Abstract
Engineering of the constant Fc part of monoclonal human IgG1 (hIgG1) Abs is an approach to improve effector functions and clinical efficacy of next-generation IgG1-based therapeutics. A main focus in such development is tailoring of in vivo half-life and transport properties by engineering the pH-dependent interaction between IgG and the neonatal Fc receptor (FcRn), as FcRn is the main homeostatic regulator of hIgG1 half-life. However, whether such engineering affects binding to other Fc-binding molecules, such as the classical FcγRs and complement factor C1q, has not been studied in detail. These effector molecules bind to IgG1 in the lower hinge–CH2 region, structurally distant from the binding site for FcRn at the CH2–CH3 elbow region. However, alterations of the structural composition of the Fc may have long-distance effects. Indeed, in this study we show that Fc engineering of hIgG1 for altered binding to FcRn also influences binding to both the classical FcγRs and complement factor C1q, which ultimately results in alterations of cellular mechanisms such as Ab-dependent cell-mediated cytotoxicity, Ab-dependent cellular phagocytosis, and Ab-dependent complement-mediated cell lysis. Thus, engineering of the FcRn–IgG1 interaction may greatly influence effector functions, which has implications for the therapeutic efficacy and use of Fc-engineered hIgG1 variants.
Introduction
Antibodies of the IgG class have rapidly become clinically important drugs for treatment of cancer as well as inflammatory diseases, and a range of Abs are under evaluation in clinical trials (1). Two such blockbusters used in therapy are trastuzumab (Herceptin), a humanized IgG1 with specificity for human epidermal growth factor receptor 2, for treatment of breast cancer and metastatic gastric or gastroesophageal junction cancer, and rituximab (Rituxan), a chimeric mouse-human IgG1 Ab against CD20 expressed on B cells, which is approved for treatment of non–Hodgkin lymphoma, chronic lymphocytic leukemia, vasculitis, and rheumatoid arthritis (1, 2). However, there is a need for improved therapeutic efficacy and patient convenience, which drives the industry toward development of next-generation Abs. Already available Abs as well as new Abs are therefore optimized with regard to improved serum half-life and tissue distribution or modulated effector functions such as Ab-dependent cell-mediated cytotoxicity (ADCC), Ab-dependent cellular phagocytosis (ADCP), and Ab-dependent complement-mediated cell lysis (ADCML) (1). For instance, enhancement of antitumor properties can be achieved by engineering the Fc region for stronger binding to activating FcγRs and C1q, or weaker binding to the inhibitory FcγRIIb to increase ADCC and ADCML effector activity (3–8). Alternatively, decreased ADCC and ADCML may be beneficial in terms of minimizing toxicity in other therapeutic settings (9–13). In either case, engineering is accomplished through alterations of amino acids in the Fc part, either by selection from display libraries of Ab fragments or by structure-guided design. Additionally, tailoring of the antennary glycans attached to the Fc part offers a powerful method to modify binding to the classical FcγRs for enhancement or elimination of ADCC activity (14–16). In general, Fc engineering for desired effector functions may be combined with any specificity of interest.
Most of the Abs approved or in clinical trials are built on the IgG class of Abs (1). In humans, IgG1, IgG2, and IgG4 all have a serum half-life of ∼3 wk, whereas IgG3 has a half-life of 1 wk, comparable with the short half-life of other Ab classes (1–2 d) (17, 18). The long half-life of IgG1 and its ability to bind FcγRs as well as C1q have made it attractive as an Ab therapeutic. The long half-life relies on its interaction with the neonatal Fc receptor (FcRn) (19, 20).
FcRn is widely expressed in different cell types and tissues, as for instance the vascular endothelium that covers the interior of blood and lymphatic vessels and in hematopoietic cells such as dendritic cells, predominantly within endosomal compartments (21–25). Similar to other circulating proteins, IgG is continuously pinocytosed by these and other cells. Subsequently, the low pH in the endosomes triggers binding to FcRn, followed by recycling to the cell surface and release upon exposure to the nearly neutral pH of the blood (24, 26, 27). Unbound material is directed from endosomes to lysosomes for degradation. Thus, the pH dependence of the interaction with FcRn is essential for efficient receptor-mediated recycling and rescue from degradation, which results in extended circulatory half-life. The serum half-life of therapeutic IgG Abs may be decreased or increased by engineering of the constant Fc domains for altered binding to human FcRn (hFcRn) (28–35). Such design has great potential for improving pharmacokinetics, as increased serum half-life offers the benefits of greater efficacy, less frequent dosing, and lower cost.
FcRn is an MHC class I–related molecule consisting of a unique transmembrane H chain that is noncovalently associated with β2-microglobulin (36). Site-directed mutagenesis studies and crystal structure analysis of the FcRn/IgG Fc complex show that the Fc amino acid residues at positions 252–256 in the CH2 domains and at 310, 433, 434, and 435 in the CH3 domains are at the core or in close proximity to the FcRn interaction site, and that the conserved histidine residues H310 and possibly H435 are responsible for the pH dependence (32, 36–38). These and other residues may be targeted by mutagenesis to alter the binding kinetics toward FcRn.
A challenge when designing IgGs with extended half-life is to maintain the pH dependence of the FcRn interaction. An IgG engineered to bind FcRn with higher affinity and reduced pH dependence potently inhibits the interaction of FcRn with endogenous IgG and thus rapidly lowers the concentration of circulating IgG, and the mutant itself is also rapidly degraded. This has been demonstrated to be the case for a human IgG1 (hIgG1) mutant with five substitutions in the Fc (M252Y/S254T/T256E/H433K/N434F; MST/HN), denoted as Abs for degradation. However, the mutant has shown promise as an FcRn blocker that accelerates the clearance of pathogenic IgG in mouse models (34, 39, 40).
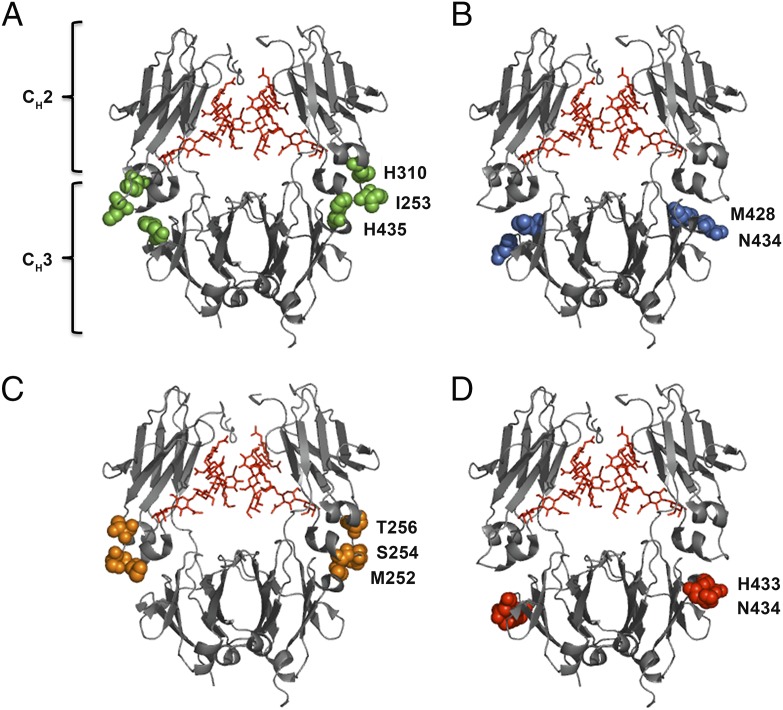
A handful of hIgG Abs have been developed that show favorable binding kinetics toward hFcRn, improved affinity at acidic pH with no or moderate binding at neutral pH (28–35). One such hIgG1 variant, with two mutations within the CH3 domains (M428L/N434S; MN) engineered into the vascular endothelial growth factor–specific Ab bevacizumab (Avastin), has demonstrated 4-fold longer half-life in hFcRn transgenic mice, and 3-fold in cynomolgus monkeys (41). Importantly, extended serum persistence was shown to translate into enhanced antitumor activity in a xenograft study in mice (41). A second hIgG1 variant with anti-lysozyme specificity with two mutations in the CH3 domain (H433K/N434F; HN) showed a 4-fold reduced half-life in wild-type (WT) mice, but enhanced FcRn-mediated transport in an ex vivo human placenta model (42). A third example is an hIgG1 variant with anti-respiratory syncytial virus specificity (motavizumab) and three mutations into the CH2 domain (M252Y/S254T/T246E) that improves affinity for hFcRn at acidic pH, resulting in 4-fold extended half-life in cynomolgus monkeys (35). Alternatively, it may be desirable to tune the half-life to achieve intermediate or shorter persistence as a means to optimize the target/nontarget localization ratios of Abs used for tumor imaging. This can be accomplished by targeting Fc residues directly involved in binding to FcRn (I253, H310, and H435), and such IgG mutants have been shown to have a range of distinct half-life profiles (43, 44). The positions of the amino acid residues are highlighted in the crystallographic illustrations of hIgG1 Fc shown in Fig. 1.
FIGURE 1.
Crystal structure illustrations of hIgG1 Fc variants. Amino acid residues targeted to modulate binding to hFcRn are highlighted. (A) The key Fc residues involved in binding to FcRn (I253, H310, and H435) at the CH2–CH3 interface are highlighted in green spheres. (B) M428 and N434 of the CH3 domains are highlighted in blue spheres. (C) The Fc residues M252, S254, and T256 of the CH2 domains are highlighted in orange spheres. (D) H433 and N434 of the CH3 domains are highlighted in red spheres. In all crystal structure illustrations the biantennary glycans attached to the N297 residues of the CH2 domains are shown in red. The figures were designed using PyMOL (http://www.pymol.org) with the crystallographic data of hIgG1 (Protein Data Bank accession code 1HZH) (70).
However, a highly relevant question with regard to such engineering is whether substitutions of residues at or near the FcRn binding site could lead to structural perturbations that affect Fc-mediated effector functions. This is essential, as some Abs require potent effector functions for optimal clinical activity. In this study, we examine how engineering for altered binding to FcRn affects binding to the classical FcγRs and complement factor C1q. We compare the panel of Fc-engineered hIgG1 variants, produced side by side and with the same specificity, for binding to human C1q (hC1q) and all human FcγRs (hFcγRs). In most cases, we found binding to be reduced. However, one of the hIgG1 variants, hIgG1-HN, showed improved binding to hC1q. Importantly, the differences in binding translated into reduction of hFcγR- and hC1q-mediated cellular processes such as ADCC, ADCP and ADCML, except in the case of hIgG1-HN. These findings have implications for tailoring of recombinant hIgG1 Abs with extended half-life and desired effector functions.
Materials and Methods
Cell culture
The adherent human embryonic kidney cell line 293E (HEK293E) was obtained from American Type Culture Collection and maintained in DMEM or RPMI 1640 (BioWhittaker) supplemented with 10% heat-inactivated FCS (Sigma-Aldrich), 2 mM l-glutamine, 25 μg/ml streptomycin, and 25 U/ml penicillin (all from BioWhittaker). The cells were incubated at 37°C in a humidified 5% CO2, 95% air incubator.
Construction and production of anti–5-iodo-4-hydroxy-3-nitro-phenacetyl hIgG1 variants
A vector cassette system (pLNOH2-NIPhIgG1-oriP) for subcloning of Fc fragments into the framework encoding WT hIgG1 has previously been described (45, 46). The vector contains the constant H chain gene from hIgG1 with specificity for the hapten 5-iodo-4-hydroxy-3-nitro-phenacetyl (NIP), and it was used as a template for subcloning of DNA fragments encoding (synthesized by GenScript) a panel of Fc mutants using the restriction sites AgeI and SfiI (CH2 mutations) or SfiI and BamHI (CH3 mutations): M252Y/S254T/T256E (MST), M428L/N434S (MN), H433K/N434F (HN), and M252Y/S254T/T256E/H433K/N434F (MST/HN). Vectors encoding the constant H chain gene from hIgG1 with the mutations P329A, L234A, and L235A have previously been described (25, 47). The Abs were produced using HEK293E cells by transient cotransfection of the vectors together with a vector encoding the mouse λ L chain with NIP specificity (pLNOH2-NIPλLC-oriP) using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Construction of a plasmid encoding anti-NIP hIgG1 with the three point mutations I253A/H310A/H435A (IHH) has been described, and the Ab was purified from a stable transfected J558L cell line (25). The Abs were purified from collected supernatants using a column coupled with 4-hydroxy-3-nitrophenyl acetyl followed by size exclusion chromatography using a Superdex 200 10/300 column (GE Healthcare) to obtain monomeric IgG1 variants. Fractions were up-concentrated by using Amicon Ultra columns (Millipore).
Production of hFcRn and FcγRs
Vectors containing truncated versions of hFcγRI, hFcγRIIa, hFcγRIIb, hFcγRIIIa, and hFcγRIIIb cDNAs encoding their three ectodomains genetically fused to a cDNA encoding the Schistosoma japonicum glutathione S-transferase (GST) have been described (43, 46). The vectors also contain a DNA segment corresponding to the EBV origin of replication (oriP). The FcγRs were produced in HEK293E cells, and secreted receptors were purified using a GSTrap FF column as described (46).
Truncated monomeric His-tagged hFcRn was produced using a baculovirus expression vector system, as previously described (48, 49). The viral stocks were gifts from Dr. Sally Ward (University of Texas, Southwestern Medical Center, Dallas, TX). Secreted receptor was purified using a HisTrap HP column supplied with Ni2+ ions (GE Healthcare) and pre-equilibrated with PBS with 0.05% sodium azide. The pH of the supernatant was adjusted with PBS/0.05% sodium azide (pH 10.9) to pH 7.2 and then applied to the column (5 ml/min) before being washed using 200 ml PBS followed by 50 ml 25 mM imidazole/PBS (pH 7.2–7.4). Bound hFcRn was eluted with 50 ml 250 mM imidazole/PBS (pH 7.2–7.4), and the collected protein was up-concentrated and buffer-changed to PBS using Amicon Ultra-10 columns (Millipore). Subsequently, the protein was loaded onto a HiLoad 26/600 Superdex 200 prep grade column (GE Healthcare) for isolation of monomeric receptor. Eluted fractions were concentrated using Amicon Ultra-10 columns (Millipore) and stored at 4°C.
Quantification of anti-NIP hIgG1 Abs
ELISA was performed using 96-well MaxiSorp plates (Nunc). Plates were coated with 100 μl BSA conjugated to NIP (BSA-NIP) diluted to 1 μg/ml in PBS. After incubation overnight at 4°C, the plates were blocked with PBS containing 4% skim milk (Sigma-Aldrich) for 2 h and washed four times with PBS containing 0.05% Tween 20 (PBST; Sigma-Aldrich). Purified anti-NIP hIgG1 variants were diluted in 4% skim milk/PBST and incubated for 1 h at room temperature (RT). The plates were washed four times as described above before alkaline phosphatase–conjugated polyclonal goat anti–hIgG Fc Ab (Sigma-Aldrich), diluted (1:4000) in 4% skim milk/PBST, was added for 1 h at RT. The plates were then washed as above before 100 μl p-nitropenylphospate substrate (Sigma-Aldrich) diluted to 10 μg/ml in diethanolamine buffer was added. The absorbance was measured at 405 nm using a Sunrise spectrophotometer (Tecan). Alternatively, a monoclonal HRP-conjugated goat anti-mouse λ L chain Ab (SouthernBiotech) was used followed by visualization using the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution (Merck), and the reactions were stopped by adding 50 μl HCl. The absorbance was measured at 450 nm using the Sunrise spectrophotometer (Tecan).
ELISA measurements of FcRn binding
Serial dilution of anti-NIP hIgG1 variants (0.08–10 μg/ml) diluted in 4% skim milk/PBST were added to BSA-NIP–coated plates (2 μg/ml) and incubated for 1 h at RT, prior to washing using PBST (pH 6.0). Then, 10 μg/ml hFcRn was diluted in 4% skim milk/PBST (pH 6.0) and added for 1 h at RT before washing as above. Bound receptor was detected using a biotinylated monoclonal mouse anti-hFcRn Ab (ADM11) (50) followed by alkaline phosphatase–conjugated streptavidin (GE Healthcare). After washing, 100 μl p-nitropenylphospate substrate (Sigma-Aldrich) was added to each well. The absorbance was measured at 405 nm using a Sunrise spectrophotometer (Tecan). The same set-up was performed at pH 7.4.
ELISA measurements of FcγRs and C1q binding
Ninety-six–well plates were coated with 100 μl BSA-NIP at 1 μg/ml. After incubation overnight at 4°C, the plates were blocked with 4% skim milk/PBS for 2 h and washed four times with PBST. Serial dilution of anti-NIP hIgG1 variants (0.3–1 μg/ml) diluted in 4% skim milk/PBST were added and incubated for 1 h at RT prior to washing using PBST (pH 7.4). Then, 1 μg/ml GST-tagged hFcγRI, hFcγRIIa, hFcγRIIb, and hFcγRIIIa were diluted in 4% skim milk/PBST and incubated for 1 h at RT before being washed as above. Bound receptors were detected using an HRP-conjugated goat anti-GST Ab (GE Healthcare) diluted 1:5000 in 4% skim milk/PBST. After washing, 100 μl TMB substrate (SouthernBiotech) was added to each well and reactions were stopped by adding 50 μl 1 M HCl. The absorbance was measured at 450 nm using a Sunrise spectrophotometer (Tecan). The same set-up (without non-fat dried milk powder) was performed by adding pure hC1q (0.366 μg/ml) (Quidel), serum from a human volunteer (1:200 dilution), or pure hC1q (0.366 μg/ml) mixed with C1q-depleted human serum (1:200 dilution; Calbiochem) to the captured anti-NIP Abs. Bound hC1q was detected using a rabbit anti-hC1q Ab (Dako) followed by biotinylated sheep anti-rabbit IgG (locally produced) and subsequently alkaline phosphatase–conjugated streptavidin (GE Healthcare). Detection of deposition of C3 and C5 was measured using a rabbit anti-human C3c Ab and a rabbit anti-human C5 Ab (both from Dako). The absorbance was measured at 405 nm using a Sunrise spectrophotometer (Tecan).
Quantification of C1q in human serum
Ninety-six–well ELISA plates were coated with 100 μl polyclonal chicken anti-hC1q Ab (Abcam) diluted to 0.5 μg/ml in PBS and incubated overnight at 4°C. Serial dilutions of pure hC1q (111.0–0.006 ng/ml) in parallel with human serum from a volunteer (1:200–1:102,400) were diluted into PBST and incubated for 1.5 h at RT. After washing with PBST, bound hC1q was detected using a rabbit anti-hC1q Ab (Dako) followed by an HRP-conjugated donkey anti-rabbit Ab (GE Healthcare). After washing, 100 μl TMB substrate was added to each well and reactions were stopped by adding 50 μl 1 M HCl. The absorbance was measured at 450 nm using a Sunrise spectrophotometer (Tecan).
Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were carried out using a Biacore 3000 instrument (GE Healthcare). CM5 sensor chips (GE Healthcare) were coupled with NeutrAvidin (Pierce) (2000 resonance units) using amine coupling as described by the manufacturer. Soluble site-specific biotinylated FcγRIIa-H131 (Sino Biological) was captured on the coupled chip, and titrated amounts of anti-NIP WT hIgG1 (8,000.0–31.0 nM), MN, MST, and MST/HN (16,000.0–62.5 nM) were injected using buffer (pH 7.4) (GE Healthcare). Regeneration of the flow cells were done by injecting 10 mM NaOH. All experiments were performed at 25°C with a flow rate of 10 μl/min. Binding responses were zero adjusted, and the reference cell value was subtracted. Binding analyses were performed using the BIAevaluation 4.1 Wizard software (GE Healthcare).
Liquid chromatographic tandem mass spectrometry analysis
Fifty microliters of each IgG1 variant (1 mg/ml) was spun down for 10 min at 13,000 × g before 1 μg trypsin dissolved in 100 μl 50 mM ammonium bicarbonate (pH 7.8) was added and incubated overnight at 37°C. The centrifugal devices were spun down at 13,000 × g for 10 min, and the flow-through was transferred to an Eppendorf tube and dried in a SpeedVac (Heto Maxi dry). Dried samples were dissolved in 20 μl 0.1% formic acid, sonicated for 30 s, and centrifuged for 10 min at 16,100 × g. Subsequently, 10 μl of each sample was transferred to new vials, and reverse phase (C18) nano online liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis of proteolytic peptides was performed using a system consisting of two Agilent 1200 HPLC binary pumps (nano and capillary) with an autosampler, column heater, and integrated switching valve. This system was coupled via a nanoelectrospray ion source to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific). For the analyses, 6 μl peptide solution was injected into the 5 × 0.3-mm extraction column filled with Zorbax 300SB-C18 of 5-μm (diameter) particle size (Agilent Technologies). After washing for 5 min with 0.1% formic acid (v/v) and 3% acetonitrile (v/v) at a flow rate of 10 μl/min, the integrated switching valve was activated and peptides were eluted in the back-flush mode from the extraction column onto a 150 × 0.075-mm C18, 3-μm resin column (GlycproSIL C18–80Å, Glycpromass). The mobile phase consisted of acetonitrile and mass spectroscopy–grade water, both containing 0.1% formic acid. Chromatographic separation was achieved using a binary gradient from 5 to 55% of acetonitrile in water for 60 min with a flow rate of 0.2 μl/min. Mass spectra were acquired in the positive ion mode applying a data-dependent automatic switch between survey scan and MS/MS acquisition. Peptide samples were analyzed with a high-energy collisional dissociation (HCD) fragmentation method with normalized collision energy at 25 and 41, acquiring one Orbitrap survey scan in the mass range of m/z 300–2000 followed by MS/MS of the three most intense ions in the Orbitrap (R7500). The target value in the LTQ-Orbitrap was 1 million for survey scan at a resolution of 30,000 at m/z 400 using lock masses for recalibration to improve the mass accuracy of precursor ions. Ion selection threshold was 500 counts. Selected sequenced ions were dynamically excluded for 180 s. Data analysis was performed on Xcalibur v2.0. MS/MS spectra for all glycopeptides and were extracted by oxonium ion search; 204.086 (N-acetylhexosamine) and 366.1388 (N-acetylhexosamine-hexose) were used. HCD fragmentation with normalized collision energy at 25 was used to detect the glycans, and the peptide mass was detected for the IgG glycopeptides. Extracted ion chromatogram for target glycopeptides (EEQYNSTYR and the miscleaved TKPREEQYNSTYR with all different glycan masses) were extracted with 10 ppm accuracy and MS/MS spectra were manually verified. HCD fragmentation with normalized collision energy at 41 was used to detect the peptide sequence and to verify that the peptide mass corresponded to the correct peptide sequence. The area under the curve for all extracted glycopeptides was calculated and the percentage ratio for each glycoform was determined.
Ab-dependent complement-mediated cell lysis
The ADCML assay was done essentially as described (51). Briefly, target cells were prepared by mixing 10 μl packed sheep RBCs (SRBCs) with 16 μl PBS and 60 μCi sodium chromate ([51Cr]; PerkinElmer) and incubating for 1 h at 37°C. Subsequently, NIP-conjugated anti-SRBC rabbit Fab fragments (2.5 μg NIP4-Fab or 13.3 μg NIP60-Fab) were added and incubated for 1 h at 37°C. The sensitized and labeled SRBCs were washed three times and diluted to 2–4 × 107 cells/ml in veronal buffer (pH 7.2; 1% BSA, 0,25 mM CaCl2, and 0.8 mM MgCl3). The assay was performed in round-bottom microtiter plates (Costar). Serial dilutions of anti-NIP IgG1 Abs (900.0–3.7 ng/ml) (50 μl) were mixed with SRBCs (50 μl) and incubated for 10 min at RT before complement source (50 μl), diluted 1:30 in veronal buffer, was added. All mixtures were incubated for 30 min at 37°C and then spun down at 1200 rpm for 3 min before the supernatants were collected with harvesting filters/devices (Skatron). Release of [51Cr] was measured using a gamma counter (Packard). The cytotoxic index (CI %) was calculated using the formula: CI % = {[cpm (test) − cpm (spontaneous)]/[cpm (maximum) − cpm (spontaneous)]} × 100. Maximum [51Cr] release was determined as radioactivity released when the SRBCs were lysed with water containing 0.1% Tween 20. Spontaneous [51Cr] release was measured by incubating SRBCs with complement source.
Ab-dependent cell-mediated cytotoxicity
The ADCC assay was done essentially as previously described (51). Briefly, 50 μl 5-fold dilution of anti-NIP hIgG1 variants (200.0–0.06 ng/ml) and 50 μl haptenated, 51Cr-labeled SRBCs (3 × 105 cells/ml) were added into round-bottom microtiter plates (Costar) and incubated for 10 min at RT before 3 × 106 cells/ml were added. PBMCs were obtained from healthy donors using Lymphoprep (Axis-Shield), and NK cells and monocytes were isolated by negative selection using Dynabeads untouched human NK cells and Dynabeads untouched human monocyte kits (Invitrogen), respectively, following the manufacturer’s instructions. Cells were diluted into DMEM/HEPES/1% FCS. The microtiter plate was spun down at 300 × g for 2 min and incubated for 4 h at 37°C. The plates were centrifuged for 3 min at 300 × g, and supernatants were collected with harvesting filters/devices (Skatron). Maximum [51Cr] release and spontaneous [51Cr] release were measured as described above.
Ab-dependent cellular phagocytosis
The phagocytosis activity was measured as respiratory burst, as previously described (52). Briefly, NIP-haptenated meningococci killed by ethanol were used as target cells whereas dihydrorhodamine 123 (Sigma-Aldrich)–primed polymorphonuclear neutrophils (PMNs) from a health donor were used as effector cells. Serial dilutions of anti-NIP hIgG1 variants were prepared in DMEM/HEPES/1% FCS (50 μl), mixed with 5 μl meningococci (109 ml), and incubated for 30 min at 37°C in microtiter plates. Then, 5 μl hIgG-depleted human serum was added as a complement source and incubated for 10 min at 37°C. Furthermore, 50 μl 4 × 106 dihydrorhodamine 123–primed effector cells were added and incubated for 10 min at 37°C. Flow cytometry was performed gated on the PMNs, and the percentage of respiratory burst was estimated. ADCP was also performed without the complement source.
Protein stability by differential scanning fluorimetry
Protein stability of monomeric IgG1 variants was measured by protein stability by differential scanning fluorimetry (DSF) using a LightCycler RT-PCR instrument (Roche). SYPO Orange (Sigma-Aldrich) was used at a 1:1000 dilution, and the protein concentration was 0.1 mg/ml in a final volume of 25 μl. All samples were run in triplicate in 96-well LightCycler 480 multiwell plates. The peaks of excitation and emission of SYPO Orange are 490 and 580 nm, respectively. The 450-nm excitation and 568-nm emission filters on the RT-PCR instrument were used. The RT-PCR instrument was programmed to raise the temperature from 25°C to 95°C after a stabilization period of 10 min at 25°C. Two measurements at each temperature were recorded, and data were collected every 0.5°C. Data transformation and analysis were performed using the DSF analysis protocol (53).
Statistical analysis
All graphs were generated and statistical analyses were performed using GraphPad Prism version 5.03 from GraphPad Software (San Diego, CA). All experiments were repeated at least three times and error bars represent SEM. A one-way ANOVA test (Dunnett test) was used to calculate the statistical significance. A p value < 0.05 was considered significant.
Results
Design of human Fc-engineered IgG1 variants
To construct recombinant IgG variants, we used two plasmids, one encoding the hIgG1 H chain and one encoding the λ mouse L chain, which together form Abs with specificity for the hapten NIP. In addition to the WT, four Fc-engineered variants were made by substitution of amino acid residues that have previously been shown to modulate binding to FcRn, transplacental transport, and serum half-life, namely hIgG1-HN, hIgG1-MN, hIgG1-MST, and hIgG1-MST/HN (34, 35, 41, 42) (Fig. 1B–D).
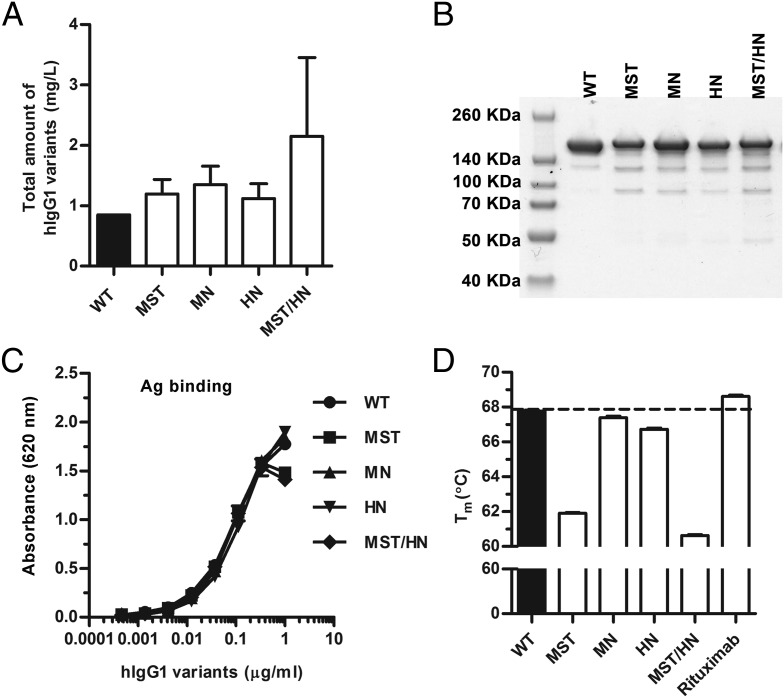
The hIgG1 variants were produced by transient transfection of HEK293E cells, and medium fractions were collected every second day for up to 2 wk, pooled, and filtrated prior to purification on a 4-hydroxy-3-nitrophenyl acetyl–coupled column followed by gel filtration. The total concentrations were in the range of 3–0.9 mg/l culture, where hIgG1-MST/HN was produced with a 3-fold higher yield than that of the others (Fig. 2A). Small fractions of the hIgG1 Abs were run on a nonreducing SDS-PAGE followed by Coomassie staining (Fig. 2B), and the migration profiles for the mutants were found to be similar to those of the WT with major bands at ∼150 kDa. Additionally, ELISA showed that the Abs bound equally well to NIP conjugated to BSA (Fig. 2C). Finally, the stability of the Abs was compared, and the melting temperature (tm) value for WT anti-NIP hIgG1 was found to be 68°C, equal to that of the commercial anti-CD20 Ab rituximab, whereas the tm values for the mutants varied from 61°C to 67.5°C (Fig. 2D).
FIGURE 2.
Production and integrity of WT hIgG1 and Fc-engineered variants. (A) Total amounts of anti-NIP hIgG1 variants produced from transient transfection of HEK293E cells. (B) Nonreducing SDS-PAGE of WT hIgG1 and Fc-engineered variants. (C) Binding of titrated amounts (1000.0–0.5 ng/ml) of anti-NIP WT hIgG1 and Fc-engineered variants to NIP-conjugated BSA. (D) A DSF thermal stability histogram showing the tm values for WT hIgG1 and Fc-engineered variants. Data are presented as mean ± SEM of experiments performed in triplicate (n = 3).
Mapping of glycans attached to the IgG1 variants
To characterize the profile of the glycans attached to N297 on the CH2 domain of the hIgG1 variants, we performed LC-MS/MS analysis on glycopeptides after trypsination. In total, we identified 32 different glycans (Supplemental Table I), where four forms dominated and comprised 77–93% of the total glycan pool, namely 4GlcNac-1Fuc-3Man, 4GlcNac-1Fuc-3Man-1Gal, 4GlcNac-1Fuc-3Man-2Gal, and 2GlcNac-5Man. The relative amount of each glycan was similar, but not identical, between variants and the WT (Supplemental Table I). In particular, hIgG1-WT had 15 and 8% less of the 4GlcNac1Fuc3Man and 4GlcNac1Fuc3Man1Gal forms, respectively, than did the mutants (Supplemental Table I). Thus, there were some differences in fucosylated oligosaccharides attached to the WT compared with the engineered variants and minor differences between the mutant variants. Because the Abs were produced in the same system side by side, our data suggest that the mutations introduced a slight shift in the profile of glycan attached to position N297.
Binding of the Fc-engineered hIgG1 to hFcRn
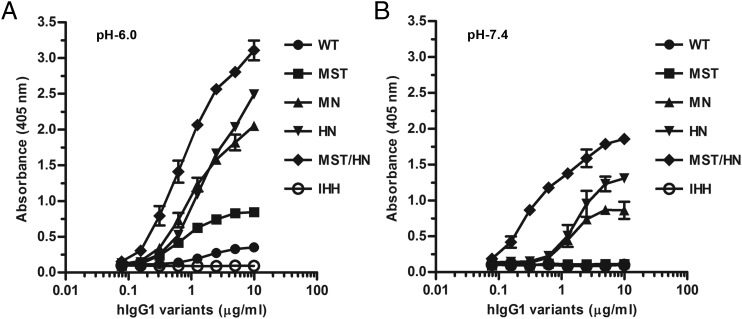
To investigate how the hIgG1 variants bound soluble hFcRn, we established an ELISA where titrated amounts of the Abs were captured on NIP-conjugated BSA coated in wells followed by addition of hFcRn at pH 6.0 or 7.4. Bound receptor was detected using an mAb (ADM11) directed toward hFcRn, the binding of which does not interfere with IgG binding (50). As expected, hFcRn was shown to bind WT hIgG1 in a strictly pH-dependent manner, binding at pH 6.0 and not binding at pH 7.4, whereas hIgG1-IHH did not bind (Fig. 3). Furthermore, a side-by-side comparison of the four Fc-engineered hIgG1 mutants revealed that they all bound more strongly than did the WT at acidic pH with a hierarchy from strongest to weakest as follows: MST/HN > HN > MN > MST > WT > IHH (Fig. 3A). At neutral pH, detectable binding was measured for MST/HN followed by intermediate binding to HN and MN whereas no binding was detected for the WT, MST, and IHH (Fig. 3B). Thus, the Fc-engineered anti-NIP hIgG1 variants showed a range of distinct FcRn binding properties, in accordance with reported data (34, 35, 41, 42).
FIGURE 3.
Measurements of pH-dependent binding to hFcRn. ELISA binding of titrated amounts (1000.0–0.5 ng/ml) of WT hIgG1 and Fc-engineered variants to hFcRn at (A) pH 6.0 and (B) pH 7.4. Data are mean ± SEM of one representative experiment out of three.
Binding of the Fc-engineered hIgG1 variants to classical hFcγRs
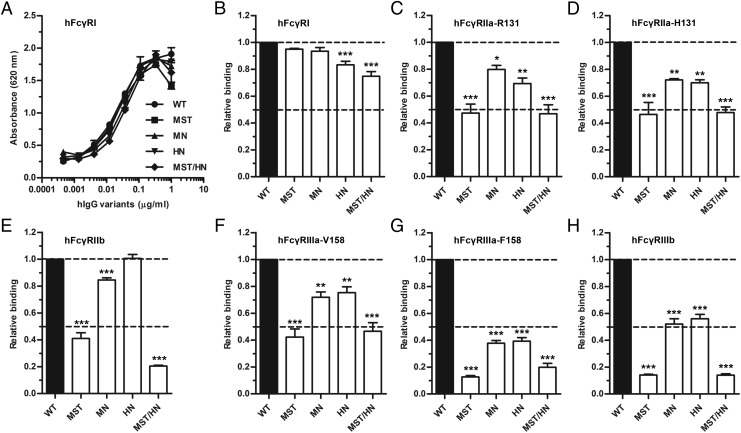
To assess whether the mutations affected binding to the five classical hFcγRs, ELISA was performed using soluble GST-tagged hFcγRs. Importantly, all recombinant receptors showed expected binding selectivity toward the four hIgG subclasses (Supplemental Fig. 1A–G), and thus they were used for screening of titrated amounts of hIgG1 variants captured on BSA-NIP.
The binding responses obtained toward hFcγRI showed that all variants bound equally as well as did the WT, with the exception of HN and MST/HN, which showed a slight reduction in binding (Fig. 4A). The titrated binding curves (Supplemental Fig. 1H–M) were used to calculate the relative binding response for each of the FcγRs toward the hIgG variants, where binding of WT hIgG1 was set to 1.0. Slightly reduced binding (5–20%) was observed between the high-affinity FcγRI and the variants (Fig. 4B). Furthermore, the two allotypes of hFcγRIIa (H/R131) showed reduced binding (20–50%) to the variants, with the most pronounced effect detected being toward MST and MST/HN (Fig. 4C, 4D). Similarly, reduced binding (20–90%) was detected toward the two hFcγRIIIa allotypes (F/V158), where again the largest negative effect was detected for MST and MST/HN followed by MN and HN (Fig. 4F, 4G). The negative effect of the Fc substitutions was most pronounced for binding to the phenylalanine-containing allotype (F158), and a similar trend was seen as for binding to hFcγRIIIb, with 40–90% reduction in binding (Fig. 4H). Furthermore, despite high homology between the hFcγRIIa allotypes and the inhibitory hFcγRIIb, hFcγRIIb showed even poorer binding to MST and MST/HN than did hFcγRIIa. Binding of HN and MN was less affected (Fig. 4E).
FIGURE 4.
Binding properties of Fc-engineered hIgG1 variants toward classical hFcγRs. ELISA binding of titrated amounts (1000.0–0.5 ng/ml) of WT hIgG1 and the Fc-engineered variants to (A) hFcγRI, and calculation of relative binding of WT hIgG1 and the Fc-engineered variants to (B) hFcγRI, (C) hFcγRIIa-R131, (D) hFcγRIIa-H131, (E) hFcγRIIb, (F) hFcγRIIIa-V158, (G) hFcγRIIIa-F158, and (H) hFcγRIIIb. The experiments were performed at pH 7.4, and obtained data are shown as mean ± SEM of one experiment performed in triplicate (A), and as mean ± SEM of three independent experiments performed in triplicate (B–H). *p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA test.
Lastly, we selected the H131 allotype of hFcγRIIa to quantify the binding affinity toward the WT and three of the hIgG1 variants (MN, MST, and MST/HN) using SPR (Supplemental Fig. 2). Fitting the equilibrium binding responses to a steady-state affinity model revealed that MST and MST/HN bound with 4- to 5-fold reduced equilibrium binding affinity compared with the WT, whereas MN only showed a slight reduction. In summary, we demonstrate that Fc engineering of hIgG1 for improved hFcRn binding may markedly reduce binding to the classical FcγRs.
ADCP
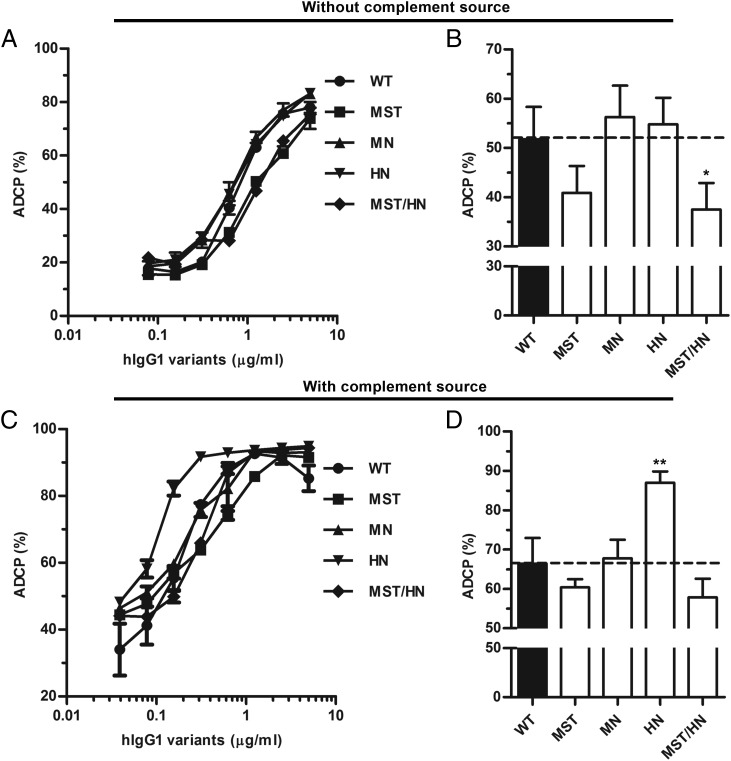
To investigate how Fc engineering affected ADCP, meningococci were directly labeled with the hapten NIP followed by addition of titrated amounts of the anti-NIP Abs. Portions of human PMNs from a volunteer were added and phagocytic activity was measured as respiratory burst. The results show that all variants induced ADCP (Fig. 5) with only minor differences between the variants, where MST/HN showed the most pronounced reduction. Furthermore, we investigated how ADCP was affected by the addition of human serum as a complement source, and we found that the phagocytic activity increased by ∼20% for all variants, but most strikingly by a further 20% for IgG1-HN (Fig. 5C, 5D). Thus, Fc engineering of hIgG1 for altered binding to FcRn may have a great impact on complement activation ability.
FIGURE 5.
ADCP activity of hIgG1 Fc-engineered variants. (A) ADCP activity measured using PMNs and titrated amounts (5000.0–0.04 ng/ml) of WT hIgG1 and Fc-engineered variants. (B) Histogram showing the average of ADCP activity. (C) ADCP activity measured using PBMCs and titrated amounts (5000.0–0.04 ng/ml) of WT hIgG1 and Fc-engineered variants in the presence of human serum as complement source. (D) Histogram showing the average of ADCP activity in the presence of complement source. Data are shown as mean ± SEM of one experiment performed in triplicate (A and C) and as mean ± SEM of three independent experiments performed in triplicate (B and D). *p < 0.05, **p < 0.01 by one-way ANOVA test.
ADCC
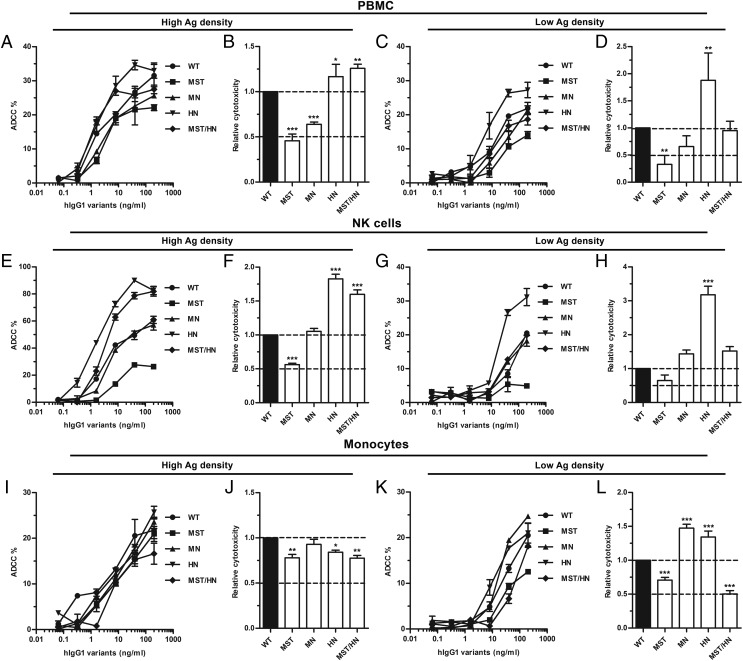
Next, an ADCC assay was conducted where 51Cr-labeled SRBCs were coated with rabbit anti-SRBC Fab fragments that were conjugated to the hapten NIP at low or high density, prior to addition of hIgG1 variants and freshly isolated PBMCs. In both cases, ADCC activity was measured as release of [51Cr], and the results obtained show hIgG1-MST to be a rather poor inducer of cytotoxicity, 50% or less efficient than the WT, followed by MN, whereas HN and MST/HN were similar or better than the WT. At low Ag density, the HN variant showed a 2-fold improved ADCC activity over the WT (Fig. 6A–D).
FIGURE 6.
ADCC activity of hIgG1 Fc-engineered variants. (A) ADCC activity measured using PBMCs and titrated amounts (200.0–0.06 ng/ml) of WT hIgG1 and Fc-engineered variants against high-haptenated SRBCs (NIP60Fab). (B) Histogram showing the relative ADCC activity of the hIgG1 variants against high-haptenated SRBCs. (C) ADCC activity measured using PBMCs and titrated amounts (200.0–0.06 ng/ml) of WT hIgG1 and Fc-engineered variants against low-haptenated SRBCs (NIP4Fab). (D) Histogram showing the relative ADCC activity of the hIgG1 variants against low-haptenated SRBCs. (E) ADCC activity measured using pure NK cells and titrated amounts (200.0–0.06 ng/ml) of WT hIgG1 and Fc-engineered variants against high-haptenated SRBCs. (F) Histogram showing the relative ADCC activity of hIgG1 variants against high-haptenated SRBCs. (G) ADCC activity measured using pure NK cells and titrated amounts (200.0–0.06 ng/ml) of WT hIgG1 and Fc-engineered variants against against low-haptenated SRBCs. (H) Histogram showing the relative ADCC activity of hIgG1 variants against low-haptenated SRBCs. (I) ADCC activity measured using pure monocytes and titrated amounts (200.0–0.06 ng/ml) of WT hIgG1 and Fc-engineered variants against high-haptenated SRBCs. (J) Histogram showing the relative ADCC activity of hIgG1 variants against high-haptenated SRBCs. (K) ADCC activity measured using pure monocytes and titrated amounts (200.0–0.06 ng/ml) of WT hIgG1 and Fc-engineered variants against low-haptenated SRBCs. (L) Histogram showing the relative ADCC activity of hIgG1 variants against at low-haptenated SRBCs. Data are presented as mean ± SEM of one experiment performed in triplicate (A, C, E, G, I, and K), and mean ± SEM of three independent experiments performed in triplicate (B, D, F, H, J, and L). *p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA test.
Next, we addressed which effector cells were responsible for the ADCC activities by repeating the assay with NK cells and monocytes isolated by negative selection using magnetic beads. Enrichment of NK cells resulted in overall enhanced ADCC activity at high Ag density, and the relative activity of the variants followed the same trend as that measured using PBMCs, with the exception of MN, which was now equally as good as the WT (Fig. 6E–H). Additionally, the cytotoxicity induced by MST/HN was more pronounced at high Ag density, whereas HN showed improved activity at both low and high Ag densities (Fig. 6E–H). Furthermore, the hIgG1 variants also induced ADCC by monocytes, but not to the same magnitude as that of NK cells at high Ag density (Fig. 6I–L). Interestingly, only minor differences were detected between the hIgG1 variants at high Ag density whereas more pronounced differences were measured at low density (Fig. 6I–L). Specifically, both MN and HN showed 1.5-fold improved ADCC activity over WT, whereas MST and MST/HN showed a reduction of 25 and 50%, respectively (Fig. 6K, 6L).
Binding of the Fc-engineered hIgG1 variants to hC1q
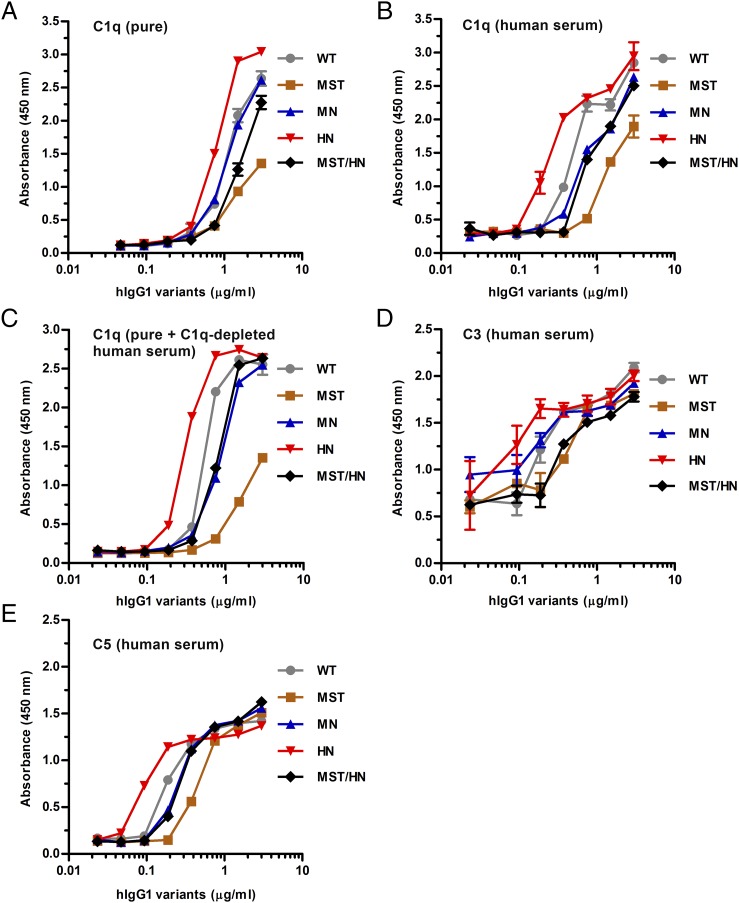
Besides FcγR-mediated effector functions, the classical complement cascade is important for humoral immunity. Because the binding sites for the classical FcγRs overlap with that of C1q (3), the key residues involved are again located far from the amino acid substitutions of the Fc-engineered hIgG1 variants. Nevertheless, we measured binding to hC1q by ELISA using two sources of complement, that is, purified hC1q and human serum. Initially, the amount of hC1q present in serum was quantified and found to be 73.2 μg/ml by ELISA (data not shown). Equal amounts of both sources of hC1q were added to titrated amounts of the Abs captured on BSA-NIP.
All hIgG1 variants bound pure hC1q and hC1q in serum with almost identical hierarchies as HN > WT ≥ MN ≥ MST/HN > MST, but the binding responses were stronger when hC1q was provided in serum, and in both cases hIgG1-HN bound more hC1q than did the WT (Fig. 7A, 7B). Notably, the WT performed equally well as MN when pure hC1q was added whereas it bound better in serum (Fig. 7A, 7B). Next, when pure hC1q was added to C1q-depleted human serum, the same hierarchy was measured (Fig. 7C). Furthermore, an even more pronounced effect of the HN mutations was detected when a constant amount of the Abs was directly coated in wells or captured on BSA-NIP followed by adding titrated amounts of hC1q from both sources (Supplemental Fig. 3A–D). Finally, the same hierarchy was found when C3 or C5 deposition was monitored (Fig. 7D, 7E). As controls, we included hIgG1 with a single point mutation (P329A) within the CH2 domain at the core of the C1q binding site (47), which completely eliminated hC1q binding, as well as hIgG1 with mutation of either of two leucine residues in the lower hinge (L234A and L235A), crucial for FcγR binding (54), which moderately reduced binding (Supplemental Fig. 3E–H).
FIGURE 7.
Binding properties of hIgG1 Fc-engineered variants toward C1q and complement factors. ELISA results showing binding of WT hIgG1 and Fc-engineered variants to (A) pure hC1q, (B) C1q present in human serum, and (C) pure C1q present in C1q-depleted human serum and activation of complement factors (D) C3 and (E) C5. Experiments were performed at pH 7.4 and results are shown as mean ± SEM of one representative experiment out of three.
ADCML
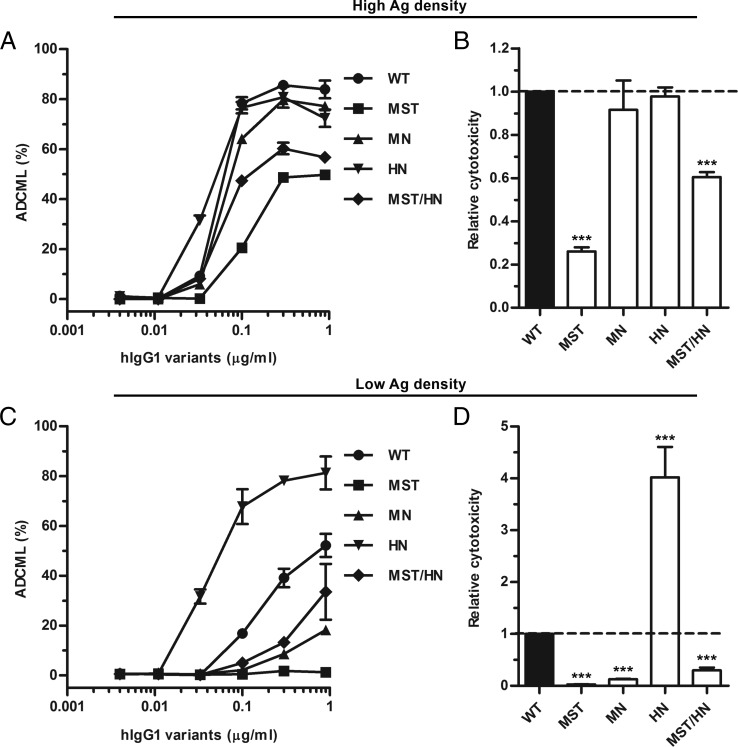
To investigate whether the complement binding data would translate into altered ADCML activity, we captured the anti-NIP Abs on 51Cr-labeled SRBCs, again coated with anti-SRBC Fab fragments from rabbit conjugated to NIP at low or high density. After addition of the complement source, release of [51Cr] was measured as ADCML activity, which gave rise to the same hierarchy as that measured in the C1q binding assay (Fig. 8). A similar trend was detected at both Ag densities, whereas the effect of the mutations was most pronounced at low Ag density. In this study, the HN variant showed a 4-fold enhancement of ADCML compared with WT, in stark contrast to the other variants, which induced very little ADCML (Fig. 8C, 8D).
FIGURE 8.
ADCML activity of hIgG1 Fc-engineered variants. (A) ADCML activity measured against high-haptenated SRBCs in the presence of titrated amounts (900.0–3.7 ng/ml) of WT hIgG1 and Fc-engineered variants. (B) Histogram showing relative ADCML activity of the hIgG1 variants against high-haptenated SRBCs. (C) ADCML activity against low-haptenated SRBCs in the presence of titrated amounts (900.0–3.7 ng/ml) of WT hIgG1 and Fc-engineered variants. (D) Histogram showing relative ADCML activity of the hIgG1 variants against low-haptenated SRBCs. Experiments were performed at pH 7.4 and results are shown as mean ± SEM of one experiment performed in triplicate (A and C), and as mean ± SEM of three independent experiments performed in triplicate (B and D). ***p < 0.001 by one-way ANOVA test.
Discussion
In the present study, we produced a panel of Fc-engineering hIgG1 variants designed to have increased binding to FcRn, and we investigated how the Fc amino acid substitutions modulate binding to the classical hFcγRs and hC1q, as well as the accompanying effector functions ADCC, ADCP, and ADCML. Such knowledge is of crucial importance prior to therapeutic use of FcRn-engineered Abs.
To compare their binding abilities and cellular effector functions, all hIgG1 variants were produced with the same specificity, side by side using the same vector and cell system. The Fc-engineered IgG variants were secreted in similar amounts upon expression in HEK293E cells, except for hIgG1-MST/HN, which was produced in higher yields. This may suggest that the mutations resulted in increased intrinsic protein stability. However, when stability was measured by DSF, a slight reduction in tm value was observed for MST/HN, which was solely due to the M252Y/S254T/T246E substitutions in the CH2 domains. Additionally, as the nature of the biantennary glycans attached to N297 of Fc plays a major role in modulating binding to the FcγRs, but not FcRn, we mapped the glycans of the hIgG1 variants by LC-MS/MS analysis and found that the dominating forms were present in both the WT and the mutant fractions. Interestingly, a difference in fucosylation was found between the WT and mutant variants, even when mutations are introduced distally from N297. Such an example was also previously described, where a CH3 mutation (Y407E) was introduced into hIgG1 and hIgG4 that caused great alterations of the glycosylation profile of N297 in the CH2 domains (55).
To assess the binding properties of the produced anti-NIP hIgG1 variants toward hFcγRs, we screened the Abs in ELISA for binding to GST-tagged hFcγRs. The variants were compared with the WT and showed distinct binding profiles. Whereas binding to the high-affinity FcγRI was only slightly affected, the mutations in all variants had a large impact on binding to the low-affinity receptors, with the MST and MST/HN variants being the most strongly affected (50–90% reduction), with a similar trend for all receptors. The most dramatic effect was detected for the MST variant with substitutions within the CH2 domain. Using SPR, we found that the MST and MST/HN variants bound with a 5-fold weaker affinity toward hFcγRIIa-H131. Interestingly, the inhibitory hFcγRIIb was not affected by the H433K/N434F mutations. Because the glycosylation profiles were similar for the mutant variants, the differences in binding are most likely due to the specific amino acid substitutions. The three mutations within the CH2 domains are in the same domain and in proximity to the binding site for the hFcγRs. Our data indicate that the mutations alter the local structure and/or dynamics of the CH2 domain, which again modulate binding to effector molecules.
Furthermore, when the MST and MST/HN variants were compared with hIgG1-WT in ADCP and ADCC assays, MST showed that both had reduced ability to induce phagocytosis and cytotoxicity. The MST/HN variant showed reduced ability to induce ADCP and ADCC by monocytes at low Ag density, whereas the ADCC inducing activity was enhanced using PBMC or NK cells at both low and high Ag density. The effect seen for MST/HN is probably due to the fact that the HN mutations had a positive effect on ADCC, although in vitro interaction analysis showed that it bound less well to the FcγRIIIa allotypes compared with the WT. Furthermore, the HN variant performed overall better than did the WT and remaining variants at high and low Ag density for PBMCs, NK cells, and monocytes.
With regard to ADCC and the effect of the M252Y/S254T/T246E mutations, the result is in line with a previous study that shows this variant, with anti-RSV specificity (MEDI-522), to have reduced binding to hFcγRIIIa (F156) and more than 2-fold reduction in ADCC. Interestingly, the ability to induce ADCC was restored by introduction of three additional mutations (S239D/A330L/I332E) (35). These mutations may also compensate for loss of ADCP activity.
The differences in ADCP activity between the hIgG1 variants were minor, except for the HN variant, which showed an enhanced ability to induce phagocytosis at low Ag density. One reason may be due to the fact that FcRn has been shown to take an active part in enhancing processing and elimination of IgG-coated bacteria in neutrophils upon engulfment via cell surface–expressed FcγRs (56). Furthermore, two recent studies suggest that phagocytosis is affected by the ratio of the hFcγRIIa and hFcγRIIIb affinities (57, 58). Because the HN and MN variants demonstrate stronger FcγRIIIb binding than do the MST and MST/HN variants, this points to these two having a favorable ratio for phagocytosis.
Classical complement activation was initially studied by measuring binding of the hIgG1 variants to pure hC1q and C1q present in human serum, which revealed that all variants bound with almost similar hierarchy to both hC1q sources. At both conditions, the hIgG1-HN variant showed greatly enhanced hC1q binding, whereas the other mutants bound less efficiently than did the WT. Increased hC1q binding to HN also resulted in higher deposition of C3 and C5, which translated into a 4-fold increased ADCML activity at low Ag density and, additionally, more efficient ADCP in the presence of serum as a complement source. Again, the MST variant was the least efficient. Interestingly, no activity was detected for any variants other than HN at lower Ag density. This finding demonstrates that ADCML activity may well be modulated by mutations in the CH3 domain, structurally distant from the core site for C1q in the hinge and upper part of the CH2 domains. This is particularly interesting in light of recent work by Diebolder et al. (59) showing that IgG1 forms hexamers via noncovalent interactions between CH2 and CH3 segments upon Ag binding on cells. Introduction of single amino acid substitutions at this site could thus result in reduced or enhanced binding to C1q, and ADCML activity could be modulated. However, in our case the effect of the HN mutations did not require capturing on the Ag, as an even more pronounced effect on hC1q binding was detected when the hIgG1 variants were coated directly in wells.
Although crystal structures of FcγRs in complex with the IgG1 Fc show that the core binding site is located at the lower hinge and CH2 regions (54, 60), no cocrystal structure exists of the C1q/Fc complex, but mutagenesis and atomic resolution modeling have shown that the binding sites for the FcγRs and C1q largely overlap (61). This is in line with our findings that mutating two leucine residues (L234 and L235), which are at the core of the FcγR site, strongly decreased C1q binding as well as C3 and C5 deposition (62).
Solving of a crystal structure of an hIgG1 Fc fragment containing the MST substitutions has revealed that the mutations do not induce large confirmation changes, but instead mediate local alterations of the targeted amino acids such that they favor binding to hFcRn (63). This is confirmed by a recently published cocrystal structure of the Fc fragment in complex with hFcRn, where the T256E mutation creates two additional salt bridges with Gln2 of β2-microglobulin (63). It now appears that the Fc region of IgG is far more flexible and dynamic than has previously been appreciated. A consequence of this is that changes introduced in one part of the Fc may well have a large impact on the binding properties to Fc effector molecules with distally located binding sites. The consequence of individual mutations is not readily predicted. Thus, as the crystal structure of the MST containing Fc fragment is very similar to that of WT Fc, it is hard to predict why these mutations have such a negative impact on binding of FcγR and C1q, other than by inducing minor conformational changes or alterations in the dynamics within the CH2 domains prior to or upon binding.
Regarding the effect of the MN and HN mutations, our data suggest that amino acid substitutions in the CH3 domains induce changes in the Fc that are manifest even at the CH2 domains. A previous report also describes the effect of two amino acid substitutions (E382V/M428I) that result in high-affinity binding to hFcγRI in the CH3 domain of an aglycosylated hIgG1 molecule (64). Interestingly, crystallography and small-angle x-ray scattering studies of this Fc fragment show that it has a more closed CH2–CH2 conformation than does a similarly aglycosylated WT fragment (65). A recent study (built on crystallization, model building, and simulation of Fc fragments) highlights that the Fc is flexible and can adapt a range of different conformations that are distinct from those observed in FcγR complexes (66). The same study also predicts that mutations at the CH2–CH3 interface may have a large impact on the flexibility of the domains, which may fit well with the data presented in the current study. Therefore, our study highlights that substitution of amino acids in the CH2 and CH3 domains, which are distally from C1q and FcγRs binding sites, may induce conformational changes or position shift of CH2 domain in glycosylated IgG, which may directly affect interaction with C1q and FcγRs, leading to unpredictable changes in the effector functions. Thus, engineering of FcRn–IgG interaction may influence effector functions, which has implications for the therapeutic efficacy and use of Fc-engineered hIgG1 variants.
In the present study, the recombinant hIgG1 variants were produced in the HEK293E cell line, which is extensively used by numerous academic laboratories for generation of Abs for structural and functional studies. However, for manufacturing of mAbs for clinical use, only the CHO cell line is approved as a host cell system whereas HEK293E is so far only approved for manufacturing of hIgG1 Fc-fused coagulation factors (67–69). The future will show whether HEK293E will be accepted for manufacturing of full-length Abs, and whether the results in the present study also hold true for Abs produced in CHO cells.
Supplementary Material
Acknowledgments
We are grateful to Sathiaruby Sivaganesh and Jan Haug Anonsen for excellent technical assistance. We also are thankful to Gregory Christianson and Derry Roopenian (The Jackson Laboratory) for providing the ADM11 Ab.
This work was supported in part by Research Council of Norway Grant 179573 through its Centres of Excellence funding scheme. J.T.A. was supported by Research Council of Norway Grants 230526/F20 and 179573/V40.
The online version of this article contains supplemental material.
- ADCC
- Ab-dependent cellular cytotoxicity
- ADCML
- Ab-dependent complement-mediated cell lysis
- ADCP
- Ab-dependent phagocytosis
- DSF
- differential scanning fluorimetry
- FcRn
- neonatal Fc receptor
- GST
- glutathione S-transferase
- HCD
- high-energy collisional dissociation
- HEK293E
- adherent human embryonic kidney cell line 293E
- hFcRn
- human FcRn
- hIgG1
- human IgG1
- HN
- H433K/N434F
- IHH
- I253A/H310A/H435A
- LC-MS/MS
- liquid chromatography–tandem mass spectrometry
- MN
- M428L/N434S
- MS/MS
- tandem mass spectrometry
- MST
- M252Y/S254T/T256E
- MST/HN
- M252Y/S254T/T256E/H433K/N434F
- NIP
- 5-iodo-4-hydroxy-3-nitro-phenacetyl
- PBST
- PBS containing 0.05% Tween 20
- PMN
- polymorphonuclear neutrophil
- RT
- room temperature
- SPR
- surface plasmon resonance
- SRBC
- sheep RBC
- tm
- melting temperature
- TMB
- 3,3′,5,5′-tetramethylbenzidine
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Chan A. C., Carter P. J. 2010. Therapeutic antibodies for autoimmunity and inflammation. Nat. Rev. Immunol. 10: 301–316. [DOI] [PubMed] [Google Scholar]
- 2.Weiner L. M., Surana R., Wang S. 2010. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat. Rev. Immunol. 10: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Idusogie E. E., Presta L. G., Gazzano-Santoro H., Totpal K., Wong P. Y., Ultsch M., Meng Y. G., Mulkerrin M. G. 2000. Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. J. Immunol. 164: 4178–4184. [DOI] [PubMed] [Google Scholar]
- 4.Lazar G. A., Dang W., Karki S., Vafa O., Peng J. S., Hyun L., Chan C., Chung H. S., Eivazi A., Yoder S. C., et al. 2006. Engineered antibody Fc variants with enhanced effector function. Proc. Natl. Acad. Sci. USA 103: 4005–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natsume A., In M., Takamura H., Nakagawa T., Shimizu Y., Kitajima K., Wakitani M., Ohta S., Satoh M., Shitara K., Niwa R. 2008. Engineered antibodies of IgG1/IgG3 mixed isotype with enhanced cytotoxic activities. Cancer Res. 68: 3863–3872. [DOI] [PubMed] [Google Scholar]
- 6.Richards J. O., Karki S., Lazar G. A., Chen H., Dang W., Desjarlais J. R. 2008. Optimization of antibody binding to FcγRIIa enhances macrophage phagocytosis of tumor cells. Mol. Cancer Ther. 7: 2517–2527. [DOI] [PubMed] [Google Scholar]
- 7.Shields R. L., Namenuk A. K., Hong K., Meng Y. G., Rae J., Briggs J., Xie D., Lai J., Stadlen A., Li B., et al. 2001. High resolution mapping of the binding site on human IgG1 for FcγRI, FcγRII, FcγRIII, and FcRn and design of IgG1 variants with improved binding to the FcγR. J. Biol. Chem. 276: 6591–6604. [DOI] [PubMed] [Google Scholar]
- 8.Hogarth P. M., Pietersz G. A. 2012. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat. Rev. Drug Discov. 11: 311–331. [DOI] [PubMed] [Google Scholar]
- 9.Alegre M. L., Peterson L. J., Xu D., Sattar H. A., Jeyarajah D. R., Kowalkowski K., Thistlethwaite J. R., Zivin R. A., Jolliffe L., Bluestone J. A. 1994. A non-activating “humanized” anti-CD3 monoclonal antibody retains immunosuppressive properties in vivo. Transplantation 57: 1537–1543. [PubMed] [Google Scholar]
- 10.Woodle E. S., Xu D., Zivin R. A., Auger J., Charette J., O’Laughlin R., Peace D., Jollife L. K., Haverty T., Bluestone J. A., Thistlethwaite J. R., Jr. 1999. Phase I trial of a humanized, Fc receptor nonbinding OKT3 antibody, huOKT3γ1(Ala-Ala) in the treatment of acute renal allograft rejection. Transplantation 68: 608–616. [DOI] [PubMed] [Google Scholar]
- 11.Bolt S., Routledge E., Lloyd I., Chatenoud L., Pope H., Gorman S. D., Clark M., Waldmann H. 1993. The generation of a humanized, non-mitogenic CD3 monoclonal antibody which retains in vitro immunosuppressive properties. Eur. J. Immunol. 23: 403–411. [DOI] [PubMed] [Google Scholar]
- 12.Reddy M. P., Kinney C. A., Chaikin M. A., Payne A., Fishman-Lobell J., Tsui P., Dal Monte P. R., Doyle M. L., Brigham-Burke M. R., Anderson D., et al. 2000. Elimination of Fc receptor-dependent effector functions of a modified IgG4 monoclonal antibody to human CD4. J. Immunol. 164: 1925–1933. [DOI] [PubMed] [Google Scholar]
- 13.Rother R. P., Rollins S. A., Mojcik C. F., Brodsky R. A., Bell L. 2007. Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nat. Biotechnol. 25: 1256–1264. [DOI] [PubMed] [Google Scholar]
- 14.Jefferis R. 2009. Glycosylation as a strategy to improve antibody-based therapeutics. Nat. Rev. Drug Discov. 8: 226–234. [DOI] [PubMed] [Google Scholar]
- 15.Shields R. L., Lai J., Keck R., O’Connell L. Y., Hong K., Meng Y. G., Weikert S. H., Presta L. G. 2002. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem. 277: 26733–26740. [DOI] [PubMed] [Google Scholar]
- 16.Umaña P., Jean-Mairet J., Moudry R., Amstutz H., Bailey J. E. 1999. Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat. Biotechnol. 17: 176–180. [DOI] [PubMed] [Google Scholar]
- 17.Morell A., Terry W. D., Waldmann T. A. 1970. Metabolic properties of IgG subclasses in man. J. Clin. Invest. 49: 673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stapleton N. M., Andersen J. T., Stemerding A. M., Bjarnarson S. P., Verheul R. C., Gerritsen J., Zhao Y., Kleijer M., Sandlie I., de Haas M., et al. 2011. Competition for FcRn-mediated transport gives rise to short half-life of human IgG3 and offers therapeutic potential. Nat. Commun. 2: 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roopenian D. C., Akilesh S. 2007. FcRn: the neonatal Fc receptor comes of age. Nat. Rev. Immunol. 7: 715–725. [DOI] [PubMed] [Google Scholar]
- 20.Ward E. S., Ober R. J. 2009. Chapter 4: multitasking by exploitation of intracellular transport functions the many faces of FcRn. Adv. Immunol. 103: 77–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akilesh S., Christianson G. J., Roopenian D. C., Shaw A. S. 2007. Neonatal FcR expression in bone marrow-derived cells functions to protect serum IgG from catabolism. J. Immunol. 179: 4580–4588. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi K., Qiao S. W., Yoshida M., Baker K., Lencer W. I., Blumberg R. S. 2009. An FcRn-dependent role for anti-flagellin immunoglobulin G in pathogenesis of colitis in mice. Gastroenterology 137: 1746–1756.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montoyo H. P., Vaccaro C., Hafner M., Ober R. J., Mueller W., Ward E. S. 2009. Conditional deletion of the MHC class I-related receptor FcRn reveals the sites of IgG homeostasis in mice. Proc. Natl. Acad. Sci. USA 106: 2788–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ober R. J., Martinez C., Vaccaro C., Zhou J., Ward E. S. 2004. Visualizing the site and dynamics of IgG salvage by the MHC class I-related receptor, FcRn. J. Immunol. 172: 2021–2029. [DOI] [PubMed] [Google Scholar]
- 25.Qiao S. W., Kobayashi K., Johansen F. E., Sollid L. M., Andersen J. T., Milford E., Roopenian D. C., Lencer W. I., Blumberg R. S. 2008. Dependence of antibody-mediated presentation of antigen on FcRn. Proc. Natl. Acad. Sci. USA 105: 9337–9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ober R. J., Martinez C., Lai X., Zhou J., Ward E. S. 2004. Exocytosis of IgG as mediated by the receptor, FcRn: an analysis at the single-molecule level. Proc. Natl. Acad. Sci. USA 101: 11076–11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prabhat P., Gan Z., Chao J., Ram S., Vaccaro C., Gibbons S., Ober R. J., Ward E. S. 2007. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc. Natl. Acad. Sci. USA 104: 5889–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dall’Acqua W. F., Woods R. M., Ward E. S., Palaszynski S. R., Patel N. K., Brewah Y. A., Wu H., Kiener P. A., Langermann S. 2002. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: biological consequences. J. Immunol. 169: 5171–5180. [DOI] [PubMed] [Google Scholar]
- 29.Ghetie V., Popov S., Borvak J., Radu C., Matesoi D., Medesan C., Ober R. J., Ward E. S. 1997. Increasing the serum persistence of an IgG fragment by random mutagenesis. Nat. Biotechnol. 15: 637–640. [DOI] [PubMed] [Google Scholar]
- 30.Hinton P. R., Johlfs M. G., Xiong J. M., Hanestad K., Ong K. C., Bullock C., Keller S., Tang M. T., Tso J. Y., Vásquez M., Tsurushita N. 2004. Engineered human IgG antibodies with longer serum half-lives in primates. J. Biol. Chem. 279: 6213–6216. [DOI] [PubMed] [Google Scholar]
- 31.Hinton P. R., Xiong J. M., Johlfs M. G., Tang M. T., Keller S., Tsurushita N. 2006. An engineered human IgG1 antibody with longer serum half-life. J. Immunol. 176: 346–356. [DOI] [PubMed] [Google Scholar]
- 32.Kim J. K., Firan M., Radu C. G., Kim C. H., Ghetie V., Ward E. S. 1999. Mapping the site on human IgG for binding of the MHC class I-related receptor, FcRn. Eur. J. Immunol. 29: 2819–2825. [DOI] [PubMed] [Google Scholar]
- 33.Petkova S. B., Akilesh S., Sproule T. J., Christianson G. J., Al Khabbaz H., Brown A. C., Presta L. G., Meng Y. G., Roopenian D. C. 2006. Enhanced half-life of genetically engineered human IgG1 antibodies in a humanized FcRn mouse model: potential application in humorally mediated autoimmune disease. Int. Immunol. 18: 1759–1769. [DOI] [PubMed] [Google Scholar]
- 34.Vaccaro C., Zhou J., Ober R. J., Ward E. S. 2005. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat. Biotechnol. 23: 1283–1288. [DOI] [PubMed] [Google Scholar]
- 35.Dall’Acqua W. F., Kiener P. A., Wu H. 2006. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). J. Biol. Chem. 281: 23514–23524. [DOI] [PubMed] [Google Scholar]
- 36.Simister N. E., Mostov K. E. 1989. Cloning and expression of the neonatal rat intestinal Fc receptor, a major histocompatibility complex class I antigen homolog. Cold Spring Harb. Symp. Quant. Biol. 54: 571–580. [DOI] [PubMed] [Google Scholar]
- 37.Burmeister W. P., Huber A. H., Bjorkman P. J. 1994. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature 372: 379–383. [DOI] [PubMed] [Google Scholar]
- 38.West A. P., Jr., Bjorkman P. J. 2000. Crystal structure and immunoglobulin G binding properties of the human major histocompatibility complex-related Fc receptor. Biochemistry 39: 9698–9708. [DOI] [PubMed] [Google Scholar]
- 39.Patel D. A., Puig-Canto A., Challa D. K., Perez Montoyo H., Ober R. J., Ward E. S. 2011. Neonatal Fc receptor blockade by Fc engineering ameliorates arthritis in a murine model. J. Immunol. 187: 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Challa D. K., Bussmeyer U., Khan T., Montoyo H. P., Bansal P., Ober R. J., Ward E. S. 2013. Autoantibody depletion ameliorates disease in murine experimental autoimmune encephalomyelitis. MAbs 5: 655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zalevsky J., Chamberlain A. K., Horton H. M., Karki S., Leung I. W., Sproule T. J., Lazar G. A., Roopenian D. C., Desjarlais J. R. 2010. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 28: 157–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaccaro C., Bawdon R., Wanjie S., Ober R. J., Ward E. S. 2006. Divergent activities of an engineered antibody in murine and human systems have implications for therapeutic antibodies. Proc. Natl. Acad. Sci. USA 103: 18709–18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andersen J. T., Foss S., Kenanova V. E., Olafsen T., Leikfoss I. S., Roopenian D. C., Wu A. M., Sandlie I. 2012. Anti-carcinoembryonic antigen single-chain variable fragment antibody variants bind mouse and human neonatal Fc receptor with different affinities that reveal distinct cross-species differences in serum half-life. J. Biol. Chem. 287: 22927–22937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenanova V., Olafsen T., Crow D. M., Sundaresan G., Subbarayan M., Carter N. H., Ikle D. N., Yazaki P. J., Chatziioannou A. F., Gambhir S. S., et al. 2005. Tailoring the pharmacokinetics and positron emission tomography imaging properties of anti-carcinoembryonic antigen single-chain Fv-Fc antibody fragments. Cancer Res. 65: 622–631. [PMC free article] [PubMed] [Google Scholar]
- 45.Norderhaug L., Olafsen T., Michaelsen T. E., Sandlie I. 1997. Versatile vectors for transient and stable expression of recombinant antibody molecules in mammalian cells. J. Immunol. Methods 204: 77–87. [DOI] [PubMed] [Google Scholar]
- 46.Berntzen G., Lunde E., Flobakk M., Andersen J. T., Lauvrak V., Sandlie I. 2005. Prolonged and increased expression of soluble Fc receptors, IgG and a TCR-Ig fusion protein by transiently transfected adherent 293E cells. J. Immunol. Methods 298: 93–104. [DOI] [PubMed] [Google Scholar]
- 47.Michaelsen T. E., Sandlie I., Bratlie D. B., Sandin R. H., Ihle O. 2009. Structural difference in the complement activation site of human IgG1 and IgG3. Scand. J. Immunol. 70: 553–564. [DOI] [PubMed] [Google Scholar]
- 48.Firan M., Bawdon R., Radu C., Ober R. J., Eaken D., Antohe F., Ghetie V., Ward E. S. 2001. The MHC class I-related receptor, FcRn, plays an essential role in the maternofetal transfer of γ-globulin in humans. Int. Immunol. 13: 993–1002. [DOI] [PubMed] [Google Scholar]
- 49.Popov S., Hubbard J. G., Kim J., Ober B., Ghetie V., Ward E. S. 1996. The stoichiometry and affinity of the interaction of murine Fc fragments with the MHC class I-related receptor, FcRn. Mol. Immunol. 33: 521–530. [DOI] [PubMed] [Google Scholar]
- 50.Christianson G. J., Sun V. Z., Akilesh S., Pesavento E., Proetzel G., Roopenian D. C. 2012. Monoclonal antibodies directed against human FcRn and their applications. MAbs 4: 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aase A., Michaelsen T. E. 1991. The use of a hapten-Fab conjugate to sensitize target cells for antibody-dependent complement-mediated lysis and antibody-dependent cell-mediated cytotoxicity. J. Immunol. Methods 136: 185–191. [DOI] [PubMed] [Google Scholar]
- 52.Aase A., Michaelsen T. E. 1994. Opsonophagocytic activity induced by chimeric antibodies of the four human IgG subclasses with or without help from complement. Scand. J. Immunol. 39: 581–587. [DOI] [PubMed] [Google Scholar]
- 53.Niesen F. H., Berglund H., Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2: 2212–2221. [DOI] [PubMed] [Google Scholar]
- 54.Radaev S., Motyka S., Fridman W. H., Sautes-Fridman C., Sun P. D. 2001. The structure of a human type III Fcγ receptor in complex with Fc. J. Biol. Chem. 276: 16469–16477. [DOI] [PubMed] [Google Scholar]
- 55.Rose R. J., van Berkel P. H., van den Bremer E. T., Labrijn A. F., Vink T., Schuurman J., Heck A. J., Parren P. W. 2013. Mutation of Y407 in the CH3 domain dramatically alters glycosylation and structure of human IgG. MAbs 5: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidarsson G., Stemerding A. M., Stapleton N. M., Spliethoff S. E., Janssen H., Rebers F. E., de Haas M., van de Winkel J. G. 2006. FcRn: an IgG receptor on phagocytes with a novel role in phagocytosis. Blood 108: 3573–3579. [DOI] [PubMed] [Google Scholar]
- 57.Derer S., Glorius P., Schlaeth M., Lohse S., Klausz K., Muchhal U., Desjarlais J. R., Humpe A., Valerius T., Peipp M. 2014. Increasing FcγRIIa affinity of an FcγRIII-optimized anti-EGFR antibody restores neutrophil-mediated cytotoxicity. MAbs 6: 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Golay J., Da Roit F., Bologna L., Ferrara C., Leusen J. H., Rambaldi A., Klein C., Introna M. 2013. Glycoengineered CD20 antibody obinutuzumab activates neutrophils and mediates phagocytosis through CD16B more efficiently than rituximab. Blood 122: 3482–3491. [DOI] [PubMed] [Google Scholar]
- 59.Diebolder C. A., Beurskens F. J., de Jong R. N., Koning R. I., Strumane K., Lindorfer M. A., Voorhorst M., Ugurlar D., Rosati S., Heck A. J., et al. 2014. Complement is activated by IgG hexamers assembled at the cell surface. Science 343: 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sondermann P., Huber R., Oosthuizen V., Jacob U. 2000. The 3.2-Å crystal structure of the human IgG1 Fc fragment-FcγRIII complex. Nature 406: 267–273. [DOI] [PubMed] [Google Scholar]
- 61.Schneider S., Zacharias M. 2012. Atomic resolution model of the antibody Fc interaction with the complement C1q component. Mol. Immunol. 51: 66–72. [DOI] [PubMed] [Google Scholar]
- 62.Hessell A. J., Hangartner L., Hunter M., Havenith C. E., Beurskens F. J., Bakker J. M., Lanigan C. M., Landucci G., Forthal D. N., Parren P. W., et al. 2007. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449: 101–104. [DOI] [PubMed] [Google Scholar]
- 63.Oganesyan V., Damschroder M. M., Woods R. M., Cook K. E., Wu H., Dall’acqua W. F. 2009. Structural characterization of a human Fc fragment engineered for extended serum half-life. Mol. Immunol. 46: 1750–1755. [DOI] [PubMed] [Google Scholar]
- 64.Jung S. T., Reddy S. T., Kang T. H., Borrok M. J., Sandlie I., Tucker P. W., Georgiou G. 2010. Aglycosylated IgG variants expressed in bacteria that selectively bind FcγRI potentiate tumor cell killing by monocyte-dendritic cells. Proc. Natl. Acad. Sci. USA 107: 604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Borrok M. J., Jung S. T., Kang T. H., Monzingo A. F., Georgiou G. 2012. Revisiting the role of glycosylation in the structure of human IgG Fc. ACS Chem. Biol. 7: 1596–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frank M., Walker R. C., Lanzilotta W. N., Prestegard J. H., Barb A. W. 2014. Immunoglobulin G1 Fc domain motions: implications for Fc engineering. J. Mol. Biol. 426: 1799–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peters R. T., Toby G., Lu Q., Liu T., Kulman J. D., Low S. C., Bitonti A. J., Pierce G. F. 2013. Biochemical and functional characterization of a recombinant monomeric factor VIII-Fc fusion protein. J. Thromb. Haemost. 11: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shapiro A. D., Ragni M. V., Kulkarni R., Oldenberg J., Srivastava A., Quon D. V., Pasi K. J., Hanabusa H., Pabinger I., Mahlangu J., et al. 2014. Recombinant factor VIII Fc fusion protein: extended-interval dosing maintains low bleeding rates and correlates with von Willebrand factor levels. J. Thromb. Haemost. 12: 1788–1800. [DOI] [PubMed] [Google Scholar]
- 69.Thompson C. A. 2014. New hemophilia A treatment allows less-frequent prophylactic doses. Am. J. Health Syst. Pharm. 71: 1153. [DOI] [PubMed] [Google Scholar]
- 70.Saphire E. O., Parren P. W., Pantophlet R., Zwick M. B., Morris G. M., Rudd P. M., Dwek R. A., Stanfield R. L., Burton D. R., Wilson I. A. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293: 1155–1159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.