling Technology (Danvers, MA). All primary antibodies were diluted (1:1000) in TBS-T buffer containing 5% BSA. Membranes were developed by ECL Western blot detection reagents (GE Healthcare and Thermo Fisher Scientific). Images for the ECL-developed membranes were taken by the Fuji Image Reader LAS1000 or ChemiDoc Touch Imaging System (Bio-Rad).
MTS Cell Proliferation Assay
1000 cells (DU-145 and MEF-UC-773587 treated cells), 2000 cells (MEF-UC-857993 treated cells), or 500 cells (NIH/3T3, Panc-1, MIA-PaCa-2, BxPC-3, and DU-145 cells) per well in triplicate or quadruplicate were seeded into a 96-well plate (Corning Glass) as 100-μl cell suspensions in complete DMEM. After culturing the cells for 24 h, cells were treated with the indicated concentrations of UC-773587, UC-857993, or DMSO for 24, 48, or 72 h. The MTS cell proliferation assay was carried out following the manufacturer's instructions (Promega, Madison, WI; catalog no. G3580). After a 2-h incubation at 37 °C and 5% CO2 with substrate, the metabolic activity of the cells was measured by absorbance at 490 nm with a VMax Kinetic ELISA microplate reader (Molecular Devices, Sunnyvale, CA).
RESULTS
Combined Virtual Screening and High Throughput Experimental Screening to Identify Inhibitors of GEF Catalytic Activity
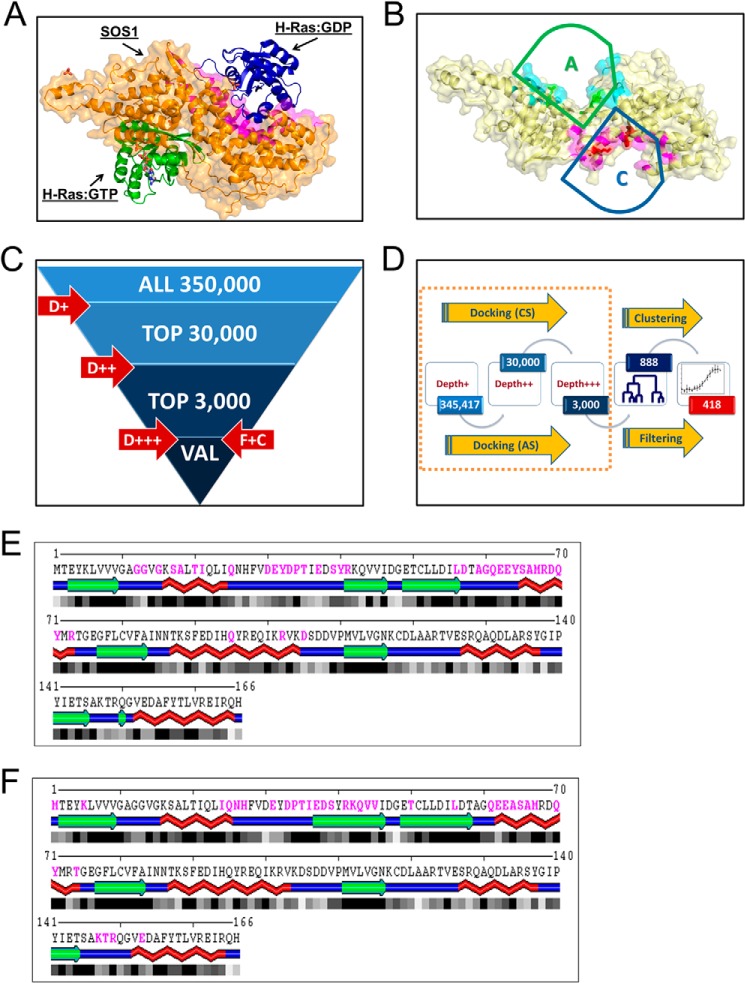
Multiple crystal structures of the Ras GEF, SOS1, and its complexes (PDB code 1XD2 and 1XD4) (Fig. 1A) were used for virtual screening to guide an ensemble docking approach targeting both the catalytic and allosteric sites within SOS1 to identify candidate inhibitors (19). Detailed structural analysis of the SOS1-Ras complexes revealed structural similarity between Ras binding interfaces. In particular, similar conformations of the switch I region of Ras are involved in the interactions with SOS1 at both the catalytic and allosteric sites (Fig. 1, E and F). Additionally, ProBis structural alignment (26, 27) identified equivalent motifs involving residues Ile-932, Leu-934, Asn-936, and Lys-939 within the catalytic site and the corresponding Leu-687, Leu-690, Asn-691, and Arg-694 residues in the allosteric site (Table 1). Thus, despite differences in the overall topology, the catalytic and allosteric sites on SOS1 share some elements of structural similarity. Therefore, both Ras-interaction interfaces (Fig. 1B) were targeted in virtual screening. Utilizing ensemble docking with multiple (bound versus unbound) structures and comparing predicted docking poses at structurally equivalent docking sites allowed us to improve sensitivity, while reducing false-positive rates. In addition, integrating multiple structures into the virtual screening protocol allowed us to partially alleviate the limitation of rigid body docking.
FIGURE 1.
Ensemble docking based virtual screening for candidate small molecule inhibitors of SOS1. A, crystal structure of the REM and Cdc25 catalytic domains of SOS1 in complex with or without both inactive and active H-Ras at the catalytic and allosteric sites of SOS1, respectively (PDB code 1XD2) (19). SOS1 is depicted in orange; inactive H-Ras at the catalytic site is depicted in blue, and active H-Ras at the allosteric site is depicted in green. B, catalytic targeted pocket in SOS1 is highlighted in magenta and indicated by a blue arc-topped triangle labeled with C. The allosteric targeted pocket in SOS1 is highlighted in turquoise and indicated by a green arc-topped triangle labeled with A. Residues at structurally equivalent positions in both pockets are further highlighted by their side chains. C, multistage virtual screening approach is as follows: initial docking performed with limited sampling (D+ or depth 1) to identify top 30,000 compounds, followed by depth 2 (D++) docking with increased sampling to identify top 3000, followed up by docking with extensive sampling (D+++), filtering using predicted inhibition constants and entropy of docking, and clustering based on chemical similarity to identify a set of about 300 compounds in each run. D, overall flow of virtual screening, starting from multiple runs of docking simulations as shown in C and targeting catalytic (CS) or allosteric (AS) sites, using bound or unbound structures of SOS1, followed by selection (filtering) of compounds with best median predicted inhibition constants and low entropy of clustering poses, and by clustering of chemically structurally similar candidates to select a final set of 418 candidates for experimental screening. E and F, SOS1 interaction interface on Ras for both the catalytic and allosteric sites. Ras residues buried upon complex formation with both the SOS1 catalytic and allosteric sites are shown in magenta, as identified by Sppider using default settings. SOS1 interaction interface on Ras for the catalytic site (PDB code 1XD2 and chain B) (E) and SOS1 interaction interface on Ras for the allosteric site (PDB code 1XD2 and chain A) (F). The large overlapping interacting regions on Ras for both the catalytic and allosteric sites, in particular within the switch I region, should be noted.
TABLE 1.
SOS1 residues involved in the formation of the interaction interfaces with Ras at the catalytic and allosteric sites, respectively
Residues at the SOS1 interfaces were identified by using the Sppider server with default settings. Residue numbers are based upon the SOS1 crystal structure (PDB code 1XD2), which was used for analysis.
| Catalytic Ras (1XD2, chain B) interface with SOS1 (chain C); residues on Sos1 at right | Glu-589, Lys-602, Trp-809, Thr-810, Lys-814, Leu-822, Ile-825, Arg-826, Thr-828, Thr-829, Thr-832, Leu-833, Glu-836, Val-875, Ser-876, Asn-879, Ser-880, Ser-881, Tyr-884, His-905, Ser-908, Asp-910, His-911, Tyr-912, Lys-913, Phe-929, Gly-931, Leu-934, Thr-935, Asn-936, Leu-938, Lys-939, Thr-940, Glu-942, Gly-943, Asn-944, Pro-945, Leu-948, Arg-950, Ser-959, Lys-963, Glu-966, Ile-967, Glu-1002, Lys-1003, Thr-1006, Asp-1007, Phe-1010, Arg-1019 |
| Allosteric Ras (1XD2, chain A) interface with SOS1 (chain C); residues on Sos1 at right | Gly-597, Ile-598, His-616, Met-617, Ala-619, Asp-620, Pro-621, Asn-622, Val-624, Lys-679, Gln-683, Pro-684, Leu-687, Arg-688, Asn-691, Arg-694, His-695, Ala-725, Lys-728, Trp-729, Ser-732, Arg-739, His-750, Asn-751, Ile-752, Thr-753, Phe-754, Gln-755, Arg-920, Ser-921, Ile-922, Asn-923, Pro-924, Gln-973, Tyr-974, Asn-976, Gln-977, Pro-978 |
Beginning with the University of Cincinnati-Drug Discovery Center's 350,000 compound collection, a multistage virtual screening protocol was used to identify the top 30,000 hits with limited sampling. Subsequently, using better sampling, the 30,000 hits were reduced to 3000 top hits (Fig. 1C) (20). At each stage, alternative conformers of Ras interaction at both the catalytic and allosteric sites on SOS1 were used to perform docking simulations to partially account for conformational changes related to SOS1-Ras complex formation and to provide consistency-based filtering along with improved ranking of candidate inhibitors. Top hits for each site on SOS1, rank base on high predicted binding affinity and consistent binding to a specific site in a dominant pose within the simulation box, as captured by low entropy of clustering poses acquired in multiple docking runs (Fig. 1D), were combined and clustered based upon chemical similarity. This resulted in a set of 418 candidate compounds, of which 394 compounds were available, for experimental screening in our high throughput screening platform (Figs. 1D and 2E).
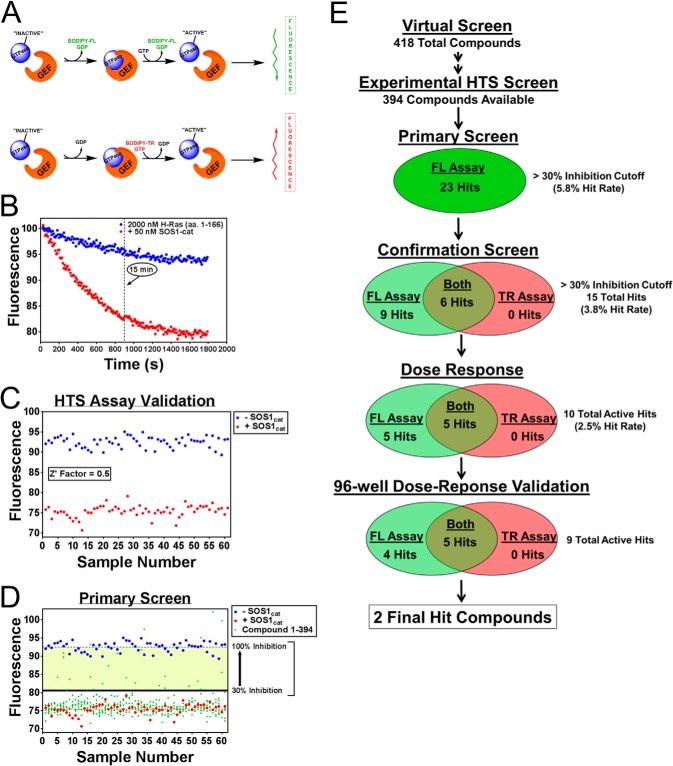
FIGURE 2.
High throughput screening platform for small molecule inhibitors of GEF catalytic activity. A, schematic of the methodology of the two complementary fluorescent (fluorescein and Texas Red) intensity guanine nucleotide exchange assays used for experimental screening. B, characterization of the BODIPY-FL-GDP dissociation primary screening assay. 50 nm purified His6-SOS1cat (red circle)-stimulated BODIPY-FL-GDP nucleotide exchange for unlabeled GTP on purified 2 μm His6-H-Ras (residues 1–166) (blue circle). C, validation of the GDP dissociation primary screening assay. −SOS1cat (blue circle) and +SOS1cat (red circle) controls from the primary screening assay plates are plotted using a 15-min time point and subsequently used to calculate the statistical coefficient, Z′-factor (22), to validate the assay for HTS. D, HTS results of the primary screen. −SOS1cat (blue circle) and +SOS1cat (red circle) DMSO controls along with the chemical compounds (green circle) tested for their ability to inhibit SOS1 catalytic activity are plotted using a 15-min time point. The >30% inhibition cutoff used is indicated. E, HTS platform experimental screening scheme used to identify inhibitors of SOS1 catalytic activity. 418 candidate compounds were identified from the virtual screen as described in Fig. 1. 394 of these 418 compounds were available for experimental screening. 23 of the 394 compounds were identified as hits from the primary screen. 15 of these 23 hits were confirmed using two complementary fluorescent intensity assays. Of these 15 hits, 10 hits were validated by a 10-point dose response (2-fold serial dilution; 0.195–100 μm), and subsequently 9 of these 10 hits were validated in 96-well format dose response (0, 2, 5, 10, 20, 50, and 100 μm). Five of the nine hits showed activity in both complementary fluorescent assays. Two of these five hits were selected for further validation and characterization. Data in B–D are expressed as percent change of fluorescence units normalized to the initial time point. Data in B were performed in triplicate. C, three outliers were removed from both the −SOS1cat and +SOS1cat control datasets based upon visual inspection.
For our high throughput screening platform, two complementary fluorescent intensity guanine nucleotide exchange assays were developed (28–30). One assay utilized the BODIPY-FL-GDP nucleotide to measure SOS1cat-mediated GDP dissociation on H-Ras that results in a loss of fluorescence, and the other assay utilized a BODIPY-TR-GTP nucleotide to measure SOS1cat-mediated GTP loading to H-Ras resulting in an increase in TR fluorescence (Fig. 2A). Integrating these two complementary assays into our high throughput screening platform scheme enabled us to identify inhibitors of SOS1 catalysis that mechanistically could disrupt two distinct steps of the GTPase cycle. Additionally, using two different fluorescent channels (i.e. fluorescein and Texas Red) as a read-out for each complementary guanine nucleotide exchange assay reduced the possibility of identifying false-positives as a result of fluorescent artifact. Because of the reproducibility and robustness of the BODIPY-FL-GDP dissociation guanine nucleotide exchange assay, it was selected to perform the primary screening stage of the multiple tier high throughput screening platform scheme (Fig. 2E). Using a time point of 15 min, the fluorescein-based dissociation assay produced a combined Z′ factor of 0.5 using the positive and negative controls (−SOS1cat and +SOS1cat) from each primary screening assay plate for calculation, indicating it to be an excellent assay for primary high throughput screening (Fig. 2, B and C) (22, 28). Using the dissociation assay, 394 candidate compounds at a dose of 100 μm were tested for their ability to inhibit SOS1cat catalytic activity. Using a cutoff of >30% inhibition, 23 hits were identified for a 5.8% hit rate (Fig. 2, D and E). Typically, one would expect a hit rate of ≤1% for high throughput screens using diverse chemical libraries (28, 31). However, our computer-aided virtual screen was able to enhance the screening hit rate to 5.8%, thus indicating the efficiency of combining computer-guided virtual screens with experimental high throughput screening platform campaigns. Subsequently, the 23 hit compounds at a dose of 100 μm were tested for their ability to inhibit SOS1 catalytic activity using a cutoff of >30% inhibition in both the dissociation and GTP loading assays. 15 of the 23 hit compounds were confirmed as true hits reducing the hit rate to 3.8%. Nine of these 15 hit compounds were active in the dissociation assay, and six of the 15 hit compounds were active in both the dissociation and GTP loading assays. These 15 hit compounds were further tested for their ability to inhibit SOS1cat catalytic activity in a 10-point dose-response assay using both fluorescent intensity guanine nucleotide exchange assays. 10 of the 15 hit compounds dose-dependently inhibited SOS1cat catalytic activity further reducing the overall hit rate to 2.5%. Of these 10 compounds, five compounds were only active in the dissociation assay, and the remaining five compounds were active in both the dissociation and GTP loading assays. At this stage, fresh powder of the 10 hit compounds was obtained and manually tested for their ability to dose-dependently inhibit SOS1cat catalytic activity in both fluorescent assays in 96-well format. Nine of the 10 compounds were validated to dose-dependently inhibit SOS1cat catalytic activity with four compounds only displaying activity in the dissociation assay and five compounds displaying activity in both the dissociation and GTP loading assays (Fig. 3). Two of the five validated compounds were selected for follow-up characterization due to their experimental efficacy and selectivity. Three of the five validated compounds were ruled out due to their lack of selectivity (UC-376097) for direct binding to SOS1 versus H-Ras, poor efficacy (UC-104460), and/or poor solubility (UC-992652).
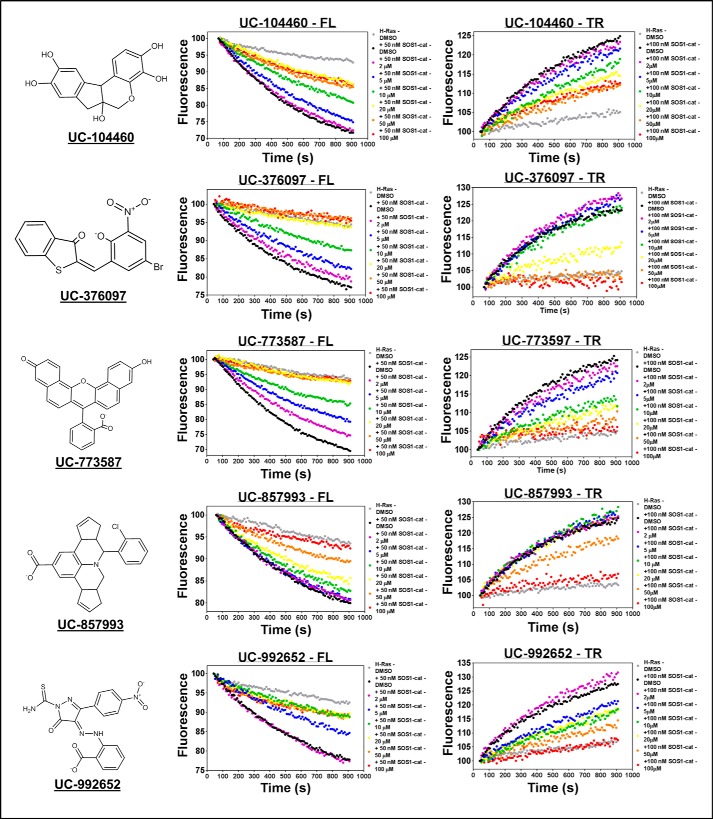
FIGURE 3.
Representative hit validation in 96-well plates. Dose-dependent inhibition of 50 nm purified His6-SOS1cat (black circle)-mediated GDP for GTP nucleotide exchange upon 2 μm His6-H-Ras (residues 1–166) (gray circle) in both the 96-well BODIPY-FL-GDP dissociation and BODIPY-TR-GTP loading assays by the five validated hit compounds that displayed activity in both complementary experiments. Chemical structures of the five hit compounds are displayed on the left side of their respective experimental data. Data are expressed as percent change of fluorescence units normalized to the initial time point. Data were performed in triplicate and represent the mean.
Biochemical Characterization of Two Chemically Distinct Inhibitors of SOS1 Catalytic Activity
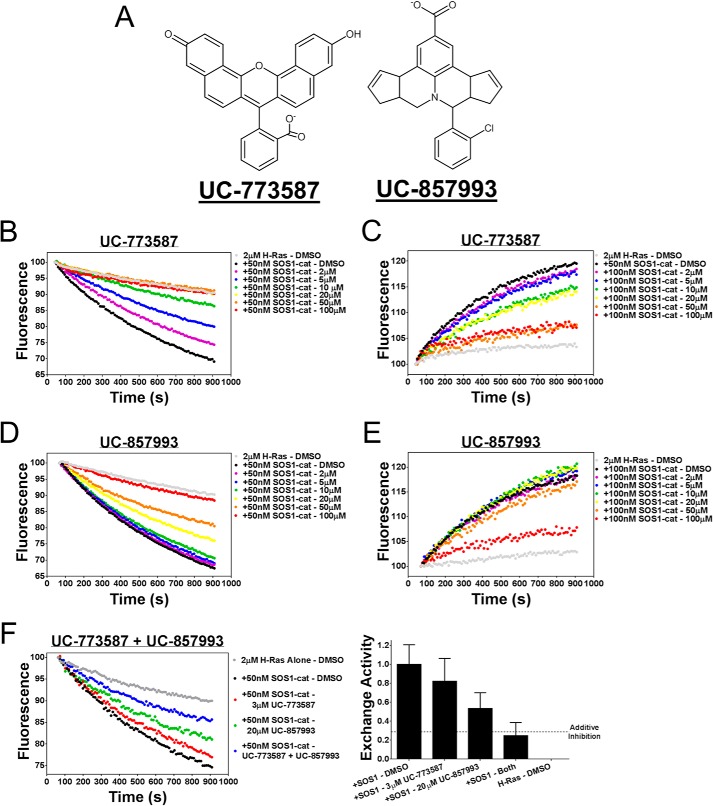
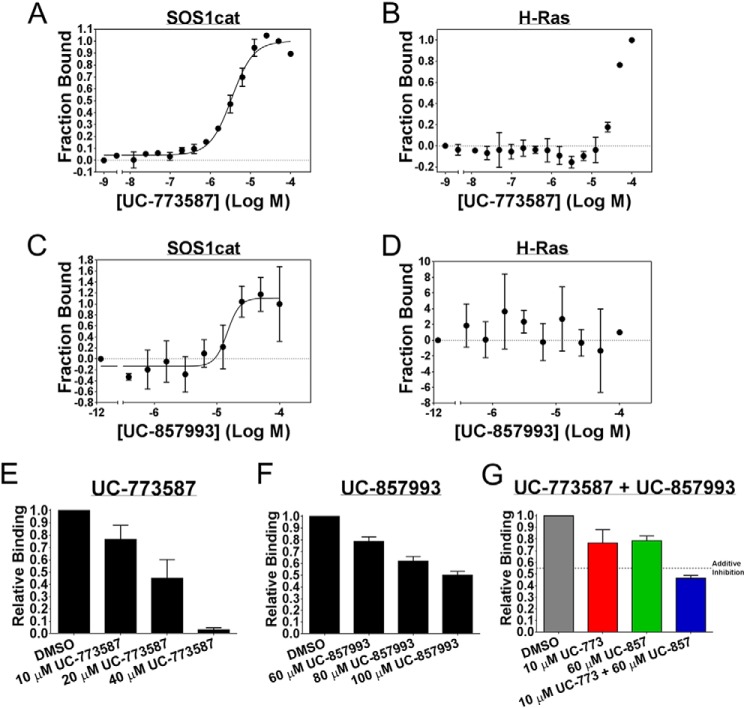
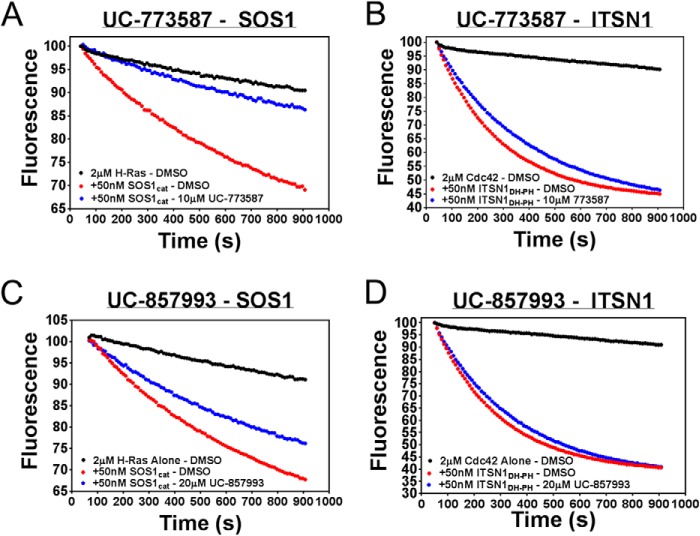
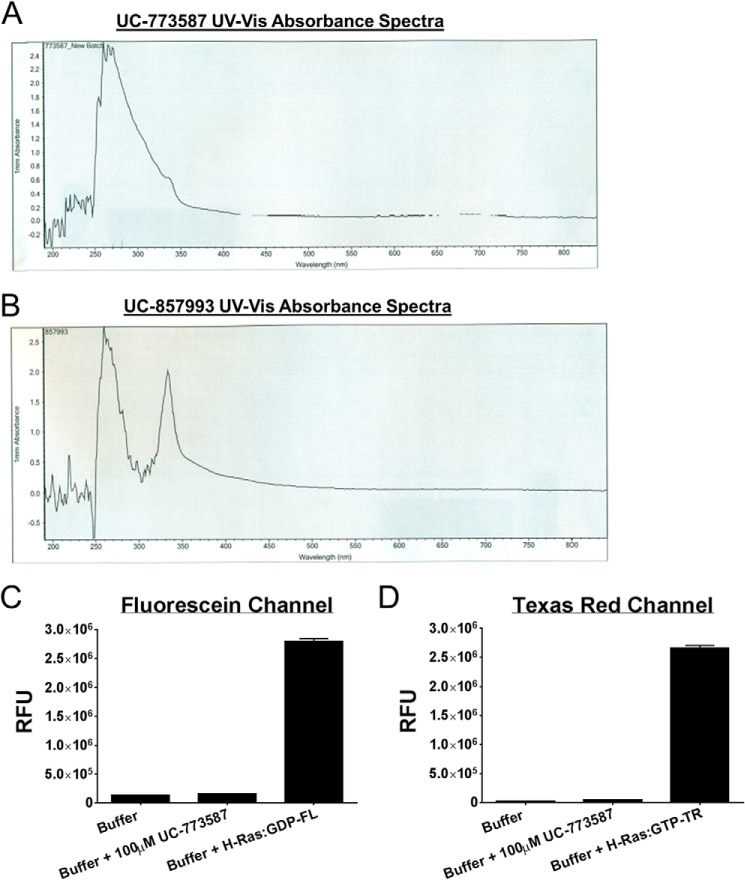
Two chemically distinct inhibitors (Fig. 4A) of SOS1 catalytic activity were further characterized as inhibitors of SOS1 in a series of follow-up biochemical experiments. First, UC-773587 and UC-857993 were tested for their ability to inhibit SOS1cat catalytic activity in our two complementary fluorescent intensity guanine nucleotide exchange assays. Both UC-773587 and UC-857993 dose-dependently inhibited SOS1cat-mediated GDP nucleotide exchange and GTP loading on H-Ras (Fig. 4, B–E). Interestingly, combination treatment of UC-773587 and UC-857993 additively inhibited SOS1cat-mediated GDP nucleotide exchange on H-Ras (Fig. 4F) suggesting that both compounds may target the same site in SOS1. Second, consistent with this result, combination treatment of UC-773587 and UC-857993 additively disrupted the interaction between SOS1cat and GDP bound H-Ras (Fig. 5G). To confirm that UC-773587 and UC-857993 inhibited both SOS1 catalytic activity and the SOS1-Ras interaction by binding to the expected target SOS1 and not H-Ras, direct binding experiments to SOS1 and H-Ras were carried out using microscale thermophoresis. As predicted, UC-773587 and UC-857993 were found to directly bind to SOS1cat (UC-773587 − Kd = 3.4 μm; UC-857993 − Kd = 14.7 μm) but not H-Ras with micromolar affinity (Fig. 5, A–D). Also, consistent with our virtual screening model, UC-773587 and UC-857993 dose-dependently disrupted the SOS1-Ras interaction by competitively inhibiting the binding of SOS1cat to GDP-bound H-Ras in a microscale thermophoresis assay (Fig. 5, E and F). Third, to test whether our compounds were selective for the Ras GEF, SOS1, versus the distantly related Rho GEF, ITSN1, UC-773587, and UC-857993, was tested for the ability to inhibit SOS1cat-mediated and the DH-PH catalytic module of Intersectin 1 (ITSN1DH-PH)-mediated GDP nucleotide exchange on H-Ras and Cdc42, respectively. Interestingly, both UC-773587 and UC-857993 showed selective inhibition of SOS1cat versus ITSN1DH-PH catalytic activity (Fig. 6). Given the fluorescent basis of these biochemical experiments, UC-773587 and UC-857993 were tested for their UV-visible absorbance spectrum and/or emission spectrum in the fluorescein and Texas Red channels to rule out spectral interference. Both compounds displayed no absorbance peaks at the wavelengths used for the biochemical experiments, and furthermore, UC-773587 did not show autofluorescence in either the fluorescein or Texas Red channels (Fig. 7).
FIGURE 4.
UC-773587 and UC-857993 inhibit GEF catalytic activity. A, chemical structures of the two hit compounds, UC-773587 and UC-857993. UC-773587 and UC-857993 dose-dependently inhibit (B and D) 50 nm His6-SOS1cat (black circle)-mediated BODIPY-FL-GDP for GTP nucleotide exchange on 2 μm His6-H-Ras (residues 1–166) (gray circle) or (C and E) 50 nm His6-SOS1cat (black circle)-mediated BODIPY-TR-GTP loading on 2 μm His6-H-Ras (residues 1–166) (gray circle) in a 96-well format at the indicated compound concentrations. F, UC-773587 (3 μm) and UC-857993 (20 μm) additively inhibit 50 nm His6-SOS1cat (black circle)-mediated BODIPY-FL-GDP for GTP nucleotide exchange on 2 μm His6-H-Ras (residues 1–166) (gray circle) in a 96-well format. Data in B–F are expressed as percent change in fluorescence units normalized to the initial time point and were performed in triplicate. Data in B–E represent the mean of three independent experiments.
FIGURE 5.
UC-773587 and UC-857993 selectively bind to SOS1 and disrupt the interaction between SOS1 and H-Ras. Direct binding of UC-773587 and UC-857993 to His6-SOS1cat or to His6-H-Ras (residues 1–166) as measured by microscale thermophoresis. A and C, 0 to 100 μm UC-773587 or UC-857993 was titrated to a constant amount of NT-647-labeled His6-SOS1cat (100 nm). UC-773587 and UC-857993 bound His6-SOS1cat with Kd values of −5.469 ± 0.02533 log M (3.4 μm) and −4.833 ± 0.09217 log M (14.7 μm), respectively. B and D, 0 to 100 μm of UC-773587 or UC-857993 was titrated to a constant amount of NT-647-labeled His6-H-Ras (100 nm) resulting in no binding. Competition of His6-H-Ras (residues 1–166) binding to NT-647-labeled His6-SOS1cat (50 nm) is shown. UC-773587 (E) and UC-857993 (F) dose-dependently inhibited titration of His6-H-Ras (amino acids 1–166) (0.2–100 μm) binding to NT-647 cysteine-labeled His6-SOS1cat (50 nm) at the indicated compound concentrations as expressed by the bar graphs. G, UC-773587 (10 μm) and UC-857993 (60 μm) additively inhibited titration of His6-H-Ras (amino acids 1–166) (0.2–100 μm) binding to NT-647 cysteine-labeled His6-SOS1cat (50 nm) as expressed by the bar graph. Dissociation constants (Kd) in A and C were determined using nonlinear regression (GraphPad Prism 6, La Jolla, CA). Data in E–G are expressed as relative binding and represent normalized binding values to the DMSO control at a nonsaturating concentration H-Ras (12.5 μm). Data in all panels represent the mean ± S.E. of three independent experiments.
FIGURE 6.
UC-773587 and UC-857993 selectively inhibit SOS1cat-mediated versus ITSN1DH-PH-mediated guanine nucleotide exchange activity. 10 μm UC-773587 (A) and 20 μm UC-857993 (C) inhibited 50 nm purified His6-SOS1cat (red circle)-mediated GDP for GTP nucleotide exchange upon 2 μm His6-H-Ras (residues 1–166) (black circle) in the 96-well BODIPY-FL-GDP dissociation exchange assay. However, 10 μm UC-773587 (B) and 20 μm UC-857993 (D) did not inhibit 50 nm purified His6-ITSN1DH-PH (red circle)-mediated GDP for GTP nucleotide exchange upon 2 μm His6-Cdc42 (residues 1–182) (black circle) in the 96-well BODIPY-FL-GDP dissociation exchange assay. Data are expressed as percent change of fluorescence units normalized to the initial time point. Data were performed in triplicate and represent the mean.
FIGURE 7.
UC-773587 and UC-857993 UV-visible absorbance spectra and UC-773587 fluorescence emission. The UV-visible absorbance spectrum for UC-773587 (A) and UC-857993 (B) was measured by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The fluorescence emission of UC-773587 in both the FL channel (C) and TR channel (D) in comparison with the fluorescence emission of exchange buffer alone and H-Ras pre-loaded with BODIPY-FL-GDP or initially loaded with BODIPY-TR-GTP as measured in the EnVision plate reader (PerkinElmer Life Sciences) (fluorescein channel: excitation, 485 nm; emission, 535 nm; Texas Red channel: excitation, 560 nm; emission, 635 nm).
Mapping the Sites of Action for UC-773587 and UC-857993
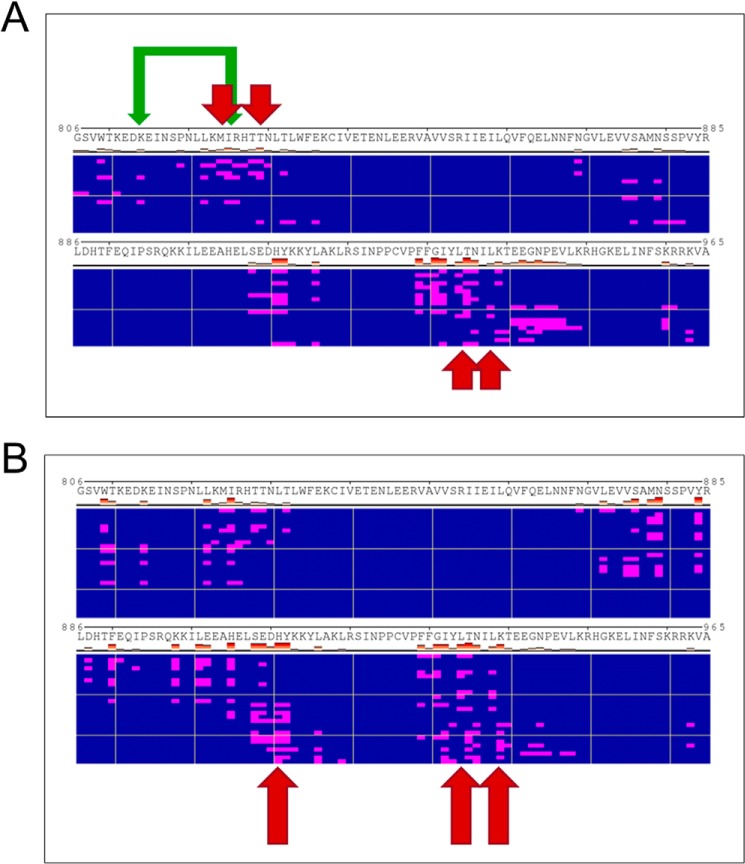
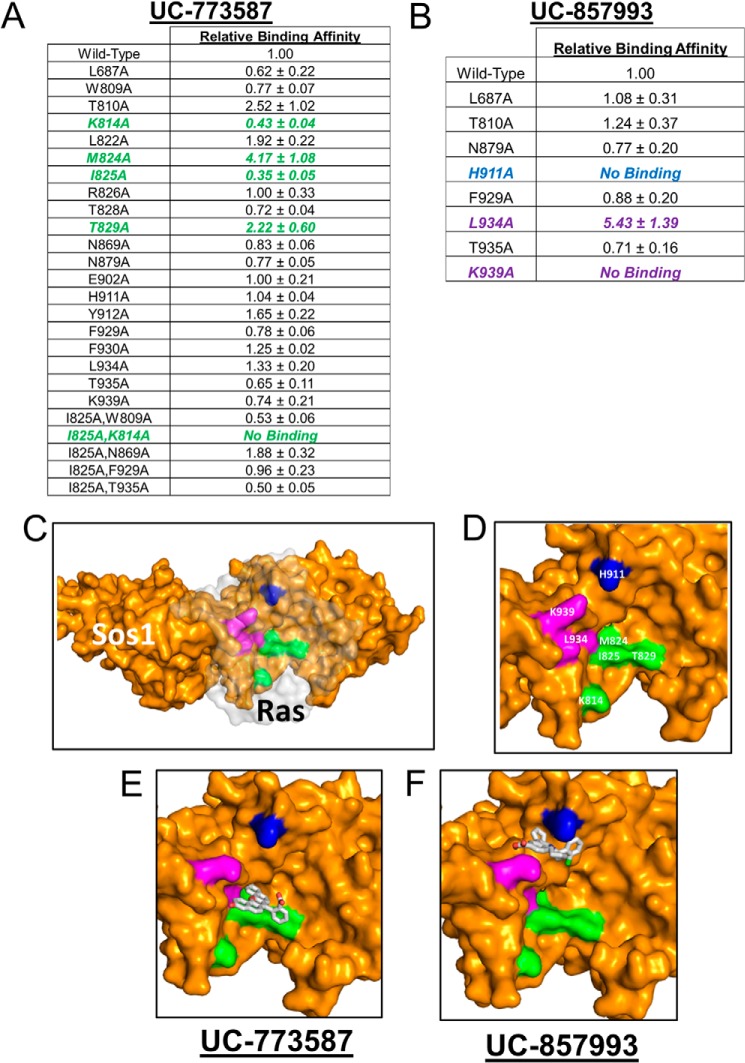
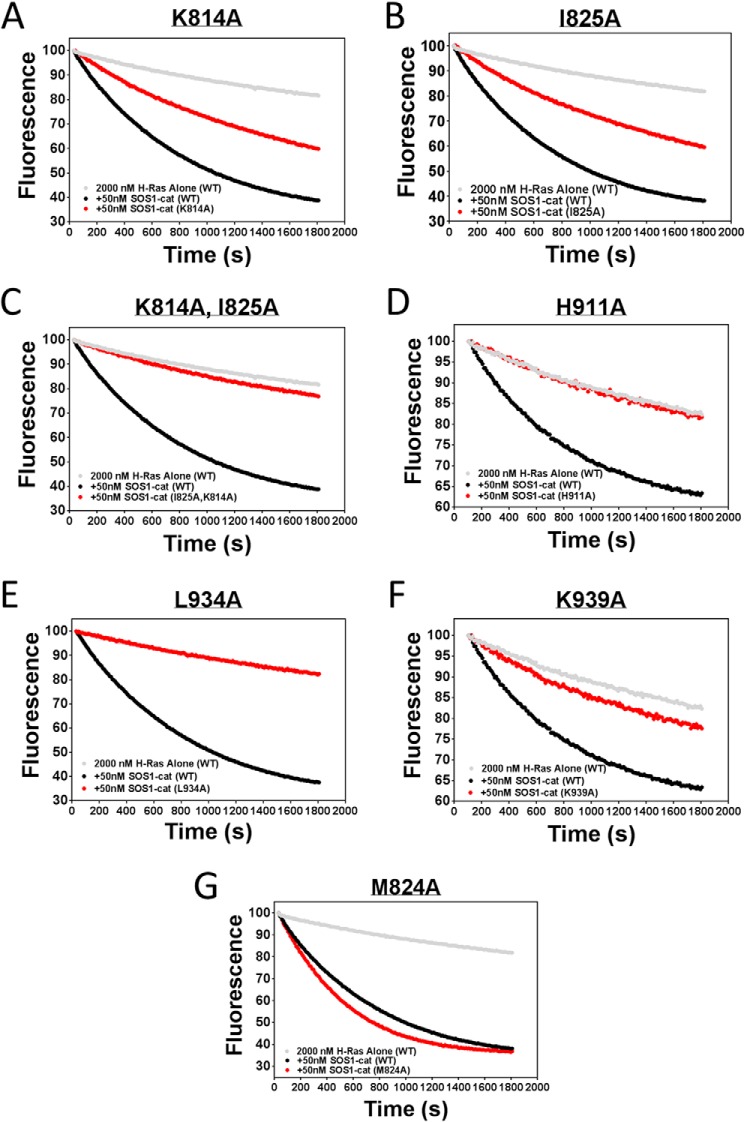
Based on our docking models, strong efficacy in our GEF exchange assays, additive inhibition of UC-773587 and UC-857993 in our biochemical experiments, and the observation that UC-773587 and UC-857993 disrupt the interaction between SOS1 and GDP-bound H-Ras, we hypothesized that UC-773587 and UC-857993 mapped to the catalytic site on SOS1. Therefore, we performed alanine scanning mutagenesis studies of residues predicted to interact with our compounds at the catalytic site (Fig. 8, A and B) by generating 19 single alanine mutants and five double alanine mutants. Additionally, we generated a single alanine mutant at the allosteric site (L687A), which has been shown previously to be critical for Ras binding (19, 32), as a negative control. Of the single point mutants, two (K939A and H911A) completely disrupted binding and one mutant (L934A) enhanced binding of UC-857993 to SOS1 (Fig. 9, B–D and F). Conversely, for UC-773587, two single point mutants (I825A and K814A) reduced binding and the double mutant (I825A/K814A) completely disrupted binding to SOS1, and the single point mutant of a residue at the bottom of the predicted binding pocket (M824A) slightly enhanced binding (Fig. 9, A and C–E). For both compounds, mutating one of the critical residues, Leu-687, at the allosteric site to alanine did not significantly affect compound binding (Fig. 9, A and B). The lack of binding, reduced binding, or enhanced binding was unlikely due to improper protein folding as four of the mutants (K814A, M824A, I825A, and K939A) were catalytically active, although the three other mutants (H911A, L934A, and K814A/I825A) were catalytically dead (Fig. 10), but the inactive mutants have been shown previously to be critical for the SOS1-Ras interaction (33, 34). Interestingly, it has been shown that His-911 and Lys-939 residues of SOS1 are critical for the interaction of SOS1 with the switch I region of Ras, whereas the Ile-825 residue of SOS1 is critical for the interaction of SOS1 with the switch II region of Ras (34). Based upon the lack of binding of UC-857993 to the H911A and K939A mutants, UC-857993 maps to the switch I Ras interaction region at the catalytic site of SOS1. The enhanced binding of UC-857993 to the L934A mutant and UC-773587 to the M824A mutant is likely due to removal of steric hindrance allowing for a better binding pocket. Based upon reduced binding of UC-773587 to the I825A single mutant and complete inhibition of binding to the K814A/I825A double mutant, UC-773587 may map to the switch II Ras interaction region at the catalytic site of SOS1. The reduced binding of UC-773587 to the K814A single mutant and subsequent complete inhibition of binding to the K814A/I825A double mutant may be attributed to an indirect conformational effect coupling the two mutated sites.
FIGURE 8.
Heat maps of the predicted docking sites for UC-773587 and UC-857993. Heat maps of predicted contacts with SOS1 made by the ligand in the predicted docking poses for UC-773587 (A) and UC-857993 (B), respectively. Polyview-MM representation of the ensemble of predicted docking poses is used, with individual rows corresponding to clusters of distinct poses, and the residues in contact with the compound in each pose highlighted in magenta. Residues that were found to contribute to binding by mutagenesis are further highlighted by arrows. Mutations that blocked or significantly reduced binding are highlighted using long or short arrows, respectively. The double mutant K814A/I825A is indicated by green arrows.
FIGURE 9.
Mutagenesis studies of the site of action for UC-773587 and UC-857993. Identification of the site of action for UC-773587 (A) and UC-857993 (B) at the catalytic site on SOS1 through mutagenesis analysis is shown. Single or double alanine mutants were generated in His6-SOS1cat as described under “Experimental Procedures.” Direct binding of UC-773587 and UC-857993 to wild-type and mutant His6-SOS1cat was measured by microscale thermophoresis. 0 to 100 μm of UC-773587 or UC-857993 was titrated to a constant amount of NT-647-labeled wild-type or mutant His6-SOS1cat (100 nm). Key interacting residues for UC-773587 and UC-857993 are highlighted in color in the tables (UC-773587, green; UC-857993, blue and magenta). C, structural view of the site of actions for both UC-773587 and UC-857993 in the catalytic site of SOS1 (yellow) in complex with Ras (transparent gray). D, residue-labeled site of action for both UC-773587 and UC-857993 in the catalytic site of SOS1. UC-857993 (E) and UC-773587 (F) docked in the catalytic pocket of SOS1. Based upon mutagenesis studies, key interacting residues are highlighted using the same colors as in C and D; note that whereas UC-773587 and UC-857993 share one site of action, they each interact with distinct residues in blue, magenta, and green patches, respectively. Data in A and B represent the mean ± S.E. of three independent experiments. Data in A and B are expressed as relative binding affinities normalized to the binding affinity of wild-type SOS1cat as determined by nonlinear regression (GraphPad Prism 6). A and B, binding affinity of >1 indicates higher affinity, and a binding affinity of <1 indicates a lower affinity with respect to wild-type SOS1cat (Note: if a binding affinity was unable to be determined due to the lack of a binding event, the data are expressed as no binding in the tables.)
FIGURE 10.
Guanine nucleotide exchange of H-Ras catalyzed by selected mutant SOS1cat. 50 nm wild-type or mutant SOS1cat (black or red circles)-mediated GDP for GTP nucleotide exchange upon 2 μm H-Ras (amino acids 1–166) (gray circles) in the 96-well BODIPY-FL-GDP dissociation assay as described under “Experimental Procedures.” The data are expressed as percent change of relative fluorescence units normalized to the initial time point over 30 min. Data were performed in triplicate and represent the mean.
SAR Studies for UC-773587 and UC-857993
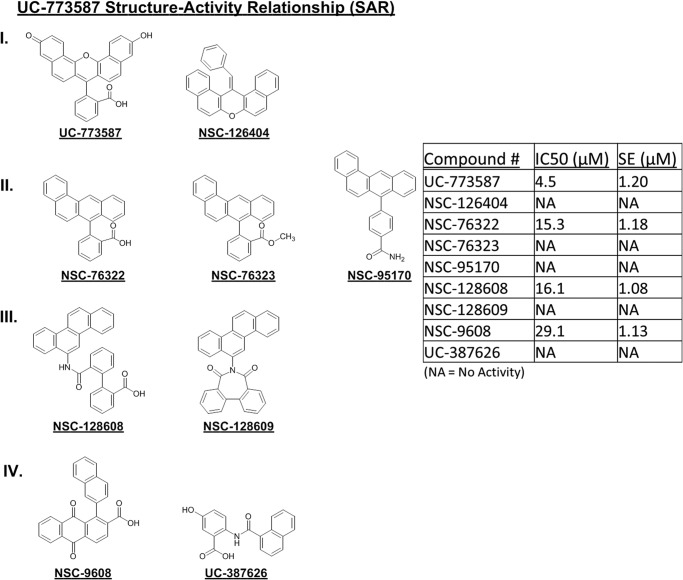
To further understand the structure-activity relationship of our two inhibitors of SOS1, computational searches for structural analogs of UC-773587 and UC-857993 were undertaken. Subsequently, the structural analogs were tested for their ability to inhibit SOS1cat-mediated GDP nucleotide exchange on H-Ras. Limited availability of closely related analogs allows only limited SAR conclusions. A carboxyl function appears to be essential because all of the activities (IC50 <50 μm) contain this group, most clearly demonstrated by the loss of activity from the carboxylate NSC-76322 to its methyl ester NSC-76323. H-bonding groups present at the distal ends of the molecule (as in UC-773587) also improve potency (Fig. 11). Interestingly, appending the carboxylate via a linkage that can adopt a perpendicular conformation (e.g. UC-773587 and NSC-76322) to the polycyclic system affords greater activity than the direct appending of the carboxylate as in NSC-9608.
FIGURE 11.
Structure-activity relationship for UC-773587. UC-773587 and related structural analogs were assayed for their ability to dose-dependently inhibit 50 nm His6-SOS1cat-mediated BODIPY-FL-GDP for GTP nucleotide exchange on 2 μm His6-H-Ras (residues 1–166) in a 96-well format. The UC-773587 analogs were grouped into four groups (I, II, III, and IV) based on their chemical structure as displayed. IC50 values were calculated based upon nonlinear regression analysis (GraphPad Prism 6) at a single time point of 900 s and are displayed in the table. IC50 values represent the mean ± S.E. of three independent experiments.
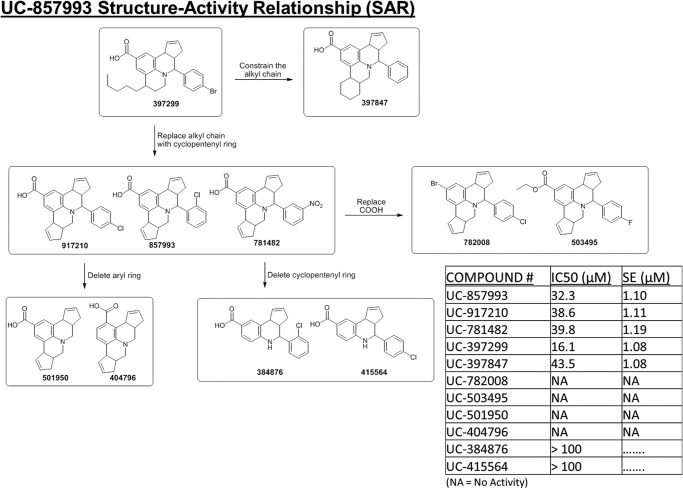
For UC-857993, analog availability allowed a more targeted range of modifications to the UC-857993 scaffold to be explored. Modification of the annulated cyclopentene ring modulated activity in the following manner: (i) deletion of the ring as in UC-384876 and UC-415564 eliminated all activity; (ii) expansion of the ring to a six-membered saturated ring as in UC-397847 led to a slight loss in activity; but (iii) replacing the ring with a flexible n-pentyl group (UC-397299) increased activity 2-fold. The appended aryl system is essential for activity (note: the loss of activity in analogs UC-501950 and UC-404796). Minor changes in substitutions as in UC-917210 and UC-781482 show little effect on activity. The carboxylate function, which most likely interacts with the Lys-939 residue in SOS1, is critical as shown by the complete loss of activity when converted to a methyl ester as in UC-503495 or replacement with bromine in UC-782008 (Fig. 12).
FIGURE 12.
Structure-activity relationship for UC-857993. UC-857993 and related structural analogs were assayed for their ability to dose-dependently inhibit 50 nm His6-SOS1cat-mediated BODIPY-TR-GTP loading on 2 μm His6-H-Ras (residues 1–166) in the 96-well format. The structural relationships of UC-857993 and related analogs are displayed as a flow diagram based upon experimental activity and chemical structure. IC50 values were calculated based upon nonlinear regression analysis (GraphPad Prism 6) at a single time point of 900 s and are displayed in the table. IC50 values represent the mean ± S.E. of three independent experiments. NA, no activity.
UC-773587 and UC-857993 Inhibit Ras Signaling and Cancer Cell Growth
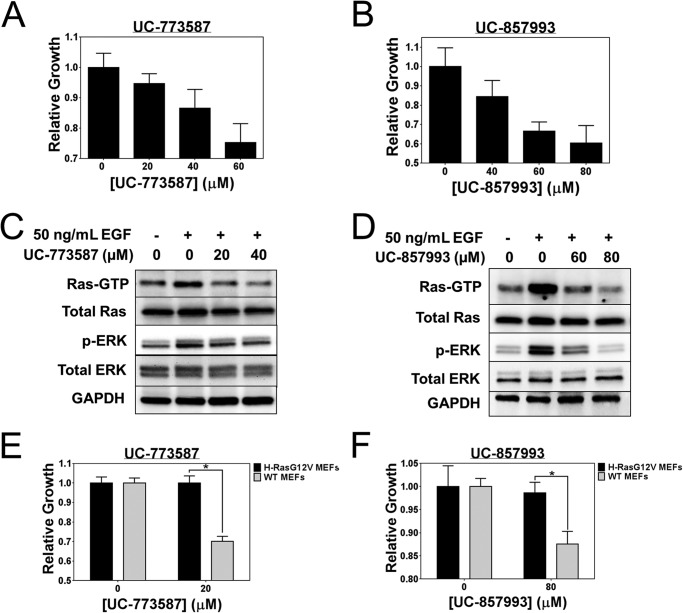
To assess cellular activity, the two SOS1 inhibitors (UC-773587 and UC-857993) were tested in cells at inhibiting epidermal growth factor receptor (EGFR)-mediated Ras activation and downstream ERK activation (35, 36). Therefore, we serum-starved NIH/3T3 mouse fibroblasts and pretreated them with either UC-773587 or UC-857993. Subsequently, we stimulated the cells with EGF to initiate Ras signaling. Both UC-773587 and UC-857993 dose-dependently inhibited (<50% inhibition) growth of NIH/3T3 cells (Fig. 13, A and B). Both UC-773587 and UC-857993 dose-dependently inhibited Ras activation and downstream ERK activation mediated through EGFR-SOS1-Ras-Raf1-MEK-ERK signaling (Fig. 13, C and D). Furthermore, to test the selectivity of both compounds for SOS1-Ras signaling, a GTPase-activating protein activity-deficient H-Ras(G12V) mutant expressing the mouse embryonic fibroblast cell line was produced (3, 25). Therefore, we tested the ability of UC-773587 and UC-857993 to inhibit growth of wild-type mouse embryonic fibroblasts versus activated mutant H-Ras(G12V)-transformed mouse embryonic fibroblasts. Interestingly, under the experimental conditions tested, UC-773587 and UC-857993 specifically inhibited wild-type mouse embryonic fibroblast growth but not the growth of oncogenic H-Ras(G12V)-expressing mouse embryonic fibroblasts (Fig. 13, E and F). As a result, this suggests that the compounds characterized here are selective for SOS1-Ras signaling.
FIGURE 13.
UC-773587 and UC-857993 inhibit Ras signaling and selective growth of mouse fibroblasts. UC-773587 (A) and UC-857993 (B) were tested for their ability to inhibit 3-day NIH/3T3 cell growth. Cells were grown in the presence of DMSO or indicated concentrations of UC-773587 or UC-857993 for 3 days and subsequently assayed for their growth by MTS as described under “Experimental Procedures.” C and D, NIH/3T3 cells were serum-starved for 24 h and pretreated with the indicated concentrations of UC-773587 for 16 h or UC-857993 for 2 h. Cells were stimulated with 50 ng/ml EGF for 5 min and then assayed for pan-Ras activation by GST-Raf1 effector domain pulldown experiments and activation of phospho-ERK by Western blotting. UC-773587 (E) and UC-857993 (F) were tested for their ability to inhibit 3-day wild-type or H-RasG12V-expressing MEF cell growth. Cells were grown in the presence of DMSO or indicated concentrations of UC-773587 or UC-857993 for 3 days and subsequently assayed for their growth by MTS as described under “Experimental Procedures.” Cell growth data were performed in triplicate and represent the mean ± S.E. Cell growth data are plotted as relative growth on day 3 normalized to the DMSO control. Western blot data are representative of n = 3 experiments. Statistical t test analysis was performed on data in E and F using GraphPad Prism 6 (E, p value = 0.006; F, p value = 0.037).
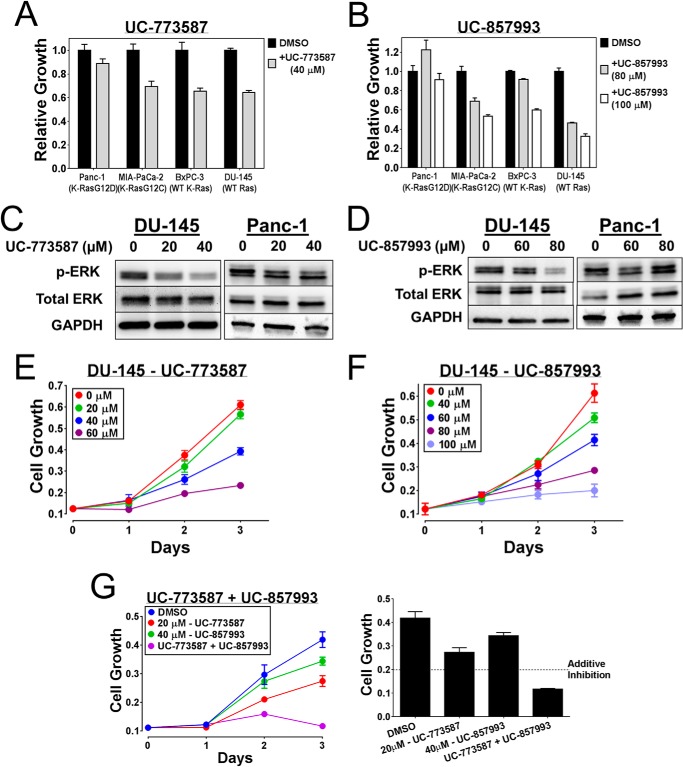
To further explore the selectivity of these inhibitors for SOS1-Ras signaling in a more translational model, UC-773587 and UC-857993 were tested for their effect upon cell growth on a panel of human cancer cell lines with wild type or activated mutant Ras status. Under the experimental conditions tested, both compounds specifically inhibited growth of prostate (DU-145) and pancreatic (BxPC-3) cancer cells containing the wild-type Ras status but not that of pancreatic (Panc-1) cancer cells containing the activated mutant K-Ras status (Fig. 14, A and B). Additionally, UC-773587 and UC-857993 inhibited downstream ERK activation in DU-145 prostate cancer cells with the wild-type Ras status but not in Panc-1 pancreatic cancer cells with the activated mutant K-Ras status (Fig. 14, C and D). Therefore, consistent with the mouse embryonic fibroblast results, these human cancer cell line results further suggest that the UC-773587 and UC-857993 are selective for SOS1-Ras signaling. Interestingly, there was an exception in which cell growth of MIA-PaCa-2 pancreatic cancer cells containing activated mutant K-Ras was suppressed by both UC-773587 and UC-857993 (Fig. 14, A and B), but these cells have previously been shown to be dependent upon SOS1 for their growth through a unique autocrine loop dependent upon wild-type H- and N-Ras (37, 38).
FIGURE 14.
UC-773587 and UC-857993 inhibit Ras signaling and growth of pancreatic and prostate cancer cells. UC-773587 (A) and UC-857993 (B) were tested for their ability to inhibit a 3-day cell growth of a panel of pancreatic and prostate cancer cell lines. Cells were grown in the presence of DMSO or indicated concentrations of UC-773587 or UC-857993 for 3 days and subsequently assayed for their growth by MTS as described under “Experimental Procedures.” C and D, DU-145 cells and Panc-1 cells were serum-starved for 24 h and pretreated with indicated concentrations of UC-773587 or UC-857993 for 16 h (DU-145-UC-773587), 2 h (DU-145-UC-857993), or 1 h (Panc-1-UC-773587 and UC-857993). Activation of phospho-ERK was measured by Western blot. Cell growth of DU-145 cells was measured in the presence of DMSO or indicated concentrations of UC-773587 (E) or UC-857993 (F) individually or in combination (G, left panel) over 72 h. Every 24 h of growth of the cells was assayed by MTS. G, right panel, combination treatment growth inhibition is represented as a bar graph at 72 h to assess additivity. Cell growth data were performed in triplicate or quadruplicate and represent the mean ± S.E. Data in A and B are plotted as relative growth on day 3 normalized to the DMSO control. Western blot data are representative of n = 3 experiments.
It has been shown previously that SOS1 is differentially expressed in prostate and breast cancer populations (7, 8) and is required for Ras signaling and growth of human prostate cancer cells (7). Given the additive effects of combination treatment of UC-773587 and UC-857993 in our biochemical experiments, we used the SOS1-dependent human DU-145 prostate cancer cells as a model to examine whether combination treatment of our SOS1 inhibitors could additively inhibit growth of the DU-145 cancer cells. As expected, both UC-773587 and UC-857993 were able to individually inhibit DU-145 cell growth in a dose-dependent manner (Fig. 14, E and F). From this result, modest inhibitory doses for UC-773587 and UC-857993 were selected to assess additivity of the compounds in the DU-145 cancer cell growth model. Consistent with the in vitro biochemical results, combination treatment of UC-773587 and UC-857993 inhibited growth of the DU-145 cells (Fig. 14G). Taken together, these cellular results suggest that both SOS1 inhibitors map to the catalytic site on SOS1 but can work additively or synergistically in suppressing SOS1 activity.
DISCUSSION
Currently, available pharmacological tools to dissect small GTPase signaling mechanisms remain limited. Although there has been significant efforts in targeting post-translational modifications (i.e. farnesyltransferases and geranylgeranyltransferases) of small GTPases (39–43) and small GTPase effector proteins (e.g. Raf, PI3K, ROCK, and PAK) (1, 44), these lead chemical inhibitors lack selectivity for upstream GTPase signaling. We and others have attempted to directly target small GTPases (i.e. Rac1, RhoA, Cdc42, and Ras) themselves (14–16, 45–49), but these chemical inhibitors tend to be of low affinity/efficacy due to the difficulty of targeting globular GTPase structures. With the exception of the Rho GEF inhibitors ITX3 for Trio and Y16 for LARG and the recently identified SOS1 inhibitor and activator (17, 50), there have been no other published reports successfully targeting the enzymatic activators of small GTPases, GEFs. The potentially structurally and functionally druggable GEFs are attractive targets for inhibition as they link upstream signaling to small GTPases and subsequent cellular signaling. Here, we present an affordable and efficient screening platform combining structure-based virtual screening and an experimental high throughput screening platform in designing and discovery of GEF inhibitors and an application to the Ras GEF, SOS1. Given the conserved mechanism of GEF action, we expect this screening platform can readily be adapted to studies of other small GTPase GEFs with available structural information.
Using our combinatorial screening platform, we were able to identify two chemically distinct inhibitors (UC-773587 and UC-857993) of the catalytic activity of SOS1. As predicted by our virtual screen, both inhibitors could bind directly to SOS1 and inhibited two distinct mechanistic steps of the GTPase cycle, including GEF-mediated GDP dissociation and GTP loading by interfering with the interaction between GDP-bound Ras and SOS1. Mutation analysis mapped both compounds to the catalytic pocket of SOS1 with UC-773587 binding to the Ras switch II interaction region and UC-857993 binding to the Ras switch I interaction region of SOS1. Interestingly, combination treatment additively inhibited SOS1 catalysis, blocked the SOS1-Ras interaction, and inhibited SOS1-dependent human prostate cancer cell growth. This suggests a potential new paradigm for dual targeting of catalytic pockets on enzymes with relatively large drug-accessible surfaces. In addition, recent studies have implicated SOS1 in oncogenic K-Ras tumorigenesis in pancreatic, colon, and breast cancer models by mediating H-Ras and N-Ras signaling through an autocrine loop (37, 38, 51), suggesting an interesting future utility of SOS1 inhibitors in K-Ras-mediated oncogenicity, which is supported here by the inhibition of MIA-PaCa-2 pancreatic cancer cell growth with both SOS1 inhibitors.
A recent review article (52) discussed the potential pitfalls of pan-assay interference compounds that often show up as false-positive hit compounds and trick chemical biologists into believing that they are true hits. Given the fluorescein-like structures of the hit compounds characterized here, we were concerned that they could be pan-assay interference compounds. To this end, we performed a series of careful validations and control experiments to confirm that this was not the case. First, in our experimental high throughput screening platform, we have built in a control for potential fluorescent artifact compounds by following up on hits using two complementary fluorescent-based biochemical exchange assays in both the fluorescein and Texas Red channels. Our hit compounds were validated to work in both exchange assays and fluorescence channels. Additionally, both compounds did not absorb light in the UV-visible spectra at wavelengths used for our biochemical experiments, and UC-773587 did not auto-fluoresce in the fluorescein or Texas Red channels, ruling out potential artifacts due to compound intrinsic fluorescence. Second, as suggested by the Baell and Walters (52), we performed follow-up validations with fresh powder compounds from our own source along with commercial sources to ensure the purity of our compounds and to show that they can directly bind to the target protein, SOS1, competitively disrupt the SOS1-Ras interaction, and selectively inhibit SOS1 catalytic activity specifically. Third, mutagenesis studies show that the hit compounds do not covalently bind to the target protein and do not bind to several mutants, and a preliminary SAR analysis shows several chemical structural analogs were inactive. Finally, a cellular test shows that the hits are capable of dose-dependently inhibiting EGFR-SOS1-Ras-Raf1-MEF-ERK signaling and proliferation of SOS1-addicted prostate and pancreatic cancer cells. Despite these collective efforts, there are limitations to our biochemical studies of the hit compounds; more detailed structural work by crystallization of the hits bound to SOS1 would help guide future medicinal chemical development. Overall, these studies provide a proof-of-concept for multiple platform screening for inhibitors of a Ras GEF, and we expect the approach can be adapted and applied to identifying lead inhibitors of other GEF proteins in academically feasible laboratories.
Acknowledgments
We thank Dr. Zhongyun Dong (Dept. of Internal Medicine, University of Cincinnati) for providing the DU-145 prostate cancer cell line.
This work was supported, in whole or in part, by the National Institutes of Health.
- GEF
- guanine nucleotide exchange factor
- PH
- pleckstrin homology
- MEF
- mouse embryonic fibroblast
- PDB
- Protein Data Bank
- FL
- fluorescein
- TR
- Texas Red
- MTS
- microscale thermophoresis
- SAR
- structure-activity relationship
- EGFR
- epidermal growth factor receptor
- DH
- Dbl homology domain
- HTS
- high throughout screening
- uHTS
- ultra-HTS
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt.
REFERENCES
- 1. Vigil D., Cherfils J., Rossman K. L., Der C. J. (2010) Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng Y. (2001) Dbl family guanine nucleotide exchange factors. Trends Biochem. Sci. 26, 724–732 [DOI] [PubMed] [Google Scholar]
- 3. Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. (2011) RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer 11, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cox A. D., Der C. J. (2010) Ras history: The saga continues. Small GTPases 1, 2–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roberts A. E., Araki T., Swanson K. D., Montgomery K. T., Schiripo T. A., Joshi V. A., Li L., Yassin Y., Tamburino A. M., Neel B. G., Kucherlapati R. S. (2007) Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 39, 70–74 [DOI] [PubMed] [Google Scholar]
- 6. Tartaglia M., Pennacchio L. A., Zhao C., Yadav K. K., Fodale V., Sarkozy A., Pandit B., Oishi K., Martinelli S., Schackwitz W., Ustaszewska A., Martin J., Bristow J., Carta C., Lepri F., et al. (2007) Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 39, 75–79 [DOI] [PubMed] [Google Scholar]
- 7. Timofeeva O. A., Zhang X., Ressom H. W., Varghese R. S., Kallakury B. V., Wang K., Ji Y., Cheema A., Jung M., Brown M. L., Rhim J. S., Dritschilo A. (2009) Enhanced expression of SOS1 is detected in prostate cancer epithelial cells from African-American men. Int. J. Oncol. 35, 751–760 [PMC free article] [PubMed] [Google Scholar]
- 8. Field L. A., Love B., Deyarmin B., Hooke J. A., Shriver C. D., Ellsworth R. E. (2012) Identification of differentially expressed genes in breast tumors from African American compared with Caucasian women. Cancer 118, 1334–1344 [DOI] [PubMed] [Google Scholar]
- 9. Tanizaki R., Katsumi A., Kiyoi H., Kunishima S., Iwasaki T., Ishikawa Y., Kobayashi M., Abe A., Matsushita T., Watanabe T., Kojima T., Kaibuchi K., Kojima S., Naoe T. (2008) Mutational analysis of SOS1 gene in acute myeloid leukemia. Int. J. Hematol. 88, 460–462 [DOI] [PubMed] [Google Scholar]
- 10. Sahai E., Marshall C. J. (2002) RHO-GTPases and cancer. Nat. Rev. Cancer 2, 133–142 [DOI] [PubMed] [Google Scholar]
- 11. Spiegel J., Cromm P. M., Zimmermann G., Grossmann T. N., Waldmann H. (2014) Small molecule modulation of Ras signaling. Nat. Chem. Biol. 10, 613–622 [DOI] [PubMed] [Google Scholar]
- 12. Patgiri A., Yadav K. K., Arora P. S., Bar-Sagi D. (2011) An orthosteric inhibitor of the Ras-Sos interaction. Nat. Chem. Biol. 7, 585–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schöpel M., Jockers K. F., Düppe P. M., Autzen J., Potheraveedu V. N., Ince S., Yip K. T., Heumann R., Herrmann C., Scherkenbeck J., Stoll R. (2013) Bisphenol A binds to ras proteins and competes with guanine nucleotide exchange: implications for GTPase-selective antagonists. J. Med. Chem. 56, 9664–9672 [DOI] [PubMed] [Google Scholar]
- 14. Zimmermann G., Papke B., Ismail S., Vartak N., Chandra A., Hoffmann M., Hahn S. A., Triola G., Wittinghofer A., Bastiaens P. I., Waldmann H. (2013) Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 497, 638–642 [DOI] [PubMed] [Google Scholar]
- 15. Maurer T., Garrenton L. S., Oh A., Pitts K., Anderson D. J., Skelton N. J., Fauber B. P., Pan B., Malek S., Stokoe D., Ludlam M. J., Bowman K. K., Wu J., Giannetti A. M., Starovasnik M. A., et al. (2012) Small molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. U.S.A. 109, 5299–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ostrem J. M., Peters U., Sos M. L., Wells J. A., Shokat K. M. (2013) K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evelyn C. R., Duan X., Biesiada J., Seibel W. L., Meller J., Zheng Y. (2014) Rational design of small molecule inhibitors targeting the Ras GEF, SOS1. Chem. Biol. 21, 1618–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sondermann H., Soisson S. M., Boykevisch S., Yang S. S., Bar-Sagi D., Kuriyan J. (2004) Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell 119, 393–405 [DOI] [PubMed] [Google Scholar]
- 20. Biesiada J., Porollo A., Velayutham P., Kouril M., Meller J. (2011) Survey of public domain software for docking simulations and virtual screening. Hum. Genomics 5, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Porollo A., Meller J. (2010) POLYVIEW-MM: web-based platform for animation and analysis of molecular simulations. Nucleic Acids Res. 38, W662–W666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang J. H., Chung T. D., Oldenburg K. R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 [DOI] [PubMed] [Google Scholar]
- 23. Jerabek-Willemsen M., Wienken C. J., Braun D., Baaske P., Duhr S. (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 9, 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wienken C. J., Baaske P., Rothbauer U., Braun D., Duhr S. (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 100. [DOI] [PubMed] [Google Scholar]
- 25. Stengel K. R., Zheng Y. (2012) Essential role of Cdc42 in Ras-induced transformation revealed by gene targeting. PLoS One 7, e37317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Konc J., Janezic D. (2012) ProBiS-2012: web server and web services for detection of structurally similar binding sites in proteins. Nucleic Acids Res. 40, W214–W221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Konc J., Janežič D. (2014) ProBiS-ligands: a web server for prediction of ligands by examination of protein binding sites. Nucleic Acids Res. 42, W215–W220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evelyn C. R., Ferng T., Rojas R. J., Larsen M. J., Sondek J., Neubig R. R. (2009) High throughput screening for small molecule inhibitors of LARG-stimulated RhoA nucleotide binding via a novel fluorescence polarization assay. J. Biomol. Screen. 14, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lenzen C., Cool R. H., Prinz H., Kuhlmann J., Wittinghofer A. (1998) Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry 37, 7420–7430 [DOI] [PubMed] [Google Scholar]
- 30. McEwen D. P., Gee K. R., Kang H. C., Neubig R. R. (2002) Fluorescence approaches to study G protein mechanisms. Methods Enzymol. 344, 403–420 [DOI] [PubMed] [Google Scholar]
- 31. Roman D. L., Talbot J. N., Roof R. A., Sunahara R. K., Traynor J. R., Neubig R. R. (2007) Identification of small molecule inhibitors of RGS4 using a high throughput flow cytometry protein interaction assay. Mol. Pharmacol. 71, 169–175 [DOI] [PubMed] [Google Scholar]
- 32. Margarit S. M., Sondermann H., Hall B. E., Nagar B., Hoelz A., Pirruccello M., Bar-Sagi D., Kuriyan J. (2003) Structural evidence for feedback activation by Ras. GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 [DOI] [PubMed] [Google Scholar]
- 33. Boriack-Sjodin P. A., Margarit S. M., Bar-Sagi D., Kuriyan J. (1998) The structural basis of the activation of Ras by Sos. Nature 394, 337–343 [DOI] [PubMed] [Google Scholar]
- 34. Hall B. E., Yang S. S., Boriack-Sjodin P. A., Kuriyan J., Bar-Sagi D. (2001) Structure-based mutagenesis reveals distinct functions for Ras switch 1 and switch 2 in Sos-catalyzed guanine nucleotide exchange. J. Biol. Chem. 276, 27629–27637 [DOI] [PubMed] [Google Scholar]
- 35. Buday L., Downward J. (2008) Many faces of Ras activation. Biochim. Biophys. Acta 1786, 178–187 [DOI] [PubMed] [Google Scholar]
- 36. Pierre S., Bats A. S., Coumoul X. (2011) Understanding SOS (Son of Sevenless). Biochem. Pharmacol. 82, 1049–1056 [DOI] [PubMed] [Google Scholar]
- 37. Grabocka E., Pylayeva-Gupta Y., Jones M. J., Lubkov V., Yemanaberhan E., Taylor L., Jeng H. H., Bar-Sagi D. (2014) Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell 25, 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jeng H. H., Taylor L. J., Bar-Sagi D. (2012) Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat. Commun. 3, 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Appels N. M., Beijnen J. H., Schellens J. H. (2005) Development of farnesyl transferase inhibitors: a review. Oncologist 10, 565–578 [DOI] [PubMed] [Google Scholar]
- 40. Hara M., Akasaka K., Akinaga S., Okabe M., Nakano H., Gomez R., Wood D., Uh M., Tamanoi F. (1993) Identification of Ras farnesyltransferase inhibitors by microbial screening. Proc. Natl. Acad. Sci. U.S.A. 90, 2281–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Epifano F., Curini M., Genovese S., Blaskovich M., Hamilton A., Sebti S. M. (2007) Prenyloxyphenylpropanoids as novel lead compounds for the selective inhibition of geranylgeranyl transferase I. Bioorg. Med. Chem. Lett. 17, 2639–2642 [DOI] [PubMed] [Google Scholar]
- 42. Peterson Y. K., Kelly P., Weinbaum C. A., Casey P. J. (2006) A novel protein geranylgeranyltransferase-I inhibitor with high potency, selectivity, and cellular activity. J. Biol. Chem. 281, 12445–12450 [DOI] [PubMed] [Google Scholar]
- 43. Peterson Y. K., Wang X. S., Casey P. J., Tropsha A. (2009) Discovery of geranylgeranyltransferase-I inhibitors with novel scaffolds by the means of quantitative structure-activity relationship modeling, virtual screening, and experimental validation. J. Med. Chem. 52, 4210–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van der Meel R., Symons M. H., Kudernatsch R., Kok R. J., Schiffelers R. M., Storm G., Gallagher W. M., Byrne A. T. (2011) The VEGF/Rho GTPase signalling pathway: a promising target for anti-angiogenic/anti-invasion therapy. Drug. Discov. Today 16, 219–228 [DOI] [PubMed] [Google Scholar]
- 45. Gao Y., Dickerson J. B., Guo F., Zheng J., Zheng Y. (2004) Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. U.S.A. 101, 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun Q., Burke J. P., Phan J., Burns M. C., Olejniczak E. T., Waterson A. G., Lee T., Rossanese O. W., Fesik S. W. (2012) Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl. 51, 6140–6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shang X., Marchioni F., Sipes N., Evelyn C. R., Jerabek-Willemsen M., Duhr S., Seibel W., Wortman M., Zheng Y. (2012) Rational design of small molecule inhibitors targeting RhoA subfamily Rho GTPases. Chem. Biol. 19, 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shutes A., Onesto C., Picard V., Leblond B., Schweighoffer F., Der C. J. (2007) Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J. Biol. Chem. 282, 35666–35678 [DOI] [PubMed] [Google Scholar]
- 49. Hong L., Kenney S. R., Phillips G. K., Simpson D., Schroeder C. E., Nöth J., Romero E., Swanson S., Waller A., Strouse J. J., Carter M., Chigaev A., Ursu O., Oprea T., Hjelle B., et al. (2013) Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J. Biol. Chem. 288, 8531–8543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Burns M. C., Sun Q., Daniels R. N., Camper D., Kennedy J. P., Phan J., Olejniczak E. T., Lee T., Waterson A. G., Rossanese O. W., Fesik S. W. (2014) Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange. Proc. Natl. Acad. Sci. U.S.A. 111, 3401–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hocker H. J., Cho K. J., Chen C. Y., Rambahal N., Sagineedu S. R., Shaari K., Stanslas J., Hancock J. F., Gorfe A. A. (2013) Andrographolide derivatives inhibit guanine nucleotide exchange and abrogate oncogenic Ras function. Proc. Natl. Acad. Sci. U.S.A. 110, 10201–10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baell J., Walters M. A. (2014) Chemistry: Chemical con artists foil drug discovery. Nature 513, 481–483 [DOI] [PubMed] [Google Scholar]