Abstract
Plasmodium falciparum merozoites use diverse alternative erythrocyte receptors for invasion and variably express cognate ligands encoded by the erythrocyte binding antigen (eba) and reticulocyte binding-like homologue (Rh) gene families. Previous analyses conducted on parasites from single populations in areas of endemicity revealed a wide spectrum of invasion phenotypes and expression profiles, although comparisons across studies have been limited by the use of different protocols. For direct comparisons within and among populations, clinical isolates from three different West African sites of endemicity (in Ghana, Guinea, and Senegal) were cryopreserved and cultured ex vivo after thawing in a single laboratory to assay invasion of target erythrocytes pretreated with enzymes affecting receptor subsets. Complete invasion assay data from 67 isolates showed no differences among the populations in the broad range of phenotypes measured by neuraminidase treatment (overall mean, 40.6% inhibition) or trypsin treatment (overall mean, 83.3% inhibition). The effects of chymotrypsin treatment (overall mean, 79.2% inhibition) showed heterogeneity across populations (Kruskall-Wallis P = 0.023), although the full phenotypic range was seen in each. Schizont-stage transcript data for a panel of 8 invasion ligand genes (eba175, eba140, eba181, Rh1, Rh2a, Rh2b, Rh4, and Rh5) were obtained for 37 isolates, showing similar ranges of variation in each population except that eba175 levels tended to be higher in parasites from Ghana than in those from Senegal (whereas levels of eba181 and Rh2b were lower in parasites from Ghana). The broad diversity in invasion phenotypes and gene expression seen within each local population, with minimal differences among them, is consistent with a hypothesis of immune selection maintaining parasite variation.
INTRODUCTION
The major human malaria parasite Plasmodium falciparum uses diverse ligand-receptor interactions in merozoite invasion of erythrocytes (1). Parasite ligands include proteins belonging to the erythrocyte binding antigen (EBA) and reticulocyte binding protein-like homologue (Rh) families, including EBA175, EBL-1, EBA140, and Rh4, which bind, respectively, to glycophorin A (GpA), GpB, GpC, and complement receptor 1 (CR1) on erythrocytes (1–6). These ligand-receptor interactions are variably used by different P. falciparum lines, while interaction between merozoite Rh5 and erythrocyte surface basigin is apparently used by all lines (7, 8). Other members of these protein families expressed in P. falciparum merozoites for which cognate erythrocyte receptors have not yet been identified are EBA181 (9) and Rh1 (10, 11) and the closely related proteins Rh2a and Rh2b (12). Variation in invasion phenotypes has been widely characterized by assessing the ability of parasites to invade erythrocytes treated with enzymes to selectively remove parts of the receptor repertoire, following pioneering studies in the 1980s (13, 14). In particular, neuraminidase treatment removes sialic acids from glycophorins and other erythrocyte receptors, whereas trypsin treatment cleaves the peptide backbone of several receptors (including GpA, GpC, and CR1), and chymotrypsin cleaves others (including GpB and CR1) (1, 2) (see Table S1 in the supplemental material). Polymorphism and plasticity of invasion phenotypes may be adaptive for the parasite due to selection by acquired immunity to individual merozoite ligands (7, 15) or possibly due to diversity in the structure and abundance of receptors on erythrocytes. This is of applicable importance, as particular ligands are being developed as vaccine antigens, with EBA175 and Rh5 being the lead candidates among these (7), and some components may be best considered in combinations (16).
If acquired immune responses to parasites can inhibit different ligand-receptor interactions, a diversity of enzyme-sensitive erythrocyte invasion phenotypes is expected to be maintained within each endemic population by frequency-dependent immune selection. This may be influenced by levels of endemicity, such that a low incidence of infection and minimal acquired immunity may allow many parasites to use a favored primary pathway, while a broader range of alternative invasion phenotypes may be selected for in areas of greater endemicity. Studies on clinical isolates have indicated a high diversity of invasion phenotypes in India (17), Brazil (18, 19), Peru (19), Colombia (19), The Gambia (20, 21), Kenya (22, 23), Tanzania (24), and Senegal (25, 26). However, apart from a small number of isolates from Peru and Colombia that were cultured together in a single laboratory (19), all other samples from each country were analyzed in different laboratories at different times, using a variety of assay protocols. Therefore, these data do not enable a standardized analysis of variation within and between populations.
We report the first comparative analysis of erythrocyte invasion and merozoite ligand gene expression by population samples of malaria parasites from different countries in regions of endemicity, assayed in the first parasite cycle ex vivo in a single laboratory. Clinical isolates of P. falciparum from three different countries in West Africa were sampled and cryopreserved, prior to assaying the isolates together with identical protocols in a blind manner. Virtually all the diversity in invasion phenotypes and gene expression was seen within each local population, as expected from a hypothesis of immune selection maintaining parasite variation at all of these sites of endemicity.
MATERIALS AND METHODS
Plasmodium falciparum clinical isolates from three populations in areas of endemicity.
Malaria patients attending local health facilities at three different sites in West Africa (Fig. 1) who tested positive for P. falciparum malaria by immunochromatic rapid diagnostic testing and who had reported not taking antimalarial drugs during the preceding 3 days were invited to participate in the study. Patients attending Kintampo Municipal Hospital in the Brong-Ahafo region of central Ghana were recruited in 2011 and 2012; 63 patients with P. falciparum parasitemia of >0.5% infected erythrocytes were included in this study, with a mean patient age of 5.3 years (standard deviation [SD], 3.5 years). Patients attending the Tiro and Banian local health centers near Faranah in the Republic of Guinea were recruited in 2012; 23 patients with P. falciparum parasitemia of >0.5% were included here (mean patient age, 12.9 years; SD, 10.1 years). Patients attending Pikine health center in Dakar, Senegal, were recruited in 2013; 44 of the patients with P. falciparum parasitemia of >0.5% were included here (mean patient age, 8.9 years; SD, 3.8 years). Written informed consent was obtained from patients or their parents or guardians, and additional assent was received from children who were 10 years of age or older. Antimalarial treatment was provided according to the relevant national and local guidelines. Immediately prior to treatment, venous blood samples of up to 5 ml in volume were collected into heparinized Vacutainer tubes in Ghana and Guinea and into EDTA anticoagulant tubes in Senegal. At each site, blood samples were centrifuged, plasma and leukocyte buffy coats were removed, and erythrocytes were cryopreserved in glycerolyte and stored frozen at −80°C or in liquid nitrogen until shipment on dry ice to the London School of Hygiene and Tropical Medicine for assay. Approval of the study was granted by the Ethics Committees of the Ghana Health Service, the Kintampo Health Research Centre, and the Noguchi Memorial Institute for Medical Research (University of Ghana), the National Ethics Committee for Health Research in the Republic of Guinea, the Ethics Committee of the Ministry of Health in Senegal, and the Ethics Committee of the London School of Hygiene and Tropical Medicine.
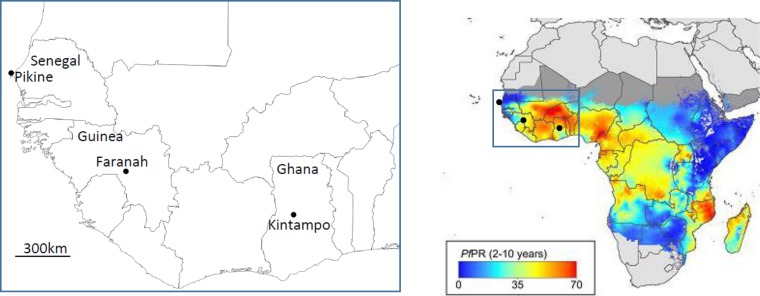
FIG 1.
Locations of the three sites in West Africa from which P. falciparum clinical isolates were sampled for assays of erythrocyte invasion and gene expression. The map on the left shows the locations of Pikine in Senegal (44 isolates), Faranah in Guinea (23 isolates), and Kintampo in Ghana (63 isolates) and utilizes graphics from d-maps.com (http://d-maps.com/carte.php?num_car=753&lang=en). Of the 130 isolates tested, 67 yielded invasion phenotype data. On the right, the area is shown within a map of Africa which indicates the estimated community prevalence of P. falciparum slide positivity in children aged between 2 and 10 years in 2010 (reprinted from the Malaria Journal [40]).
Parasite culture and erythrocyte invasion assays.
A total of 130 isolates that each had parasitemia represented by at least 0.5% infected erythrocytes were thawed and given serial codes for culture by one laboratory investigator at the London School of Hygiene and Tropical Medicine. Prior to parasite culture being initiated by the same investigator, erythrocytes from each sample were washed 4 times in total to ensure that no anticoagulant or plasma components from the samples were included. The schedule typically involved processing between 6 and 10 isolates from a mixture of the different source population samples each week, and blind processing was ensured, as the subsequent invasion assays were conducted by a different laboratory investigator who had no role in the sample collection, storage, thawing, or culture initiation procedures. Parasites were cultured at 2% hematocrit in RPMI 1640 medium containing 2% human AB serum (GE Healthcare, Amersham, United Kingdom) and 0.3% Albumax II (Life Technologies, Paisley, United Kingdom) under an atmosphere of 1% O2, 3% CO2, and 96% N2.
Erythrocytes (blood group A, rhesus positive) for each invasion assay were labeled by incubation at 4% hematocrit in 5 μM 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) succinimidyl ester (DDAO-SE) fluorescent dye (Life Technologies, Paisley, United Kingdom)–RPMI 1640 with rotation in the dark for 2 h at 37°C. The erythrocytes were then washed twice with RPMI 1640 and suspended at 4% hematocrit into separate tubes for alternative enzyme treatments. Neuraminidase (Vibrio cholerae; Sigma, United Kingdom) was used at a final concentration of 100 mU/ml, trypsin (TPCK [tosylsulfonyl phenylalanyl chloromethyl ketone] treated, bovine) (Sigma, United Kingdom) at 104 U/ml, and chymotrypsin (TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone] treated) (Worthington, USA) at 45 U/ml. All enzyme treatments were performed at 37°C for 1 h, following which erythrocytes were pelleted, washed 3 times in RPMI 1640, and suspended at 4% hematocrit. To check that the enzyme treatments were effective, 20 μl of each of the cell preparations was added to 20 μl of anti-M monoclonal antibody (MAb) (successful trypsin digestion of GpA leads to loss of agglutination), anti-S MAb (successful chymotrypsin digestion of GpB leads to loss of agglutination), or peanut lectin (successful neuraminidase digestion of sialic acids leads to gain of agglutination). The efficacy of enzyme treatment was also confirmed in one batch of erythrocytes by flow cytometric analysis, showing the expected loss of GpA (CD235a), GpC (CD236), and CR-1 (CD35), as detected by monoclonal antibodies (anti-CD235a MAb clone E4 [diluted 1/100], anti-CD236 MAb clone E5 [diluted 1/1,000], and anti-CD35 MAb clone J3D3 [diluted 1/100]; all antibodies from Santa Cruz Biotechnology Inc.). Effects of these enzyme treatments on different erythrocyte receptors as defined by previous studies are listed in Table S1 in the supplemental material.
Invasion assays were performed using a protocol based on a previously described method (27), with each cell preparation tested in triplicate wells of round-bottom 96-well plates, in a culture volume of 200 μl per well at 1% hematocrit. Plates were gassed with 1% O2, 3% CO2, and 96% N2 in a hypoxia chamber (Billups Rothberg Inc., USA) and incubated at 37°C. Assay plates were removed from the incubator after 48 h and centrifuged at 450 × g for 1 min to pellet the cells, and the supernatant was removed. To each assay well, 200 μl of phosphate-buffered saline (PBS) containing 2× SYBR green I was added. The plates were incubated for 1 h at 37°C and then removed from the incubator and centrifuged as before, and the supernatant was removed. The cells were washed a further 3 times in PBS before being suspended in PBS at a final hematocrit of 0.15% for flow cytometry on a FACSCalibur cell analyzer (BD Biosciences, CA, USA). Erythrocytes were gated using forward-scatter and side-scatter characteristics, and DDAO-SE-positive cells were determined by emission at 645 to 677 nm following excitation at ∼635 nm, while levels of SYBR green I-positive cells were determined by emission at 500 to 560 nm following excitation at 488 nm. Invasion of enzyme-treated erythrocytes was determined by analysis of the proportion of 50,000 counted DDAO-SE-positive cells that were also SYBR green I positive, and data were analyzed using FlowJo (Tree Star Inc.). Percent inhibition of invasion of each type of each treated cell preparation (TC) was calculated in comparison with negative-control cells (NC [no enzyme treatment]) and positive-control cells (PC [treated with all the enzymes, i.e., trypsin, chymotrypsin, and neuraminidase]) as follows:
| (1) |
An isolate was determined to have passed quality control if the difference between the mean percentages of infected cells (from triplicate well counts) in positive controls (PC) and negative controls (NC) was >0.2, corresponding to a difference of more than 100 parasites for 50,000 counted erythrocytes from each well.
RNA extraction and quantitative transcript analysis.
For parasite RNA preparation, the equivalent of at least 100 μl of packed erythrocytes from each isolate was cultured in complete RPMI 1640 at 2% hematocrit and 37°C for up to 48 h in separate flasks until most parasites were at the schizont stage of parasite development, following which erythrocytes were resuspended at 50% hematocrit and immediately mixed with a 4× volume of TRIzol reagent (Life Technologies). Aliquots were stored at −80°C for subsequent RNA extraction using an RNeasy Micro kit (Qiagen). The extracted RNA was quantified using Qubit fluorometric quantitation (Life Technologies), and samples below the detection limit for this assay were not carried forward for transcript analysis. The mRNA was reverse transcribed (RT) with oligo(dT) using TaqMan reagents (Life Technologies), and cDNA was quantified in a fluorogenic 5′-nuclease assay on a Rotor-Gene 3000 system (Corbett Life Sciences/Qiagen). Gene-specific TaqMan primer and probe sets for each gene were exactly as described previously for eba140, eba175, and eba181 in reference 28, for Rh1, Rh2a, Rh2b, and Rh4 in reference 29, and for Rh5 in reference 21. All primers and probes were designed to match unique sequences that had no polymorphisms in P. falciparum, and this was rechecked with all additional sequence data available at www.plasmodb.org on 1 May 2013, revealing no polymorphisms in any of the matching sequences except for a single nucleotide polymorphism (SNP) in 1 of 71 isolates in the sequence corresponding to the reverse primer for eba181. These protocols were used to allow direct comparisons with results from a previous study conducted in The Gambia, and as in that study, the ebl-1 gene which contains a stop codon in many isolates was not analyzed here. Real-time PCRs were performed using a Kapa Probe Fast quantitative PCR (qPCR) kit (Kapa Biosystems) in 10-μl volumes with 250 nM concentrations of each primer and 250 nM concentrations of each probe at 95°C for 3 min followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. Each run included controls and 3D7 genomic DNA standards, with each sample assayed in duplicate and standard curves generated during each run. The threshold fluorescence value was determined automatically by Rotor-Gene 3000 software for each run. The relative transcript level for each individual gene determined by RT-qPCR was normalized as the proportion of the sum of the transcript levels for the 8 genes assayed in each isolate.
Statistical analysis.
Nonparametric statistics were used to analyze the distribution of culture phenotypes. The Kruskal-Wallis test was applied to test for statistical significance with respect to heterogeneity across the three sampled populations. Where this was significant, Mann-Whitney tests were applied to investigate pairwise differences between different populations, with Bonferroni correction for open testing of multiple pairwise interpopulation comparisons. Correlations between the distributions of different enzyme inhibition phenotypes were tested by Spearman's ρ (rho).
RESULTS
Erythrocyte invasion phenotype variation.
The first ex vivo cycle invasion phenotypes for all of the different enzyme pretreatments were successfully obtained for 67 (52%) of the 130 clinical parasite isolates that had been thawed for short-term culture, including 34 (54%) of 63 isolates from Ghana, 12 (52%) of 23 isolates from Guinea, and 21 (48%) of 44 isolates from Senegal. Those that did not yield reliable results had either poor parasite growth or fewer than 100 postinvasion parasitized erythrocyte counts that differed between the untreated and triple-enzyme-treated erythrocytes. The proportions from each of the three populations that had successful growth were not significantly different from those obtained in previous assays of fresh West African clinical isolates without any cryopreservation step, including the largest study which yielded results for 163 (63%) of 263 isolates assayed in The Gambia (21).
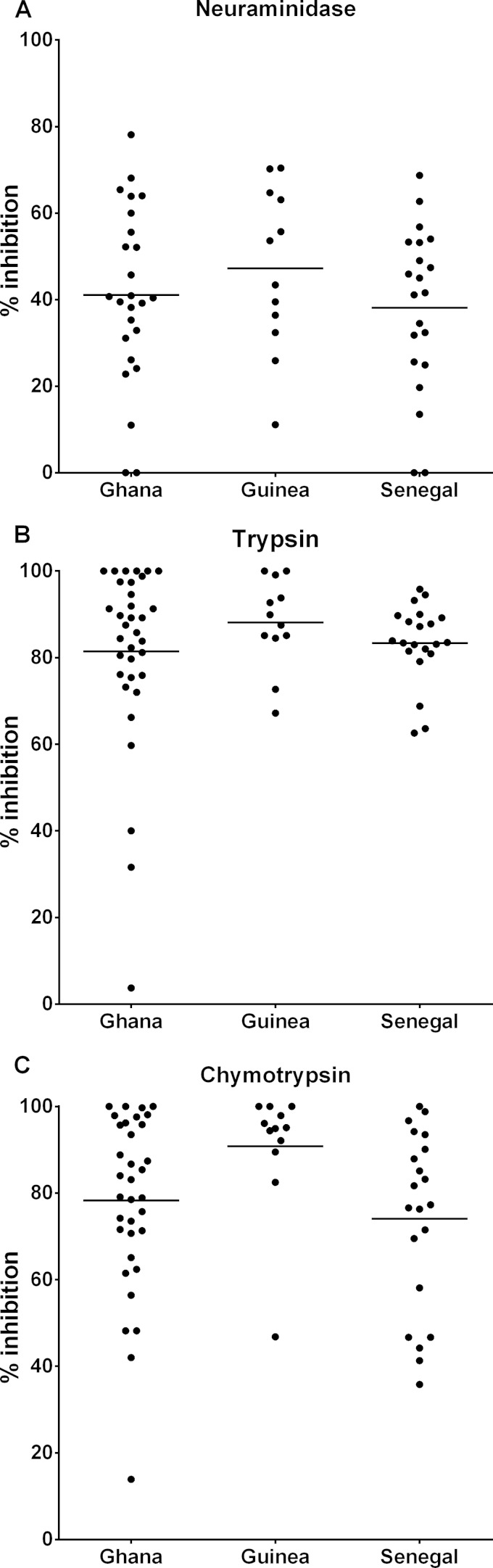
There was wide variability among the isolates in the ability to invade erythrocytes that had been treated with each of the enzymes compared to the control erythrocytes (Fig. 2). The overall mean level of invasion inhibition by neuraminidase treatment was 40.6% (range, 0 to 78.1%), and the mean levels were similar across samples from the three different countries (Kruskal-Wallis test, P = 0.441) (Fig. 2A). Trypsin treatment of erythrocytes led to greatly reduced invasion by most isolates, with an overall mean inhibition level of 83.3% (range, 3.7% to 100%), and the mean levels were similar across the different countries (Kruskal-Wallis test, P = 0.338) (Fig. 2B). Chymotrypsin treatment of erythrocytes caused an overall mean invasion inhibition level of 79.2% (range, 13.9% to 100%), with significant heterogeneity across the samples from different countries (Kruskal-Wallis test, P = 0.023) (Fig. 2C). Isolates from Guinea were inhibited more by chymotrypsin treatment (mean inhibition of 90.8%) than isolates from Ghana (mean of 78.3%; Mann-Whitney uncorrected P = 0.033, P corrected for multiple pairwise comparisons = 0.10) or Senegal (mean of 74.1%; Mann-Whitney uncorrected P = 0.004, P corrected for multiple pairwise comparisons = 0.012).
FIG 2.
Invasion phenotypes of P. falciparum isolates from Ghana (n = 34), Guinea (n = 12), and Senegal (n = 21), determined by the reduction in invasion of erythrocytes treated with enzymes compared to untreated control erythrocytes. Horizontal bars show the mean values within each group. (A) Percentages of inhibition for all countries following neuraminidase treatment. (B) Percentages of inhibition for all countries following trypsin treatment. (C) Percentages of inhibition following chymotrypsin treatment. Pairwise Mann-Whitney tests for differences between population samples in the chymotrypsin treatment phenotypes showed two differences: for Ghana versus Guinea, uncorrected P = 0.033, P corrected for multiple pairwise comparisons = 0.10; for Guinea versus Senegal, uncorrected P = 0.004, P corrected for multiple pairwise comparisons = 0.012. Invasion data values for all individual isolates are listed in Dataset S1 in the supplemental material.
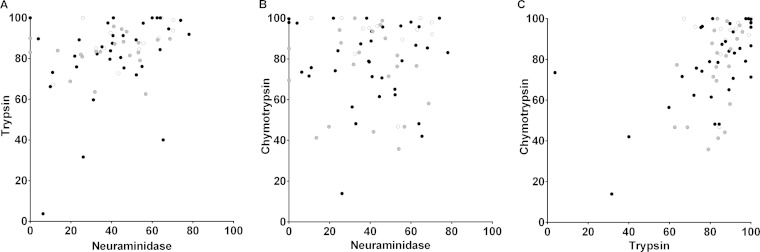
Across all isolates, the inhibition by neuraminidase treatment of erythrocytes was positively correlated with inhibition by trypsin treatment (Spearman's rho = 0.319, P = 0.009) (Fig. 3A) but not with inhibition by chymotrypsin treatment (Spearman's rho = −0.016, P = 0.9) (Fig. 3B). Invasion inhibition by chymotrypsin was positively correlated with inhibition by trypsin (Spearman's rho = 0.447, P < 0.001) (Fig. 3C).
FIG 3.
Pairwise correlations of invasion phenotypes across isolates as defined by different enzyme pretreatments of erythrocytes. Sources of individual isolates are identified by different shaded symbols as follows: Ghana, black; Guinea, white; and Senegal, gray. (A) Significant positive correlation between neuraminidase treatment and trypsin treatment (Spearman's rho = 0.319, P = 0.009). (B) No significant correlation between neuraminidase treatment and chymotrypsin treatment (Spearman's rho = −0.016, P = 0.9). (C) Significant positive correlation between trypsin treatment and chymotrypsin treatment (Spearman's rho = 0.497, P < 0.001).
Analysis of transcript levels.
In parallel with the invasion assay cultures, ex vivo parasites were cultured for up to 48 h until they reached stages consisting predominantly of schizonts, which yielded sufficient RNA to allow relative transcript levels of 8 eba and Rh genes to be successfully assayed for 37 isolates of the 67 that had had their invasion phenotypes determined (26 from Ghana, 3 from Guinea, and 8 from Senegal). The remaining 30 isolates had too low a yield of overall RNA for reliable quantification after extraction from the limited amount of schizont-stage culture available.
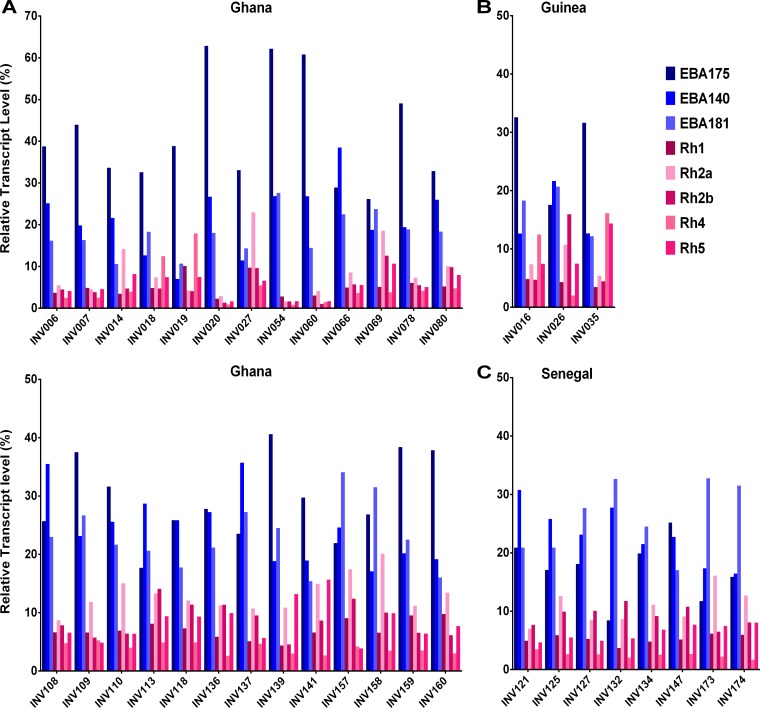
Overall, the eba gene transcripts were more abundant than the Rh gene transcripts, with eba175 being most abundant (mean of 28% of the total for the 8 genes in each isolate) followed by eba140 and eba181 (means of 21% and 20%, respectively) (Fig. 4). The profiles were broadly similar across the samples from different countries. However, the relative levels of eba175 transcripts tended to be lower in isolates from Senegal than in those from Ghana (respective means of 17% and 32% of the total transcripts across all 8 genes; Mann-Whitney uncorrected P = 0.0002, Bonferroni corrected P = 0.005); conversely, the relative levels of eba181 were higher in isolates from Senegal than in those from Ghana (respective means of 26% and 18%; uncorrected P = 0.0012, P corrected for multiple comparisons = 0.03). The relative levels of Rh2b transcripts were also higher in isolates from Senegal than in those from Ghana, although the differences were not statistically significant after correction for multiple comparisons (respective means of 9% and 6%; uncorrected P = 0.008, P corrected for multiple comparisons = 0.19) (Fig. 4). Profiling across the 8 transcripts for individual isolates showed that most had higher levels of eba175 transcripts than of transcripts of any of the other genes, although a proportion of isolates (including 6 of the 8 Senegalese isolates) had higher levels of eba181 transcripts (Fig. 5). Among the isolates from Ghana, tests were performed for correlations between the 8 individual gene transcript levels and the 3 different enzyme inhibition measurements. Of the 24 correlations tested, two were marginally significant without correction for multiple comparisons (neuraminidase inhibition versus eba140 relative levels, r = 0.47, uncorrected P = 0.017; chymotrypsin inhibition versus eba181 relative levels, r = −0.39, uncorrected P = 0.047), although these were not significant after Bonferroni correction.
FIG 4.
Relative transcript levels of 3 eba genes and 5 Rh genes in ex vivo schizont-stage cultures of P. falciparum clinical isolates. (A) Isolates from Ghana (n = 26). (B) Isolates from Guinea (n = 3). (C) Isolates from Senegal (n = 8). Each dot shows the percentage of transcript for each gene in a single clinical isolate as a proportion of the total for all 8 genes in the isolate. Horizontal lines denote the mean for each transcript across all isolates sampled in each population. Levels of eba175 were higher in isolates from Ghana than in isolates from Senegal (Mann-Whitney uncorrected P = 0.0002, P corrected for multiple comparisons = 0.005); conversely, the levels of eba181 were higher in isolates from Senegal than in isolates from Ghana (Mann-Whitney uncorrected P = 0.0012, P corrected for multiple comparisons = 0.03), as were the relative levels of Rh2b (Mann-Whitney uncorrected P = 0.008), although these data were not statistically significant after correction for multiple comparisons (P = 0.19). The results of all other pairwise tests comparing transcript levels between countries were not statistically significant. Transcript data values for all individual isolates are listed in Dataset S1 in the supplemental material.
FIG 5.
Individual isolate expression profiles for 8 eba and Rh genes in ex vivo schizont-stage cultures. (A) Isolates from Ghana (n = 26 [two panels]). (B) Isolates from Guinea (n = 3). (C) Isolates from Senegal (n = 8). The percentage of each transcript is normalized as a proportion of the sum of all 8 gene transcripts. To assist visual clarity, the plotted order of the 8 genes for each isolate is the same as the order of the genes in the legend.
DISCUSSION
This first direct comparison of data reflecting erythrocyte invasion by clinical P. falciparum isolates from three different countries in regions of endemicity shows that almost all of the overall phenotypic diversity is contained within each local population. There were no significant differences among the populations in the invasion inhibition of clinical isolates by neuraminidase or trypsin treatment of target erythrocytes. A slightly greater effect of chymotrypsin treatment on the invasion of parasites from Guinea than on those from Ghana and Senegal is not likely to reflect differences in endemicity, as the disease is more endemic in the sampled sites in both Guinea and Ghana than in the site in Senegal. The correlation between the levels of inhibition by trypsin and chymotrypsin seen here was not seen in previous studies on West African parasites, conducted in Senegal or The Gambia (21, 25). This might reflect differences in protocols among laboratories or variations in the activities of commercial enzyme batches or receptor properties of target erythrocytes used for the assays, illustrating the value of conducting comparisons within a single laboratory where possible.
Expression of genes encoding P. falciparum merozoite ligands involved with invasion is tightly regulated as part of the parasite developmental cycle (30, 31), and variation between parasites is known to be under epigenetic regulation (32–34), but the selective determinants of transcript profiles seen in vivo are not well understood. A diversity of eba and Rh gene expression profiles has been previously seen in separate studies of clinical isolates from individual populations, but epidemiological correlations with function or host immunity remain to be established (21, 24, 25, 29). In a large study of Gambian children, there were no differences in expression of these genes between isolates from patients with severe malaria and those from patients with mild malaria, although expression of eba175 correlated negatively with patient age, whereas expression of eba181 correlated positively (21). It was suggested that use of EBA175 might be favored by parasites in nonimmune hosts but that acquired immune responses to EBA175 may select for parasites using alternative receptors such as EBA181 in older individuals. In the present study, isolates from Ghana had slightly higher levels of eba175 gene expression, and concomitantly lower levels of eba181, than isolates from Senegal. Although the Ghanaian patients were slightly younger on average than those in Senegal, the endemicity of malaria was lower overall at the study site in Senegal, so this observed difference in expression does not correlate with a simple prediction based on likely differences in immunity. The expression profiles seen for each of the samples here fall within the range of those reported in the previous large study from The Gambia (21).
The phenotypes and expression profiles described here that correspond to the first replicative cycle ex vivo necessarily refer to total parasite populations sampled from each infected patient rather than to isolated individual parasite types. Most clinical P. falciparum infections in these areas of West Africa contain a mixture of different parasite genotypes (35, 36), so analysis of individual genotypes and replication of assays on parasite lines would require cloning and longer-term growth, with a potential for changes in phenotypes during culture adaptation. Slight differences in sampling dates between the countries over a 2-year period are unlikely to have caused variation here, as previous studies in The Gambia showed similar distributions of invasion phenotypes in population samples of mild clinical malaria isolates taken up to 6 years apart (20, 21), and similar profiles have been seen in Senegal in successive years (25, 26). It is possible that changes might be seen over longer periods if levels of endemicity and acquired immunity in particular populations are reduced by enhanced malaria treatment interventions. The current study was not designed to directly analyze components of immunity such as antibodies, as samples from each of the patients were taken at only a single time point, and antibodies to parasite antigens are generally elicited during clinical malaria infection in most individuals (37). Analysis of samples collected at multiple time points from patients or by random sampling from communities (38) is necessary to characterize immune response profiles informatively for comparisons over distance or time.
These results are consistent with a hypothesis of selection of parasites to maintain invasion phenotype diversity within each local area of endemicity. Frequency-dependent selection by acquired immune responses to each of the alternative parasite ligands would potentially maintain a diverse range of allelic forms and expression profiles. This is consistent with observed signatures of balancing selection previously seen with respect to the coding sequences of some ligands (particularly EBA175 and Rh2), translational phase variation (due to a polymorphic frameshift mutation in the ebl-1 gene), and variation in ligand expression levels (1, 2, 7, 15, 21). It should be noted that this study involved comparisons of West African sites, and parasite populations are extensively connected in this large region of endemicity (35, 39), so it is possible that parasites in other parts of the world might have different phenotypes, particularly in areas where there is little acquired immunity. However, the observed variation in each of the populations studied here covered most of the range seen in a nonstandardized comparison of data from different studies reported separately on more-diverse individual populations, including those from Asia and South America (17–26). A vaccine based on known parasite ligands involved in invasion would need to incorporate a broad range of alternative EBA and Rh proteins in order to be effective for use in any local population, unless an effective vaccine can be based on a ligand such as Rh5 which appears to be consistently required for invasion (7).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all the malaria patients and clinical staff for their participation in the study. The sample collection was facilitated by staff at Kintampo Health Research Centre, at the National Institute for Public Health in Guinea, and at Pikine Health Centre in Senegal. We appreciate the support of many colleagues at the Medical Research Council Unit in The Gambia, the Laboratory of Bacteriology and Virology, Le Dantec Hospital in Senegal, Noguchi Memorial Institute for Medical Research and the University of Ghana, and the London School of Hygiene and Tropical Medicine in enabling this work. We thank Amy Bei, Michel Theron, and Julian Rayner for helpful advice and discussion on invasion assay protocols.
This study was funded by an ERC Advanced Award (grant AdG-2011-294428 to D.J.C.) and a Leverhulme-Royal Society Africa Award (grant AA110050 to G.A.A. and D.J.C.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03009-14.
REFERENCES
- 1.Bei AK, Duraisingh MT. 2012. Functional analysis of erythrocyte determinants of Plasmodium infection. Int J Parasitol 42:575–582. doi: 10.1016/j.ijpara.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tham WH, Healer J, Cowman AF. 2012. Erythrocyte and reticulocyte binding-like proteins of Plasmodium falciparum. Trends Parasitol 28:23–30. doi: 10.1016/j.pt.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Sim BKL, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. 1994. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 4.Mayer DC, Cofie J, Jiang L, Hartl DL, Tracy E, Kabat J, Mendoza LH, Miller LH. 2009. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci U S A 106:5348–5352. doi: 10.1073/pnas.0900878106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maier AG, Duraisingh MT, Reeder JC, Patel SS, Kazura JW, Zimmerman PA, Cowman AF. 2003. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat Med 9:87–92. doi: 10.1038/nm807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tham WH, Wilson DW, Lopaticki S, Schmidt CQ, Tetteh-Quarcoo PB, Barlow PN, Richard D, Corbin JE, Beeson JG, Cowman AF. 2010. Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand. Proc Natl Acad Sci U S A 107:17327–17332. doi: 10.1073/pnas.1008151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright GJ, Rayner JC. 2014. Plasmodium falciparum erythrocyte invasion: combining function with immune evasion. PLoS Pathog 10:e1003943. doi: 10.1371/journal.ppat.1003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, Mboup S, Ndir O, Kwiatkowski DP, Duraisingh MT, Rayner JC, Wright GJ. 2011. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature 480:534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilberger T-M, Thompson JK, Triglia T, Good RT, Duraisingh MT, Cowman AF. 2003. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J Biol Chem 278:14480–14486. doi: 10.1074/jbc.M211446200. [DOI] [PubMed] [Google Scholar]
- 10.Triglia T, Duraisingh MT, Good RT, Cowman AF. 2005. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol Microbiol 55:162–174. doi: 10.1111/j.1365-2958.2004.04388.x. [DOI] [PubMed] [Google Scholar]
- 11.Rayner JC, Vargas-Serrato E, Huber CS, Galinski MR, Barnwell JW. 2001. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant invasion pathway. J Exp Med 194:1571–1581. doi: 10.1084/jem.194.11.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duraisingh MT, Triglia T, Ralph SA, Rayner JC, Barnwell JW, McFadden GI, Cowman AF. 2003. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J 22:1047–1057. doi: 10.1093/emboj/cdg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell GH, Hadley TJ, McGinniss MH, Klotz FW, Miller LH. 1986. Invasion of erythrocytes by Plasmodium falciparum malaria parasites: evidence for receptor heterogeneity and two receptors. Blood 67:1519–1521. [PubMed] [Google Scholar]
- 14.Hadley TJ, Klotz FW, Pasvol G, Haynes JD, McGinniss MH, Okubo Y, Miller LH. 1987. Falciparum malaria parasites invade erythrocytes that lack glycophorin A and B (MkMk). Strain differences indicate receptor heterogeneity and two pathways for invasion. J Clin Invest 80:1190–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persson KE, Fowkes FJ, McCallum FJ, Gicheru N, Reiling L, Richards JS, Wilson DW, Lopaticki S, Cowman AF, Marsh K, Beeson JG. 2013. Erythrocyte-binding antigens of Plasmodium falciparum are targets of human inhibitory antibodies and function to evade naturally acquired immunity. J Immunol 191:785–794. doi: 10.4049/jimmunol.1300444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopaticki S, Maier AG, Thompson J, Wilson DW, Tham WH, Triglia T, Gout A, Speed TP, Beeson JG, Healer J, Cowman AF. 2011. Reticulocyte and erythrocyte binding-like proteins function cooperatively in invasion of human erythrocytes by malaria parasites. Infect Immun 79:1107–1117. doi: 10.1128/IAI.01021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okoyeh JN, Pillai CR, Chitnis CE. 1999. Plasmodium falciparum field isolates commonly use erythrocyte invasion pathways that are independent of sialic acid residues of glycophorin A. Infect Immun 67:5784–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobo CA, de Frazao K, Rodriguez M, Reid M, Zalis M, Lustigman S. 2004. Invasion profiles of Brazilian field isolates of Plasmodium falciparum: phenotypic and genotypic analyses. Infect Immun 72:5886–5891. doi: 10.1128/IAI.72.10.5886-5891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Perez M, Villasis E, Machado RL, Povoa MM, Vinetz JM, Blair S, Gamboa D, Lustigman S. 2012. Plasmodium falciparum field isolates from South America use an atypical red blood cell invasion pathway associated with invasion ligand polymorphisms. PLoS One 7:e47913. doi: 10.1371/journal.pone.0047913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baum J, Pinder M, Conway DJ. 2003. Erythrocyte invasion phenotypes of Plasmodium falciparum in The Gambia. Infect Immun 71:1856–1863. doi: 10.1128/IAI.71.4.1856-1863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Escobar N, Amambua-Ngwa A, Walther M, Okebe J, Ebonyi A, Conway DJ. 2010. Erythrocyte invasion and merozoite ligand gene expression in severe and mild Plasmodium falciparum malaria. J Infect Dis 201:444–452. doi: 10.1086/649902. [DOI] [PubMed] [Google Scholar]
- 22.Deans AM, Nery S, Conway DJ, Kai O, Marsh K, Rowe JA. 2007. Invasion pathways and malaria severity in Kenyan Plasmodium falciparum clinical isolates. Infect Immun 75:3014–3020. doi: 10.1128/IAI.00249-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awandare GA, Spadafora C, Moch JK, Dutta S, Haynes JD, Stoute JA. 2011. Plasmodium falciparum field isolates use complement receptor 1 (CR1) as a receptor for invasion of erythrocytes. Mol Biochem Parasitol 177:57–60. doi: 10.1016/j.molbiopara.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bei AK, Membi CD, Rayner JC, Mubi M, Ngasala B, Sultan AA, Premji Z, Duraisingh MT. 2007. Variant merozoite protein expression is associated with erythrocyte invasion phenotypes in Plasmodium falciparum isolates from Tanzania. Mol Biochem Parasitol 153:66–71. doi: 10.1016/j.molbiopara.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Jennings CV, Ahouidi AD, Zilversmit M, Bei AK, Rayner J, Sarr O, Ndir O, Wirth DF, Mboup S, Duraisingh MT. 2007. Molecular analysis of erythrocyte invasion in Plasmodium falciparum isolates from Senegal. Infect Immun 75:3531–3538. doi: 10.1128/IAI.00122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lantos PM, Ahouidi AD, Bei AK, Jennings CV, Sarr O, Ndir O, Wirth DF, Mboup S, Duraisingh MT. 2009. Erythrocyte invasion profiles are associated with a common invasion ligand polymorphism in Senegalese isolates of Plasmodium falciparum. Parasitology 136:1–9. doi: 10.1017/S0031182008005167. [DOI] [PubMed] [Google Scholar]
- 27.Theron M, Hesketh RL, Subramanian S, Rayner JC. 2010. An adaptable two-color flow cytometric assay to quantitate the invasion of erythrocytes by Plasmodium falciparum parasites. Cytometry A 77:1067–1074. doi: 10.1002/cyto.a.20972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blair PL, Witney A, Haynes JD, Moch JK, Carucci DJ, Adams JH. 2002. Transcripts of developmentally regulated Plasmodium falciparum genes quantified by real-time RT-PCR. Nucleic Acids Res 30:2224–2231. doi: 10.1093/nar/30.10.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nery S, Deans A-M, Mosobo M, Marsh K, Rowe JA, Conway DJ. 2006. Expression of Plasmodium falciparum genes involved in erythrocyte invasion varies among isolates cultured directly from patients. Mol Biochem Parasitol 149:208–215. doi: 10.1016/j.molbiopara.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balu B, Maher SP, Pance A, Chauhan C, Naumov AV, Andrews RM, Ellis PD, Khan SM, Lin JW, Janse CJ, Rayner JC, Adams JH. 2011. CCR4-associated factor 1 coordinates the expression of Plasmodium falciparum egress and invasion proteins. Eukaryot Cell 10:1257–1263. doi: 10.1128/EC.05099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Llinás M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. 2006. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res 34:1166–1173. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang L, Lopez-Barragan MJ, Jiang H, Mu J, Gaur D, Zhao K, Felsenfeld G, Miller LH. 2010. Epigenetic control of the variable expression of a Plasmodium falciparum receptor protein for erythrocyte invasion. Proc Natl Acad Sci U S A 107:2224–2229. doi: 10.1073/pnas.0913396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Comeaux CA, Coleman BI, Bei AK, Whitehurst N, Duraisingh MT. 2011. Functional analysis of epigenetic regulation of tandem RhopH1/clag genes reveals a role in Plasmodium falciparum growth. Mol Microbiol 80:378–390. doi: 10.1111/j.1365-2958.2011.07572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rovira-Graells N, Gupta AP, Planet E, Crowley VM, Mok S, Ribas de Pouplana L, Preiser PR, Bozdech Z, Cortes A. 2012. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res 22:925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mobegi VA, Loua KM, Ahouidi AD, Satoguina J, Nwakanma DC, Amambua-Ngwa A, Conway DJ. 2012. Population genetic structure of Plasmodium falciparum across a region of diverse endemicity in West Africa. Malar J 11:223. doi: 10.1186/1475-2875-11-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owusu-Agyei S, Asante KP, Adjuik M, Adjei G, Awini E, Adams M, Newton S, Dosoo D, Dery D, Agyeman-Budu A, Gyapong J, Greenwood B, Chandramohan D. 2009. Epidemiology of malaria in the forest-savanna transitional zone of Ghana. Malar J 8:220. doi: 10.1186/1475-2875-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akpogheneta OJ, Dunyo S, Pinder M, Conway DJ. 2010. Boosting antibody responses to Plasmodium falciparum merozoite antigens in children with highly seasonal exposure to infection. Parasite Immunol 32:296–304. doi: 10.1111/j.1365-3024.2009.01193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cook J, Kleinschmidt I, Schwabe C, Nseng G, Bousema T, Corran PH, Riley EM, Drakeley CJ. 2011. Serological markers suggest heterogeneity of effectiveness of malaria control interventions on Bioko Island, equatorial Guinea. PLoS One 6:e25137. doi: 10.1371/journal.pone.0025137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Duong S, Nguon C, Chuor CM, Saunders D, Se Y, Lon C, Fukuda MM, Amenga-Etego L, Hodgson AV, Asoala V, Imwong M, Takala-Harrison S, Nosten F, Su XZ, Ringwald P, Ariey F, Dolecek C, Hien TT, Boni MF, Thai CQ, Amambua-Ngwa A, Conway DJ, Djimde AA, Doumbo OK, Zongo I, Ouedraogo JB, Alcock D, Drury E, Auburn S, Koch O, Sanders M, Hubbart C, Maslen G, Ruano-Rubio V, Jyothi D, Miles A, O'Brien J, Gamble C, Oyola SO, et al. . 2013. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gething PW, Patil AP, Smith DL, Guerra CA, Elyazar IR, Johnston GL, Tatem AJ, Hay SI. 2011. A new world malaria map: Plasmodium falciparum endemicity in 2010. Malar J 10:378. doi: 10.1186/1475-2875-10-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.