Abstract
Mutations in GJB2 are a major cause of autosomal recessive non-syndromic hearing loss (ARNSHL) in many populations. A single mutation of this gene (35delG) accounts for approximately 70% of GJB2 mutations that are associated with ARNSHL in Caucasians in many European countries and also in Iranian. In this study, we used PCR and restriction digestion to genotype five single nucleotide polymorphisms (SNPs) that define the genetic background of the 35delG mutation over an interval of 98Kbp that includes the coding and flanking regions of GJB2. Two microsatellite markers, D13S175 and D13S141, were also analyzed in patients and controls. These data suggest that the 35delG mutation originated in north and northwest of Iran.
Keywords: GJB2, 35delG, Iran, ARNSHL
INTRODUCTION
One in every 1000 neonates is born with profound congenital deafness, often the result of autosomal recessive inheritance (autosomal recessive non-syndromic hearing loss, ARNSHL). Although more than 77 genetic loci have been implicated in ARNSHL (known as DFNB loci), mutations in one gene – GJB2 (MIM *121011) – are the most frequent genetic cause of congenital deafness in many populations. Previous studies have demonstrated that the most frequent GJB2 mutation in the northern European population is the 35delG mutation, which originated on a specific genetic background consistent with an ancestral founder. [Zelante et al, 1997 ; Chaib et al, 1994 ; Denoyelle et al, 1997 ; Kelsell et al, 1997 ; Morell et al, 1998 ; Park et al, 2000 ; Maw et al, 1995].
The 35delG mutation in GJB2 is also the most common pathogenic allele in persons with ARNSHL in Iran, especially in provinces that include Turkish and Gilaki ethnicities where in aggregate, mutations in GJB2 account for 22.2% and 38.3% of ARNSHL, respectively. In these two populations, homozygosity for the 35delG mutation is estimated at 32% and 17% respectively.[Najmabadi et al, 2005; Hashemzadeh Chaleshtori et al, 2007].
Based on the historical route of early human migration from Siberia to the Iran plateau and Europe, a report postulating a possible 35delG founder in the Middle East [Van Laer et al, 2001], and the low prevalence of this mutation in Pakistan and Oman (3.7 % and 0%, respectively).[Brown et al, 1996; Simsek et al, 2001a Simsek et al, 2001b] we conducted this study to test the hypothesis that the GJB2 35delG mutation may have originated in an Iranian population several thousand years ago.
MATERIALS AND METHODS
PATIENTS
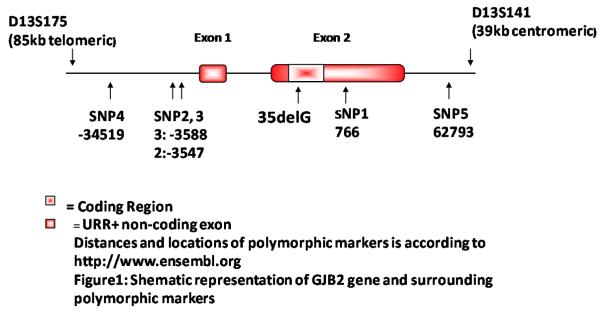
Fifty probands (20 males, 30 females) 16.3 ±11 years of age were selected from unrelated families originating from the Turkish (East Azerbayjan) and Gilaki (Gilan Province) ethnic groups of Iran for inclusion in this study. All persons were homozygous for the GJB2 35delG mutation. Controls with wild type GJB2 sequence were selected from healthy blood donors and matched to the case group by sex, age and ethnicity. Five single nucleotide polymorphisms (SNPs) and two short tandem repeat (STR) markers flanking GJB2 were used to define the genetic background (Fig 1).
Figure 1.
SNPs and STRs located near and within GJB2.
Informed consent was obtained from cases and controls after the study had been approved by the local ethics committee of the Genetics Research Center, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran.
MICROSATELLITE ANALYSIS
Two dinucleotide repeat microsatellite markers – D13S141(UniSTS:147652) and D13S175(UniSTS:19888) – flanking GJB2 were amplified in a volume of 20 µl containing 100 ng of template DNA, 1x PCR buffer (F. Hoffmann–La Roche Lt –Applied), 200 mM each dNTP(Roche -Applied), one unit of Taq DNA polymerase (F. Hoffmann–La Roche Lt –Applied) and 10 pmol of each primer. Amplification of D13S141 was performed using primers: forward, 5'-ACC ACG GAG CAA AGA ACA GA-3' and reverse, 5'-GTC CTC CCG GCC TAG TCT TA-3'. D13S175 was amplified using primers: forward, 5'-TAT TGG ATA CTT GAA TCT GCT G-3' and reverse, 5'-CAC TTA AAA TCT ACT CTC TCA GCA G-3'. Amplification conditions included a 30-cycle three-step PCR (denaturation at 94 °C for 30 s, annealing 58 °C for 30 s, extension at 72 °C for 30 s) with an initial step at 94° for 5 min and an extra extension time at 72 °C for 3 min. PCR products were resolved using 8% polyacrylamide gel electrophoresis and silver nitrate staining.
SNP GENOTYPING
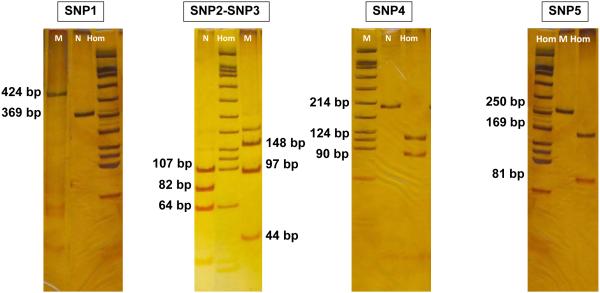
The five SNPs flanking the 35delG mutation were genotyped by conventional PCR and restriction digestion. Amplification of SNP1(rs3751385) was performed as previously described [Tekin et al, 2005] with genotyping completed by digestion of the amplification product using MaeI (F. Hoffmann–La Roche Lt –Applied). SNP2(rs9552101 or SNP 1285) and SNP3(SNP 1245) were included in the same amplicon and resolved by digestion with BanI (New England Biolabs) and HaeIII (F. Hoffmann–La Roche Lt –Applied), respectively. SNP4(rs877098) and SNP5(rs747931) were also amplified using a single primer pair and resolved by digestion with EcoRI and PstI, respectively [Tekin et al, 2005] (Fig 2).
Figure 2.
Restriction digests to resolve SNPs 1-5 (Hom: homozygous for the mentioned SNP; N: SNP wild type; M: marker).
PCR was completed using genomic DNA amplified (120 ng) and 1 U of FastStart Taq polymerase (Roche-Applied), 1.5 mM of MgCl2, dNTP (0.2 mM) and 10 pmol of the appropriate primers. For amplification, hot start PCR with 40 cycles of 3 steps including: 94 °C, 30 s; 58 °C, 30 s; and 72 °C, 45s was performed with an initial denaturation for 5 min and a final extension of 7 min.
RESULTS
Results of genotyped SNPS and STR markers and final analysis in patients and controls by SPSS version 13.0 are demonstrated in Table 1. With the exception of SNP5, which is located far from 35delG, allele distribution is significantly different between cases and controls. The 35delG mutation segregates most commonly on a T-C-G-T genetic background (SNP4-SNP3-SNP2-SNP1, respectively) ; in contrast, genotypes from control samples are in Hardy-Weinberg equilibrium (PLINK software) (Table 1).
Table I.
Results of genotyping analysis by SPSS version 13.0
| Marker | Alleles | Patients homozygous for 35delG mutation (Chr=100) |
Controls (Chr=100) |
P value | |
|---|---|---|---|---|---|
| D13S175(84785 bp telomeric) | 1(107) | 1 4 | 4 | 0.00000 | ☆ |
| 2(109) | 1 | 14 | |||
| 3(111) | 94 | 51 | |||
| 4(113) | 1 | 11 | |||
| 5(115) | 0 | 14 | |||
| 6(117) | 0 | 6 | |||
| SNP4(34519 bp telomeric) | C | 7 | 59 | 0.00000 | ☆ |
| T | 93 | 41 | |||
| SNP3(3588 bp telomeric) | C | 100 | 24 | 0.00000 | ☆ |
| T | 0 | 76 | |||
| SNP2(3547 bp telomeric) | A | 0 | 6 | 0.029 | ☆ |
| G | 100 | 94 | |||
| SNP1(765 bp centromeric) | C | 8 | 84 | 0.00000 | ☆ |
| T | 92 | 16 | |||
| D13S141(39262 bp centromeric) | 1(116) | 0 | 0 | 0.00000 | ☆ |
| 2(118) | 0 | 2 | |||
| 3(120) | 2 | 6 | |||
| 4(122) | 4 | 76 | |||
| 5(124) | 86 | 10 | |||
| 6(126) | 8 | 6 | |||
| 7(130) | 0 | 0 | |||
| SNP5(62793 bp centromeric) | C | 35 | 39 | 0.661 | |
| T | 65 | 61 |
shows the significant difference ( P value < 0.05) of the variable between cases and controls for those specific alleles.
Eighty five percent of Gilaki cases carry the T-C-G-T haplotype, as do 80% of Turkish cases. Eight of 50 individuals(16%) did not carry the most common block, consistent with a recombination event after the 35del mutation arose [Rothrock et al. 2003] (Table 2).
Table II.
Eight of 50 individuals (16%) did not carry the most common block, consistent with a recombination event after the 35delG mutation arose
| SNP4 | SNP3 | SNP2 | SNP1 | Ethnicity | Frequency |
|---|---|---|---|---|---|
| C | C | G | T | Turk–Gilaki | (3/50)6% |
| T | C | G | C | Turk–Gilaki | (5/50)10% |
DISCUSSION
Although mutations in GJB2 are a frequent cause of hearing loss in many populations, there are some populations – like the Pakistani and Arab – in which GJB2 – related deafness is rare [Brown et al, 1996; Simsek et al, 2001a Simsek et al, 2001b]. In this study, we investigated the origins of the 35delG mutation in GJB2 by studying flanking polymorphisms and found that 41 of 50 probands with severe-to-profound deafness and homozygous for the 35delG mutation shared a common haplotype T-C-G-T for SNP4-SNP3-SNP2-SNP1, respectively. These results are consistent with the presence of a 35delG founder in northern Iran. As SNP5 is more distant from the 35delG mutation, recombination is more possible [Rothrock et al, 2003].
In multiple northern European populations reported by Van Laer and colleagues, significant differences between the genotypes of patients and controls were found for five SNPs close to GJB2, with nearly complete association of one SNP allele (SNP1 in their paper) with the 35delG mutation. They concluded that the 35delG mutation may have originated somewhere in the Middle East and then spread throughout Europe [Van Laer et al, 2001]. This hypothesis is in agreement of another study which presented- by application of different markers - evidence for a founder in Caucasian and Middle Eastern population.[Rothrock et al. 2003]
Based on this statement, and reported carrier frequencies for the 35delG mutation of 1/35.2 and 1.78% in the Turkish population, Tekin and colleagues sought to determine whether the 35delG mutation has single origin in Anatolia. They found the prevalence of haplotype (T-C-G-T) to be 43% and postulated a single 35delG origin in Anatolia [Gasparini et al. and Tekin et al. 2001 and Tekin et al. 2004]
Our analysis of 35delG mutation-carrying chromosomes - using the same STR and SNP markers shows that 42 of 50 probands (84%) share the common haplotype (T-C-G-T), in agreement with data in the Anatolian population [Tekin et al, 2004]. The comparison of these haplotypes in different populations is shown in Table 3.
Table III.
Comparison of haplotypes in different populations (according to the last published data in the references)
| Allele | Iran | Turkey | Italy | Brazil | Us | Ashkenazi | Greece | Belgian | British | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||
| Deaf | Normal | Deaf | Normal | ||||||||||
| SNP1 (rs3751385) | C T |
8 92 |
84 16 |
0 89 |
89 29 |
— — |
— — |
0 45 |
— — |
1 117 |
0 33 |
0 27 |
|
| SNP2 (rs9552101 or SNP1285) |
G A |
100 0 |
94 6 |
91 0 |
87 3 |
100 3 |
94.6 5.5 |
100 0 |
100 0 |
100 0 |
— — |
— — |
— — |
| SNP3 (SNP1245) | C T |
100 0 |
24 76 |
92 0 |
5 67 |
99.3 0.7 |
100 0 |
100 0 |
100 0 |
— — |
— — |
— — |
|
| SNP4 (rs877098 or Hcv1813042) |
T C |
93 7 |
41 59 |
86 6 |
55 69 |
95.4 4.6 |
89.2 10.7 |
— — |
— — |
— — |
— — |
— — |
|
| SNP5 (rs747931) | C T |
35 65 |
39 61 |
35 57 |
49 87 |
— — |
— — |
— — |
— — |
39 71 |
— — |
— | |
Lucotte et al. compared 35delG frequencies in different European and Middle Eastern populations and found a lower-to-higher gradient from northern to southern Europe, wth the highest 35delG frequencies in southern Europe and the Mediterranean regions. Based on the high 35delG carrier frequencies in southern European countries (3.19%) such as Italy, Greece and south of France compared to the relatively lower carrier frequency in Middle East, they concluded that ancient Greece might be the centre of origin for 35delG mutation and not Turkey. [Lucotte et al. 2005 and Belguith et al 2005]
In another study, by using the same markers as used by Van Laer (2001), Kokota et al. found a common haplotype in the Greek population. Based on the high carrier frequency, a common haplotype, and some historical documents, they concluded that ancient Greece might be the origin of the GJB2 35delG mutation. [Kokota et al. 2008 and 2010]
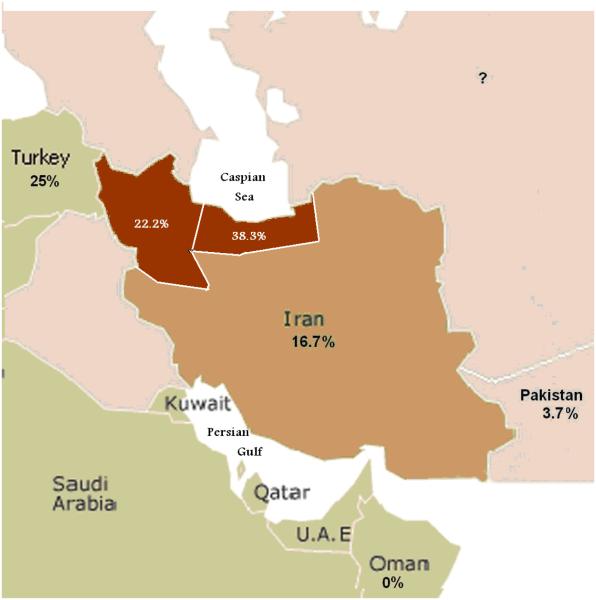
The 35delG mutation is one of the most prevalent ARNSHL-causing mutations in the north and northwest provinces of Iran amongst the Gilaki and Turkish populations [Najmabadi et al, 2005 ; Hashemzadeh Chaleshtori et al, 2007](Fig 3). These populations (Turk and Gilaki) are bounded by the Caspian Sea in the North and remain relatively isolated by mountains from other parts of Iran. [Najmabadi et al, 2005]
Figure 3.
GJB2-related deafness of studied in the Middle East.
A gradual decrease in the frequency of the 35delG mutation can be seen as we move from the northwest-to-south and east through the Persian Gulf countries.[Najmabadi, 2005]. It is noteworthy that the frequency of the 35delG mutation in resident populations in the Middle East varies, while more eastward, in Pakistan (southeast of Iran), the frequency of GJB2-related deafness is very low (3.7%)[Brown et al, 1996]. Southward in Oman, no GJB2 pathogenic alleles have been reported [Simsek et al, 2001a Simsek et al, 2001b].
Our result (82% for a common haplotype) supports the presence of founder in northern Iran (as compared to 43% in Anatolia). Based on the population frequencies of the 35delG mutation and migration routes from central Asia to Anatolia 10 centuries ago, we suspect that the 35delG mutation arose in or near northwest Iran [Tekin et al, 2004].
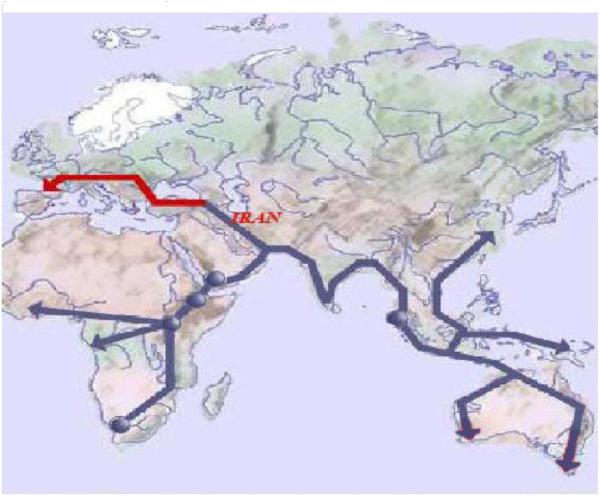
In ancient Iran Aryans were, at one time, the only residents of the Iran Plateau. The route of presumed migration of ancient humans from this area was to Europe. Consistent with this migratory route, as one moves from Iran to the east or south, the spectrum and prevalence of GJB2 mutations changes. In aggregate, our data place the origin of the 35delG mutation in the Middle East and more specifically in northern Iran.
Figure 4.
Journey of mankind presented by Bradshow Foundation (http://www.bradshawfoundation.com/journey/). The red color shows the migration path from Iran to Europe.
Acknowledgments
We would like to thank our patients and their families for their collaboration in this research. This project was sponsored by the Iranian National Science Foundation grant numbers 85073/23 and 85033/10. R. Smith is the Sterba Hearing Research Professor, University of Iowa College of Medicine, who supported the project with National Institutes of Health (NIH)-NIDCD grants RO1 DCOO2842 and RO1 DCO3544.
REFERENCES
- Belguith H, Hadjji S, Salem N, et al. Analysis of GJB2 mutation: evidence for a Mediterranean ancestor for the 35delG mutation. Clin. Genet. 2005;68:188–189. doi: 10.1111/j.1399-0004.2005.00474.x. [DOI] [PubMed] [Google Scholar]
- Brown KA, Janjua AH, Karbani G, Parry G, Noble A, Crockford G, Bishop DT, Newton VE, Markham AF, Mueller RF. Linkage studies of non-syndromic recessive deafness (NSRD) in a family originating from the Mirpur region of Pakistan maps DFNB1 centromeric to D13S175. Hum Mol Genet. 1996;5:169–173. doi: 10.1093/hmg/5.1.169. [DOI] [PubMed] [Google Scholar]
- Chaib H, Lina-Granade G, Guilford P, Plauchu H, Levilliers J, Morgon A, Petit C. A gene responsible for a dominant form of neurosensory nonsyndromic deafness maps to the NSRD1 recessive deafness gene interval. Hum Mol Genet. 1994;3:2219–2222. doi: 10.1093/hmg/3.12.2219. [DOI] [PubMed] [Google Scholar]
- Denoyelle F, Weil D, Maw MA, Wilcox SA, Lench NJ, Allen-Powell DR, Osborn AH, Dahl HH, Middleton A, Houseman MJ, et al. Prelingual deafness: High prevalence of a 30delG mutation in the connexin 26 gene. Hum Mol Genet. 1997;6:2173–2177. doi: 10.1093/hmg/6.12.2173. [DOI] [PubMed] [Google Scholar]
- Gasparini P, Rabionet R, Barbujani G, Melçhionda S, Petersen M, Brøndum-Nielsen K, Metspalu A, Oitmaa E, Pisano M, Fortina P, Zelante L, Estivill X. Eur J Hum Genet. 2000;8(1):19–23. doi: 10.1038/sj.ejhg.5200406. [DOI] [PubMed] [Google Scholar]
- Hashemzadeh Chaleshtori M, Farrokhi E, Shahrani M, Kheiri S, Dolati M, Hoghooghi Rad L, Pour-Jafari H, Ghatreh Samani K, Safa Chaleshtori K, Crosby AH. High carrier frequency of the GJB2 mutation(35delG) in the north of Iran. International Journal of Pediatric Otorhinolaryngology. 2007;71:863–867. doi: 10.1016/j.ijporl.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, Mueller RF, Leigh IM. Connexin 26 mutations in hereditary nonsyndromic sensorineural deafness. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- Kokotas H, Van Laer V, Grigoriadou M, Iliadou V, Economides J, Pomoni S, Pampanos A, Eleftheriades N, Ferekidou E, Korres S, Giannoulia-Karantana A, Van Camp G, Petersen M. Strong Linkage Disequilibrium for the Frequent GJB2 35delG Mutation in the Greek Population. American Journal of Medical Genetics Part A. 2008;146A:2879–2884. doi: 10.1002/ajmg.a.32546. [DOI] [PubMed] [Google Scholar]
- Kokotas H, Grigoriadou M, Villamar M, Giannoulia-Karantana A, Del Castillo I, Petersen M. Hypothesizing an ancient Greek origin of the GJB2 mutation:can science meet history? Genetic Testing and molecular biomarkers. 2010;14:183–187. doi: 10.1089/gtmb.2009.0146. [DOI] [PubMed] [Google Scholar]
- Lucotte G, Diéterlen F. The 35delG Mutation in the Connexin 26 Gene (GJB2) Associated with Congenital Deafness: European Carrier Frequencies and Evidence for Its Origin in Ancient Greece. GENETIC TESTING. 2005;9:20–25. doi: 10.1089/gte.2005.9.20. © Mary Ann Liebert, Inc. [DOI] [PubMed] [Google Scholar]
- Maw MA, Allen-Powell DR, Goodey RJ, Stewart IA, Nancarrow DJ, Hayward NK, Gardner RJ. The contribution of the DFNB1 locus to neurosensory deafness in a Caucasian population. Am J Hum Genet. 1995;57:629–635. [PMC free article] [PubMed] [Google Scholar]
- Morell RJ, Kim HJ, Hood LJ, et al. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med. 1998;339:1500–1505. doi: 10.1056/NEJM199811193392103. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Nishimura C, Kahrizi K, Riazalhosseini Y, Malekpour M, Daneshi A, Farhadi M, Mohseni M, Mahdieh N, Ebrahimi A, et al. GJB2 Mutations: Passage through Iran. American Journal of Medical Genetics. 2005;133A:132–137. doi: 10.1002/ajmg.a.30576. [DOI] [PubMed] [Google Scholar]
- Park HJ, Hahn SH, Chun YM, Park K, Kim HN. Connexin26 mutations associated with nonsyndromic hearing loss. Laryngoscope. 2000;110:1535–1538. doi: 10.1097/00005537-200009000-00023. [DOI] [PubMed] [Google Scholar]
- Rothrock CR, Murgia A, Sartorato EL, Leonardi E, Wei S, Lebeis SL, Yu LE, Elfenbein JL, Fisher RA, Friderici KH. Connexin 26 35delG does not represent a mutational hotspot. Hum Genet. 2003;113:18–23. doi: 10.1007/s00439-003-0944-2. [DOI] [PubMed] [Google Scholar]
- Simsek M, Al-Wardy N, Al-Khabory M. A seminested PCR test for simultaneous detection of two common mutations (35delG and 167delT) in the connexin-26 gene. Mol Diagnostic. 2001a;6:63–67. doi: 10.1054/modi.2001.22119. [DOI] [PubMed] [Google Scholar]
- Simsek M, Al-Wardy N, Al-Khayat A, Shanmugakonar M, Al-Bulushi T, Al-Khabory M, Al-Mujeni S, Al-Harthi S. Absence of deafness associated connexin-26 (GJB2) gene mutations in the Omani population. 2001b. [DOI] [PubMed]
- Tekin M, Akar N, Cin S, Blanton SH, Xia XJ, Liu XZ, Nance WE, Pandya A. Connexin 26 (GJB2) mutations in the Turkish population: implications for the origin and high frequency of the 35delG mutation in Caucasians. Hum Genet. 2001;108(5):385–9. doi: 10.1007/s004390100507. [DOI] [PubMed] [Google Scholar]
- Tekin M, Boğoclu G, Arican ST, Orman MN, Tastan H, Elsobky E, Elsayed S, Akar N. Evidence for single origins of 35delG and delE120 mutations in the GJB2 gene in Anatolia. Clin Genet. 2004;67(3):273. doi: 10.1111/j.1399-0004.2004.00334.x. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Coucke P, Mueller RF, Caethoven G, Flothmann K, Prasad SD, Chamberlin GP, Houseman M, Taylor GR, Van de Heyning CM. A common founder for the 35delG GJB2 gene mutation in connexin 26 hearing impairment. J Med Genet. 2001;38:515–518. doi: 10.1136/jmg.38.8.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante L, Gasparini P, Estivill X, Melchionda S, D’Agruma L, Govea N, Mila M, Monica MD, Lutfi J, Shohat M, et al. Connexin 26 mutations associated with the most common form of nonsyndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet. 1997;6:1605–160. doi: 10.1093/hmg/6.9.1605. [DOI] [PubMed] [Google Scholar]