Abstract
Whole-genome sequencing analysis of ∼800 strains of group A Streptococcus (GAS) found that the gene encoding the multiple virulence gene regulator of GAS (mga) is highly polymorphic in serotype M59 strains but not in strains of other serotypes. To help understand the molecular mechanism of gene regulation by Mga and its contribution to GAS pathogenesis in serotype M59 GAS, we constructed an isogenic mga mutant strain. Transcriptome studies indicated a significant regulatory influence of Mga and altered metabolic capabilities conferred by Mga-regulated genes. We assessed the phosphorylation status of Mga in GAS cell lysates with Phos-tag gels. The results revealed that Mga is phosphorylated at histidines in vivo. Using phosphomimetic and nonphosphomimetic substitutions at conserved phosphoenolpyruvate:carbohydrate phosphotransferase regulation domain (PRD) histidines of Mga, we demonstrated that phosphorylation-mimicking aspartate replacements at H207 and H273 of PRD-1 and at H327 of PRD-2 are inhibitory to Mga-dependent gene expression. Conversely, non-phosphorylation-mimicking alanine substitutions at H273 and H327 relieved inhibition, and the mutant strains exhibited a wild-type phenotype. The opposing regulatory profiles observed for phosphorylation- and non-phosphorylation-mimicking substitutions at H273 extended to global gene regulation by Mga. Consistent with these observations, the H273D mutant strain attenuated GAS virulence, whereas the H273A strain exhibited a wild-type virulence phenotype in a mouse model of necrotizing fasciitis. Together, our results demonstrate phosphoregulation of Mga and its direct link to virulence in M59 GAS strains. These data also lay a foundation toward understanding how naturally occurring gain-of-function variations in mga, such as H201R, may confer an advantage to the pathogen and contribute to M59 GAS pathogenesis.
INTRODUCTION
Group A Streptococcus (GAS) is a versatile human pathogen that colonizes a broad spectrum of anatomical sites within the host and causes a variety of clinical manifestations (1, 2). Given that each host microenvironment presents unique challenges to the pathogen in terms of nutrient availability, anatomical barriers, and host defense responses, it is critical that GAS possess sophisticated sensing systems and elicit potent adaptive responses. Such sensory systems are coordinated in GAS by a combination of global transcription regulators and two-component signaling systems (3–7). Global transcription regulators sense specific environmental cues present at the colonization surface and transduce those signals into a tailored transcriptional response that confers a survival advantage to the bacteria within the host (5, 8–12). Consistent with their central role in bacterial in vivo fitness and pathogenesis, these regulators are, as revealed by genomic sequencing studies, under strong selective pressure during human infection (6, 8, 13–16).
Genomic sequencing of ∼800 serotype M59 GAS strains isolated from an epidemic of invasive infections across North America revealed that the highest frequency of single nucleotide polymorphisms (SNPs) resulting in nonsynonymous amino acid replacements occurs in the gene encoding the global regulator multiple virulence gene regulator of GAS (mga) (13, 14). Multiple polymorphisms in mga occur independently, suggesting that the mga locus is under strong evolutionary pressure during the course of this outbreak (13, 14, 17). This is in contrast to observations made in similar analyses of serotype M1 and M3 strains, in which SNPs occur predominantly in genes encoding the global regulators ropB and covRS (6, 15). These observations led us to hypothesize that Mga and its gene regulation are critical for the survival of the organism in vivo and contribute to the pathogenesis of infection caused by GAS serotype M59 strains in a serotype-specific fashion.
Mga is a global transcription regulator that controls the expression of genes involved in host cell attachment, immune evasion, and carbohydrate metabolism during early stages of GAS infection (3, 18, 19). However, the Mga regulon in GAS has significant interserotype variation in the composition and number of genes (3). For example, a transcriptome analysis of Mga in GAS serotype M1 and M4 strains indicated that it controls ∼10% of genes, whereas it influences only ∼2% of genes in a serotype M6 strain (3). Regardless, the core Mga regulon, as defined by the genes with the highest degree of regulation and containing Mga-binding sites in their promoters, consists of proven virulence factors, including emm, scpA, sclA, sic, fba, and sof (3). Consistent with its critical role in GAS gene regulation and pathogenesis, upregulation of mga in M59 GAS increases virulence, and conversely, inactivation of mga in several tested serotypes attenuates GAS virulence (10, 20, 21).
Mga is a 536-amino-acid-long protein that belongs to a group of transcription regulators with phosphoenolpyruvate:carbohydrate phosphotransferase (PTS) regulation domains (PRDs). Mga is predicted to have a multidomain architecture (22, 23). The amino terminus has a conserved Mga domain 1 (CMD-1) and two helix-turn-helix (HTH) motifs that are critical for interaction of Mga with its target promoters (22–24). The central region of the protein has two PRDs, and the carboxy terminus has an oligomerization domain (10, 25). As in other bacterial PRD-containing antiterminators and transcription activators, the regulatory activity of Mga is controlled by PTS-dependent phosphorylation of conserved histidine residues in the PRDs (10).
Despite the wealth of information available about Mga-dependent gene regulation, knowledge of the influence of Mga on gene regulation in serotype M59 strains is lacking. Given that mga polymorphisms are prevalent in serotype M59 disease isolates, it is important to understand the molecular mechanism of gene regulation and the adaptive responses controlled by Mga in this group of organisms. To this end, we defined the mga regulon in serotype M59 by transcriptome sequencing (RNA-seq) experiments. Although Mga phosphorylation has been demonstrated in vitro by using recombinant PTS components, here we provide evidence that Mga is phosphorylated in vivo in GAS cells grown in laboratory medium. Finally, using isogenic mutant strains with single-amino-acid replacements at conserved histidines mimicking phosphorylated and nonphosphorylated states of Mga, we demonstrated the global influence of phosphorylation on gene regulation by Mga and its contribution to GAS virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strain MGAS15249 was used as the reference strain because its genome has been sequenced and it has the wild-type sequence for all major regulators, including mga (14). MGAS15249 is genetically representative of serotype M59 strains that cause invasive infections (14). All GAS growth was performed routinely on Trypticase soy agar containing 5% sheep blood (SA; Becton Dickinson) or in Todd-Hewitt broth containing 0.2% yeast extract (THY; Difco) at 37°C with 5% CO2. When required, ampicillin or chloramphenicol was added to a final concentration of 80 μg/ml or 8 μg/ml, respectively. Ampicillin was used for Escherichia coli growth, whereas chloramphenicol was used for both E. coli and GAS cultures. When appropriate, spectinomycin was added to a final concentration of 100 μg/ml for GAS growth. E. coli strain DH5α was used as the host for plasmid constructions, and strain BL21(DE3) was used for recombinant protein overexpression. E. coli strains were grown in Luria-Bertani (LB) broth (EMD chemicals). All GAS growth experiments were done in triplicate on three separate occasions. Cultures grown overnight were inoculated into fresh medium to achieve an initial A600 of <0.1. Growth was monitored by measuring the optical density at 600 nm. Bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material.
Creation of the mga-inactivated mutant strain.
An isogenic mutant strain lacking the majority of the coding region (codons 6 through 533) of mga (Δmga) in parental strain MGAS15249 was generated by insertional inactivation with a spectinomycin cassette, as described previously (21).
Construction of an mga trans-complementation plasmid.
To complement the isogenic Δmga mutant, the coding sequence of the full-length mga gene along with its native promoter (676 bp upstream of the start codon) was cloned into the E. coli-GAS shuttle vector pDC123 (26). The DNA fragment was amplified by PCR from GAS genomic DNA, digested with BglII and NdeI, and ligated into digested vector pDC123 to generate complementation plasmid pDC-mga. The inserts were verified by DNA sequencing and electroporated into the isogenic Δmga mutant strain to create the Δmga:pDC-mga strain. Primers used for the construction and sequencing of the trans-complementation plasmid are listed in Table S2 in the supplemental material.
Site-directed mutagenesis of mga.
Plasmid pDC-mga containing the wild-type mga coding region was used as a template for site-directed mutagenesis. A QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce single-amino-acid substitutions into Mga that resulted in amino acid changes of histidines to either alanine (non-phosphorylation mimicking) or aspartate (phosphorylation mimicking). Substitutions were confirmed by DNA sequencing, and the plasmids with single-amino-acid substitutions were subsequently introduced into the isogenic Δmga mutant strain. The primers used for site-directed mutagenesis are listed in Table S2 in the supplemental material.
Construction of hexahistidine-tagged Mga.
Plasmid pDC-mga containing the wild-type mga coding region was used as a template. Using site-directed mutagenesis, we introduced a hexahistidine tag at the C-terminal end of the mga coding region. The presence of the histidine tag was confirmed by DNA sequencing, and the plasmid containing mga-His6 (pDC-mga-His6) was subsequently electroporated into the isogenic Δmga mutant strain, to create the wild-type–His6 strain. The primers used to introduce the histidine tag are listed in Table S2 in the supplemental material.
RNA-seq analysis.
GAS strains were grown in THY broth to the late exponential growth phase (A600 of ∼1.0), and cells were collected by centrifugation. RNA isolation and purification were performed by using an RNeasy minikit (Qiagen), and cells were treated with DNase by using the Turbo DNA-free kit (Ambion). Purified RNA was evaluated for quality and concentration by using an Agilent 2100 bioanalyzer and fluorometric quantitation (Qubit RNA assay kit; Life Technologies). The rRNA was then removed by using a Ribo-Zero treatment kit (Epicenter) and further purified by using the Min-Elute RNA purification kit (Qiagen). The ribosomally depleted RNA was then used to synthesize cDNA libraries containing adaptor-tagged libraries by using the ScriptSeq V2 RNA-seq library preparation kit (Epicentre). The directional cDNA library was prepared and indexed at a single end by using reverse primers (Epicentre) and amplified by 15 cycles of PCR. The purified cDNA libraries were then run on a MiSeq instrument using the MiSeq v2 reagent kit (Illumina). Approximately 2 million reads were obtained per sample, and the reads were mapped to the MGAS15252 genome by using CLC-Biosystems WorkBench, version 7 (CLC Bio). The expression values were obtained as RPKM (reads per kilobase of exon model per million mapped reads) and normalized by using quantile normalization. Differential expression was computed by using Baggerly's test on proportions, and P value correction for multiple testing was performed by using the Bonferroni correction. Genes were considered differentially expressed if the fold change was >1.5 and the Bonferroni-corrected P value was <0.05.
Phos-tag SDS-PAGE analysis of GAS cell lysates for phosphorylated Mga.
GAS strains were grown to the late exponential phase of growth (A600 of ∼1.0). Cell pellets were resuspended in buffer A (20 mM Tris-HCl [pH 8.0] supplemented with protease inhibitor set III [Calbiochem]) and lysed by using the FastPrep cell disruptor (MP Bio). Cleared cell lysates were subjected to methanol precipitation, and the resulting pellet was air dried at room temperature. The pellet was suspended in 20 mM Tris-HCl (pH 8.0), and the total protein concentration was measured by a Bradford assay (27). To remove the phosphoryl modification of histidines in Mga, cell lysates of the Δmga:pDC-mga-His6 strain were treated with 500 mM hydroxylamine-HCl for 1 h at 37°C. Typically, the lysates contain ∼1.3 μg/μl of total protein content. Samples (∼26 μg of total protein/sample) were subsequently diluted with loading buffer (15% 2-mercaptoethanol, 5% SDS, 50% glycerol, 0.01% bromophenol blue) and resolved on a 10% Bis-Tris-buffered Zn2+–Phos-tag SDS-PAGE gel (Wako Pure Chemicals) for 3 h at 4°C. The gel was washed once in transfer buffer (running buffer containing 20% methanol and 1 mM EDTA) and again in transfer buffer without EDTA. Samples were transferred onto a nitrocellulose membrane and probed with antihexahistidine monoclonal antibodies conjugated to horseradish peroxidase (Clontech). Mga was visualized by chemiluminescence using a SuperSignal West Femto IgG detection kit (Thermo Scientific).
Transcript analysis by quantitative RT-PCR.
GAS strains were grown to late exponential phase (A600 of ∼1.0) and incubated with 2 volumes of RNAprotect (Qiagen) for 10 min at room temperature. Bacteria were harvested by centrifugation, and the cell pellets were snap-frozen with liquid nitrogen. RNA isolation and purification were performed by using an RNeasy kit (Qiagen). Purified RNA was analyzed for quality and concentration by using an Agilent 2100 bioanalyzer. cDNA was synthesized from the purified RNA by using Superscript III (Invitrogen), and reverse transcription-quantitative PCR (qRT-PCR) was performed with an ABI 7500 Fast system (Applied Biosystems). Comparison of transcript levels was done by using the ΔCT method of analysis, using tufA as the endogenous control gene (see Fig. S1 in the supplemental material) (28). The TaqMan primers and probes used are listed in Table S2 in the supplemental material.
Virulence studies with mouse models of infection.
Virulence of the isogenic mutant strains of M59 GAS was assessed by using a mouse model of necrotizing fasciitis (8). Outbred immunocompetent CD1 mice (Charles River) were inoculated intramuscularly in the right hind limb with 5 × 107 CFU of each GAS strain. Near-mortality (n = 20 mice/strain) was determined by using previously defined criteria. Results were graphically displayed as a Kaplan-Meier survival curve, with a P value of <0.05 by the log rank test being considered to be statistically significant. Visual and microscopic examinations of infected limbs collected on day 5 postinoculation were performed as previously described (8). Tissues were examined independently by two pathologists blind to treatment groups. Representative micrographs were obtained with a BX5 microscope fitted with a DB70 digital camera (Olympus, Tokyo, Japan). GAS burden (n = 20 mice/strain) in infected limbs was determined as previously described (8, 14). Results were graphically displayed as the mean number of CFU recovered per gram of tissue, with a P value of <0.05 by the Mann-Whitney log rank test being considered to be statistically significant. Mouse experiments were performed according to protocols approved by the Houston Methodist Research Institute (HMRI) Institutional Animal Care and Use Committee (Office of Laboratory Animal Welfare [OLAW] assurance no. A4555-01; USDA assurance no. 740R-0192).
RNA-seq accession number.
The RNA sequencing data discussed in this publication have been deposited in NCBI's GENE Expression Omnibus and are accessible through GEO Series accession number GSE67988.
RESULTS
Growth phase-dependent gene regulation by Mga in M59 serotype strains.
Inasmuch as gene regulation in GAS is growth phase dependent (6, 29) and the kinetics of mga expression and mga-dependent gene regulation in serotype M59 strains are not known, we first measured the transcript levels of mga and its well-characterized regulatory target, the emm gene, at different stages of growth by qRT-PCR. Consistent with strains of other M protein serotypes, transcript levels of mga were maximal during exponential phases of growth and decreased during the stationary growth phase (Fig. 1). The emm transcript level mirrored the mga expression level, as it reached a peak during the mid- and late exponential phases of growth, whereas it decreased during the stationary phase (Fig. 1). These results are consistent with a regulatory influence of mga on emm expression in serotype M59 GAS. Collectively, these data indicate that Mga exerts a greater influence on the expression of its target genes during the exponential growth phase. Thus, analysis of gene regulation by Mga at this growth phase may help identify the regulatory targets of Mga in serotype M59 GAS.
FIG 1.
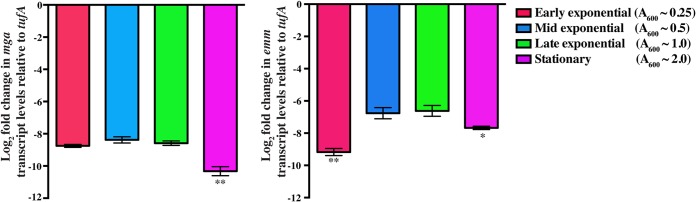
mga and emm are expressed at maximal levels during exponential phases of growth. Transcript levels of mga and emm in strain MGAS15249 were measured by qRT-PCR. Samples were collected at the indicated growth phases for transcript analysis. Three biological replicates were grown and analyzed in triplicate. Data graphed are means ± standard deviations. Values with single asterisks (P < 0.01) and double asterisks (P < 0.001) indicate statistically significantly different transcript levels compared to the late exponential phase of growth.
Gene regulation by Mga in serotype M59 strains.
Transcriptome analyses to identify Mga-regulated genes in GAS serotypes M1, M4, and M6 have been previously reported (3). The general finding is that the composition of the mga regulon and the degree of gene regulation by Mga showed significant variation among the tested serotypes (3). The mga regulon in serotype M59 has not been defined. Given that the regulatory function of Mga is under strong selective pressure in serotype M59 during natural infection (13, 14), determination of the mga regulon in this genetic background might shed light on Mga-regulated genes that contribute to GAS pathogenesis in these organisms. We constructed an insertionally inactivated isogenic mga mutant strain (Δmga) from reference serotype M59 strain MGAS15249. In this construct, the majority of the mga coding region (codons 6 through 533) is replaced with a spectinomycin cassette. Next, we complemented this isogenic mutant strain in trans with plasmid pDC-mga, containing the entire coding region of mga fused with its native promoter. Successful complementation of the mutant was verified by analyzing mga transcript levels in the MGAS15249:pDC, Δmga:pDC, and Δmga:pDC-mga strains by qRT-PCR (see Fig. S2 in the supplemental material). Target gene regulation by the trans-complemented strain was also confirmed by analysis of the transcript levels of emm by qRT-PCR, indicating that mga expressed from the complementation plasmid is functional in vivo (see Fig. S3 in the supplemental material). The isogenic Δmga mutant strain trans-complemented with wild-type mga (Δmga:pDC-mga) was used as a control. Using the wild-type and Δmga strains, we performed comparative genome-wide transcription profiling by RNA-seq analysis. Briefly, cells were grown to late exponential phase (A600 of ∼1.0) in THY medium, and RNA purification was performed and cDNA libraries were generated by using commercially available kits. Genes were considered differentially regulated between the two strains if the fold change was >1.5 and the Bonferroni-corrected P value was <0.05.
We found that Mga influences the expression of 144 genes (∼7% of the 1,951 total predicted genes in the genome), with 70 genes being upregulated and 74 genes being downregulated (Fig. 2A to D; see also Table S3 in the supplemental material). Consistent with data reported previously for strains of other M protein serotypes, the differentially regulated genes belonged to 3 major categories: virulence factors, nutrient metabolism and transport, and uncharacterized hypothetical proteins (see Table S3 in the supplemental material). Virulence genes upregulated by Mga include components of the core mga regulon identified in other serotypes, such as the emm, mrp, scpA, enn, sclA, sof, and sfbX genes (Fig. 2D; see also Table S3 in the supplemental material). However, a key difference was that the magnitude of the Mga regulatory influence was more pronounced in serotype M59 than in other tested serotypes (3). Additional virulence factors controlled by Mga in this serotype included the operon encoding NAD-glycohydrolase (NADase) (nga), the intracellular inhibitor of nga (ifs), streptolysin O (SLO) (slo), and 3 small, putative secreted peptides (MGAS15252_0178, MGAS15252_0179, and MGAS15252_0181) (Fig. 2B; see also Table S3 in the supplemental material). Uncharacterized hypothetical small peptides and proteins form one of the major groups of genes activated by Mga. Consistent with previously reported observations of strains of other serotypes, many genes involved in nutrient metabolism and transport were downregulated by Mga (see Table S4 in the supplemental material). Taken together, these data show that Mga is a regulator of global gene expression in serotype M59 GAS.
FIG 2.
Genome-wide transcript analysis of the Δmga strain compared to the wild type. (A) Gene expression in the MGAS15249 Δmga strain. The relative gene expression level in the Δmga:pDC strain compared to the Δmga:pDC-mga strain is shown. Mean fold changes in transcript levels of differentially regulated genes and their respective genomic coordinates are shown (P < 0.05 compared to the wild type, determined by Baggerly's test after applying Bonferroni's correction for multiple comparisons). (B to D) Genetic arrangement of significantly regulated genes in the Δmga:pDC strain (genes with a >10-fold change in transcript levels).
Mga is phosphorylated in vivo.
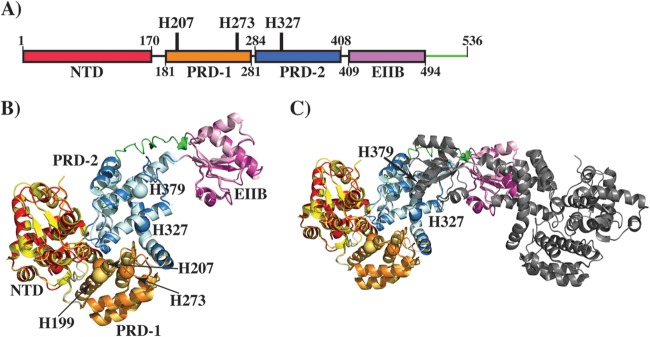
To deduce the molecular mechanism of gene regulation by Mga, we first performed modeling studies with I-TASSER (30–32). Mga is predicted to share a high degree of structural homology with AtxA, a PRD-containing activator from Bacillus anthracis (PDB accession number 4R6I) (10, 33, 34). The predicted model has a high template modeling (TM) score of 0.62 ± 0.14 (a score of >0.5 is reliable), suggesting that the model has the correct topology (30–32). Furthermore, the model superimposed well with the AtxA structure, with a root mean square deviation of 0.8 Å over 420 amino acids, indicating structural similarity between the two proteins (Fig. 3A and B). In light of this new information, we redefined the domain boundaries within Mga and attempted to elucidate the mode of gene regulation. The model shares 4 domains with AtxA: an N-terminal DNA-binding domain with two HTH motifs (amino acids 1 to 170), PRD-1 (amino acids 181 to 281), PRD-2 (amino acids 284 to 408), and the EIIB domain (amino acids 409 to 492) (Fig. 3A and B). In addition, the amino acid sequence of Mga has an additional 44 amino acids in its C terminus (Fig. 3B). Of note, the side chain of H207 aligns well structurally with the phosphorylatable histidine (H199) of AtxA PRD-1, suggesting that Mga function may be influenced by phosphorylation of H207 (Fig. 3B and C).
FIG 3.
Modeling studies of Mga. (A) Schematic representation of the domain architecture of Mga. Individual domains are labeled, and the domain boundaries are indicated by the amino acid numbering at the bottom. The locations of phosphorylatable histidines are indicated and labeled. (B) Ribbon representation of the Mga model, as predicted by I-TASSER, superimposed on the structure of AtxA from B. anthracis (PDB accession number 4R6I). Individual domains of Mga are color-coded as shown in panel A. Individual domains of AtxA are color-coded as follows: the N-terminal domain (NTD) is in yellow, PRD-1 is in brown, PRD-2 is in light blue, and the EIIB domain is in light pink. An additional C-terminal fragment in Mga is indicated in green. Phosphorylatable histidines, H207 and H273 of PRD-1 and H327 of PRD-2, are shown as spheres and marked by arrows. A phosphorylatable histidine (H199) in PRD-1 of AtxA is shown as an orange sphere. (C) Ribbon representation of the model of the Mga dimer. One subunit is color-coded as described above for panel B, and the second subunit is shown in gray. The orientation of the model in panel C is tilted and rotated on the x axis relative to that in panel B for a better representation of the dimer.
We next investigated if Mga in the serotype M59 strain undergoes phosphorylation at histidines. Although recent data showed that recombinant purified Mga is phosphorylated in vitro by recombinant PTS components (10), evidence regarding the phosphorylation status of Mga in vivo is lacking. To investigate if Mga is phosphorylated in vivo, we conducted Phos-tag SDS-PAGE and immunoblot analysis of GAS cell lysates. The Phos-tag compound specifically forms a complex with phosphorylated amino acids in the presence of divalent cations (35–37). Phosphorylated proteins complexed with the Phos-tag compound migrate more slowly in Phos-tag SDS-PAGE gels than do their nonphosphorylated counterparts (11, 36–38). To detect Mga in GAS cell lysates, we constructed C-terminally hexahistidine-tagged Mga (Mga-His6). To ensure that modified Mga retained regulatory activity, we measured the transcript levels of the emm gene in strains containing either native Mga or Mga-His6 by qRT-PCR. The modification did not affect the regulatory function of Mga-His6 (see Fig. S3 in the supplemental material). Furthermore, we tested the ability of a monoclonal anti-His6 antibody to detect Mga in cell lysates by immunoblotting. The results demonstrated that the cell lysates of the Δmga:pDC or Δmga:pDC-mga (native) strain lacked an immunoreactive signal, whereas the lysates of the Δmga:pDC-mga-His6 strain had a reactive band corresponding to the predicted molecular weight of Mga (Fig. 4A). This result shows that the antibody can detect Mga in bacterial cell lysates (Fig. 4A).
FIG 4.
Mga is phosphorylated in vivo. The isogenic mga mutant strain trans-complemented with an empty vector (Δmga:pDC), the wild-type mga strain (Δmga:pDC-mga), or the hexahistidine-tagged wild-type mga strain (Δmga:pDC-mga-His6) was grown in THY medium to the late exponential growth phase (A600 of ∼1.0). Cells were lysed, and cell lysates were clarified by methanol precipitation. (A) Equal concentrations of cell lysates of the Δmga:pDC (lane 1), Δmga:pDC-mga (lane 2), and Δmga:pDC-mga-His6 strains were resolved on a 10% SDS-PAGE gel and probed by monoclonal antihexahistidine antibodies. (B) Equal concentrations of cell lysates of the Δmga:pDC strain (lane 1), the Δmga:pDC-mga-His6 strain (lane 2), and the Δmga:pDC-mga-His6 strain treated with hydroxylamine (lane 3) were resolved on a 10% SDS-PAGE gel containing 50 μM Phos-tag and ZnSO4 and probed by monoclonal antihexahistidine antibodies. Bands corresponding to nonphosphorylated Mga (Mga-His6) and phosphorylated Mga (Mga-P-His6) are labeled and indicated by arrows.
To test the hypothesis that Mga is phosphorylated in vivo, isogenic Δmga mutant strains trans-complemented with either an empty vector (Δmga:pDC) or Mga-His6 (Δmga:pDC-mga-His6) were grown to late exponential phase in THY medium. Subsequently, we subjected the cell lysates to Phos-tag SDS-PAGE to investigate the presence of phosphorylated Mga. Consistent with our hypothesis, we observed two bands corresponding to Mga, a slow-migrating species likely arising from phosphorylated Mga and a fast-migrating band corresponding to nonphosphorylated Mga (Fig. 4B). A similar analysis of cell lysates on SDS-PAGE gels without Phos-tag revealed only one band, indicating that retarded migration of Mga occurs only in Phos-tag gels, likely due to Mga phosphorylation (Fig. 4B). Since Mga is hypothesized to be phosphorylated at conserved histidine residues and histidine phosphorylation is acid labile, we performed Phos-tag analysis of cell lysates treated with hydroxylamine-HCl (39). In accordance with histidine phosphorylation of Mga, the slow-migrating band was absent in the hydroxylamine-HCl-treated samples, suggesting that the shifted species corresponds to Mga phosphorylated at one or more its conserved PRD histidines (Fig. 4B). Together, these data provide the first evidence that Mga is phosphorylated at PRD histidine residues in vivo.
Phosphorylation- and non-phosphorylation-mimicking amino acid substitutions at conserved PRD-1 histidines exert a significant influence on the regulatory activity of Mga.
We next investigated the influence of phosphorylation events on the Mga-mediated regulation of its target genes by introducing amino acid substitutions at conserved PRD-1 histidines that mimic phosphorylated (His-to-Asp) or nonphosphorylated (His-to-Ala) states. PRD-1 (amino acids 181 to 281) of Mga has 6 histidines (H201, H207, H234, H242, H260, and H273), and PRD-2 (amino acids 284 to 408) has 3 histidines (H303, H327, and H358). Previous analyses of M1 and M4 serotypes identified H207 and H273 of PRD-1 (numbered H204 and H270 in the previous study) as the sites for PTS-mediated phosphorylation (10). Furthermore, whole-genome sequencing analysis of serotype M59 strains identified H201 to be highly polymorphic, and a naturally occurring histidine-to-arginine replacement at H201 (H201R) caused increased GAS virulence (13, 14, 21). Thus, we performed mutational analysis of H201, H207, and H273 of PRD-1 and characterized the mutants for target gene regulation.
We used complementation plasmid pDC-mga as the template to introduce the single-amino-acid replacements at 3 PRD-1 histidines (H201, H207, and H273), and the resulting plasmids with phosphorylation- or non-phosphorylation-mimicking substitutions were subsequently introduced into the Δmga mutant strain. The Δmga mutant strain complemented with empty vector pDC123 (Δmga:pDC) was used as a negative control, and the Δmga:pDC-mga strain was used as a wild-type control. When grown in laboratory medium, growth characteristics of the wild type, the Δmga mutant strain (Δmga:pDC), and strains with substitutions at PRD-1 histidines did not differ significantly (data not shown). SDS-PAGE analysis of the mutant recombinant proteins indicated that the solubility of the mutant proteins was similar to that of wild-type Mga, suggesting that the mutations do not cause drastic structural defects (see Fig. S4 in the supplemental material).
To determine the regulatory consequences of phosphorylation or nonphosphorylation at PRD-1 histidines, we measured the transcript levels of mga and its regulatory targets, emm, scpA, and sclA, by qRT-PCR. Although an arginine substitution at H201 resulted in the upregulation of the target genes (21), neither an alanine nor an aspartate substitution at this position caused an alteration in the regulatory activity of Mga (Fig. 5). Consistent with this, it was proposed that the H201R mutation likely either interferes with the phosphorylation event indirectly or impedes the phosphorylation-dependent conformational changes in Mga and the resulting alterations in gene regulation (21). Together, these results suggest that H201 is an unlikely candidate for phosphorylation in Mga.
FIG 5.

Phosphorylation- and non-phosphorylation-mimicking substitutions at H201 of PRD-1 of Mga do not alter transcript levels of Mga-regulated genes. Transcript levels of mga, emm, scpA, and sclA were measured by qRT-PCR. Four biological replicates were grown and analyzed in triplicate. Data graphed are means ± standard deviations.
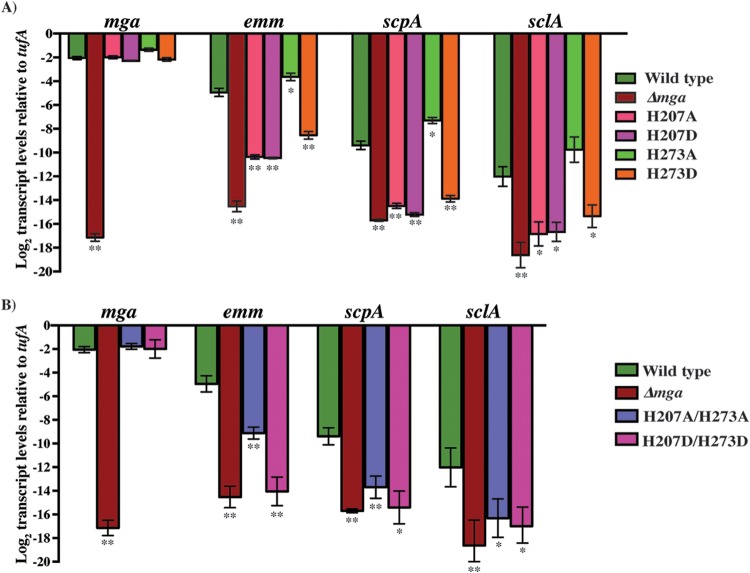
However, analysis of the transcript levels of phosphorylation- or non-phosphorylation-mimicking mutations at either H207 or H273 showed significantly altered gene regulation by Mga. Interestingly, depending on the histidine at which the substitutions were made, two distinct phenotypes were observed. First, regardless of the nature of the substitution, alterations at H207 resulted in a drastic decrease of Mga regulatory activity (Fig. 6A). Compared to the wild type, an alanine or aspartate substitution at H207 caused 32- to 48-fold reductions in the transcript levels of emm, scpA, and sclA (Fig. 6A). On the other hand, phosphorylation-mimicking (H273D) or non-phosphorylation-mimicking (H273A) substitutions at H273 had opposing effects on gene regulation by Mga. Importantly, the alanine substitution at H273 resulted in slightly elevated (2- to 4-fold) levels of regulatory activity, whereas the aspartate substitution caused a 12- to 24-fold reduction in the transcript levels of target genes compared to the wild type (Fig. 6A). Furthermore, with the exception of the H207A substitution, the regulatory profile observed for the mga mutants in our study is reminiscent of the phenotypes that are typically observed in PRD-containing proteins; i.e., phosphorylation- and non-phosphorylation-mimicking substitutions at phosphorylatable PRD histidines result in opposing regulatory outcomes (40). We next investigated whether the introduction of double-alanine (A/A) or double-aspartate (D/D) substitutions at H207 and H273 augments the regulatory effect of single-alanine or -aspartate substitutions. The phenotype of mutant strains with either A/A or D/D substitutions resembled that of single-alanine or -aspartate mutants of H207, indicating that substitutions at H207 of Mga have a dominant-negative phenotype (Fig. 6B). Collectively, these data indicate that phosphomimetic substitution at H207 or H273 inhibits Mga-dependent gene regulation, whereas nonphosphomimetic substitution at H273 relieves this inhibition.
FIG 6.
Phosphorylation- and non-phosphorylation-mimicking substitutions at H207 and H273 of PRD-1 of Mga alter transcript levels of Mga-regulated genes. Isogenic mutant strains with single (A) or double (B) phosphorylation- and non-phosphorylation-mimicking substitutions at H207 and H273 of PRD-1 were grown to the exponential phase of growth. Transcript levels of mga, emm, scpA, and sclA in the indicated strains were measured by qRT-PCR. Four biological replicates were grown and analyzed in triplicate. Data graphed are means ± standard deviations. Values with single asterisks (P < 0.05) and double asterisks (P < 0.0005) indicate statistically significantly different transcript levels compared to the wild type.
Phosphorylation- and non-phosphorylation-mimicking substitutions at conserved PRD-2 histidines play a lesser role in the regulation of Mga activity.
Since Mga is predicted to contain two PRDs and PRD-2 has two conserved histidines (H327 and H358), we sought to determine if these two histidines played a role in gene regulation. Single-alanine or -aspartate substitutions were introduced at the two histidines (H327 and H358), and the resulting strains were characterized for target gene regulation by qRT-PCR. Substitutions at H358 did not alter the transcript levels of emm, scpA, and sclA, indicating that H358 is an unlikely target for phosphoregulation in Mga (Fig. 7). However, single-alanine or -aspartate substitutions at H327, H327A, and H327D caused opposing effects, albeit to a lesser extent than the analogous substitutions at H207 and H273, on gene transcripts (Fig. 7). Although the non-phosphorylation-mimicking substitution (H327A) had transcript levels comparable to those of the wild type, the phosphorylation-mimicking aspartate mutation resulted in a modest 2- to 3-fold decrease in the transcript levels of target genes (Fig. 7). Consistent with these observations, analogous studies of serotype M1/M4 identified H327 as a phosphorylation site in Mga (10). Together, these data indicate that phosphorylation and nonphosphorylation events at H327 of PRD-2 mediate a lesser influence on Mga-mediated gene regulation than the phosphorylation events at PRD-1 histidines.
FIG 7.

Phosphorylation- and non-phosphorylation-mimicking substitutions at H327 of PRD-2 of Mga alter transcript levels of Mga-regulated genes. Transcript levels of mga, emm, scpA, and sclA in the indicated strains were measured by qRT-PCR. Four biological replicates were grown and analyzed in triplicate. Data graphed are means ± standard deviations. Values with single asterisks (P < 0.05) and double asterisks (P < 0.0005) indicate statistically significantly different transcript levels compared to the wild type.
Phosphorylation/non-phosphorylation-mimicking substitutions at PRD-1 histidines affect Mga-dependent gene regulation at the global level.
Given the greater influence of phosphorylation/non-phosphorylation-mimicking mutations at PRD-1 histidines than at PRD-2 histidines on Mga-dependent gene regulation, we investigated the effect of these mutations on global gene regulation by Mga. Relevant strains were grown to late exponential phase in laboratory medium, and genome-wide transcriptome analysis was performed by RNA-seq analysis. In accordance with the qRT-PCR results, 3 distinct profiles of gene expression were observed (Fig. 8A and B). First, both substitutions at H207 resulted in an altered expression profile, with the core mga regulon being significantly downregulated (Fig. 8A and B; see also Tables S5 and S6 in the supplemental material). Second, depending on the nature of substitutions at H273, an opposing pattern of gene regulation was observed. Specifically, the non-phosphorylation-mimicking substitution at H273 (H273A) had a pattern comparable to that of the wild type, whereas the phosphorylation-mimicking mutation (H273D) caused a significant downregulation of Mga-regulated genes (Fig. 8A and B; see also Tables S7 and S8 in the supplemental material). Finally, the phosphomimetic substitutions at either H207 or H273 resulted in similar genome-wide transcription profiles, with the H207D substitution causing a more pronounced effect on Mga-dependent gene regulation (Fig. 8A and B; see also Tables S5 and S6 in the supplemental material). Together, these results demonstrated that phosphorylation-mimicking substitutions of PRD-1 histidines downregulate Mga-dependent gene expression at the global level, whereas a non-phosphorylation-mimicking substitution at H273 of PRD-1 results in upregulation of Mga regulatory activity.
FIG 8.

Phosphorylation/non-phosphorylation-mimicking substitutions at H207 and H273 of PRD-1 of Mga alter the global mga transcriptional profile. (A) Heat map depicting the alterations in the gene expression pattern at the global level in the indicated strains. (B) Heat map of transcript levels of selected genes in the indicated strains relative to the wild type. The scheme for the color values is shown at the bottom. Red in the key represents downregulated genes, black represents genes not detected/not differentially expressed, and green represents upregulated genes in the indicated strains relative to the wild type.
Given the lack of opposing phenotypes for phosphorylation- and non-phosphorylation-mimicking substitutions at H207, we investigated whether the phosphorylation of H207 occurs in vivo. By using C-terminally hexahistidine-tagged mga (pDC-mga-His6) as the template, single- or double-alanine substitutions were introduced at H273 of PRD-1 and H327 of PRD-2. Cells were grown to late exponential phase in THY medium, and cell lysates were subjected to Phos-tag SDS-PAGE. Consistent with our hypothesis that H207 is phosphorylated in vivo, a slow-migrating species corresponding to phosphorylated Mga was observed with both single- and double-alanine substitutions at H273 and H327 (Fig. 9). Thus, results from Phos-tag analysis indicated that single- or double-alanine substitutions at H273 and H327 do not prevent Mga phosphorylation and that additional phosphorylation events occur in Mga, presumably at H207.
FIG 9.
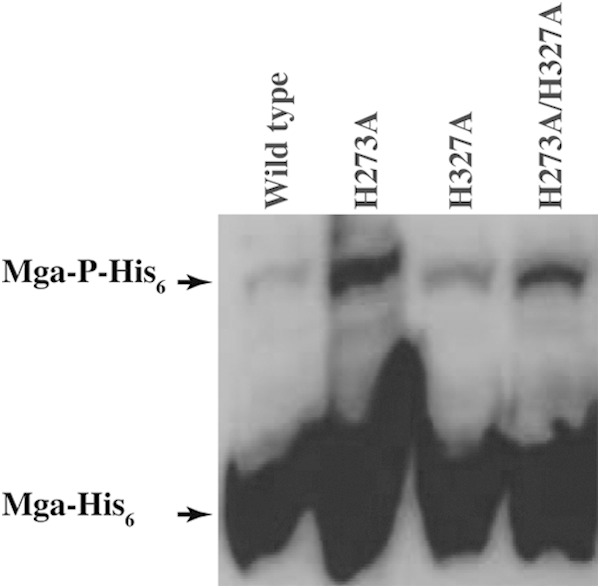
In vivo phosphorylation status of the wild type and various alanine mutants of Mga, as assessed by Phos-tag immunoblot analysis. Equal concentrations of cell lysates from the indicated strains were resolved on a 10% SDS-PAGE gel containing 50 μM Phos-tag and ZnSO4 and probed by monoclonal antihexahistidine antibodies. Bands corresponding to nonphosphorylated Mga (Mga-His6) and phosphorylated Mga (Mga-P-His6) are labeled and indicated by arrows.
Phosphorylation/non-phosphorylation-mimicking substitutions at H273 alter virulence in a mouse model of necrotizing fasciitis.
To test the hypothesis that modulation of Mga regulatory activity by phosphorylation/non-phosphorylation-mimicking substitutions at H273 contributes to GAS virulence, we compared the Δmga strain and the Δmga strain trans-complemented with either wild-type mga or individual mutants in an intramuscular mouse model of infection. Consistent with the results from RNA-seq studies, the wild-type and non-phosphorylation-mimicking H273A mutant strains were more virulent than the Δmga and phosphorylation-mimicking H273D mutant strains (P < 0.05) (Fig. 10A). We hypothesized that the increased virulence of the wild-type and H273A mutant strains would manifest as a significantly altered tissue phenotype. To test this hypothesis, we performed visual and microscopic examinations of the tissue lesions. Consistent with the near-mortality data, mice infected with the wild-type and H273A mutant strains exhibited larger lesions with more severe tissue damage and local dissemination than did mice infected with the Δmga and H273D mutant strains (Fig. 10B). Given that target genes regulated by Mga are implicated in host cell attachment and immune evasion, we next hypothesized that phosphorylation/non-phosphorylation-mimicking mutants would alter the bacterial burden in vivo. To this end, we compared the GAS CFU recovered from infected limbs. The results demonstrated that significantly more CFU per gram were recovered from the lesions of mice infected with the wild type than from the lesions of mice infected with the Δmga mutant (P < 0.05), suggesting that Mga-dependent gene regulation is critical for GAS survival during infection (Fig. 10C). Furthermore, an increased bacterial burden was observed for the lesions from mice infected with the non-phosphorylation-mimicking H273A mutant compared to the phosphorylation-mimicking H273D mutant (P < 0.05) (Fig. 10C). Together, these data demonstrate that the regulatory activity of Mga is critical for GAS survival in vivo and that modulation of Mga function by phosphorylation/non-phosphorylation-mimicking substitutions at H273 contributes significantly to M59 GAS virulence.
FIG 10.

Phosphorylation- and non-phosphorylation-mimicking substitutions at H273 of PRD-1 of Mga alter M59 GAS virulence. (A) Twenty mice were infected with each indicated strain intramuscularly, and near-mortality was graphed as a Kaplan-Meier survival curve. Statistical significance between strains, as assessed by a log rank test, is indicated. (B) Histological examination of muscular lesions from mice infected with the indicated strains. The confined, less destructive lesions are indicated by arrowheads, and the areas of damaged muscle tissue with more destructive lesions are boxed. (C) Twenty mice were infected intramuscularly, and mean CFU recovered from the infected muscle tissue are shown, with P values being determined by a t test.
Both phosphorylation- and non-phosphorylation-mimicking substitutions at H207 attenuate GAS virulence in a mouse model of necrotizing fasciitis.
Unlike the opposing effects of phosphorylation- and non-phosphorylation-mimicking substitutions at H273 on Mga-dependent gene regulation, both substitutions at H207 resulted in a downregulation of Mga-controlled target genes in vitro. To investigate whether the observed in vitro phenotype of H207 substitutions correlates with GAS pathogenicity in vivo, we compared the Δmga strain and the Δmga strain trans-complemented with either wild-type mga or individual H207 mutants for near-mortality in an intramuscular mouse model of infection. Consistent with the results from RNA-seq studies and analyses of transcript levels, both alanine and aspartate substitutions at H207 caused a significant attenuation of GAS virulence compared to the strain trans-complemented with wild-type mga (P < 0.05) (Fig. 11). Together, these data indicate that both phosphomimetic and nonphosphomimetic substitutions at H207 significantly alter M59 GAS virulence.
FIG 11.
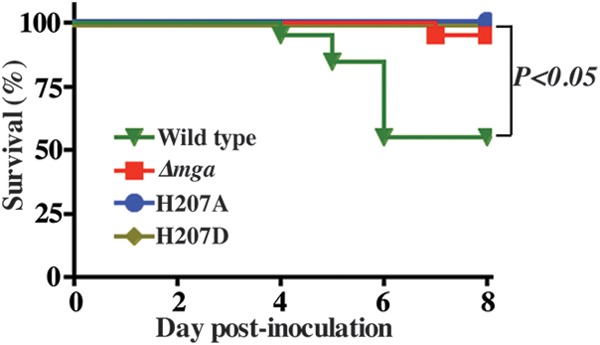
Phosphorylation- and non-phosphorylation-mimicking substitutions at H207 of PRD-1 of Mga attenuate M59 GAS virulence. Twenty mice were infected with each indicated strain intramuscularly, and near-mortality was graphed as a Kaplan-Meier survival curve. Statistical significance between strains, as assessed by a log rank test, is indicated.
DISCUSSION
Population genomic analyses of the bacterial isolates recovered from epidemics have provided unprecedented insights into the evolution of the pathogens, genetic changes in the bacterial genome that contribute to altered virulence, and loci that are under strong selection during natural infection (14–16, 41–46). Whole-genome sequencing analyses of ∼800 strains of GAS serotype M59 from invasive epidemics revealed that the mga locus is under strong selection pressure, suggesting a critical role for the regulatory function of Mga in the pathogenesis of serotype M59 GAS (13, 14, 17). Consistent with this, a naturally occurring single-amino-acid replacement in Mga, H201R, caused upregulation of various Mga-regulated toxins and conferred a hypervirulence phenotype compared to the wild type (21). These data prompted us to investigate the molecular mechanism of gene regulation by Mga and the serotype-specific contribution of Mga to M59 GAS pathogenesis.
Our findings demonstrated that Mga exerts a significant regulatory influence on GAS gene expression in the M59 serotype. Depending on the composition of the core mga regulon and the amino acid sequence of Mga, it has been classified into two distinct alleles, mga-1 and mga-2 (18, 19, 47). To obtain insights into the serotype-specific contribution of Mga to GAS pathogenesis, we compared the transcriptomes controlled by mga-2 alleles from two different GAS serotypes, M59 from our study and M4 from a previous study. As expected, the core regulon that is composed of the emm, mrp, scpA, enn, sclA, sof, and sfbX genes is highly regulated in both serotypes (3, 18). In addition, Mga in M59 GAS influences the expression of an operon encoding streptolysin O (SLO) and NAD-glycohydrolase (NADase) and its intracellular inhibitor immunity factor for Streptococcus pyogenes NADase (IFS). Both SLO and NADase are proven virulence factors and have been implicated in intracellular survival and GAS resistance to macrophage-mediated cytotoxicity (48–52). Beyond the core regulon, genes regulated by Mga in the two serotypes diverge significantly and point toward differing metabolic capabilities of the two serotypes. Importantly, Mga-regulated genes in the M4 serotype involved the uptake of maltose (malE-malG operon involved in maltose transport and metabolism [malE-G]), glucose (ptsG), and mannose (ptsA-ptsD operon [ptsA-D]) as well as regulatory proteins (ccpA and licT) involved in carbohydrate metabolism (3). In contrast, the transcriptional response controlled by Mga in the M59 serotype involved nucleotide metabolism (pyrP, pyrB, MGAS15252_0417, MGAS15252_0325, mutY, rnhC, and the pur operon), amino acid metabolism (glnP, glnQ, glnA, arcDABC, and carAB), fatty acid biosynthesis (fab and acc operons), and mannose uptake systems (manXZ) (see Tables S3 and S4 in the supplemental material). Furthermore, Mga controls the expression of the iron uptake system (siuADBG) in the M4 serotype, whereas it upregulates zinc acquisition systems (lmb and phtD) in the M59 serotype (see Tables S3 and S4 in the supplemental material) (3). Together, these data indicate the delicate differences in the serotype-specific regulation of adaptive responses by Mga and highlight the important metabolic differences between the two serotypes. Although these results support the hypothesis that Mga mediates gene regulation in a serotype-specific fashion, further investigations into the contribution of these Mga-controlled metabolic pathways to GAS serotype M59 pathogenesis will be required to validate this hypothesis.
We have demonstrated that the regulatory activity of Mga is modulated by phosphorylation- and non-phosphorylation-mimicking substitutions at each of the 3 conserved histidines, H207 and H273 of PRD-1 and H327 of PRD-2 of Mga. Although the phenotypes of alanine and aspartate substitutions at the conserved PRD histidines of Mga are consistent with the mode of phosphoregulation observed for other previously characterized PRD-containing transcription regulators, alternative possibilities cannot be ruled out. These possibilities include potential direct or indirect roles for the PRD histidines in DNA binding and multimerization, structural changes in Mga required to mediate gene regulation, and protein-protein interactions with transcription machinery. However, similar studies of GAS serotype M4 demonstrated that the 3 histidines identified in our study, H207 and H273 of PRD-1 and H327 of PRD-2, are the target sites for PTS-dependent Mga phosphorylation in vitro (10). Thus, findings from studies of serotype M4 lend further support to the conclusion that alanine and aspartate substitutions at the conserved PRD histidines of Mga mimic nonphosphorylated and phosphorylated states, respectively.
Phosphoryl modification at each of the 3 histidines inhibits Mga-dependent gene regulation, whereas the lack of phosphorylation at either H273 or H327 positively influences Mga regulatory activity. The phenotype of the H207A mutant was an exception to the opposing modes of phosphoregulation, as the H207A mutation failed to reverse the phosphorylation-induced loss of Mga regulatory function. To understand the underlying cause for the observed H207A phenotype, we analyzed the local environment of H207 in the Mga model (Fig. 3A to C). The location of H207 in the Mga model corresponds to phosphorylatable H199 in the crystal structure of AtxA (33). The side chain of H199 in AtxA is involved in an H-bonding network, with the amino acids in the loop connecting PRD-1 and the N-terminal DNA-binding domain. Thus, it was proposed that the side chain of H199 in its nonphosphorylated state interacts with the interdomain loop and locks the DNA-binding domain of AtxA in an apo state. Conversely, phosphorylation of H199 disrupts the interactions between PRD-1 and the DNA-binding domain, which releases the DNA-binding domain to interact with its cognate DNA (33). However, phosphorylations of H199 in AtxA and of H207 in Mga result in opposing regulatory outcomes, with the phosphoryl modification of H199 causing the activation of AtxA regulatory activity and H207 resulting in an inhibition of Mga regulatory activity. Given the structural homology between AtxA and Mga, it is likely that H207 in Mga is also involved in similar critical interdomain interactions. However, contrary to the proposed mechanism of phosphoregulation in AtxA, the locked conformation favored by the interactions between nonphosphorylated H207 and the DNA-binding domain might represent the DNA-bound conformation of Mga. Consequently, phosphorylation of H207 might disrupt these interactions and inhibit DNA binding and transcription activation by Mga. In this model, both phosphomimetic and nonphosphomimetic substitutions at H207 might disrupt the interdomain contacts and result in inhibition of Mga regulatory activity. Failure of the mutants to mimic their respective phosphorylated states occurs and has been documented for other PRD-containing regulators (34, 53, 54).
Despite the similarities between studies on phosphorylation of Mga in two GAS serotypes, the regulatory consequences of Mga phosphorylation in serotype M4 are quite different from our observations of serotype M59 (10). First, single-aspartate or -alanine substitutions at conserved histidines failed to alter Mga regulatory activity in a serotype M4 strain (10). In contrast, single-amino-acid substitutions at PRD histidines in serotype M59 GAS markedly altered the regulatory phenotype. Second, the phenotypes of non-phosphorylation-mimicking substitutions at the histidines are indistinguishable from those of the respective phosphorylation-mimicking mutations, as both substitutions reduced Mga activity in a serotype M4 strain (10). Consistent with the existing model of phosphoregulation, non-phosphorylation-mimicking substitutions at either H273 or H327 exhibited opposing phenotypes of H273D or H327D in serotype M59 GAS. Finally, phosphorylation at PRD-2 H327 was proposed to induce Mga activity in a serotype M4 strain (10). Contrary to this, phosphorylation at H327 negatively impacts Mga regulation in serotype M59 GAS. Comparison of the Mga model and the AtxA crystal structure provided key insights into the differing structural and functional consequences of phosphorylation of the PRD-2 histidines. The side chain of phosphorylatable PRD-2 H379 in AtxA is located in the interface between PRD-2 and the EIIB dimerization domain (Fig. 3C). Thus, phosphorylation of H379 was proposed to weaken the dimer interface and inhibit AtxA-dependent gene regulation by causing defective dimerization (10). Contrarily, the side chain of H327 is not involved in dimerization interactions and is situated at the interface between PRD-1 and PRD-2 (Fig. 3C). As a result, phosphorylation of H327 is likely to have a lesser influence on Mga multimerization but might impact Mga-DNA interactions indirectly by its interference with the interactions between PRD-1 and DNA-binding domains. The observed regulatory differences between the two studies were further extended to the virulence phenotype. In the previous study, both the phosphomimetic and nonphosphomimetic substitutions attenuated GAS virulence in mouse models of infection (10). However, in accordance with the transcription profiling and qRT-PCR results for serotype M59 GAS, a strain with a non-phosphorylation-mimicking substitution at H273 exhibited the wild-type virulence phenotype, whereas the phenotype of the H273D mutant was comparable to that of the Δmga strain. Although these observations underscore the increased sensitivity of the Mga regulatory network to its cognate environmental stimuli in serotype M59, the genetic elements that account for the differential response of Mga to phosphoregulation in different GAS serotypes remain unknown. The genome of strain MGAS15249 used in this study is fully sequenced and devoid of function-altering polymorphisms in major GAS regulators. However, the genome sequence of the serotype M4 strain used in previous studies has not been reported.
PTS-mediated phosphoregulation has been well documented for several PRD-containing bacterial antiterminators and transcription activators (55, 56). The regulatory proteins often contain two PRDs, of which one PRD is modified by the phosphorylated E1-HPr system and the second PRD is phosphorylated by the sugar-specific EIIB enzyme (56). Phosphorylation of each PRD has opposing effects on the regulatory activity of PRD-containing proteins (56). However, variations of the general principles exist in the number of phosphorylatable histidines in each PRD, the regulatory effect of the phosphorylation of individual PRDs, and the role of PTS components in the phosphorylation of PRDs (33, 34, 40, 53, 57–59). Intriguingly, phosphorylation at both PRD-1 and PRD-2 histidines negatively influences the regulatory activity of Mga, which is contrary to the opposing effect of phosphorylation of PRD1 and PRD2 observed in the majority of the PRD-containing regulators (34, 53, 59, 60). An exception to this phenotype also occurs in the regulation of the antiterminator LicT from Bacillus subtilis, in which the phosphorylation events at both domains result in the same regulatory outcome (58). Additionally, the magnitude of regulation exerted by each phosphorylation event in Mga varies, with the modification of H207 displaying a drastic effect, followed by an intermediate effect of H273 mutations and a modest effect of H327 mutations. This raises the possibility that each phosphorylation event is additive toward the inhibition of Mga regulatory activity and that the incremental inhibition of Mga occurs in concert with altering environmental cues by sequential Mga phosphorylation. Given that phosphorylation of the PRD histidines inhibits transcription activation by Mga, the possible mechanism of Mga activation remains unknown. Possibilities include activation by dephosphorylation and the consequent relief of inhibition, as occurs in LicT from B. subtilis (58), or additional unknown genetic elements that contribute to Mga induction. The observation that induction in MtlR from B. subtilis occurs by direct phosphorylation of a cysteine (C419) located in the EIIB domain lends support to the latter hypothesis, as Mga has a cysteine, C444, located in its EIIB domain (40).
Although it is evident that phosphorylation of Mga controls its regulatory properties, the molecular mechanism behind how this signaling event is translated to gene regulation is poorly understood. However, analyses of Mga from GAS and pneumococci and AtxA from B. anthracis provide clues to the likely mechanism. The binding sites of Mga in the target promoters lack a consensus sequence and display low sequence identity, suggesting that sequence-independent mechanisms dictate Mga-DNA interactions (24). Furthermore, phosphorylation of Mga does not alter DNA binding but affects its ability to multimerize (10), indicating that Mga oligomerization may be the regulatory target of phosphorylation. A similar regulatory role for phosphorylation-dependent oligomerization was observed for AtxA (33, 61). Consistent with this, pneumococcal Mga (Mga-Spn) binds to target promoters in a sequence-independent fashion and mediates gene regulation by polymerization of Mga-Spn on the target promoters (62). Based on these observations, we propose a speculative model for gene regulation by Mga. Regardless of the phosphorylation state, Mga recognizes the operator sequences by indirect readout and binds to the target promoters. Phosphorylation of PRD histidines induces structural changes in Mga, which negatively influence Mga multimerization and gene regulation. Conversely, nonphosphorylated Mga binds and multimerizes on DNA to form a transcriptionally productive complex and mediate the upregulation of target genes.
In conclusion, using genetic, biochemical, modeling, and animal infection studies, we have demonstrated that Mga is phosphorylated in vivo and that phosphorylation events at conserved histidines of PRD-1 and PRD-2 constitute the regulatory switch that controls Mga regulatory activity. Phosphoregulation influences Mga regulatory activity at the global level and contributes significantly to the virulence of serotype M59 GAS. Further structural and biochemical characterizations of Mga will be necessary to elucidate the phosphorylation-induced molecular events occurring in Mga that lead to altered regulatory activity.
Supplementary Material
ACKNOWLEDGMENTS
M.K. was supported in part by NIH grants 1R21AI103708-01 and 1R01AI109096-01A1. M.S. was supported in part by the Escuela de Medicina y Ciencias de la Salud, Tecnológico de Monterrey y Consejo Nacional de Ciencia y Tecnología (CONACyT) (grant 421460/263885). J.M.M. was supported in part by the Fondren Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03023-14.
REFERENCES
- 1.Cunningham MW. 2000. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev 13:470–511. doi: 10.1128/CMR.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musser JM, Shelburne SA III. 2009. A decade of molecular pathogenomic analysis of group A Streptococcus. J Clin Invest 119:2455–2463. doi: 10.1172/JCI38095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ribardo DA, McIver KS. 2006. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A Streptococcus. Mol Microbiol 62:491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 4.McIver KS. 2009. Stand-alone response regulators controlling global virulence networks in Streptococcus pyogenes. Contrib Microbiol 16:103–119. doi: 10.1159/000219375. [DOI] [PubMed] [Google Scholar]
- 5.Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. 2015. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res 43:418–432. doi: 10.1093/nar/gku1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll RK, Shelburne SA III, Olsen RJ, Suber B, Sahasrabhojane P, Kumaraswami M, Beres SB, Shea PR, Flores AR, Musser JM. 2011. Naturally occurring single amino acid replacements in a regulatory protein alter streptococcal gene expression and virulence in mice. J Clin Invest 121:1956–1968. doi: 10.1172/JCI45169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horstmann N, Sahasrabhojane P, Suber B, Kumaraswami M, Olsen RJ, Flores A, Musser JM, Brennan RG, Shelburne SA III. 2011. Distinct single amino acid replacements in the control of virulence regulator protein differentially impact streptococcal pathogenesis. PLoS Pathog 7:e1002311. doi: 10.1371/journal.ppat.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen RJ, Sitkiewicz I, Ayeras AA, Gonulal VE, Cantu C, Beres SB, Green NM, Lei B, Humbird T, Greaver J, Chang E, Ragasa WP, Montgomery CA, Cartwright J, McGeer A, Low DE, Whitney AR, Cagle PT, Blasdel TL, DeLeo FR, Musser JM. 2010. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci U S A 107:888–893. doi: 10.1073/pnas.0911811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makthal N, Rastegari S, Sanson M, Ma Z, Olsen RJ, Helmann JD, Musser JM, Kumaraswami M. 2013. Crystal structure of peroxide stress regulator from Streptococcus pyogenes provides functional insights into the mechanism of oxidative stress sensing. J Biol Chem 288:18311–18324. doi: 10.1074/jbc.M113.456590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hondorp ER, Hou SC, Hause LL, Gera K, Lee CE, McIver KS. 2013. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A Streptococcus. Mol Microbiol 88:1176–1193. doi: 10.1111/mmi.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horstmann N, Saldana M, Sahasrabhojane P, Yao H, Su X, Thompson E, Koller A, Shelburne SA III. 2014. Dual-site phosphorylation of the control of virulence regulator impacts group A streptococcal global gene expression and pathogenesis. PLoS Pathog 10:e1004088. doi: 10.1371/journal.ppat.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shelburne SA III, Olsen RJ, Makthal N, Brown NG, Sahasrabhojane P, Watkins EM, Palzkill T, Musser JM, Kumaraswami M. 2011. An amino-terminal signal peptide of Vfr protein negatively influences RopB-dependent SpeB expression and attenuates virulence in Streptococcus pyogenes. Mol Microbiol 82:1481–1495. doi: 10.1111/j.1365-2958.2011.07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fittipaldi N, Tyrrell G, Low D, Martin E, Lin D, Kumar LH, Musser JM. 2013. Integrated whole-genome sequencing and temporospatial analysis of a continuing group A Streptococcus epidemic. Emerg Microbes Infect 2:e13. doi: 10.1038/emi.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fittipaldi N, Beres SB, Olsen RJ, Kapur V, Shea PR, Watkins ME, Cantu CC, Laucirica DR, Jenkins L, Flores AR, Lovgren M, Ardanuy C, Linares J, Low DE, Tyrrell GJ, Musser JM. 2012. Full-genome dissection of an epidemic of severe invasive disease caused by a hypervirulent, recently emerged clone of group A Streptococcus. Am J Pathol 180:1522–1534. doi: 10.1016/j.ajpath.2011.12.037. [DOI] [PubMed] [Google Scholar]
- 15.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffman S, Musser JM. 2014. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A 111:E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, Goldman TD, Feldgarden M, Birren BW, Fofanov Y, Boos J, Wheaton WD, Honisch C, Musser JM. 2010. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci U S A 107:4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fittipaldi N, Olsen RJ, Beres SB, Van Beneden C, Musser JM. 2012. Genomic analysis of emm59 group A Streptococcus invasive strains, United States. Emerg Infect Dis 18:650–652. doi: 10.3201/eid1804.111803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hondorp ER, McIver KS. 2007. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol 66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- 19.Podbielski A, Flosdorff A, Weber-Heynemann J. 1995. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun 63:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo F, Lizano S, Banik S, Zhang H, Bessen DE. 2008. Role of Mga in group A streptococcal infection at the skin epithelium. Microb Pathog 45:217–224. doi: 10.1016/j.micpath.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanson M, O'Neil BE, Kachroo P, Anderson JR, Flores AR, Valson C, Cantu CC, Makthal N, Karmonik C, Fittipaldi N, Kumaraswami M, Musser JM, Olsen RJ. 2015. A naturally occurring single amino acid replacement in multiple gene regulator of group A Streptococcus significantly increases virulence. Am J Pathol 185:462–471. doi: 10.1016/j.ajpath.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIver KS, Myles RL. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A Streptococcus. Mol Microbiol 43:1591–1601. doi: 10.1046/j.1365-2958.2002.02849.x. [DOI] [PubMed] [Google Scholar]
- 23.Vahling CM, McIver KS. 2006. Domains required for transcriptional activation show conservation in the mga family of virulence gene regulators. J Bacteriol 188:863–873. doi: 10.1128/JB.188.3.863-873.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hause LL, McIver KS. 2012. Nucleotides critical for the interaction of the Streptococcus pyogenes Mga virulence regulator with Mga-regulated promoter sequences. J Bacteriol 194:4904–4919. doi: 10.1128/JB.00809-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hondorp ER, Hou SC, Hempstead AD, Hause LL, Beckett DM, McIver KS. 2012. Characterization of the group A Streptococcus Mga virulence regulator reveals a role for the C-terminal region in oligomerization and transcriptional activation. Mol Microbiol 83:953–967. doi: 10.1111/j.1365-2958.2012.07980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaffin DO, Rubens CE. 1998. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219:91–99. doi: 10.1016/S0378-1119(98)00396-5. [DOI] [PubMed] [Google Scholar]
- 27.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Virtaneva K, Porcella SF, Graham MR, Ireland RM, Johnson CA, Ricklefs SM, Babar I, Parkins LD, Romero RA, Corn GJ, Gardener DJ, Bailey JR, Parnell MJ, Musser JM. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci U S A 102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIver KS, Scott JR. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J Bacteriol 179:5178–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy A, Yang J, Zhang Y. 2012. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res 40:W471–W477. doi: 10.1093/nar/gks372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammerstrom TG, Horton LB, Swick MC, Joachimiak A, Osipiuk J, Koehler TM. 2015. Crystal structure of Bacillus anthracis virulence regulator AtxA and effects of phosphorylated histidines on multimerization and activity. Mol Microbiol 95:426–441. doi: 10.1111/mmi.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsvetanova B, Wilson AC, Bongiorni C, Chiang C, Hoch JA, Perego M. 2007. Opposing effects of histidine phosphorylation regulate the AtxA virulence transcription factor in Bacillus anthracis. Mol Microbiol 63:644–655. doi: 10.1111/j.1365-2958.2006.05543.x. [DOI] [PubMed] [Google Scholar]
- 35.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Kinoshita E, Kinoshita-Kikuta E. 2011. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 11:319–323. doi: 10.1002/pmic.201000472. [DOI] [PubMed] [Google Scholar]
- 37.Kinoshita E, Kinoshita-Kikuta E, Kojima H, Nakano Y, Chayama K, Koike T. 2005. Reliable and cost-effective screening of inherited heterozygosity by Zn2+-cyclen polyacrylamide gel electrophoresis. Clin Chem 51:2195–2198. doi: 10.1373/clinchem.2005.051011. [DOI] [PubMed] [Google Scholar]
- 38.Boulanger A, Chen Q, Hinton DM, Stibitz S. 2013. In vivo phosphorylation dynamics of the Bordetella pertussis virulence-controlling response regulator BvgA. Mol Microbiol 88:156–172. doi: 10.1111/mmi.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciesla J, Fraczyk T, Rode W. 2011. Phosphorylation of basic amino acid residues in proteins: important but easily missed. Acta Biochim Pol 58:137–148. [PubMed] [Google Scholar]
- 40.Joyet P, Derkaoui M, Poncet S, Deutscher J. 2010. Control of Bacillus subtilis mtl operon expression by complex phosphorylation-dependent regulation of the transcriptional activator MtlR. Mol Microbiol 76:1279–1294. doi: 10.1111/j.1365-2958.2010.07175.x. [DOI] [PubMed] [Google Scholar]
- 41.Young BC, Golubchik T, Batty EM, Fung R, Larner-Svensson H, Votintseva AA, Miller RR, Godwin H, Knox K, Everitt RG, Iqbal Z, Rimmer AJ, Cule M, Ip CLC, Didelot X, Harding RM, Donnelly P, Peto TE, Crook DW, Bowden R, Wilson DJ. 2012. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A 109:4550–4555. doi: 10.1073/pnas.1113219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Liu F, Wang Q, Ge P, Woo PCY, Yan J, Zhao Y, Gao GF, Liu CH, Liu C. 2014. Genomic and transcriptomic analysis of NDM-1 Klebsiella pneumoniae in spaceflight reveal mechanisms underlying environmental adaptability. Sci Rep 4:6216. doi: 10.1038/srep06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salipante SJ, Roach DJ, Kitzman JO, Snyder MW, Stackhouse B, Butler-Wu SM, Lee C, Cookson BT, Shendure J. 2015. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 25:119–128. doi: 10.1101/gr.180190.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A 108:20142–20147. doi: 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Ligget CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci U S A 109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koser CU, Holden MTG, Ellington MJ, Cartwright EJP, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Podbielski A. 1993. Three different types of organization of the vir regulon in group A streptococci. Mol Gen Genet 237:287–300. [DOI] [PubMed] [Google Scholar]
- 48.Kimoto H, Fujii Y, Hirano S, Yokota Y, Taketo A. 2006. Genetic and biochemical properties of streptococcal NAD-glycohydrolase inhibitor. J Biol Chem 281:9181–9189. doi: 10.1074/jbc.M506879200. [DOI] [PubMed] [Google Scholar]
- 49.Meehl MA, Pinkner JS, Anderson PJ, Hultgren SJ, Caparon MG. 2005. A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathog 1:e35. doi: 10.1371/journal.ppat.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bricker AL, Cywes C, Ashbaugh CD, Wessels MR. 2002. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol Microbiol 44:257–269. doi: 10.1046/j.1365-2958.2002.02876.x. [DOI] [PubMed] [Google Scholar]
- 51.Bastiat-Sempe B, Love JF, Lomayesva N, Wessels MR. 2014. Streptolysin O and NAD-glycohydrolase prevent phagolysosome acidification and promote group A Streptococcus survival in macrophages. mBio 5(5):e01690-14. doi: 10.1128/mBio.01690-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Seaghdha M, Wessels MR. 2013. Streptolysin O and its co-toxin NAD-glycohydrolase protect group A Streptococcus from xenophagic killing. PLoS Pathog 9:e1003394. doi: 10.1371/journal.ppat.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henstra SA, Duurkens RH, Robillard GT. 2000. Multiple phosphorylation events regulate the activity of the mannitol transcriptional regulator MtlR of the Bacillus stearothermophilus phosphoenolpyruvate-dependent mannitol phosphotransferase system. J Biol Chem 275:7037–7044. doi: 10.1074/jbc.275.10.7037. [DOI] [PubMed] [Google Scholar]
- 54.Charrier V, Buckley E, Parsonage D, Galinier A, Darbon E, Jaquinod M, Forest E, Deutscher J, Claiborne A. 1997. Cloning and sequencing of two enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate-dependent, phosphotransferase system-catalyzed phosphorylation of a single histidyl residue. J Biol Chem 272:14166–14174. doi: 10.1074/jbc.272.22.14166. [DOI] [PubMed] [Google Scholar]
- 55.Van Tilbeurgh H, Declerck N. 2001. Structural insights into the regulation of bacterial signalling proteins containing PRDs. Curr Opin Struct Biol 11:685–693. doi: 10.1016/S0959-440X(01)00267-6. [DOI] [PubMed] [Google Scholar]
- 56.Joyet P, Bouraoui H, Aké FMD, Derkaoui M, Zébré AC, Cao TN, Ventroux M, Nessler S, Noirot-Gros M-F, Deutscher J, Milohanic E. 2013. Transcription regulators controlled by interaction with enzyme IIB components of the phosphoenolpyruvate:sugar phosphotransferase system. Biochim Biophys Acta 1834:1415–1424. doi: 10.1016/j.bbapap.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 57.Rothe FM, Bahr T, Stülke J, Rak B, Görke B. 2012. Activation of Escherichia coli antiterminator BglG requires its phosphorylation. Proc Natl Acad Sci U S A 109:15906–15911. doi: 10.1073/pnas.1210443109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindner C, Galinier A, Hecker M, Deutscher J. 1999. Regulation of the activity of the Bacillus subtilis antiterminator LicT by multiple PEP-dependent, enzyme I- and HPr-catalysed phosphorylation. Mol Microbiol 31:995–1006. doi: 10.1046/j.1365-2958.1999.01262.x. [DOI] [PubMed] [Google Scholar]
- 59.Martin-Verstraete I, Charrier V, Stülke J, Galinier A, Erni B, Rapoport G, Deutscher J. 1998. Antagonistic effects of dual PTS-catalysed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol Microbiol 28:293–303. doi: 10.1046/j.1365-2958.1998.00781.x. [DOI] [PubMed] [Google Scholar]
- 60.Tortosa P, Declerck N, Dutartre H, Lindner C, Deutscher J, Le Coq D. 2001. Sites of positive and negative regulation in the Bacillus subtilis antiterminators LicT and SacY. Mol Microbiol 41:1381–1393. doi: 10.1046/j.1365-2958.2001.02608.x. [DOI] [PubMed] [Google Scholar]
- 61.Hammerstrom TG, Roh JH, Nikonowicz EP, Koehler TM. 2011. Bacillus anthracis virulence regulator AtxA: oligomeric state, function, and CO(2)-signaling. Mol Microbiol 82:634–647. doi: 10.1111/j.1365-2958.2011.07843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Solano-Collado V, Lurz R, Espinosa M, Bravo A. 2013. The pneumococcal MgaSpn virulence transcriptional regulator generates multimeric complexes on linear double-stranded DNA. Nucleic Acids Res 41:6975–6991. doi: 10.1093/nar/gkt445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.