Abstract
The enteric bacterium Proteus mirabilis is associated with a significant number of catheter-associated urinary tract infections (UTIs). Strict regulation of the antagonistic processes of adhesion and motility, mediated by fimbriae and flagella, respectively, is essential for disease progression. Previously, the transcriptional regulator MrpJ, which is encoded by the mrp fimbrial operon, has been shown to repress both swimming and swarming motility. Here we show that MrpJ affects an array of cellular processes beyond adherence and motility. Microarray analysis found that expression of mrpJ mimicking levels observed during UTIs leads to differential expression of 217 genes related to, among other functions, bacterial virulence, type VI secretion, and metabolism. We probed the molecular mechanism of transcriptional regulation by MrpJ using transcriptional reporters and chromatin immunoprecipitation (ChIP). Binding of MrpJ to two virulence-associated target gene promoters, the promoters of the flagellar master regulator flhDC and mrp itself, appears to be affected by the condensation state of the native chromosome, although both targets share a direct MrpJ binding site proximal to the transcriptional start. Furthermore, an mrpJ deletion mutant colonized the bladders of mice at significantly lower levels in a transurethral model of infection. Additionally, we observed that mrpJ is widely conserved in a collection of recent clinical isolates. Altogether, these findings support a role of MrpJ as a global regulator of P. mirabilis virulence.
INTRODUCTION
Bacterial pathogens utilize a myriad of complex regulatory networks to adapt gene expression in response to environmental cues encountered in the host. Trying to walk a fine balance of avoiding detection by the host immune system, while ensuring acquisition of all essential nutrients and promoting growth, bacteria employ transcriptional and posttranscriptional strategies to combat the host's defenses. A prominent example is the specific induction of iron and zinc uptake systems during infection, regulated by Fur and Zur, respectively, which allows pathogens to gain access to these essential transition metals in the restricted host environment (1, 2).
Urinary tract infections (UTIs) are among the most common bacterial infections (3), presenting a significant public health burden amounting to annual costs of about 3.5 billion dollars in the United States alone (4). Although Proteus mirabilis causes a relatively small proportion of UTIs in healthy individuals, it is a common cause of cystitis and pyelonephritis in individuals with indwelling catheters or anatomical abnormalities of the urinary tract (4, 5).
UTIs occur almost exclusively in an ascending route, meaning that bacteria of fecal origin gain entry to the bladder via the urethra and then spread to the kidneys via the ureters (4). Initially, bacteria adhere to host epithelial cells via proteinaceous supramolecular structures referred to as fimbriae (or pili) (6), which allow pathogens to withstand the mechanical flow of urine (5). Later in infection, bacteria can move into the kidneys using the force created by rotational movement of peritrichous flagella (7).
Flagellum-mediated motility and adhesion through fimbriae are inherently opposing processes that underlie tight regulation to allow a productive infection to take place. Sequencing of the first P. mirabilis genome in 2008 (8) revealed the presence of 17 potential chaperone-usher fimbriae. To date, biological evidence for the production of seven of these has been collected (9–16). Among these, the best studied is the mannose-resistant Proteus-like (MR/P) fimbria, which has been shown to be an important virulence factor that contributes to colonization of the urinary tract in a murine model of infection (17), as well as affecting the localization of bacteria within the bladder (18). In fact, all mutants defective in MR/P fimbria assembly tested thus far are attenuated for virulence in experimental UTIs (19–22). Expression of MR/P fimbriae undergoes phase variation: the mrp promoter contains an invertible element flanked by inverted repeat sequences, and the orientation of this element strictly regulates transcription of the fimbrial operon (20, 23). Transcribed at low levels in culture, the genes of the mrp operon show the largest fold induction of all P. mirabilis genes in vivo (24). Consequently, the major fimbrial subunit MrpA, as well as the tip adhesin MrpH, have been the subjects of multiple vaccine efforts (25–31). Recent work illustrates that mrpA is present in 96% of a collection of 48 clinical P. mirabilis isolates collected in 2012, as well as all seven sequenced genomes available at the time (13).
MrpJ, a transcriptional regulator encoded by the last gene of the MR/P fimbrial operon (32), has previously been shown to repress both swarming and swimming motility in P. mirabilis, permitting the bacteria to reciprocally control motility and adherence when mrp is expressed (32, 33). This helix-turn-helix (HTH) xenobiotic response element (XRE) family member interacts directly with the promoter of the flagellar master regulator flhDC (33). PapX, a functional homolog of MrpJ associated with P fimbriae in uropathogenic Escherichia coli (UPEC) similarly represses motility (34, 35). Recently, FmrA has been identified as a novel fimbria-associated regulator of motility in Shiga toxin-producing E. coli O157:H7 (36). Homologs of mrpJ have been reported in the Enterobacteriaceae species Photorhabdus temperata (37) and Xenorhabdus nematophila (38), although their function remains to be elucidated. BLAST searches reveal mrpJ homologs in at least 20 bacterial species. They are frequently associated with fimbrial operons in Proteus and Providencia spp., and P. mirabilis reference strain HI4320 carries 14 mrpJ paralogs in addition to mrpJ itself (33).
This study closely examines the role of MrpJ as an important regulator of motility and adherence in the uropathogen P. mirabilis. Transcriptomic data reveal a complex network of genes affected by mrpJ expression, many of which have previously been associated with virulence. We demonstrate that MrpJ has positive autoregulatory function using in vitro reporter assays that allow us to define an MrpJ-responsive fragment of the mrp promoter. Utilizing chromatin immunoprecipitation (ChIP), we extend our observations to the native state of the bacterial chromosome, confirming direct interactions of MrpJ with the mrp and flhDC promoters. Coupled with the attenuation of the mrpJ mutant observed in the murine UTI challenge in vivo, our results elucidate significant novel aspects of this important transcriptional regulator with regard to P. mirabilis virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. The New York University (NYU) clinical isolate collection and 10 nursing home isolates used for the mrpJ PCR survey have been previously described (13). All strains were grown at 37°C in low-salt nonswarming Luria broth (LB) (per liter, 10 g tryptone, 5 g yeast extract, 0.5 g NaCl) or on LB medium solidified with 1.5% agar. The following antibiotics were supplied at the indicated concentrations when necessary: ampicillin (100 μg/ml), gentamicin (15 μg/ml), and kanamycin (25 μg/ml). Transformants of P. mirabilis Φ(mrpAp-lacZ) operon fusion strains with pLX3607 or pLX3805 were selected on LB agar with 25 to 50 μg/ml ampicillin but subsequently maintained using 100 μg/ml ampicillin.
To construct the arabinose-inducible His6-tagged mrpJ vector used in chromatin immunoprecipitation (ChIP) experiments, a fragment containing mrpJ was PCR amplified from pLX2501 (32) using primers QE60Nco and QE60Hind. The resulting amplicon was digested with NcoI, which cuts at the 5′ insertion site for mrpJ coding sequence, and ligated into pBAD/Myc-HisA (Life Technologies) which had been linearized with NcoI and PmeI, resulting in plasmid pMP190.
All primer sequences are described in Table S2 in the supplemental material.
Identification of mrpJ in sequenced genomes and clinical isolates.
BLAST (39) was used to identify mrpJ in the 14 P. mirabilis genomes currently available in GenBank (strains HI4320, BB2000, PR03, C05028, ATCC 29906, ATCC 7002, WGLW4, WGLW6, Pm-Oxa48, and FDA_MicroDB strains 60, 67, 86, 87, and 91) (8, 40–43). PCR with mrpJ-specific primers was used to determine the presence of mrpJ in a clinical isolate collection (13). To determine whether mrpJ was part of the mrp operon in mrpA-negative strain NYU014, we used mrpH- and mrpJ-specific primers mrpH-seqF and mrpJ-RTR.
Microarrays.
The P. mirabilis HI4320 microarray has been described previously (44). To measure MrpJ-regulated gene transcription, P. mirabilis HI4320, HI4320ΔmrpJ, vector control HI4320(pLX3607), or mrpJ in vivo mimic HI4320(pLX3805) were cultured to logarithmic phase with aeration (optical density at 600 nm [OD600] of 0.7 to 0.9). Briefly, RNA was stabilized with RNAprotect (Qiagen) and isolated from cultures using the RNeasy minikit (Qiagen) according to the manufacturer's directions, with the exception of treating stabilized bacteria with 3 mg/ml lysozyme in Tris-EDTA (TE) buffer for 15 min. RNA was used as a cDNA template and labeled with either cyanine 3 or cyanine 5 dye as previously described (44) before microarray hybridization. Slides were scanned with a ScanArray Express microarray scanner (PerkinElmer) at 10-μm resolution, and data were analyzed using MeV software (v. 4.7.4; J. Craig Venter Institute). Four independent microarrays were analyzed for each condition.
qRT-PCR.
RNA from logarithmic-phase, aerated broth cultures of P. mirabilis (microarray validation) or aerated broth cultures of P. mirabilis with an OD600 of 2 (ChIP validation) was used as the template for cDNA synthesis using the Superscript first-strand synthesis system (Life Technologies) according to the manufacturer's protocol. Quantitative reverse transcriptase PCR (qRT-PCR) was performed as previously described (44), with two modifications: the 2× PCR master mix was either SsoAdvanced Universal SYBR green (Bio-Rad; microarray validation) or Maxima SYBR green (Thermo Scientific; ChIP validation), and the thermal cycler was a CFX Connect real-time system (Bio-Rad).
Determination of the transcriptional start site.
The 5′ rapid amplification of cDNA ends (5′ RACE) system (Life Technologies) was used to identify the transcriptional start site for the mrpABCDEFGHJ and flhDC operons according to the manufacturer's protocol. Amplicons were cloned, and full-length clones were sequenced. Four (mrp) or five (flh) identical clones were used to localize the transcriptional +1 site.
Construction of transcriptional fusions to lacZ.
lacZ was amplified from the operon fusion vector pRS415 using Q5 High-Fidelity polymerase (NEB) and cloned into XhoI/KpnI-HF-digested pUC18R6K-mini-Tn7T-Gm to form pNB022. Transformants were streaked on LB-gentamicin (LB plus gentamicin) plates supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml), and isolates showing the blue color indicative of LacZ activity were selected. Sequencing (GENEWIZ Inc.) confirmed that amplified regulatory elements upstream of lacZ were maintained without error.
5′ truncated promoter fragments of mrpA were generated by PCR from P. mirabilis HI4320 genomic DNA using a common reverse primer and forward primers that anneal to various distances upstream. The resulting fragments were cloned into the XhoI and NotI sites of pNB022. Forward primers NB75 to NB78 specifically anneal to the mrp promoter invertible element (IE) in the on direction. Promoter fragments that contain the full IE (amplified using forward primers NB65, NB66, and NB90) were confirmed to be in the on orientation. All plasmids were sequenced to verify the correct sequence of the cloned inserts (using primers NB52 and NB60).
A method by Choi et al. was adapted to construct single-copy operon fusions integrated into the HI4320 attTn7 locus downstream of glmS (45). Briefly, strain HI4320 with the IE locked in the OFF orientation (MR/P L-OFF) was transformed with 100 to 200 ng of plasmid carrying a Φ(mrpAp-lacZ) operon fusion and the transposase expression vector pTNS3. Cells were recovered in LB medium for 1 h postelectroporation. Gentamicin-resistant transformants were screened for successful integration at the glmS attTn7 site by colony PCR as described previously (45). A second PCR using primers specific to the secondary attTn7 site in carA (45) was performed at all times to ensure that no double integration event had taken place. Following purification of a single colony, chromosomal DNA was extracted from a liquid overnight culture using the QIAamp DNA minikit (Qiagen) according to the instructions of the manufacturer. Using primers annealing to the flanking region of the two attTn7 loci (JS72/Tn7rev and JS73/NB46), a second PCR was performed with Taq DNA polymerase (NEB). The cycle conditions for the second PCR were as follows: (i) 5 min at 95°C; (ii) 30 cycles, with 1 cycle consisting of 30 s at 94°C, 40 s at 61°C, and 8 min at 68°C; and (iii) 10 min at 72°C. Successful integration resulted in an observable size shift of the amplified fragment, corresponding to the length of the transformed mini-Tn7 derivative.
β-Galactosidase assay.
To determine the effect of MrpJ overproduction on the expression of Φ(mrpAp-lacZ) transcriptional fusions, saturated overnight cultures were diluted to a starting OD600 of approximately 0.04 in 5 ml LB supplied with ampicillin. The cultures were incubated in a shaker incubator at 37°C and 225 rpm until mid-logarithmic growth phase (OD600 of 0.7 to 0.9), when the cells were harvested by centrifugation and washed in 0.88% NaCl prior to enzymatic assays.
Bacteria were permeabilized by the addition of chloroform and sodium dodecyl sulfate, and LacZ activity was measured as described previously (46). All assays were performed at room temperature (approximately 21°C). Activities are expressed in arbitrary Miller units (47). Individual cultures were assessed in duplicate, and graphed values represent the averages from four biologically independent experiments and the corresponding standard deviations (SD).
Immunoblot analysis.
Protein samples were separated by SDS-PAGE. Western blot analysis was conducted using penta-His antibody (anti-His5; Qiagen) (1:4,000 dilution in phosphate-buffered saline [PBS] with 3% bovine serum albumin [BSA]) and peroxide-conjugated goat anti-mouse IgG (Bio-Rad) secondary antibodies. Western blots were developed using ECL Plus Western blotting detection system (Thermo Scientific) according to the manufacturer's protocol. Each gel was run in duplicate, followed by Coomassie blue staining to control for loading.
Bacterial ChIP and promoter walk.
The bacterial ChIP protocol was developed for P. mirabilis and was guided by published protocols (48–50) (see supplemental material for the detailed method). Briefly, P. mirabilis cells were cross-linked by formaldehyde, DNA-protein complexes were sheared by sonication, and His6-tagged MrpJ was immunoprecipitated with anti-His5 antibody. Next, the immunoprecipitated targets were stringently washed to remove nonspecific background, heat treated at 65°C to reverse cross-linking and column purified. PCR was used to analyze MrpJ-DNA interaction with mrpA and flhD upstream regulatory elements.
We designed a ChIP-PCR primer set to target a previously published positive electrophoretic mobility shift assay (EMSA) probe region (33) of the flhD promoter (fragment “j”) to confirm the successful immunoprecipitation (IP) of MrpJ-bound DNA complexes. A no-antibody bead-only control, an immunoprecipitation of a vector control and rpoA PCR as a nonspecific target gene control were included in each ChIP assay and in their downstream analysis. ChIP-PCR of MrpJ-bound DNA complexes was quantified using Image Studio software (LI-COR).
Mouse model of UTI.
Five- to 6-week-old female CBA/J mice (Jackson Laboratory) were transurethrally inoculated with P. mirabilis in accordance with NYU Langone Medical Center (NYULMC) IACUC-approved animal protocol number 140204. The infection procedure was carried out with some modifications of a published protocol (24, 51). Specifically, overnight cultures of wild-type P. mirabilis HI4320 and the isogenic mrpJ mutant (32) were adjusted to an estimated density of 2 × 108 CFU/ml (OD600 of 0.2). Mice were then infected with 50 μl of this culture (1 × 107 CFU) via a transurethral sterile polyethylene catheter attached to an infusion pump (Harvard Apparatus), after being anesthetized by intraperitoneal injection of anesthetic. Seven days postinfection, urine was collected by gentle abdominal massage, and then organs (bladders, kidneys, and spleens) were harvested aseptically, weighed, and homogenized in sterile 1× PBS. Bacterial titers were determined by plating serial dilutions of homogenates or urine samples on agar plates. The limit of detection was set at 200 CFU g−1 tissue for homogenate samples with undetectable colony numbers.
Statistical analysis.
Statistical significance of qRT-PCR data was calculated using a one-sample t test and a theoretical mean of 1.0. Significance of β-galactosidase activity and ChIP complex enrichment was calculated using Student's t test. The Mann-Whitney U test was used to analyze the results from animal experiments. Graphing and analysis were performed using Prism software (v. 6.0f; GraphPad).
Microarray data accession number.
The data from the microarray experiments are available at GEO accession number GSE63321.
RESULTS
MrpJ is present across diverse P. mirabilis isolates.
We recently reported that mrpA is highly conserved in a collection of 65 P. mirabilis strains, including 7 sequenced genomes, 10 nursing home urinary catheter isolates collected in the early 1980s, and 48 recent isolates from NYU Tisch Hospital (62/65 strains [95%]) (13). To determine whether mrpJ is similarly widespread, we examined the same strains by PCR or sequence analysis. We detected mrpJ in 63/65 (97%) of these strains. All strains that were positive for mrpA were also positive for mrpJ. Interestingly, the NYU014 strain was PCR negative for mrpA but positive for mrpJ; however, an amplicon that bridged mrpH and mrpJ was obtained. As this strain is also negative for MR/P-type hemagglutination (data not shown), its mrp operon is possibly cryptic. In addition, mrpJ is located at the end of the mrp operon in all 14 P. mirabilis genomes currently searchable by BLAST (see Materials and Methods). Alignment of these 14 sequences revealed synonymous mutations in 6/333 nucleotides (1.8%), and one nonsynonymous change (P to L) found only in FDA_MicroDB_91.
MrpJ regulates virulence factors in addition to flagella.
MrpJ has previously been shown to repress swimming and swarming motility by binding the promoter of the master regulator of flagella, flhDC (32, 33). Additional evidence suggested that MrpJ autoregulates the mrp operon: a ΔmrpJ mutant produces less MrpA protein (32). Furthermore, when viewed by transmission electron microscopy, P. mirabilis overexpressing mrpJ or mrpJ paralogs produce fimbriae with distinct appearances (33). To define the breadth of the MrpJ regulon, we used microarray analysis of P. mirabilis with various levels of mrpJ expression. Two sets of microarray experiments were conducted: wild-type strain HI4320 versus HI4320ΔmrpJ and strain HI4320 carrying an empty vector (pLX3607) versus HI4320 with mrpJ under an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter (pLX3805). We expressed mrpJ in trans because the highest levels of expression measured thus far for this gene occur during UTIs, and mrpJ is expressed only at low levels in vitro. The amount of mrpJ transcript in mid-logarithmic-phase culture resulting from uninduced pLX3805 (2,178-fold median increase versus vector control) is comparable to the amount detected in the urine of mice experimentally infected with P. mirabilis 1 day postinfection (1,103-fold compared to in vitro broth control), and therefore, this strain may be considered an in vivo mimic for mrpJ (24).
We found many genes were differentially regulated at least 2-fold by MrpJ (Fig. 1). Specifically, during expression of mrpJ at levels comparable to those during infection, 70 genes were induced and 147 genes were repressed compared to the vector control (Tables 1 and 2, with complete results in Tables S3 and S4 in the supplemental material). In the ΔmrpJ strain, 13 genes were induced and 56 genes were repressed compared to the wild type (Tables S5 and S6). The amount of regulation in the ΔmrpJ strain compared to the wild type was relatively modest with the exception of mrpA, which was repressed 146.93-fold. This was not surprising, given that mrpJ is not expressed well in vitro (32).
FIG 1.
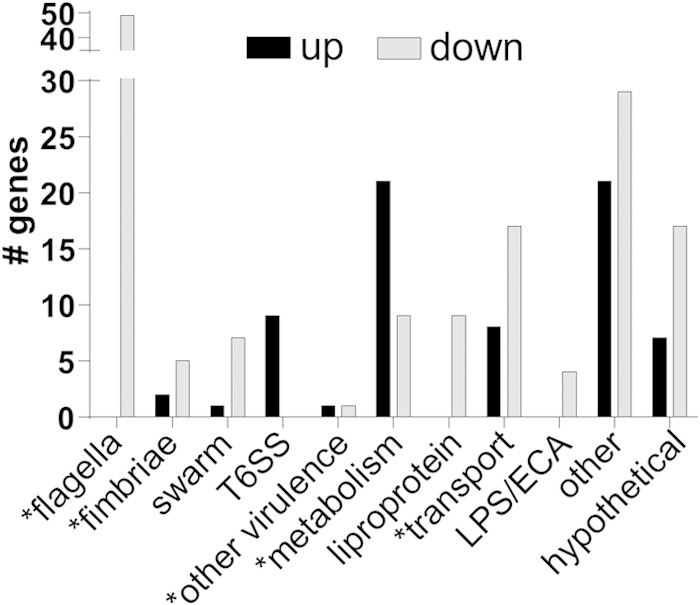
Categories of genes regulated by MrpJ. A vector control was compared to mrpJ expressed in trans at levels comparable to those during experimental UTIs, and differential transcription was measured by microarray. Gene classes that have previously been shown to contribute to P. mirabilis pathogenesis are indicated by an asterisk. ECA, enterobacterial common antigen.
TABLE 1.
Top 50 genes repressed in mrpJ overexpression strain compared to the vector control
| ORFa | Gene | Annotation | Fold change |
|---|---|---|---|
| PMI1961 | ccm | Putative membrane protein (Ccm1 protein) | −630.29 |
| PMI1635 | fliK | Flagellar hook length control protein | −509.87 |
| PMI1652 | flgD | Basal body rod modification protein | −469.29 |
| PMI1631 | fliG | Flagellar motor switch protein | −467.90 |
| PMI1647 | flgI | Flagellar P-ring protein precursor (basal body P-ring protein) | −466.93 |
| PMI1637 | fliM | Flagellar motor switch protein | −450.58 |
| PMI1639 | fliO | Flagellar protein | −441.62 |
| PMI1670 | motA | Chemotaxis protein (motility protein A) | −435.30 |
| PMI1632 | fliH | Flagellar assembly protein | −398.84 |
| PMI1660 | flhB | Flagellar biosynthetic protein | −341.98 |
| PMI1654 | flgB | Flagellar basal body rod protein | −329.89 |
| PMI1618 | fliA | RNA polymerase sigma factor for flagellar operon | −320.62 |
| PMI1659 | flhA | Flagellar biosynthesis protein | −307.03 |
| PMI1636 | fliL | Flagellar protein FliL | −302.51 |
| PMI1648 | flgH | Flagellar L-ring protein precursor | −297.81 |
| PMI1630 | fliF | Flagellar M-ring protein | −286.30 |
| PMI1638 | fliN | Flagellar motor switch protein | −279.07 |
| PMI1634 | fliJ | Flagellar protein FliJ | −276.57 |
| PMI1629 | fliE | Flagellar hook-basal body complex protein | −261.77 |
| PMI1623 | fliT | Flagellar protein | −242.66 |
| PMI1469 | fim8A | Fimbrial subunit | −234.31 |
| PMI3460 | Putative lipoprotein | −227.61 | |
| PMI3115 | umoA | Putative upregulator of flagellar operon | −222.72 |
| PMI1633 | fliI | Flagellum-specific ATP synthase | −192.53 |
| PMI1669 | motB | Chemotaxis protein (motility protein B) | −174.67 |
| PMI1617 | fliZ | FliZ protein | −161.68 |
| PMI1655 | flgA | Flagellar basal body P-ring formation protein | −152.90 |
| PMI1640 | fliP | Flagellar biosynthetic protein | −140.09 |
| PMI1621 | flaD; fliD | Flagellar hook-associated protein 2 | −127.34 |
| PMI1653 | flgC | Flagellar basal body rod protein | −122.76 |
| PMI1650 | flgF | Flagellar basal body rod protein | −98.71 |
| PMI0833 | Putative exported protein | −95.10 | |
| PMI2421 | Putative phospholipid-binding protein | −78.61 | |
| PMI0599 | modC | Molybdenum ABC transporter, ATP-binding protein | −76.09 |
| PMI1622 | fliS | Flagellar protein FliS | −70.74 |
| PMI0182 | Putative transcriptional regulator (MrpJ homolog) | −70.05 | |
| PMI1656 | flgM | Negative regulator of flagellin synthesis (anti-sigma-28 factor) | −66.60 |
| PMI1668 | cheA | Chemotaxis protein | −62.85 |
| PMI1645 | flgK | Flagellar hook-associated protein 1 | −62.65 |
| PMI1642 | fliR | Flagellar biosynthetic protein | −62.63 |
| PMI1649 | flgG | Flagellar basal body rod protein (distal rod protein) | −61.59 |
| PMI1540 | Putative transcriptional regulator | −56.75 | |
| PMI1663 | cheB | Chemotaxis response regulator protein-glutamate methylesterase | −53.46 |
| PMI1646 | flgJ | Peptidoglycan hydrolase (muramidase) | −46.92 |
| PMI1651 | flgE | Flagellar hook protein | −46.64 |
| PMI1666 | cheD | Methyl-accepting chemotaxis protein | −41.63 |
| PMI2808 | Methyl-accepting chemotaxis protein | −37.69 | |
| PMI1470 | fim8J | Fimbrial operon regulator | −37.36 |
| PMI1665 | tap | Methyl-accepting chemotaxis protein | −35.58 |
| PMI1664 | cheR | Chemotaxis protein methyltransferase | −31.00 |
ORF, open reading frame.
TABLE 2.
Top 50 genes induced in mrpJ overexpression strain compared to vector control
| ORF | Gene | Annotationa | Fold change |
|---|---|---|---|
| PMI0271 | mrpJ | Fimbrial operon regulator | 2,178.32 |
| PMI0748 | Type VI secretion system protein | 23.45 | |
| PMI0747 | Type VI secretion system protein | 20.49 | |
| PMI0347 | lysA | Diaminopimelate decarboxylase | 15.29 |
| PMI3238 | argB | Acetylglutamate kinase | 13.95 |
| PMI0749 | Type VI secretion system protein | 12.81 | |
| PMI0263 | mrpA | Major mannose-resistant/Proteus-like fimbrial protein | 12.28 |
| PMI2157 | fruK | 1-Phosphofructokinase | 12.18 |
| PMI1079 | Conserved hypothetical protein | 9.43 | |
| PMI1702 | Putative membrane protein | 8.92 | |
| PMI2292 | ptsG | PTS system, glucose-specific IIBC component | 8.70 |
| PMI0287 | Putative amidohydrolase/metallopeptidase | 8.01 | |
| PMI2528 | hyfA | Hydrogenase 4 component A | 6.21 |
| PMI2755 | lysC | Lysine-sensitive aspartokinase III | 5.68 |
| PMI2307 | argA | Amino acid acetyltransferase | 5.22 |
| PMI0744 | Putative phosphopeptide-binding protein; type VI secretion | 5.15 | |
| PMI0742 | Type VI secretion system protein | 5.12 | |
| PMI2660 | caiF | Transcriptional activator for carnitine metabolism | 4.93 |
| PMI2825 | def | Peptide deformylase | 4.92 |
| PMI0746 | Type VI secretion system protein | 4.84 | |
| PMI2523 | hyfF | Hydrogenase 4 component F | 4.66 |
| PMI3577 | fdhF | Formate dehydrogenase H, selenopolypeptide subunit | 4.30 |
| PMI0741 | Type VI secretion system protein | 4.18 | |
| PMI2821 | argD | Acetylornithine/succinyldiaminopimelate aminotransferase | 3.87 |
| PMI1777 | PTS system IIA component | 3.60 | |
| PMI2873 | Putative acetyltransferase | 3.56 | |
| PMI0667 | Putative exported protein | 3.31 | |
| PMI3225 | Putative membrane protein | 3.19 | |
| PMI3581 | hydN | Electron transport protein | 3.17 |
| PMI3457 | argI | Ornithine carbamoyltransferase chain I | 3.12 |
| PMI3553 | trkH | Trk system potassium uptake protein | 3.12 |
| PMI2769 | purD | Phosphoribosylamine-glycine ligase | 3.01 |
| PMI3125 | hipA | Putative regulatory protein | 3.00 |
| PMI2288 | dapD | 2,3,4,5-Tetrahydropyridine-2-carboxylate N-succinyltransferase | 2.96 |
| PMI2661 | fsaA | Fructose-6-phosphate aldolase | 2.90 |
| PMI0637 | moeA | Molybdopterin biosynthesis protein | 2.82 |
| PMI1263 | Putative membrane protein | 2.82 | |
| PMI0967 | Phage protein | 2.66 | |
| PMI0030 | exbD | Biopolymer transport protein | 2.57 |
| PMI2956 | chbB; celA | N,N′-diacetylchitobiose-specific PTS system, EIIb component | 2.55 |
| PMI0244 | Putative membrane protein | 2.53 | |
| PMI3698 | Putative membrane protein | 2.44 | |
| PMI2761 | eda | KHG/KDPG aldolase | 2.39 |
| PMI1023 | Putative kinase | 2.33 | |
| PMI0825 | ndpA | Nucleoid-asociated protein | 2.33 |
| PMI1313 | pyrF | Orotidine-5′-phosphate decarboxylase | 2.32 |
| PMI3141 | Putative restriction endonuclease | 2.29 | |
| PMI2360 | Adenylate cyclase-like protein | 2.29 | |
| PMI2168 | Probable short-chain dehydrogenase | 2.27 | |
| PMI1405 | pykF | Pyruvate kinase | 2.27 |
PTS, phosphotransferase; KHG, 2-keto-4-hydroxyglutarate; KDPG, 2-keto-3-deoxy-6-phosphogluconate.
As expected in the presence of mrpJ, flagellar genes were repressed: these genes included flhDC (−5.16- and −6.25-fold, respectively), which encodes the master regulator of flagella. In addition, swarming regulators umoA and umoB were repressed by MrpJ (−222- and −2.68-fold, respectively), while lrhA was induced (2.21-fold) (44, 52, 53), indicating that MrpJ not only represses motility but also interferes with swarmer cell differentiation. The cell shape-determining gene ccm, also implicated in swarming behavior (54), was the most repressed gene in the in vivo mimic strain (630-fold). The polyamine putrescine has also been found to contribute to swarming (55), and we found MrpJ repression of speF (−3.75-fold), which catalyzes the production of putrescine from ornithine.
Also consistent with previously published evidence that suggested an altered fimbrial profile when mrpJ was overexpressed (33), we found that several fimbrial operons were regulated by mrpJ: pmfA, fim8A, and fim14A were repressed, while the mrp operon itself was induced (−3.14-, −234-, −20.5-, and 12.3-fold, respectively). Three of these fimbriae, MR/P, PMF (P. mirabilis fimbria), and Fim14, have previously been shown to contribute to virulence in a mouse model of UTI (15, 19–22, 56, 57). In addition to regulation of anticipated systems, we found that MrpJ regulates other established virulence genes encoding the Pta toxin (2.25-fold) and ZapA protease (−14.3-fold) (58, 59). Furthermore, MrpJ regulated genes that could be associated with virulence or immune evasion, such as lipopolysaccharide (LPS) modifications (pagP, lpxK, and msbB; −5.01-, −2.21-, and −2.94-fold, respectively). A type VI secretion system (T6SS), including effector genes idsA and PMI0750 as well as seven genes in the operon which encodes the structural proteins of the T6SS (PMI0749-0734) (60–62), was induced by MrpJ. Thus, MrpJ regulates a variety of genes that are known or hypothesized to contribute to pathogenesis. These data, combined with the fact that mrpJ is strongly induced during experimental UTI, suggest that MrpJ is a master regulator of virulence.
Microarray data were validated by measuring transcript levels of selected genes by qRT-PCR (Fig. 2A; see Fig. S1 in the supplemental material). Even though the observed fold changes in the ΔmrpJ mutant were very modest, gene expression was generally the inverse of the in vivo mimic strain. For example, mrpA was induced by mrpJ, and flagellum- or swarming-related genes were repressed, while in the ΔmrpJ strain, transcript levels of mrpA were downregulated and those of motility-associated genes increased. Of the 15 mrpJ paralogs encoded by P. mirabilis genes, ucaJ is among the strongest in its repression of motility (33); therefore, we also tested the effect of ucaJ overexpression on mrpJ-regulated genes (Fig. 2B). As expected, flagellum-related genes were approximately 2-fold more repressed by ucaJ than by mrpJ. Interestingly, ucaJ did not induce the T6SS, and regulation of fimbrial and other virulence-associated genes varied in both direction and magnitude compared to that of mrpJ.
FIG 2.
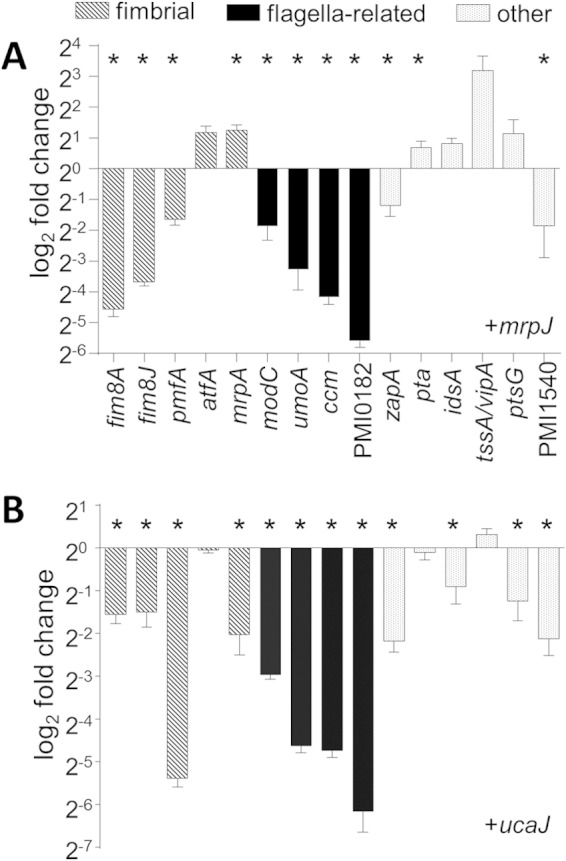
qRT-PCR validation of selected microarray data. (A) Gene expression by the mrpJ in vivo mimic strain compared to the vector control. (B) Gene expression by a ucaJ overexpression strain. Data are the results of three independent experiments, normalized to rpoA, and error bars show standard errors of the means (SEM). Statistically significant differences compared to vector control (P < 0.05) as calculated by Student's unpaired t test are indicated by an asterisk.
Promoter deletion analysis identifies an MrpJ-responsive region in the mrpA promoter.
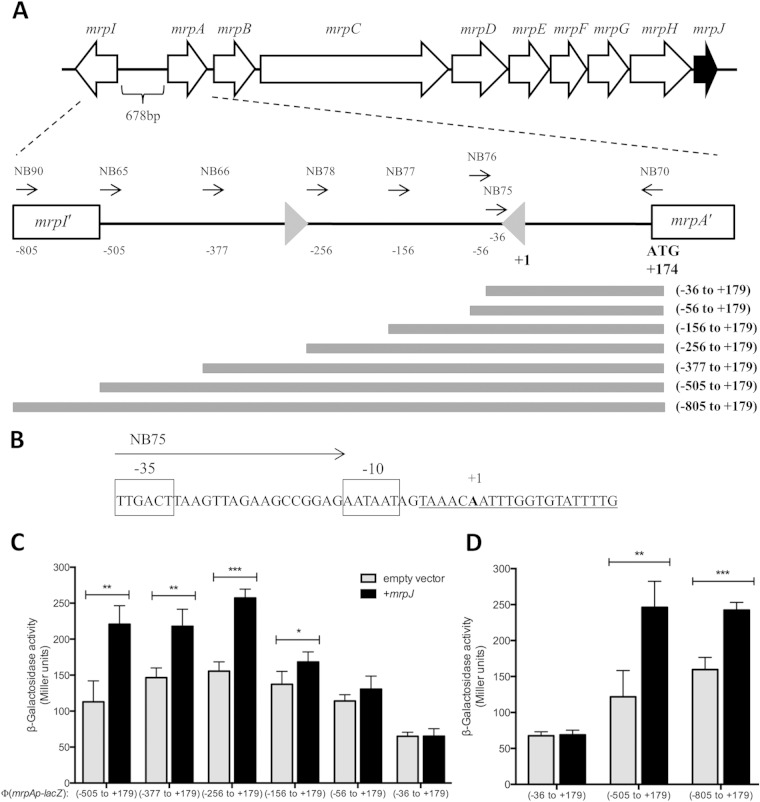
Following our transcriptomic identification of mrpJ targets, we sought to deepen our mechanistic understanding of transcriptional regulation by MrpJ. To start to address this question, we determined the promoter fragment required for mrpJ-dependent induction of the mrp operon. Expression of MR/P fimbriae is phase variable in response to environmental stimuli by means of an invertible element (23, 63) (Fig. 3A). 5′ RACE mapped the transcriptional start of mrpA to the adenine residue located 173 residues upstream of the predicted ATG start codon. This residue is located within the inverted repeat closest to mrpA. Visual inspection of the sequence immediately upstream of the start of transcription revealed the presence of putative −10 and −35 regions separated by a 17-bp spacer, both of which closely match the consensus sequence of a bacterial σ70 promoter (64) (Fig. 3B), when the invertible element is in the ON position, allowing transcription of the mrp operon. Of note, a previously reported canonical σ70 promoter sequence facing mrpA in the ON orientation (23) could not be located on the sequenced genome of strain HI4320.
FIG 3.
MrpJ-dependent activation of Φ(mrpAp-lacZ) expression. (A) Schematic showing the mrp gene locus, including mrpI and the fimbrial operon mrpABCDEFGHJ. The intergenic region contains an invertible element flanked by inverted repeats (gray triangles) here depicted in the ON orientation, resulting in MR/P fimbria production. The numbers are the 5′ positions of Φ(mrpAp-lacZ) operon fusions relative to the transcriptional start site of mrpA (+1), which was determined by 5′ RACE. Gray bars visualize Φ(mrpAp-lacZ) operon fusions with respect to the included promoter elements. (B) Nucleotide sequence surrounding the transcriptional start site of mrpA. The inverted repeat sequence is underlined; the transcriptional start site (+1) is indicated by bold type. Putative −10 and −35 regions are boxed and labeled. (C and D) Effect of promoter truncation on mrpJ-dependent activation of Φ(mrpAp-lacZ) expression in mid-logarithmic growth phase. Single-copy fusions to lacZ include specified amounts of DNA upstream of the predicted MrpA start codon. Strains were transformed with empty vector (pLX3607) or mrpJ expression plasmid (pLX3805; black bars). β-Galactosidase activity was determined as described in Materials and Methods. Values represent the means of four independent experiments plus standard deviations (SD) (error bars). Statistically significant differences comparing presence or absence of mrpJ as calculated by Student's unpaired t test are indicated by bars and asterisks as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To identify the promoter region necessary for MrpJ-dependent activation of mrpA expression, we constructed a set of single-copy Φ(mrpA-lacZ) operon fusion strains. All lacZ fusion strains contained different lengths of DNA upstream of mrpA (Fig. 3A) integrated at the attTn7 chromosomal locus downstream of glmS. The reverse primer was chosen to anneal to the beginning of the mrpA coding sequence to ensure all relevant regulatory elements are included in our transcriptional fusions. During aerobic growth, P. mirabilis exists as a mixed population of bacteria expressing MR/P fimbriae (promoter in the ON position) and those that do not (invertible element OFF). To avoid complication of our results by variation of mrpJ expression from its native promoter, we utilized a previously described insertion-disruption mutant in mrpI (32), which has the invertible element in the OFF orientation (L-OFF). MrpI is the only recombinase known to control phase variation of MR/P fimbriae (20). Hence, this mutation irreversibly locks the invertible element in the OFF position, thereby eliminating production of MR/P fimbriae. All strains were cultured to replicate the experimental conditions of the transcriptomic analysis.
An operon fusion comprising the whole intergenic region of mrpI and mrpA to lacZ (nucleotides −505 to +179) responded to mrpJ expression in trans, with β-galactosidase levels increasing approximately 2-fold compared to an empty vector control (Fig. 3C). Activities of two other constructs containing shorter fragments of the intergenic region (from −377 to +179 and from −256 to +179) showed induction levels similar to that of the full-length intergenic region, suggesting that these 249 bp do not contribute to MrpJ-dependent activation of Φ(mrpAp-lacZ) expression. The highest overall activity under Φ(mrpAp-lacZ)-inducing conditions (+mrpJ) was measured for the construct from −256 to +179. Induction of the construct from −156 to +179 by MrpJ was significantly reduced and completely abolished upon further deletion of an additional 100 nucleotides (−56 to +179). These results imply that the region between 256 and 156 bp upstream of the transcriptional start of mrpA is required for MrpJ-mediated induction of Φ(mrpAp-lacZ) expression. The small, albeit statistically significant, response to MrpJ production by the fragment from −156 to +179 may suggest the presence of a partial MrpJ binding site, although the biological significance of this result has yet to be established.
The shortest promoter fragment tested (nucleotides −36 to +179) showed low levels of β-galactosidase activity unaffected by mrpJ overexpression (Fig. 3C). This construct contains the predicted −35 and −10 region, resulting in basal expression of the Φ(mrpAp-lacZ) fusion. Including an additional 20 nucleotides upstream (−56 to +179) results in a marked increase in transcription independent of MrpJ production, demonstrating that MrpJ is most likely not the only factor activating mrpA transcription.
Validation of epitope-tagged MrpJ for use in ChIP assay.
To further dissect mechanistic aspects of MrpJ-mediated transcriptional regulation, we performed ChIP analysis to define MrpJ-DNA interactions in vivo in the native state of the bacterial chromosome. In order to conduct these experiments, we validated an expression system that was compatible with ChIP. MrpJ with a C-terminal His6 tag under the tight regulation of an arabinose-inducible promoter was expressed in trans (pMP190) in P. mirabilis HI4320 to match the mrpJ expression level to the in vivo level found during the course of UTIs in mice (24).
To confirm that the antibody for our ChIP assay recognized MrpJ protein without significant cross-reactivity, immunoblot analysis was performed on arabinose-induced pMP190 and vector control lysates harvested at an approximate OD600 of 2. As shown in Fig. 4A, probing of His6-tagged MrpJ by this anti-His5 antibody is highly specific. To verify even loading, samples were also separated on a parallel gel, and the proteins were visualized using Coomassie blue staining (see Fig. S2A in the supplemental material). Figure 4B shows the transcript level of mrpJ from pMP190 (pMrpJ) with or without arabinose induction. Quantitative RT-PCR was performed to measure mrpJ expression compared to the vector control at an OD600 of 2 (0.002% arabinose induction at an OD600 of 0.5), normalized to the level of rpoA. The induced sample [pMrpJ(I)] had 2,251-fold-more expression of mrpJ compared to the uninduced sample, similar to the level found during the progression of murine UTI (1 day postinfection, 1,103-fold induced over in vitro control [24]).
FIG 4.
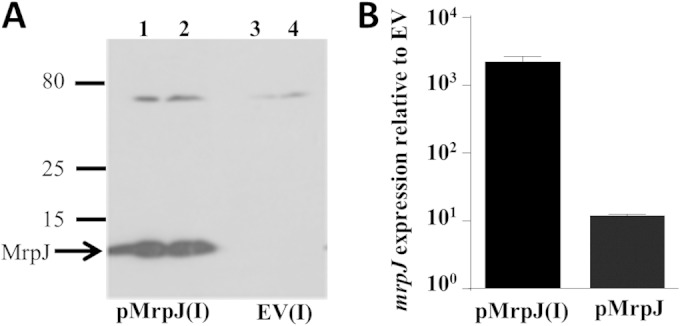
Validation of mrpJ expression system used in ChIP assay. (A) Levels of MrpJ analyzed by Western blotting. The gel was loaded with 5 μl (lanes 1 and 3) or 10 μl (lanes 2 and 4) of pMrpJ or empty vector (EV) lysate, respectively, induced (I) by 0.002% arabinose. The positions of size markers (in kilodaltons) are shown to the left of the gel. (B) mrpJ transcript level in the induced (I) or uninduced pMrpJ strain compared to EV by qRT-PCR; error bars indicate SEM of replicates, and data are from two independent experiments.
No significant difference in the growth pattern was found when growth curve analysis of P. mirabilis HI4320/pMP190 (pMrpJ) and the vector control (empty vector) with and without an arabinose induction (0.002%) was performed (see Fig. S2B and S2D in the supplemental material). Both strains were subcultured in 1:100 dilutions from an overnight aerated culture and allowed to reach stationary phase, a native state for mrp fimbrial operon induction (17). Serial dilution of this culture (OD600 of 2) resulted in no obvious distinction in CFU.
The length of the sonicated fragment of the cross-linked protein-DNA complexes is a major determinant of a successful ChIP experiment (65). For this study, we sheared the formaldehyde-cross-linked MrpJ-DNA complexes into a final range of 200- to 650-bp fragments as shown in the representative sonication profile in Fig. S2C in the supplemental material.
ChIP shows in vivo binding of the mrp promoter by MrpJ.
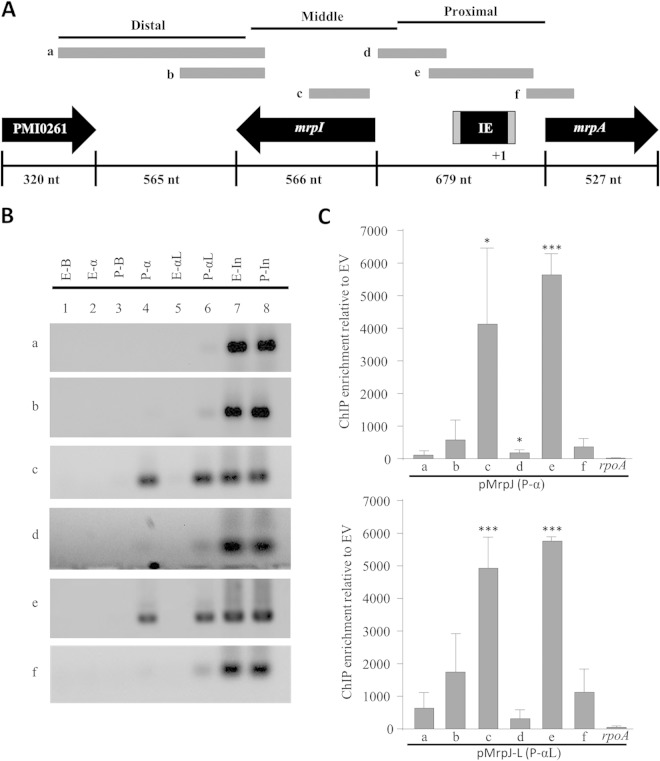
An approximately 2-kb region upstream from the translational start site of mrpA was analyzed for in vivo MrpJ binding by ChIP-PCR. Figure 5A shows a schematic organization of this upstream regulatory sequence, including the invertible element (IE) region and the mrpI gene required for ON/OFF switching of the promoter orientation. The entire region, starting with mrpA and going up to PMI0261, has been divided into base pair length landmarks for easy reference. A series of primers was designed to walk toward the translational start of mrpA in graduated steps of PCR amplicons. Amplified fragments were designated as proximal, middle, and distal based on their relative proximity to the start site. Figure 5B shows the results for one representative ChIP-PCR gel analysis, and Fig. 5C shows the average values from three independent experiments for the mrp promoter.
FIG 5.
ChIP-PCR analysis of mrpA upstream promoter element. (A) Schematic representation of mrpA genomic organization depicting fragments “a” to “f” tested by ChIP-PCR as gray bars. The invertible element (IE) is not drawn to scale. nt, nucleotides. (B) Agarose gel analysis of mrpA ChIP-PCR. Lanes 1 to 8 are as follows: E-B, empty vector bead control; E-α, empty vector His5-IP; P-B, MrpJ plasmid bead control; P-α, MrpJ plasmid His5-IP with antibody-coupled beads; E-αL, empty vector His5-IP using antibody-coupled lysate; P-αL, MrpJ plasmid His5-IP using antibody-coupled lysate; E-In, 10% pre-IP empty vector input; P-In, 10% pre-IP MrpJ plasmid input. The results of one experiment are shown. We performed three experiments with similar results. (C) Image Studio analysis of ChIP-PCR data. MrpJ-His6 ChIP signal enrichment is shown relative to the EV signal (comparing data from panel B [lanes 4 versus 2 or lanes 6 versus 5]). The means of three independent ChIP experiments are shown in the graphs; error bars represent standard deviations. Statistically significant differences compared to the rpoA enrichment value as calculated by Student's unpaired t test are indicated by asterisks as follows: *, P ≤ 0.05; ***, P ≤ 0.001.
Two primer sets were used to probe for the distal 5′ element relative to the start site; of these, the set encompassing the larger amplicon, fragment “a” (661 bp), showed little signal in all three of our ChIP assays (Fig. 5B and C). The ChIP signal increased slightly with the shorter amplicon, fragment “b” (443 bp), which had the forward primer nested 195 bp downstream, indicating a potential direct binding of MrpJ in the vicinity. Indeed, as we PCR walked into the mrpI intragenic region with fragment “c” (250 bp), the ChIP signal became highly pronounced, confirming in vivo MrpJ-DNA complex formation in this site. The approximate total length of the area flanked by fragments “b” and “c” (with a 236-bp gap in between) is about 929 bp. On the basis of our 200- to 650-bp sonicated DNA fragment range in each ChIP experiment, we believe that the gradually increasing ChIP signals from fragments “b” to “c” stem from a single MrpJ nucleoprotein complex and that the minor residual population of larger sonicated fragments, as shown in Fig. S2C in the supplemental material, could account for the slight IP signal for fragment “b.” ChIP signal was completely lost as we PCR walked further into the mrpI proximal putative promoter region, as shown by fragment “d,” further supporting the specificity of MrpJ-DNA complex formation in the mrpI intragenic region.
The 679-bp intergenic region between mrpI and mrpA contains the invertible element. As described earlier, our β-galactosidase reporter assays have shown a consistently strong transcriptional activation of mrpA with the construct from nucleotides −256 to +179 and longer promoter elements (Fig. 3), which include the IE region in its ON orientation. We extended our ChIP-PCR walk into the proximal promoter region of mrpA with fragment “e” (300 bp), which encases the IE and its flanking inverted repeat sequences. Consistent with the reporter assay data, we found the strongest in vivo MrpJ binding in this region, suggesting that the MrpJ-dependent activation of mrpA in the transcriptional reporter strains is a direct result of nucleoprotein complex formation and feedback regulation by MrpJ on this site. ChIP signal was lost as we PCR walked further downstream of the intergenic region between mrpI and mrpA and with the inclusion of mrpA intragenic sequence fragment “f” (199 bp). Robust PCR amplicons in the input samples showed that the low or negligible ChIP-PCR signal with some of our primer sets is not due to inefficient PCRs; rather, they support the specificity of the positive ChIP signals.
Our data suggest that MrpJ directly autoregulates the mrp operon in vivo and that there are two direct MrpJ interaction regions on the mrpA promoter; the distal site is located in the intragenic region of mrpI, and the proximal one is located within the invertible element. To test whether the mrpI site contributes to MrpJ regulation of mrpA, we examined this new site using the transcriptional reporter assay; including the distal binding site within mrpI (−805 to +179) did not further increase MrpJ-dependent expression of Φ(mrpAp-lacZ) (Fig. 3D). Indeed, the fold change measured for the promoter fragment including mrpI was almost identical to that observed for the transcriptional fusion restricted to the full intragenic region between mrpI and mrpA.
ChIP demonstrates in vivo MrpJ-DNA complex formation on the flhDC promoter.
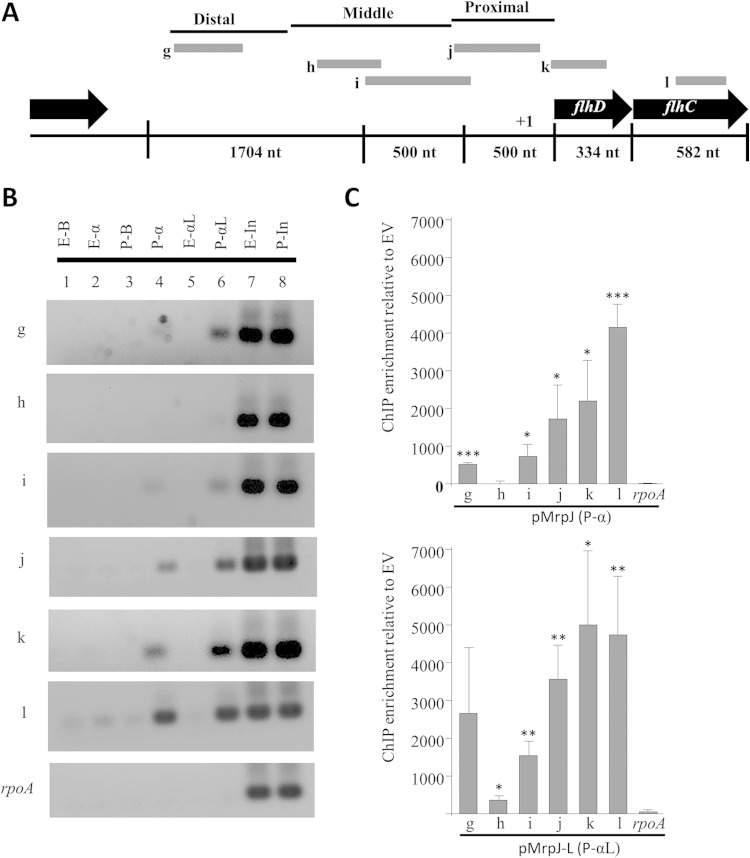
Next, we extended our ChIP-PCR analysis to the flhDC upstream promoter element, an operon previously shown to be repressed by MrpJ (32, 33). Figure 6A shows a schematic diagram of the genomic organization of this region, specifically depicting the regions amplified for our analysis. The transcriptional start we identified for flhD by 5′ RACE is in agreement with an earlier reported putative start site in another strain of P. mirabilis (66). The most distal fragment, fragment “g” (373 bp), showed positive MrpJ binding signal, especially in the antibody-coupled lysate IP for anti-His5-MrpJ. While first conjugating the antibody with the beads prior to the addition of lysate (P-α) could yield lower IP background, coupling the lysate with antibody before bead addition (P-αL) augments signal enrichment. We found that immunoprecipitation of MrpJ-His6 using antibody-coupled lysate (P-αL) provided greater efficiency than IP of MrpJ-His6 using antibody-coupled beads. The ChIP signal was reproducibly lost when we PCR walked further downstream (fragment “h” [322 bp]) into the middle section but started to enhance with the mid-proximal transition amplicon, fragment “i” (241 bp). The signal was further pronounced with the proximal “j” fragment (356 bp), corresponding to the previously reported EMSA probe for MrpJ binding in vitro. Based on our 5′ RACE data, this region also carries the transcriptional start site; this set was used as a positive control in all of our ChIP assays. The length of DNA flanked by fragments “i” and “j” is 597 bp. On the basis of the sonication fragment range maintained in our ChIP assays (200 to 650 bp), we believe the gradual increase in binding signal from these two sets (fragments “i” and “j”) originates from one MrpJ binding event. To our surprise, a fragment primarily within flhD coding sequence (fragment “k” [283 bp]) placed 183 bp downstream of fragment “j” provided a very strong positive ChIP signal in all three of our independent experiments. This was intriguing to us, as based on the earlier in vitro EMSA data, we did not expect to see flhD intragenic MrpJ binding signal. Although the total linear length of DNA encompassing fragments “j” and “k” is 822 bp, larger than the size range of our sonication fragments (200 to 650 bp), a slim chance remains for intermediate length fragments carrying the “k” amplicon on their 3′ end and the MrpJ binding region on their 5′ side. It is also possible that MrpJ forms more than one viable nucleoprotein complex on the proximal and internal regions of flhDC. To evaluate whether this is occurring, flhC intragenic PCR was performed, and a strong ChIP signal was found in that region as well (fragment “l”). The proximal promoter region and the flhD gene are potentially part of a more complex conformation than their usually perceived linear structure. This would give rise to a comparatively condensed DNA before cross-linking and sonication, generating a much longer DNA fragment after the reversal of cross-linking even if only one DNA-protein interaction had happened in vivo (67–70).
FIG 6.
ChIP-PCR analysis of flhDC upstream promoter element and intragenic region. (A) Schematic representation of flhDC genomic organization depicting the fragments “g” to “l” tested by ChIP-PCR as gray bars. (B) Agarose gel analysis of flhDC ChIP-PCR. Lane designations are the same as in Fig. 5B. The results of one representative experiment are shown (three experiments were performed). (C) Image Studio analysis of flhDC ChIP-PCR data. MrpJ-His6 ChIP signal enrichment is shown relative to the EV signal. The averages of three independent ChIP experiments are shown; error bars represent standard deviation. Statistically significant differences compared to the rpoA enrichment value as calculated by Student's unpaired t test are indicated by asterisks as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
MrpJ modulates P. mirabilis pathogenesis in an in vivo mouse UTI model.
Previously, an mrpJ mutant was found to be outcompeted by wild-type strain HI4320 in a mouse coinfection UTI model (32). Our transcriptional analyses showed a diverse virulence gene network under MrpJ, expanding its contribution beyond flagellar and mrp fimbrial regulation. Genes modulating such a broad virulence network may have a greater effect on a pathogen's fitness, and we hypothesized that deletion of mrpJ would lead to a profound decrease in virulence even when the mutant was not in direct competition with the wild-type parent strain. Hence, to address whether mrpJ deficiency can directly affect P. mirabilis virulence within the urinary tract, we challenged mice with monocultures of P. mirabilis isolate HI4320 or the isogenic ΔmrpJ mutant using a murine UTI model. As shown in Fig. 7, bladder colonization was highly compromised in the absence of functional MrpJ (approximately 10,000-fold; P = 0.0079), whereas kidney samples show a comparatively moderate effect. As previously reported (32), there was no significant deviation in the in vitro growth profile comparison between the mrpJ mutant and wild-type HI4320, indicating that MrpJ is involved in the regulation of the P. mirabilis pathogenic lifestyle in the lower urinary tract.
FIG 7.
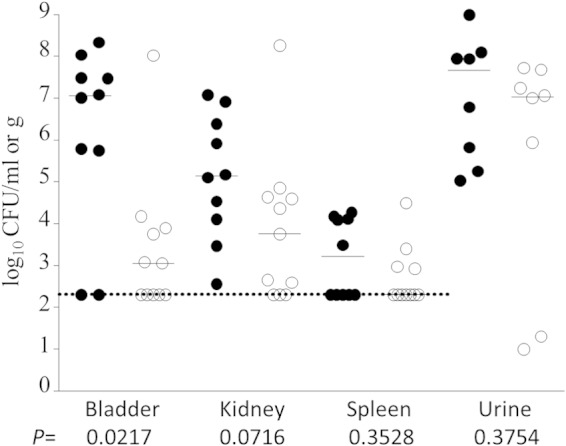
P. mirabilis HI4320ΔmrpJ challenge in a mouse model of ascending UTI. Deficiency of mrpJ compromises P. mirabilis fitness within the bladder. Female mice were infected via transurethral catheterization with either wild-type HI4320 or an isogenic mrpJ mutant in an independent challenge experiment. Each symbol represents the bacterial titer present in the bladder, kidney, spleen, or urine of an individual mouse at 7 days postinoculation. Bars denote median values for each group of mice (wild type [●] and mutant [○]). The dotted line indicates the limit of detection for homogenized organ data. Statistical significance was determined using the Mann-Whitney U test. Data depicted here are from two independent experiments.
DISCUSSION
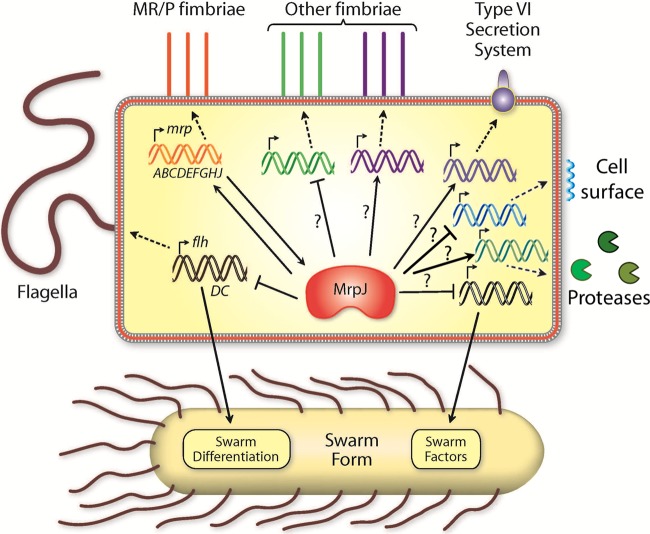
The inverse interplay between fimbria-mediated attachment and flagellar motility in P. mirabilis is critical for the establishment of a successful infection in the urinary tract (5). Earlier studies have shown that MrpJ plays an important role in this dynamic by positively regulating its own operon while repressing flagella through the flagellar master regulator flhDC (32, 33). The present study expands the role of MrpJ beyond this interplay by identifying an array of cellular processes and target genes under its influence, showing a more global regulation of virulence by this transcriptional modulator. On the basis of our microarray and qRT-PCR data, we propose that MrpJ is a master regulator of P. mirabilis virulence (Fig. 8).
FIG 8.
Model for MrpJ as a master regulator of virulence. MrpJ directly represses flhDC and induces the mrp operon. Other fimbriae (e.g., PMF, Fim8), the T6SS, cell surface modification systems (e.g., PagP, WzyE), and proteases (e.g., ZapA, Pta) are regulated by MrpJ, but this could be direct or indirect (represented by a question mark).
P. mirabilis HI4320 encodes 15 mrpJ-type genes (33), and most of these also act to repress motility. All but four are located within fimbrial operons. We propose that MrpJ is the dominant regulator of this network during UTIs. First, the highest levels of mrpJ transcript are detected in urine samples from infected mice, and the mrp operon is the most highly induced set of genes during the shift from in vitro to in vivo growth (24). Second, 13/14 of the other mrpJ paralogs are expressed during infection at or below background detection levels (24); the exception is orphan gene PMI0982 which although detectable, is not differentially expressed in vivo. Third, the data we present demonstrate that MrpJ controls numerous virulence processes, although this may occur via intermediate factors, as microarrays cannot discriminate between indirect and direct regulation. Fourth, MrpJ regulates other fimbrial operons, including fim8 and fim14, both of which encode mrpJ paralogs. Indeed, fim8J was repressed by MrpJ in our microarray analysis. The aerated broth growth condition, while beneficial for detecting regulation of flagella, is not ideal for fimbrial expression, and it is possible that MrpJ regulates additional fimbriae that were not detected in this set of microarrays. Nevertheless, we performed our analysis using mid-logarithmic-phase cultures, which allowed us to carefully adjust mrpJ expression to mimic the amount that occurs during UTI. Last, two of the four orphan mrpJ paralogs, PMI0182 and PMI3508, were repressed by MrpJ. However, an important question remains: do other MrpJ paralogs contribute to virulence? It is possible that low levels of transcription of one or more mrpJ paralogs could trigger a regulatory cascade. Expression in trans of other mrpJ paralogs leads to differing surface fimbrial appearances (33), and we now show that ucaJ represses mrpA; it will be important to decipher the complete regulatory networks of UcaJ and other MrpJ paralogs to fully understand how P. mirabilis coordinates the expression of fimbria-encoding genes. It is possible that these related genes orchestrate genetic programs beneficial to specific niches beyond the bladder.
The sequencing and annotation of prototypic P. mirabilis strain HI4320 (8) revealed the presence of 17 different potential chaperone-usher fimbriae in its genome. Our microarray analysis has now shown that a number of these fimbriae are under the control of MrpJ; while MrpJ augmented mrp gene expression, pmfA, fim8A, and fim14A were downregulated. Interestingly, pmfA and fim8A were also repressed when gene expression was monitored in mice infected with strain HI4320 (24). Fimbrial regulation is a quintessential aspect of virulence in many pathogens, including causative agents of UTIs, and in many instances, more than one fimbrial type contributes to disease progression depending on the environmental cues or tissue niches (4, 71). Although several fimbriae, including MR/P, PMF, and Fim14, have been reported to modulate P. mirabilis pathogenesis, little is known about the regulation and combinatorial implication of fimbriae in P. mirabilis-mediated UTIs (13, 15, 57, 72). To our knowledge, this is the first report describing a molecular regulator simultaneously targeting a number of fimbriae in a positive or negative manner in this uropathogen.
Among other differentially regulated targets, we identified flagellum-related genes, including the flagellar master transcriptional regulator flhDC as anticipated based on the previously reported inverse play between MR/P and flagella (32, 33). Our microarray data show that MrpJ not only regulates flagella but also regulates factors specifically involved in swarming which are not necessarily under the control of FlhD4C2. These genes include the swarming regulator genes umoA and umoB (53), as well as mrpJ paralogs, which are not associated with fimbrial operons (PMI0182 and PMI3508) (33).
A number of T6SS genes were also induced by MrpJ; among them were the effector genes idsADE as well as seven T6SS structural genes. Although T6SS is involved in virulence in a variety of pathogens (5, 73–75), its role in UTIs has not yet been investigated. Recent reports have described the role of the P. mirabilis T6SS in the competition of some strains over others (60, 62), particularly when distinct isolates encounter each other while swarming. However, during in vitro swarming, the mrp operon is repressed (44). We speculate that the T6SS may play a swarming-independent role during infection by mediating direct interaction with the host, perhaps mediated by secretion of a different set of effector proteins. It is also possible that MrpJ-mediated regulation of T6SS components facilitates P. mirabilis swarming behavior across catheter surfaces in a polymicrobial environment when signals from the host are encountered.
In addition to these factors, several other putative or known virulence genes were revealed as the targets of MrpJ; the Pta toxin and ZapA metalloprotease have already been shown to contribute to UTI in mice (58, 76, 77). MrpJ regulates expression or modification of cell surface molecules, such as lipoproteins and LPS, which have been associated with virulence by other bacterial pathogens (78–81), and we hypothesize these changes aid P. mirabilis survival in the urinary tract by bolstering bacterial defenses against the innate immune response.
We chose two candidate virulence operons to perform further molecular analyses of MrpJ-mediated transcriptional regulation: mrpABCDEFGHJ and flhDC, an activation and a repression target, respectively. We used two molecular approaches to probe the nature of the MrpJ-target gene interactions: β-galactosidase reporter assays to determine the essential promoter element and ChIP analysis to evaluate the in vivo MrpJ-chromosome interaction. Our single-copy chromosomal operon fusions identified the necessary promoter region on the mrpA upstream element (Fig. 3C). Specifically, the highest LacZ activity was seen by the promoter fragment from nucleotides −256 to +179, which includes the invertible element region (Fig. 3C), and this was consistent with our in vivo ChIP data showing direct MrpJ binding in this region (fragment “e” [Fig. 5]). Interestingly, there was a mild decrease in LacZ activity with the longer promoter fragments used in this assay, indicating possible repressor binding. ChIP data further support this hypothesis, as there was only a background level of MrpJ binding in this region (fragment “d”). Intriguingly, ChIP also showed a second MrpJ-DNA interaction upstream of the mrpA regulatory element in the intragenic region of mrpI. However, because the distal site did not contribute to transcriptional activity of our reporter constructs, we believe that proximity of MrpJ to this upstream region is influenced by the curvature and conformation of DNA in this area of the native chromosome. A previous report showed that mutation of mrpJ leads to the mrp promoter invertible element being predominantly in the OFF orientation (63). Our data suggest this could be due to either MrpJ interaction with the invertible element or the mrpI coding sequence. It remains to be determined whether MrpJ binding to the mrp promoter affects transcription of mrpI in vivo.
The presence of an upstream regulatory region in conjunction with a more proximally situated (relative to the transcriptional start site) regulatory element is seen in many instances in bacteria (69, 82), and in the case of the mrp promoter, it would provide for an elegant mechanism of regulation between the recombinase MrpI and its target operon mrp. This is especially important for genes involved in virulence, such as mrpA, to exert more stringent control over transcriptional regulation depending on various environmental cues. Although our transcriptional reporters reaffirm the notion of MrpJ-mediated autoregulation of the mrp operon, we point out that the fold changes observed in the transcriptional reporter assays are smaller than those reported for our microarray data (Fig. 3; see Tables S3 and S4 in the supplemental material). We hypothesize that the combination of integrating the reporters at a nonnative chromosomal site, as well as the lower sensitivity of β-galactosidase assays compared to qRT-PCR measurements, contributed to this outcome.
Surprisingly, we detected a consistent intragenic ChIP signal in our assay of MrpJ-mediated in vivo regulation of flhDC. Internal binding of transcriptional regulators within a gene in bacteria has been seen in many instances, and recently, it has also been observed for flhDC and fliA (83–87). In this particular case of MrpJ-flhDC regulation, these intragenic signals could originate from additional MrpJ-DNA interactions, or alternatively, they could arise from a condensed higher-order conformation of the P. mirabilis chromosome in this region (67–70, 88). The intergenic region preceding the flhDC operon is quite large (over 2.7 kb) and highly AT rich (24.7% GC versus 38.9% chromosomal GC content) in this pathogen. Although we tested flhD transcriptional reporters with MrpJ, a significant reduction in flhD promoter activity was not seen reproducibly (data not shown). We hypothesize that a much larger upstream fragment in its native state is required to replicate MrpJ-mediated flhD promoter repression. To support the critical role of flagellar motility in various stages of this bacterium's life cycle, a number of regulators (e.g., H-NS, Crp, Lrp, RcsB, LrhA) have been shown or implicated to influence the flhD upstream region (5, 66, 89–91). Furthermore, it has recently been proposed that the Salmonella enterica global regulator LeuO binds to DNA differently to initiate or impede transcription of target genes (92). Distal as well as proximal binding profiles of MrpJ were evident for both the mrp gene and the flhD upstream regulatory element in vivo, suggesting that the binding of MrpJ on separate regions preceding its target genes occurs during both gene activation as well as repression. Therefore, our results warrant further studies to define the precise regulatory mechanism and structural reference of transcriptional regulation by MrpJ.
An obvious binding sequence for MrpJ could not be identified at this time. Although a repeat sequence was found in the MrpJ binding region of the mrpA promoter, this sequence was not found in the flhDC promoter. It is not uncommon for global bacterial transcriptional regulators to lack a consensus binding motif; in fact, a low level of binding specificity has been proposed as a genomic diagnostic tool to identify factors regulating genome-wide targets versus the local transcriptional regulators with highly specific binding sites (69, 70, 82, 93).
In P. mirabilis, this is the first report of a transcriptional modulator regulating a variety of processes, including several aspects of virulence. Our independent challenge experiments further highlight the critical role of MrpJ in virulence regulation in this bacterium. While the contribution of this regulator to UTI progression has been documented earlier in a cochallenge experiment in the presence of a wild-type counterpart (32), the direct modulatory effect of MrpJ deficiency is more evident in an individual challenge. However, the ability of this mutant to ascend to the kidney in spite of a poor bladder colonization profile implies that there could be other fimbriae compensating for the loss of MR/P fimbriae or perhaps involvement by other MrpJ paralogs.
In summary, this report analyzes the molecular nature of MrpJ-mediated transcriptional regulation in P. mirabilis by extending its contribution beyond the reciprocity of adherence and motility and introduces this regulator of transcription as a multifaceted virulence regulator of UTIs.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Andrew Darwin for his input in the design of the β-galactosidase reporter assays, as well as his gift of the CC118 λpir and pRS415 strains. We thank Herbert Schweizer for sending us plasmids pTNS3 and pUC18R6K-mini-Tn7T-Gm. We appreciate Jessica Schaffer and Keith Martinez for technical assistance with experiments and Gordon Cook for artwork. We also thank Aleksandra Sikora, Victor Torres, and Jessica Schaffer for critical reading of the manuscript.
This work was supported by U.S. Public Health Service grant AI083743 (M.M.P.) and in part by a grant from the Urology Care Foundation Research Scholars Program and AUA New York Section Research Scholar Fund (I.D.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02978-14.
REFERENCES
- 1.Hood MI, Skaar EP. 2012. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Troxell B, Hassan HM. 2013. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol 3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foxman B. 2010. The epidemiology of urinary tract infection. Nat Rev Urol 7:653–660. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- 4.Nielubowicz GR, Mobley HLT. 2010. Host-pathogen interactions in urinary tract infection. Nat Rev Urol 7:430–441. doi: 10.1038/nrurol.2010.101. [DOI] [PubMed] [Google Scholar]
- 5.Armbruster CE, Mobley HLT. 2012. Merging mythology and morphology: the multifaceted lifestyle of Proteus mirabilis. Nat Rev Microbiol 10:743–754. doi: 10.1038/nrmicro2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waksman G, Hultgren SJ. 2009. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol 7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane MC, Alteri CJ, Smith SN, Mobley HLT. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Walker D, Whithead S, Thomson NR, Rather PN, Parkhill J, Mobley HLT. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J Bacteriol 190:4027–4037. doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adegbola RA, Old DC, Senior BW. 1983. The adhesins and fimbriae of Proteus mirabilis strains associated with high and low affinity for the urinary tract. J Med Microbiol 16:427–431. doi: 10.1099/00222615-16-4-427. [DOI] [PubMed] [Google Scholar]
- 10.Bahrani FK, Mobley HLT. 1994. Proteus mirabilis MR/P fimbrial operon: genetic organization, nucleotide sequence, and conditions for expression. J Bacteriol 176:3412–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bijlsma IG, van Dijk L, Kusters JG, Gaastra W. 1995. Nucleotide sequences of two fimbrial major subunit genes, pmpA and ucaA, from canine-uropathogenic Proteus mirabilis strains. Microbiology 141:1349–1357. doi: 10.1099/13500872-141-6-1349. [DOI] [PubMed] [Google Scholar]
- 12.Cook SW, Mody N, Valle J, Hull R. 1995. Molecular cloning of Proteus mirabilis uroepithelial cell adherence (uca) genes. Infect Immun 63:2082–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuan L, Schaffer JN, Zouzias CD, Pearson MM. 2014. Characterization of 17 chaperone-usher fimbriae encoded by Proteus mirabilis reveals strong conservation. J Med Microbiol 63:911–922. doi: 10.1099/jmm.0.069971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massad G, Bahrani FK, Mobley HLT. 1994. Proteus mirabilis fimbriae: identification, isolation, and characterization of a new ambient-temperature fimbria. Infect Immun 62:1989–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massad G, Lockatell CV, Johnson DE, Mobley HLT. 1994. Proteus mirabilis fimbriae: construction of an isogenic pmfA mutant and analysis of virulence in a CBA mouse model of ascending urinary tract infection. Infect Immun 62:536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wray SK, Hull SI, Cook RG, Barrish J, Hull RA. 1986. Identification and characterization of a uroepithelial cell adhesin from a uropathogenic isolate of Proteus mirabilis. Infect Immun 54:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahrani FK, Massad G, Lockatell CV, Johnson DE, Russell RG, Warren JW, Mobley HLT. 1994. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infect Immun 62:3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen AM, Lockatell V, Johnson DE, Mobley HLT. 2004. Mannose-resistant Proteus-like fimbriae are produced by most Proteus mirabilis strains infecting the urinary tract, dictate the in vivo localization of bacteria, and contribute to biofilm formation. Infect Immun 72:7294–7305. doi: 10.1128/IAI.72.12.7294-7305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Johnson DE, Mobley HLT. 1999. Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis. Infect Immun 67:2822–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Lockatell CV, Johnson DE, Mobley HLT. 2002. Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis. Mol Microbiol 45:865–874. doi: 10.1046/j.1365-2958.2002.03067.x. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Zhao H, Geymonat L, Bahrani F, Johnson DE, Mobley HLT. 1997. Proteus mirabilis mannose-resistant, Proteus-like fimbriae: MrpG is located at the fimbrial tip and is required for fimbrial assembly. Infect Immun 65:1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zunino P, Sosa V, Schlapp G, Allen AG, Preston A, Maskell DJ. 2007. Mannose-resistant Proteus-like and P. mirabilis fimbriae have specific and additive roles in P. mirabilis urinary tract infections. FEMS Immunol Med Microbiol 51:125–133. doi: 10.1111/j.1574-695X.2007.00285.x.. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Li X, Johnson DE, Blomfield I, Mobley HLT. 1997. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Mol Microbiol 23:1009–1019. doi: 10.1046/j.1365-2958.1997.2791645.x. [DOI] [PubMed] [Google Scholar]
- 24.Pearson MM, Yep A, Smith SN, Mobley HLT. 2011. Transcriptome of Proteus mirabilis in the murine urinary tract: virulence and nitrogen assimilation gene expression. Infect Immun 79:2619–2631. doi: 10.1128/IAI.05152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Erbe JL, Lockatell CV, Johnson DE, Jobling MG, Holmes RK, Mobley HLT. 2004. Use of translational fusion of the MrpH fimbrial adhesin-binding domain with the cholera toxin A2 domain, coexpressed with the cholera toxin B subunit, as an intranasal vaccine to prevent experimental urinary tract infection by Proteus mirabilis. Infect Immun 72:7306–7310. doi: 10.1128/IAI.72.12.7306-7310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Lockatell CV, Johnson DE, Lane MC, Warren JW, Mobley HLT. 2004. Development of an intranasal vaccine to prevent urinary tract infection by Proteus mirabilis. Infect Immun 72:66–75. doi: 10.1128/IAI.72.1.66-75.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellegrino R, Galvalisi U, Scavone P, Sosa V, Zunino P. 2003. Evaluation of Proteus mirabilis structural fimbrial proteins as antigens against urinary tract infections. FEMS Immunol Med Microbiol 36:103–110. doi: 10.1016/S0928-8244(03)00103-2. [DOI] [PubMed] [Google Scholar]
- 28.Scavone P, Miyoshi A, Rial A, Chabalgoity A, Langella P, Azevedo V, Zunino P. 2007. Intranasal immunisation with recombinant Lactococcus lactis displaying either anchored or secreted forms of Proteus mirabilis MrpA fimbrial protein confers specific immune response and induces a significant reduction of kidney bacterial colonisation in mice. Microbes Infect 9:821–828. doi: 10.1016/j.micinf.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 29.Scavone P, Rial A, Umpiérrez A, Chabalgoity A, Zunino P. 2009. Effects of the administration of cholera toxin as a mucosal adjuvant on the immune and protective response induced by Proteus mirabilis MrpA fimbrial protein in the urinary tract. Microbiol Immunol 53:233–240. doi: 10.1111/j.1348-0421.2009.00111.x. [DOI] [PubMed] [Google Scholar]
- 30.Scavone P, Sosa V, Pellegrino R, Galvalisi U, Zunino P. 2004. Mucosal vaccination of mice with recombinant Proteus mirabilis structural fimbrial proteins. Microbes Infect 6:853–860. doi: 10.1016/j.micinf.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Scavone P, Umpiérrez A, Maskell DJ, Zunino P. 2011. Nasal immunization with attenuated Salmonella Typhimurium expressing an MrpA-TetC fusion protein significantly reduces Proteus mirabilis colonization in the mouse urinary tract. J Med Microbiol 60:899–904. doi: 10.1099/jmm.0.030460-0. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Rasko DA, Lockatell CV, Johnson DE, Mobley HLT. 2001. Repression of bacterial motility by a novel fimbrial gene product. EMBO J 20:4854–4862. doi: 10.1093/emboj/20.17.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson MM, Mobley HLT. 2008. Repression of motility during fimbrial expression: identification of 14 mrpJ gene paralogues in Proteus mirabilis. Mol Microbiol 69:548–558. doi: 10.1111/j.1365-2958.2008.06307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiss DJ, Mobley HLT. 2011. Determination of target sequence bound by PapX, repressor of bacterial motility, in flhD promoter using systematic evolution of ligands by exponential enrichment (SELEX) and high throughput sequencing. J Biol Chem 286:44726–44738. doi: 10.1074/jbc.M111.290684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simms AN, Mobley HLT. 2008. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect Immun 76:4833–4841. doi: 10.1128/IAI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allison SE, Silphaduang U, Mascarenhas M, Konczy P, Quan Q, Karmali M, Coombes BK. 2012. Novel repressor of Escherichia coli O157:H7 motility encoded in the putative fimbrial cluster OI-1. J Bacteriol 194:5343–5352. doi: 10.1128/JB.01025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meslet-Cladiere LM, Pimenta A, Duchaud E, Holland IB, Blight MA. 2004. In vivo expression of the mannose-resistant fimbriae of Photorhabdus temperata K122 during insect infection. J Bacteriol 186:611–622. doi: 10.1128/JB.186.3.611-622.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He H, Snyder HA, Forst S. 2004. Unique organization and regulation of the mrx fimbrial operon in Xenorhabdus nematophila. Microbiology 150:1439–1446. doi: 10.1099/mic.0.26853-0. [DOI] [PubMed] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Khalid MI, Teh LK, Lee LS, Zakaria ZA, Salleh MZ. 2013. Genome sequence of Proteus mirabilis strain PR03, isolated from a local hospital in Malaysia. Genome Announc 1(3):e00327–13. doi: 10.1128/genomeA.00327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minogue TD, Daligault HE, Davenport KW, Bishop-Lilly KA, Bruce DC, Chain PS, Coyne SR, Chertkov O, Freitas T, Frey KG, Jaissle J, Koroleva GI, Ladner JT, Palacios GF, Redden CL, Xu Y, Johnson SL. 2014. Draft genome assemblies of Proteus mirabilis ATCC 7002 and Proteus vulgaris ATCC 49132. Genome Announc 2(5):e01064–14. doi: 10.1128/genomeA.01064-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi X, Zhu Y, Li Y, Jiang M, Lin Y, Qiu Y, Chen Q, Yuan Y, Ni P, Hu Q, Huang S. 2014. Genome sequence of Proteus mirabilis clinical isolate C05028. Genome Announc 2(2):e00167–14. doi: 10.1128/genomeA.00167-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan NL, Septer AN, Fields AT, Wenren LM, Gibbs KA. 2013. The complete genome sequence of Proteus mirabilis strain BB2000 reveals differences from the P. mirabilis reference strain. Genome Announc 1(5):e00024–13. doi: 10.1128/genomeA.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pearson MM, Rasko DA, Smith SN, Mobley HLT. 2010. Transcriptome of swarming Proteus mirabilis. Infect Immun 78:2834–2845. doi: 10.1128/IAI.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with secondary, non-glmS-linked attTn7 sites: example Proteus mirabilis HI4320. Nature Protoc 1:170–178. doi: 10.1038/nprot.2006.26. [DOI] [PubMed] [Google Scholar]
- 46.Maloy SR, Stewart VJ, Taylor RK. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 47.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Grainger DC, Overton TW, Reppas N, Wade JT, Tamai E, Hobman JL, Constantinidou C, Struhl K, Church G, Busby SJ. 2004. Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J Bacteriol 186:6938–6943. doi: 10.1128/JB.186.20.6938-6943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo C, Lam E. 2014. Quantitatively profiling genome-wide patterns of histone modifications in Arabidopsis thaliana using ChIP-seq. Methods Mol Biol 1112:177–193. doi: 10.1007/978-1-62703-773-0_12. [DOI] [PubMed] [Google Scholar]
- 50.Luo C, Sidote DJ, Zhang Y, Kerstetter RA, Michael TP, Lam E. 2012. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J 73:77–90. doi: 10.1111/tpj.12017. [DOI] [PubMed] [Google Scholar]
- 51.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Edén C. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clemmer KM, Rather PN. 2007. Regulation of flhDC expression in Proteus mirabilis. Res Microbiol 158:295–302. doi: 10.1016/j.resmic.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 53.Dufour A, Furness RB, Hughes C. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol Microbiol 29:741–751. doi: 10.1046/j.1365-2958.1998.00967.x. [DOI] [PubMed] [Google Scholar]
- 54.Hay NA, Tipper DJ, Gygi D, Hughes C. 1999. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol 181:2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sturgill G, Rather PN. 2004. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol Microbiol 51:437–446. doi: 10.1046/j.1365-2958.2003.03835.x. [DOI] [PubMed] [Google Scholar]
- 56.Himpsl SD, Lockatell CV, Hebel JR, Johnson DE, Mobley HLT. 2008. Identification of virulence determinants in uropathogenic Proteus mirabilis using signature-tagged mutagenesis. J Med Microbiol 57:1068–1078. doi: 10.1099/jmm.0.2008/002071-0. [DOI] [PubMed] [Google Scholar]
- 57.Zunino P, Sosa V, Allen AG, Preston A, Schlapp G, Maskell DJ. 2003. Proteus mirabilis fimbriae (PMF) are important for both bladder and kidney colonization in mice. Microbiology 149:3231–3237. doi: 10.1099/mic.0.26534-0. [DOI] [PubMed] [Google Scholar]
- 58.Alamuri P, Mobley HLT. 2008. A novel autotransporter of uropathogenic Proteus mirabilis is both a cytotoxin and an agglutinin. Mol Microbiol 68:997–1017. doi: 10.1111/j.1365-2958.2008.06199.x. [DOI] [PubMed] [Google Scholar]
- 59.Walker KE, Moghaddame-Jafari S, Lockatell CV, Johnson D, Belas R. 1999. ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol Microbiol 32:825–836. doi: 10.1046/j.1365-2958.1999.01401.x. [DOI] [PubMed] [Google Scholar]
- 60.Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HLT. 2013. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog 9:e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibbs KA, Urbanowski ML, Greenberg EP. 2008. Genetic determinants of self identity and social recognition in bacteria. Science 321:256–259. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wenren LM, Sullivan NL, Cardarelli L, Septer AN, Gibbs KA. 2013. Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. mBio 4(4):e00374–13. doi: 10.1128/mBio.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lane MC, Li X, Pearson MM, Simms AN, Mobley HLT. 2009. Oxygen-limiting conditions enrich for fimbriate cells of uropathogenic Proteus mirabilis and Escherichia coli. J Bacteriol 191:1382–1392. doi: 10.1128/JB.01550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feklistov A, Sharon BD, Darst SA, Gross CA. 2014. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 65.Browne JA, Harris A, Leir SH. 2014. An optimized protocol for isolating primary epithelial cell chromatin for ChIP. PLoS One 9:e100099. doi: 10.1371/journal.pone.0100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furness RB, Fraser GM, Hay NA, Hughes C. 1997. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J Bacteriol 179:5585–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 68.Dorman CJ. 2013. Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem Soc Trans 41:542–547. doi: 10.1042/BST20120222. [DOI] [PubMed] [Google Scholar]
- 69.Minchin SD, Busby SJ. 2009. Analysis of mechanisms of activation and repression at bacterial promoters. Methods 47:6–12. doi: 10.1016/j.ymeth.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 70.Wade JT, Struhl K, Busby SJ, Grainger DC. 2007. Genomic analysis of protein-DNA interactions in bacteria: insights into transcription and chromosome organization. Mol Microbiol 65:21–26. doi: 10.1111/j.1365-2958.2007.05781.x. [DOI] [PubMed] [Google Scholar]
- 71.Wiles TJ, Kulesus RR, Mulvey MA. 2008. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol 85:11–19. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pellegrino R, Scavone P, Umpiérrez A, Maskell DJ, Zunino P. 2013. Proteus mirabilis uroepithelial cell adhesin (UCA) fimbria plays a role in the colonization of the urinary tract. Pathog Dis 67:104–107. doi: 10.1111/2049-632X.12027. [DOI] [PubMed] [Google Scholar]
- 73.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kapitein N, Mogk A. 2013. Deadly syringes: type VI secretion system activities in pathogenicity and interbacterial competition. Curr Opin Microbiol 16:52–58. doi: 10.1016/j.mib.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 75.Russell AB, Peterson SB, Mougous JD. 2014. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Belas R, Manos J, Suvanasuthi R. 2004. Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect Immun 72:5159–5167. doi: 10.1128/IAI.72.9.5159-5167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phan V, Belas R, Gilmore BF, Ceri H. 2008. ZapA, a virulence factor in a rat model of Proteus mirabilis-induced acute and chronic prostatitis. Infect Immun 76:4859–4864. doi: 10.1128/IAI.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bishop RE. 2005. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol Microbiol 57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 79.Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect Immun 79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ranallo RT, Kaminski RW, George T, Kordis AA, Chen Q, Szabo K, Venkatesan MM. 2010. Virulence, inflammatory potential, and adaptive immunity induced by Shigella flexneri msbB mutants. Infect Immun 78:400–412. doi: 10.1128/IAI.00533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tan Y, Kagan JC. 2014. A cross-disciplinary perspective on the innate immune responses to bacterial lipopolysaccharide. Mol Cell 54:212–223. doi: 10.1016/j.molcel.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Balleza E, López-Bojorquez LN, Martínez-Antonio A, Resendis-Antonio O, Lozada-Chávez I, Balderas-Martinez YI, Encarnación S, Collado-Vides J. 2009. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol Rev 33:133–151. doi: 10.1111/j.1574-6976.2008.00145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bonocora RP, Fitzgerald DM, Stringer AM, Wade JT. 2013. Non-canonical protein-DNA interactions identified by ChIP are not artifacts. BMC Genomics 14:254. doi: 10.1186/1471-2164-14-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fitzgerald DM, Bonocora RP, Wade JT. 2014. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genetics 10:e1004649. doi: 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jungwirth B, Sala C, Kohl TA, Uplekar S, Baumbach J, Cole ST, Pühler A, Tauch A. 2013. High-resolution detection of DNA binding sites of the global transcriptional regulator GlxR in Corynebacterium glutamicum. Microbiology 159:12–22. doi: 10.1099/mic.0.062059-0. [DOI] [PubMed] [Google Scholar]
- 86.Singh SS, Singh N, Bonocora RP, Fitzgerald DM, Wade JT, Grainger DC. 2014. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev 28:214–219. doi: 10.1101/gad.234336.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wade JT, Grainger DC. 2014. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol 12:647–653. doi: 10.1038/nrmicro3316. [DOI] [PubMed] [Google Scholar]
- 88.Dorman CJ. 2013. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat Rev Microbiol 11:349–355. doi: 10.1038/nrmicro3007. [DOI] [PubMed] [Google Scholar]
- 89.Ko M, Park C. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Bacteriol 182:4670–4672. doi: 10.1128/JB.182.16.4670-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lanois A, Jubelin G, Givaudan A. 2008. FliZ, a flagellar regulator, is at the crossroads between motility, haemolysin expression and virulence in the insect pathogenic bacterium Xenorhabdus. Mol Microbiol 68:516–533. doi: 10.1111/j.1365-2958.2008.06168.x. [DOI] [PubMed] [Google Scholar]
- 91.Mouslim C, Hughes KT. 2014. The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog 10:e1003987. doi: 10.1371/journal.ppat.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guadarrama C, Medrano-Lopez A, Oropeza R, Hernandez-Lucas I, Calva E. 2014. The Salmonella enterica serovar Typhi LeuO global regulator forms tetramers: residues involved in oligomerization, DNA binding, and transcriptional regulation. J Bacteriol 196:2143–2154. doi: 10.1128/JB.01484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lozada-Chávez I, Angarica VE, Collado-Vides J, Contreras-Moreira B. 2008. The role of DNA-binding specificity in the evolution of bacterial regulatory networks. J Mol Biol 379:627–643. doi: 10.1016/j.jmb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.