Abstract
Pathogenic bacteria often need to survive in the host and the environment, and it is not well understood how cells transition between these equally challenging situations. For the human and animal pathogen Salmonella enterica serovar Typhimurium, biofilm formation is correlated with persistence outside a host, but the connection to virulence is unknown. In this study, we analyzed multicellular-aggregate and planktonic-cell subpopulations that coexist when S. Typhimurium is grown under biofilm-inducing conditions. These cell types arise due to bistable expression of CsgD, the central biofilm regulator. Despite being exposed to the same stresses, the two cell subpopulations had 1,856 genes that were differentially expressed, as determined by transcriptome sequencing (RNA-seq). Aggregated cells displayed the characteristic gene expression of biofilms, whereas planktonic cells had enhanced expression of numerous virulence genes. Increased type three secretion synthesis in planktonic cells correlated with enhanced invasion of a human intestinal cell line and significantly increased virulence in mice compared to the aggregates. However, when the same groups of cells were exposed to desiccation, the aggregates survived better, and the competitive advantage of planktonic cells was lost. We hypothesize that CsgD-based differentiation is a form of bet hedging, with single cells primed for host cell invasion and aggregated cells adapted for persistence in the environment. This allows S. Typhimurium to spread the risks of transmission and ensures a smooth transition between the host and the environment.
INTRODUCTION
Salmonella persistence, or the ability of cells to survive under stressful conditions, has traditionally been studied separately from virulence. The continued success of these pathogens, however, indicates that both traits must be inherently connected. Most of the >2,000 serovars of Salmonella collectively account for approximately 94 million annual cases of human gastroenteritis (1), a disease characterized by intestinal inflammation and diarrhea (2). The gastroenteritis-causing serovars are referred to as nontyphoidal Salmonella (NTS), and Salmonella enterica serovar Typhimurium is one of the serovars most commonly isolated in the world. Most NTS isolates are considered to be host generalists, with a cyclical lifestyle involving exposure to the environment between host colonization events (3). This is in contrast to typhoidal Salmonella serovars (i.e., Salmonella enterica serovar Typhi and Salmonella enterica serovar Paratyphi A), which are human restricted (4, 5). Despite their differences, both NTS and typhoidal Salmonella strains rely on type three secretion systems (T3SS) to stimulate invasion, uptake, and survival within human epithelial cells and macrophages (6). While our understanding of pathogenesis is improving, less is known about the strategies that pathogenic Salmonella strains employ to survive outside the host (7, 8). It is generally assumed that people become infected with Salmonella through ingestion of contaminated food and water, but there are also numerous examples of sporadic human cases where the causative source is unknown (9).
One strategy for bacterial survival is through the formation of biofilms, where cells aggregate and become embedded in a self-produced extracellular matrix, usually in contact with a surface. This has been described in Salmonella as the rdar (red, dry, and rough) morphotype (10, 11). The extracellular matrix surrounding rdar-positive cells is comprised of protein polymers (curli fimbriae) and extracellular polysaccharides (cellulose and an O-antigen capsule) that physically link cells together (10–13). The genes coding for these polymers are conserved throughout Salmonella (14, 15) and Escherichia coli (16, 17). Comparisons between rdar-positive and rdar-negative S. Typhimurium and E. coli strains have shown that embedded cells have enhanced persistence upon exposure to conditions of starvation, desiccation, and treatment with disinfectants (15, 18–21), as well as an increased ability to adhere to both biotic and abiotic surfaces (22, 23). Rdar-positive cells can also cause infections after long periods (24) but do not appear to be specifically adapted for virulence (25). Rdar-negative Salmonella and E. coli isolates tend to cause more invasive forms of disease (26, 27).
Most NTS isolates are rdar positive, and most typhoidal Salmonella isolates are rdar negative (21, 25, 26). One possible explanation for this correlation is that the rdar morphotype is an adaptation necessary for life outside the host. However, there have been recent links between the rdar morphotype, immune stimulation, and invasion (28, 29), making it difficult to fully understand this physiology. Although they are informative, a potential problem with strain-to-strain comparisons is that they could miss fluctuations that exist within individual populations of cells. It was recently demonstrated that when S. Typhimurium is grown under biofilm-inducing conditions in liquid medium, the population of cells in the fluid phase differentiates into large, multicellular aggregates and single (planktonic) cells (25, 30). This bifurcation is caused by the bistable synthesis of CsgD, the central transcriptional regulator of the rdar morphotype (31). Aggregated cells are in a CsgD-on state, whereas the nonaggregative planktonic cells are primarily in a CsgD-off state. The multicellular aggregates can be thought of as noncanonical biofilms, as the cells strongly express extracellular matrix components (25, 30) but are not in direct contact with a surface. While it is assumed that the aggregates retain the resistance properties of classic adherent biofilms, the planktonic cells that result from CsgD bistability have never been analyzed in detail, and it is unclear what role they may have in S. Typhimurium physiology.
In this study, we isolated and examined these two groups of cells and demonstrate for the first time that they could represent a critical link between the virulence and persistence of Salmonella. Bistability and the formation of specialized cell types are predicted to contribute to S. Typhimurium transmission success, with single cells adapted for host cell invasion and multicellular aggregates adapted for persistence in the environment.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. enterica serovar Typhimurium strain ATCC 14028 was the wild-type strain used in this study. In general, strains were inoculated from frozen stocks onto LB agar and grown for 20 h at 37°C. One isolated colony was used to inoculate 5 ml LB broth (containing 1% NaCl), and the culture was incubated for 12 h at 37°C with agitation at 200 rpm. To analyze growth under biofilm-inducing conditions, approximately 4 × 109 CFU was inoculated into 400 ml of 1% tryptone, pH 7.4, and the culture was distributed in 100-ml volumes into four 250-ml flasks that were incubated at 28°C for 8 h, 13 h, 18 h, or 32 h with agitation at 200 rpm. The csgD promoter (PcsgD) luxCDABE fusion plasmid used for bioluminescence assays has been previously described (21). To measure luminescence, 200-μl aliquots of culture were removed at hourly intervals in triplicate using a wide-bore pipette, transferred into individual wells of a 96-well clear-bottom black plate (product 9520; Corning Life Sciences), and assayed on a Victor X3 multilabel plate reader (Perkin-Elmer Life Sciences).
Generation of S. Typhimurium 14028 ΔcsgD and ΔSPI-1 mutant strains.
ΔcsgD and ΔSPI-1 strains were generated using the lambda red recombinase knockout procedure (32). Primers containing a 50-nucleotide sequence on either side of the entire Salmonella pathogenicity island 1 (SPI-1) region (SPI-1F and SPI-1R) (33) or csgD (csgDko sense [GCAGCTGTCAGATGTGCGATTAAAAAAAGTGGAGTTTCATCATGTTTAATGTGTAGGCTGTAGCTGCTTC] and csgDko antisense [CTCTGCTGCTACAATCCAGGTCAGATAGCGTTTCATGGCCTTACCGCCTGCCTCCTTAGTTCCTATTCCG]) were used to amplify the cat gene from pKD3 using Phusion High-Fidelity DNA polymerase (New England BioLabs). The PCR products were purified (AxyPrep Mag PCR cleanup kit; Axygen) and electroporated into S. Typhimurium 14028 cells containing pKD46. Mutant strains were first selected by growth at 37°C on LB agar supplemented with 7 μg ml−1 chloramphenicol (Cm) before restreaking onto LB agar containing 30 μg ml−1 Cm. The mutations were moved into clean wild-type backgrounds by transduction with P22 phage (34). The cat gene was resolved from the chromosome using pCP20 (32). DNA sequencing of PCR products amplified from the chromosome of the ΔcsgD or ΔSPI-1 strain confirmed the loss of the majority of csgD or SPI-1, respectively.
Generation of Knr or Cmr S. Typhimurium strains.
sig70_16 is a reporter plasmid that contains a synthetic σ70-dependent promoter driving luxCDABE expression (35). A chloramphenicol-resistant (Cmr) version of the sig70_16 plasmid was generated, and the sig70_16 kanamycin-resistant (Knr) or Cmr fragments were used for Tn7 integration (36) into the S. Typhimurium ATCC 14028 chromosome, using a modified protocol (D. Shivak, K. MacKenzie, B. Jones, and A. White, submitted for publication). To confirm that the fragments had been properly inserted into the att Tn7 site downstream of the glmS gene of S. Typhimurium, the chromosomal region was PCR amplified and the resulting products were sequenced. To ensure the absence of secondary mutations, the Knr and Cmr sig70_16 fragments were moved into a clean wild-type or ΔcsgD background by P22 transduction.
Isolation of multicellular-aggregate and planktonic-cell subpopulations.
S. Typhimurium biofilm cultures were transferred into 15-ml conical tubes and subjected to low-speed centrifugation (210 × g; 2 min; 4°C). The supernatant fractions were transferred into new 15-ml tubes, and the cell aggregates were transferred into 1.5-ml tubes using a sterile loop. After brief centrifugation (10,000 × g; 30 s), the culture medium was removed and the wet weight of the aggregates was determined. The aggregates were resuspended in 1 ml of phosphate-buffered saline (PBS) (for direct counting) or 1 ml cell-free 1% tryptone culture supernatant (for subsequent applications) and homogenized for approximately 30 s (25 Dounce homogenizations) with a glass tissue grinder (product 7727-2; Corning Life Sciences). For CFU determination, serial dilutions of planktonic cells or homogenized aggregates were plated in 4-μl drops onto LB agar prior to incubation overnight at 37°C.
Visualizing the homogenization of multicellular aggregates.
Multicellular aggregates were harvested from an S. Typhimurium 14028 reporter strain containing a csgBAC promoter-green fluorescent protein (GFP) fusion (25) and grown for 13 h (time point 2 [TP2]). Ten milligrams of aggregates was resuspended in 1 ml of cell-free culture supernatant and homogenized in a tissue grinder, and two successive 1-in-10 dilutions were used to prepare a solution of 1.2 × 107 CFU/ml. Ten 200-μl aliquots were individually concentrated onto the surfaces of 3-in. by 1-in. by 1.0-mm glass slides (Fisher Scientific; 12-550-343) using a Cytospin-4 cytocentrifuge (Thermo Shandon, Asheville, NC). Samples were centrifuged at 500 rpm for 5 min with low acceleration. Five microliters of Mowiol mounting medium (Sigma-Aldrich; 81381) was added to the cells on each slide, and each mixture was topped with a 12-mm-diameter micro-cover glass (72230-01; Electron Microscopy Sciences). The slides were air dried overnight prior to visualization on a Leica TCS SP5 confocal microscope, using a 63× oil immersion objective with the laser operating at 488 nm.
SDS-PAGE and Western blotting.
Planktonic cells were sedimented (11,000 × g; 10 min; 4°C) to concentrate the samples before suspending them in 1 ml of SDS-PAGE sample buffer (without 2-mercaptoethanol and bromophenol blue). Samples (100 mg) of multicellular aggregates were resuspended in 1 ml of water and homogenized with a glass tissue grinder. The cell material was sedimented (10,000 × g; 1 min) before resuspending it in 1 ml of SDS-PAGE sample buffer and boiling it for 5 min. Protein concentrations were determined using the DC protein assay kit (Bio-Rad Laboratories), samples were normalized to 30 μg of total protein per lane, and bromophenol blue (0.0002% final concentration) and 2-mercaptoethanol (0.2% final concentration) were added. SDS-PAGE was performed with a 5% stacking gel and a 12% resolving gel or with 10%, 12%, or 4 to 15% precast Mini-Protean TGX gels (Bio-Rad Laboratories). For immunoblots, proteins were transferred to nitrocellulose for 40 min at 25 V using a Trans-Blot SD semidry transfer cell (Bio-Rad Laboratories) in buffer recommended by the manufacturer. For detection of curli fimbriae, cell samples were boiled for 5 min in SDS-PAGE sample buffer with 2-mercaptoethanol, and the cell debris was sedimented (14,000 × g; 2 min) and washed twice in 500 μl of distilled water before suspension in 250 μl of 90% formic acid, freezing at −80°C, and lyophilization (10). The dried samples were resuspended in SDS-PAGE sample buffer and loaded directly, without boiling, into each gel lane prior to electrophoresis.
CsgD was detected using the monoclonal antibody described below. CsgA, the major subunit of curli fimbriae, was detected using rabbit polyclonal immune serum raised to whole purified curli (10). GroEL was used as a protein-loading control and was detected with rabbit polyclonal immune serum used at a 1-in-10,000 dilution (Sigma-Aldrich; G6532). SPI-1 T3SS proteins (InvG, SipD, SopE2, and SopB) were detected with rabbit polyclonal immune sera generated against each of the individual proteins (33). The secondary antibodies were goat anti-rabbit immunoglobulin G (IgG)-alkaline phosphatase or human-adsorbed goat-anti-mouse IgG-alkaline phosphatase conjugates (KPL). Proteins were detected by dye precipitation using the substrate 5-bromo-4-chloro-3-indolylphosphate with 4-nitroblue tetrazolium chloride (Sigma-Aldrich) as an enhancer.
Generation of CsgD-specific monoclonal antibodies.
csgD was PCR amplified from S. Typhimurium 14028 genomic DNA using Phusion High-Fidelity DNA polymerase (New England BioLabs) and primers csgD_expFOR (GGCTCATATGTTTAATGAAGTCCATAG) and csgD_expREV2 (ATTTCTCGAGCCGCCTGAGATTATCGTTTG). The PCR product was digested with NdeI and XhoI and ligated into NdeI/XhoI-digested pET-21a (Novagen), using T4 DNA ligase (NEB), to generate a vector encoding CsgD with a 6×His tag at the C-terminal end. E. coli BL21(DE3) plus pET21a-csgDHis was grown at 37°C with agitation at 200 rpm in LB broth supplemented with 100 μg ml−1 ampicillin until an optical density at 600 nm (OD600) of 0.7 was reached; expression of CsgD-His was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM and growing the cells for 2 h. Cells were harvested and lysed, and CsgD-His was purified using a Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen), following procedures outlined by the manufacturer (Qiagen; QIAexpressionist, protocol 9). Approximately 5 mg of purified CsgD-His was used for Rapid Prime custom monoclonal antibody development (ImmunoPrecise Antibodies Ltd., Victoria, BC, Canada). Concentrated tissue culture supernatants containing a CsgD-specific monoclonal antibody were used for Western blotting experiments.
Measurement of c-di-GMP levels inside cells.
Planktonic cells and multicellular aggregates were harvested from 100-ml S. Typhimurium biofilm cultures grown for 8 h (TP1) or 13 h (TP2). Planktonic-cell samples were prepared by centrifugation (10,000 × g; 10 min), and multicellular-aggregate samples were homogenized as described above in cell-free culture supernatant. Cyclic-di-GMP (c-di-GMP) extractions were performed with 100 μl of 40% methanol-40% acetone in 0.1 N formic acid, used for every 48 mg (wet weight) of cells (37). Cell samples were transferred into screw-cap tubes containing 0.2 g of 0.1-mm zirconia beads, and homogenization was performed in a high-speed mixer mill (Retsch; MM400) for 2 min at 30 Hz. After centrifugation (16,000 × g; 2 min), the supernatant was neutralized by the addition of 4 μl of 15% ammonium bicarbonate per 100 μl of sample. Aliquots (50 μl) of each sample were dried down in a speed vacuum concentrator, and the residues were dissolved in 300 μl of 30% methanol, followed by solid-phase extraction with Agilent OPT 30-mg/ml reversed-phase cartridges (Agilent; 5982-3013) to remove lipids and other lipophilic compounds. The flowthrough fractions were collected under a 3- to 5-lb/in2 vacuum, and the cartridges were washed with 400 μl of 30% methanol. The flowthrough fractions were collected and pooled for each sample and dried down in a speed vacuum concentrator. The residues were resuspended in 50 μl of 80% acetonitrile, and 20-μl aliquots were injected into a Dionex UPLC 3000 system coupled to an AB Sciex 4000 QTrap mass spectrometer, which was operated in the electrospray ionization and negative-ion multiple-reaction-monitoring (MRM) mode. The MRM transition for quantitation was 689.2/344.2. Ultraperformance liquid chromatography (UPLC) separation was carried out on a 10-cm-long UPLC column, which was run in a hydrophilic interaction (HILIC) mode. The mobile phase was ammonium acetate-acetonitrile for binary gradient elution. The column temperature was 30°C, and the flow rate was 0.40 ml/min. A standard curve for cyclic-di-GMP (Axxora; BLG-C057) was prepared within a concentration range of 2 to 400 fmol/ml. The concentration of cyclic-di-GMP in each sample was calculated with external calibration. Since multicellular aggregates consist of both cells and extracellular matrix, a conversion factor was applied to all c-di-GMP concentrations. One milligram (wet weight) of planktonic cells was equivalent to 6.94 × 108 CFU, whereas 1 mg (wet weight) of multicellular aggregates was equivalent to 1.73 × 108 CFU. This conversion was thought to represent a more accurate cell-to-cell comparison between samples.
RNA purification procedure.
Total RNA was isolated using a modified procedure based on the SV Total RNA Isolation kit (Promega). At TP1, cells were sedimented by centrifugation (10,000 × g; 2 min; 4°C) and resuspended in 1 ml of RNALater (Life Technologies). At later time points, cell subpopulations were first separated by low-speed centrifugation, as described above. Planktonic-cell fractions were sedimented as described above and resuspended in 1 ml of RNALater (Life Technologies). Multicellular-aggregate samples were resuspended in 1 ml of RNALater and homogenized for approximately 30 s using a tissue grinder. Cells in 1 ml of RNALater were sedimented by centrifugation (13,200 × g; 1 min) and resuspended in 700 μl of RNA lysis buffer A (RLA). This cell suspension was transferred to a 1.5-ml screw-cap tube containing 0.2 g of 0.1-mm zirconia beads and homogenized in a high-speed mixer mill (Retsch; MM400) for 5 min at 30 Hz. Approximately 300 to 400 μl RLA solution was recovered from the beads, and 600 to 800 μl of RNA dilution buffer was added, followed by 0.4 volume of 95% ethanol. Samples were centrifuged (13,200 × g; 10 min) to remove extracellular debris that precipitated upon addition of ethanol prior to loading onto the RNA purification column. This step was necessary to avoid plugging the column, especially with the multicellular-aggregate samples. The column-washing steps and on-column DNase I treatment were performed according to the manufacturer's instructions. Bound RNA was eluted from each column twice with 50 μl of RNase-free water for a final volume of 100 μl. After elution, samples were DNase treated a second time in solution using a Turbo DNA-free kit (Life Technologies; AM1907). The MicrobExpress bacterial mRNA enrichment kit (Life Technologies; AM1905) was used for mRNA enrichment. To assess the quantity, purity, and integrity of RNA, samples were analyzed using a NanoDrop ND-1000 spectrophotometer (Fisher Scientific) and an Agilent 2100 Bioanalyzer with a Prokaryote Total RNA Nano chip.
Directional transcriptome sequencing (RNA-seq).
For sequencing on the Illumina HiSeq 2000, first-strand cDNA was synthesized from RNA using random-hexamer priming and reverse transcriptase. Second-strand cDNA synthesis was performed with DNA polymerase I in the presence of dATP, dGTP, dCTP, and dUTP. Double-stranded cDNA was fragmented by sonication, and 70- to 200-bp fragments were purified with AMPure magnetic beads (Beckman-Coulter). After the standard addition of 18 μl of AMPure beads to every 10 μl of sample, 9.3 μl isopropanol was added. This step allows recovery of fragments below 100 bp, as described in the AxyPrep Mag PCR Cleanup kit protocol (Axygen Biosciences Inc.). cDNA fragments were ligated to Illumina adapters and primers containing bar-coded sequences; fragment sizes in the final library ranged from approximately 196 to 326 bp. The second cDNA strand containing UTP was selectively removed by uridine digestion (37°C; 15 min) in 5 μl of 1× Tris-EDTA (TE) buffer, pH 7.5, with 1 unit of uracil-N-glycosylase (UNG) (Applied Biosystems) prior to library amplification and sequencing (38). In total, 21 libraries were sequenced (75-bp paired-end tag [PET] sequencing), prepared from three biological replicates of planktonic cells (samples at TP1, TP2, TP3, and TP4) and multicellular aggregates (samples at TP2, TP3, and TP4).
For sequencing on the Illumina MiSeq, cDNA libraries prepared from S. Typhimurium 14028 ΔcsgD at TP2 were constructed using the NEBNext Ultra Directional RNA Library Prep kit for Illumina (New England BioLabs) according to the manufacturer's protocol. This differs from the above-mentioned protocol in that fragmentation occurs through autocatalysis of RNA before first-strand cDNA synthesis. The final library fragment sizes were 200 to 300 bp. The approach also incorporates dUTP into the second cDNA strand synthesis, followed by digestion of the second strand before PCR amplification. The 75-bp PET sequencing was performed on three libraries using the MiSeq reagent kit version 3 (Illumina).
Mapping and comparison of sequencing reads.
Sequencing reads were mapped onto the S. Typhimurium ATCC 14028 chromosome (NC_016856) and plasmid (NC_016855) sequence using Geneious v5.6.5 (Biomatters). The mapping parameters were set as follows: (i) gaps not allowed, (ii) words repeated more than twice ignored, and (iii) maximum mismatches per read set as 4%. The other parameters conformed to the default settings of Geneious. The coordinates and strand of each putative protein-encoding gene were according to genome annotations (39), and in-house perl scripts were used for counting the mapping reads to each protein-encoding gene and its antisense strand. The small-RNA (sRNA) coverage was determined in a similar way. The full set of known Salmonella sRNAs (40) were retrieved, and the orthologous counterparts in S. Typhimurium 14028 were found by direct sequence alignment.
Before comparing the gene expression levels between samples, the counts of mapped reads were normalized with edgeR, which, instead of using RPKM notation, assumes that the total RNA amount could vary among different samples and that expression of more genes should be stable among libraries (41). A negative binomial distribution was fitted for the normalized read counts for each pair of genes. Differentially expressed genes were identified as those with P values of <0.05, determined by a Benjamini and Hochberg correction for the multiple testing results (42, 43), with the false-discovery rate (FDR) set as <0.05. The normalization effect was evaluated before data analysis by observing the variance among technical and biological repeats after normalization.
Motility comparison between planktonic cells and multicellular aggregates.
Cell subpopulations were isolated from S. Typhimurium biofilm cultures at TP2 and either tested for motility directly or after homogenization for 30 s in a tissue grinder. One-microliter aliquots (approximately 5 × 106 CFU) of each cell suspension were inoculated into the middle of swim agar (lysogeny broth [Luria] plus 0.25% agar), and the plates were allowed to dry for 20 min. Migration distances from the center of the plate were recorded after incubation at 37°C for 8 h. Five technical replicates of each sample were tested, prepared from three biological replicate cultures. The statistical difference between samples was calculated using unpaired t tests.
Invasion assay using polarized Caco-2 cells.
Caco-2 cells (obtained from Brett Finlay, University of British Columbia) were grown at 37°C in the presence of 5% CO2 in complete Dulbecco modified Eagle medium (DMEM) (HyClone; Fisher Scientific) supplemented with 10% fetal bovine serum (Seracare Life Sciences) and 1% nonessential amino acids (Life Technologies). To obtain polarized monolayers, Caco-2 cells were seeded onto Transwell inserts (24-mm diameter; 0.4-μm pore size; product 3470; Corning Life Sciences) for approximately 21 days. The cells were used for invasion assays once the transepithelial resistance (TER) was 700 to 900 Ω cm−2 (33). Planktonic cells and multicellular aggregates isolated from S. Typhimurium cultures were suspended in complete DMEM and applied in 200-μl aliquots to the apical surfaces of the Caco-2 cells; invasion in three replicate wells was assessed for every sample. Wild-type and ΔSPI-1 mutant strains grown to late exponential phase at an OD600 of 0.7 in LB broth at 37°C were used as positive and negative invasion controls, respectively. After 1 h exposure to S. Typhimurium, Caco-2 cells were washed three times with 200 μl of PBS and incubated for 2 h with complete DMEM containing 1.2 mg ml−1 gentamicin to kill any remaining extracellular bacteria. The Caco-2 cells were washed two times with 200 μl of PBS and lysed by exposure to 1% Triton. Serial dilutions of the Salmonella-containing lysate were plated in 4-μl drops on LB agar and incubated overnight at 37°C. We aimed for a multiplicity of infection (MOI) of 10 based on an estimated 2.5 × 105 polarized Caco-2 cells per well. The true MOI was determined by serial dilution and plating on LB agar, followed by incubation overnight at 37°C.
Gentamicin susceptibility testing.
Samples containing approximately 5 × 107 CFU ml−1 of planktonic cells or multicellular aggregates isolated from S. Typhimurium 14028 cultures were prepared in cell-free, spent culture medium. One-milliliter aliquots were centrifuged (8,000 × g; 2 min), and the cells were resuspended in 1 ml of complete DMEM containing gentamicin at concentrations ranging from 0.2 to 1.2 mg ml−1. After 2 h exposure, the cells were sedimented, washed, resuspended in PBS, and homogenized for 30 s using a tissue grinder. Serial dilutions of each sample were plated on LB agar and grown overnight at 37°C to determine the number of CFU. Four technical replicates of each sample were tested, and the experiment was repeated three times. The statistical difference between samples was calculated using unpaired t tests.
CI infections of mice.
Female C57BL/6 mice (6 to 8 weeks old) purchased from Jackson Laboratory (Bar Harbor, ME) were assigned to cage groups using a randomization table prepared in Microsoft Excel, and individual mice were marked with ear notches. Challenge formulations A and B were prepared in 100 mM HEPES, pH 8, at a concentration of ∼107 CFU per 100 μl, consisting of an approximate 1:1 ratio of cells isolated from Cmr or Knr S. Typhimurium cultures at TP2 (see Fig. S6 in the supplemental material). Animal care technicians assigned challenges A and B to 2 cages each (i.e., 4 groups of 6 mice; n = 24) and performed all infections. The cage assignments were revealed to members of the White laboratory only after the final results were analyzed. The infected mice were weighed daily and monitored for clinical signs of infection; mice that had a >20% drop in weight were euthanized, typically 4 to 7 days postinfection. The spleen, liver, cecum, and mesenteric lymph nodes (MLN) were homogenized in Eppendorf Safe-Lock tubes containing 1 ml of PBS and a 5-mm stainless steel bead (Qiagen; 69989) using a high-speed mixer mill (Retsch; MM400). To determine the number of CFU in the initial challenges or organ homogenates, serial dilutions were plated on LB agar supplemented with 10 μg ml−1 Cm or 50 μg ml−1 Kn. The competitive index (CI) was calculated as follows: (CFU planktonic/CFU biofilm)output/(CFU planktonic/CFU biofilm)input. For statistical analysis, the normality of each set of CI data was assessed using the Shapiro-Wilk normality test, followed by the Wilcoxon signed-rank test to determine if the median CI was significantly different from 1.
Input and output ratios for CI infections.
For the initial CI trial, the input ratio for challenge A was 0.76 (7.9 × 106 CFU planktonic cells and 1.0 × 107 CFU aggregates) and that for challenge B was 0.60 (5.9 × 106 CFU planktonic cells and 9.9 × 106 CFU aggregates). To analyze the effect of homogenization, challenge mixtures were homogenized in a tissue grinder for approximately 30 s before oral gavage. The input ratio for challenge A was 1.19 (4.8 × 106 CFU planktonic cells and 4.0 × 106 CFU aggregates), and for challenge B it was 0.85 (5.6 × 106 CFU planktonic cells and 6.6 × 106 CFU aggregates). The input ratios for the trial of wild-type planktonic cells versus ΔcsgD planktonic cells were 1.07 for challenge A (6.7 × 106 CFU wild type and 6.3 × 106 CFU ΔcsgD cells) and 1.11 for challenge B (7.7 × 106 CFU wild type and 6.9 × 106 CFU ΔcsgD cells). For the trial using planktonic cells and multicellular aggregates prepared from ΔSPI-1 cultures, the input ratio for challenge A was 0.27 (4.8 × 106 CFU planktonic cells and 1.8 × 107 CFU aggregates), and for challenge B it was 3.7 (4.4 × 106 CFU planktonic cells and 1.2 × 106 CFU aggregates). For the trial using dehydrated planktonic cells and multicellular aggregates (see below), the input ratio for challenge A was 0.11 (7.2 × 105 CFU planktonic cells and 6.5 × 106 CFU aggregates), and for challenge B it was 0.07 (3.0 × 105 CFU planktonic cells and 4.2 × 106 CFU aggregates).
Desiccation of S. Typhimurium challenge formulations.
Planktonic cells and multicellular aggregates from the initial CI trial were suspended in spent culture supernatant, and 200-μl aliquots were pipetted into individual wells of 24-well tissue culture plates. The plates were placed in a biological safety cabinet, the liquid was allowed to evaporate, and then the plates were covered and left to sit on the laboratory bench. At time zero, 4 weeks, and 5.5 weeks, 500 μl of PBS was added to individual wells, and the cells were allowed to rehydrate for 30 min prior to homogenization using a tissue grinder and plating to determine the number of surviving CFU from each cell type. The statistical difference between samples was calculated using unpaired t tests. For the CI trial performed at 4 weeks, cells were resuspended in a total volume of 3 ml of 100 mM HEPES, pH 8, per 24-well plate.
Ethics statement.
All animals were cared for and used in accordance with the Guidelines of the Canadian Council on Animal Care and the Regulations of the University of Saskatchewan Committee on Animal Care and Supply. All mouse experiments were performed under Animal Use Protocol 20110057, which was approved by the University of Saskatchewan's Animal Research Ethics Board.
RNA-seq accession numbers.
RNA-seq data are available in the SRA database (study no. SRP056892) under the following accession numbers: SRX976427, SRX976344, SRX976343, SRX976341, SRX976337, SRX976336, SRX976335, and SRX974437.
RESULTS
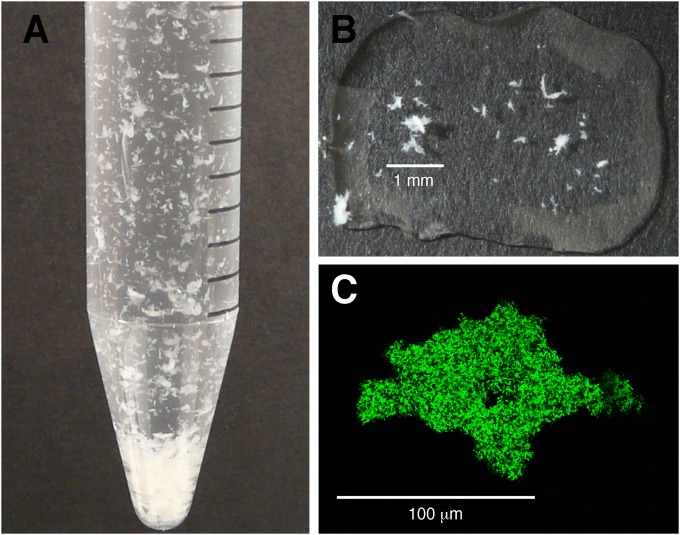
We grew S. Typhimurium 14028 in liquid culture under biofilm-inducing conditions to examine differentiation within the population. After 13 h growth, multicellular aggregates were clearly visible within the culture (Fig. 1A), ranging in size from 100 to 500 μm (Fig. 1B). The aggregates had high levels of csgBAC (curli) transcription, as measured using a GFP reporter (Fig. 1C). To monitor the dynamics of the differentiation process, we measured the real-time expression of csgD in the total population while observing the appearance of multicellular aggregates (Fig. 2A). Four time points were chosen for further analysis: TP1 was considered a preaggregation condition, when overall csgD expression was low (Fig. 2A) and the culture appeared homogeneous (data not shown); TP2 had peak csgD expression that coincided with the appearance of multicellular aggregates; TP3 had high csgD expression, and the culture appeared fully differentiated; and TP4 was visually the same as TP3, but csgD expression had dropped to near baseline levels. Low-speed centrifugation was used to sediment the multicellular aggregates, while the planktonic cells remained suspended in the medium (see Fig. S1 in the supplemental material). The formation of these two cell types was consistent, with a coefficient of variation of less than 10% between biological replicate cultures. The aggregates could be disrupted using a tissue grinder, which generated a primarily single-cell population. Using this technique, we were able to establish a correlation between the wet weight of aggregated cells and the number of CFU (see Fig. S1 in the supplemental material). In addition to the cells in the fluid phase, there were cells that formed an adherent biofilm at the air-liquid interface of the culture flask. These cells differed in terms of oxygen availability (30), and the amount of biofilm formed was highly variable (data not shown); therefore, they were excluded from further analysis.
FIG 1.
Differentiation of cells within an S. Typhimurium biofilm culture. (A) Aliquot of S. Typhimurium 14028 culture containing multicellular aggregates and planktonic cells after 13 h growth at 28°C, prior to separation of the two subpopulations. (B) Isolated aggregates resuspended in PBS to give an example of their relative sizes. (C) Aggregates isolated from an S. Typhimurium culture containing a csgBAC promoter-GFP fusion plasmid (25) visualized on a Leica TCS SP5 confocal microscope.
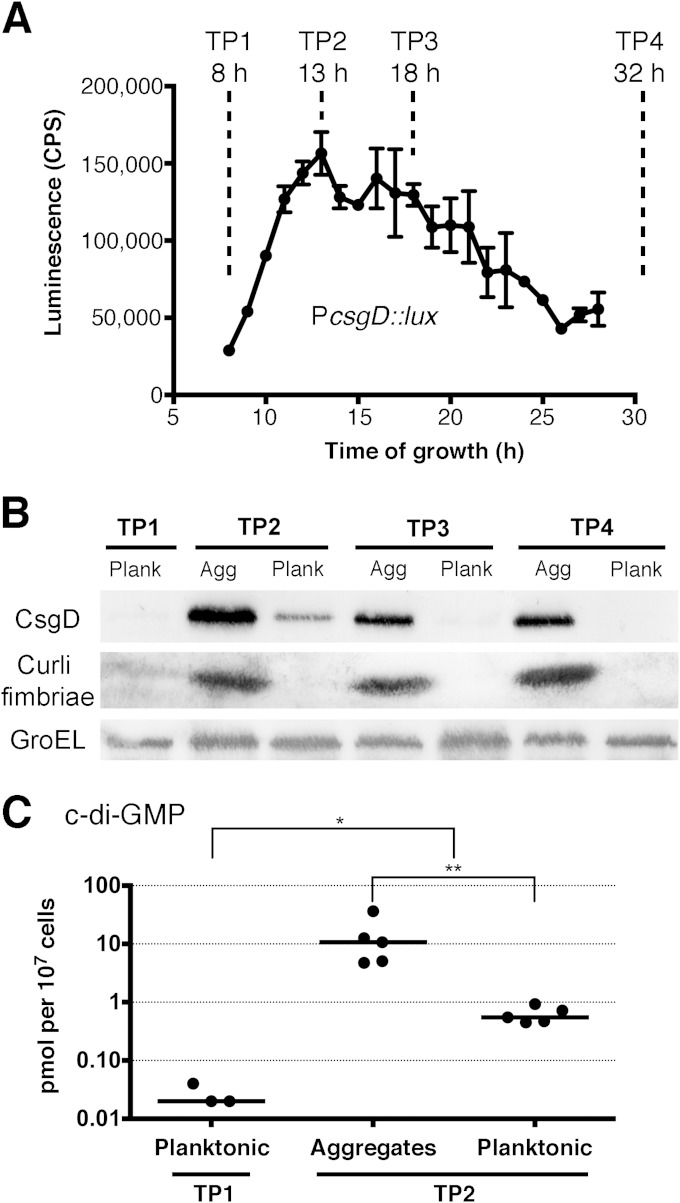
FIG 2.
Analysis of multicellular aggregates and planktonic cells isolated from S. Typhimurium biofilm cultures. (A) csgD expression was measured during growth using a promoter-luciferase (luxCDABE) fusion plasmid designed to measure gene expression by light production (CPS, counts per second). The line represents the average CPS of two biological replicate cultures measured at hourly intervals in triplicate; the error bars represent the range of expression. The time points (TP1 to TP4) were chosen to reflect the dynamics of cellular differentiation. (B) Cell fractions were isolated at each time point and analyzed by Western blotting for synthesis of CsgD, the central regulator of Salmonella biofilm formation, and CsgA, the major subunit of curli fimbriae. GroEL was detected as a loading control to ensure that equal amounts of protein were loaded into each lane. Agg, multicellular aggregates; Plank, planktonic cells. (C) Cyclic-di-GMP concentrations were calculated as pmol per 107 CFU. Each point represents the measurement from one biological replicate culture; c-di-GMP could not be detected for two samples of planktonic cells at TP1. The horizontal bars represent the median values. Statistical significance: *, P < 0.05; **, P < 0.01.
Western blot analysis of the subpopulations isolated from the fluid phase confirmed the bistable production of CsgD; multicellular aggregates displayed high levels of CsgD at all time points, whereas the planktonic cells had either low (TP2), trace (TP1 and TP3), or undetectable (TP4) levels (Fig. 2B). We also analyzed both subpopulations for production of curli fimbriae, an important biofilm extracellular matrix component and functional bacterial amyloid (29). The major subunit protein of curli, CsgA, was detected in abundance in all aggregate fractions, whereas the planktonic-cell fractions at TP2, TP3, and TP4 were negative (Fig. 2B). These results confirmed that the subpopulations had been cleanly separated by low-speed centrifugation. Trace amounts of CsgA were detected at TP1 (Fig. 2B), indicating that some cells had already begun to differentiate at this early time point.
The bacterial secondary messenger c-di-GMP is known to play a role in regulating csgD transcription and CsgD activity (30, 31). High levels of c-di-GMP are associated with biofilm formation, whereas low levels are associated with a single-cell lifestyle (44). To test if these same principles were at work in our system, we measured the intracellular concentrations of c-di-GMP in planktonic cells at TP1 and in multicellular aggregates and planktonic cells at TP2. While planktonic cells at TP1 had nearly undetectable levels of c-di-GMP, there was an order of magnitude increase for planktonic cells at TP2 and another order of magnitude increase for aggregates, which had the highest levels (Fig. 2C). The levels of c-di-GMP correlated with the levels of CsgD in these cell types.
Transcriptome divergence between planktonic and aggregate cell types.
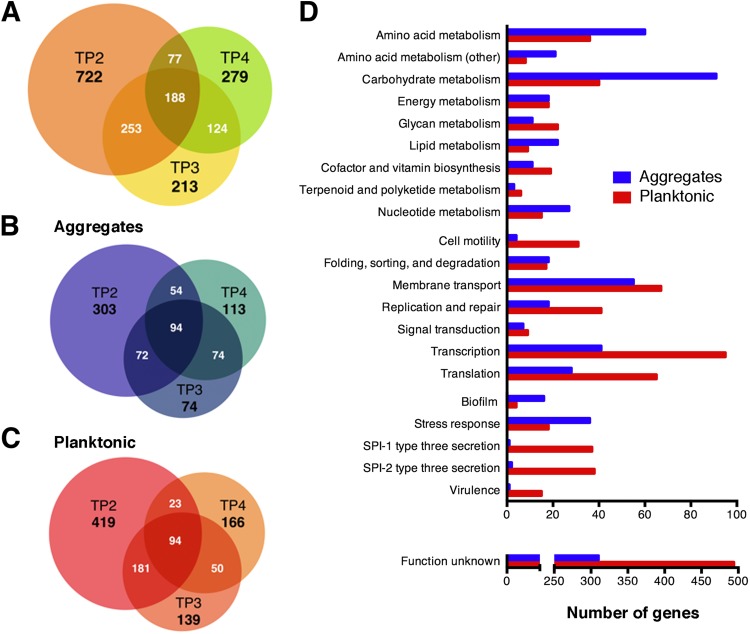
To get a comprehensive view of the transcriptional differences, RNA was purified from each cell subpopulation at each time point, and directional RNA-seq was performed (Table 1). In total, 1,856 genes, or ∼34% of the genes in the genome, were differentially expressed when comparing planktonic cells to multicellular aggregates at each time point (see Data Set S1 in the supplemental material; P < 0.05). The largest differences were identified at TP2, with 1,240 genes being differentially expressed (Fig. 3A). At TP3 and TP4, the numbers of differentially expressed genes were reduced to 778 and 668, respectively; 642 of the differentially expressed genes were shared by at least two time points, and 188 were shared by all three time points. Of the 1,856 genes that were differentially expressed, 784 were expressed at higher levels in multicellular aggregates (Fig. 3B) and 1,072 were more highly expressed in planktonic cells (Fig. 3C). We also performed a comparison between the nonaggregative ΔcsgD strain, which represents an entirely single-cell population (13, 21), and the wild-type planktonic cells at TP2. Gene expression in planktonic cells from these two strains was more similar, with only 220 genes being differentially expressed (see Data Set S2 in the supplemental material).
FIG 3.
RNA-seq-based transcriptome comparison between multicellular-aggregate and planktonic-cell subpopulations. (A) Venn diagram showing the total number of differentially expressed genes (P < 0.05) between planktonic cells and multicellular aggregates at each time point. (B and C) The total was subdivided into genes that were more highly expressed in aggregates (B) and genes that were more highly expressed in planktonic cells (C). (D) Differentially expressed genes in aggregates versus planktonic cells were categorized using the KEGG (Kyoto Encyclopedia of Genes and Genomes; http://www.genome.jp/kegg/) pathway database and are listed in Data Set S1 in the supplemental material.
TABLE 1.
RNA-seq-based transcriptome analysis of planktonic cells and multicellular aggregates isolated from S. Typhimurium culture
| Characteristic | Value |
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild-type planktonic cells |
Wild-type aggregates |
ΔcsgD planktonic (TP2) | ||||||
| TP1 | TP2 | TP3 | TP4 | TP2 | TP3 | TP4 | ||
| Total no. of readsa | 50,889,232 | 64,194,848 | 60,641,580 | 66,424,412 | 64,372,950 | 51,211,612 | 59,213,696 | 32,648,668 |
| mRNA and sRNA | 5,142,914 | 9,183,473 | 6,970,035 | 4,208,777 | 7,112,841 | 4,395,392 | 4,089,516 | 3,971,434 |
| Genome coverageb | 77.7 | 138.7 | 105.3 | 63.6 | 107.5 | 66.4 | 61.8 | 61.2 |
| Protein-coding genes (%)c | ||||||||
| Sense | 75.3 | 74.0 | 68.9 | 66.0 | 74.8 | 71.6 | 70.3 | 73.9 |
| Antisense | 5.9 | 5.4 | 6.7 | 8.9 | 2.8 | 4.1 | 4.4 | 10.4 |
| sRNAs (%)d | 8.0 | 10.6 | 14.2 | 9.5 | 14.4 | 13.9 | 8.9 | 6.5 |
| Intergenic regions (%) | 10.8 | 10.0 | 10.2 | 15.6 | 8.0 | 10.4 | 16.4 | 9.1 |
| Plasmid (%) | 0.8 | 0.8 | 0.7 | 0.6 | 0.4 | 0.4 | 0.4 | 0.8 |
Reads of three technical replicates were merged for each time point for each sample. In total, 416 million strand-specific reads of 75-nucleotide length were obtained.
The reads mapped to rRNAs and tRNAs were removed prior to statistical and comparative analyses. This yielded an average genome coverage of 89.9× (±23.2×) for each time point of each sample excluded.
There are 5,414 putative open reading frames (ORFs) in the S. Typhimurium ATCC 14028 genome, including 5,313 chromosome-located and 101 plasmid-borne ORFs.
The full set of known sRNAs in Salmonella was annotated from reference 40. Approximately 11.5% ± 2.8% of the mapped reads represented known sRNAs, and 11.8% ± 3.4% of the reads mapped to additional intergenic regions.
Closer analysis of the activated genes revealed that the planktonic cells had higher expression of numerous pathways involved in virulence, including both SPI-1 and SPI-2 T3SSs, part of the phoPQ regulon, motility and chemotaxis, and a high proportion (>40%) of function unknown (FUN) genes (Fig. 3D). In contrast, multicellular aggregates had higher expression of genes associated with extracellular matrix production in biofilms (i.e., csgDEFG, csgBAC, and yaiC [adrA]), changes in metabolism (i.e., amino acid, carbohydrates, lipids, and nucleotides), and pathways associated with stress response (i.e., osmoprotection) (Fig. 3D). The nature of the transcriptional differences between planktonic cells and multicellular aggregates suggested that they represented specialized cell types. Many of the genes listed in Data Sets S1 and S2 in the supplemental material were distributed into operons predicted from RNA-seq through a combination of transcriptional start and termination sites, intergenic distance, and coexpression information (91).
Expression of c-di-GMP enzymes and other genetic factors contributing to CsgD bistability.
We identified several differentially expressed genes associated with the complex c-di-GMP signaling system that contributes to CsgD regulation (Table 2). Two diguanylate cyclase (DGC) (i.e., c-di-GMP-generating enzymes) genes, STM1987 (45) and adrA (46), were more highly expressed in aggregates at TP2, which correlated with higher levels of c-di-GMP in these cells. yhjH, which encodes a phosphodiesterase (PDE) (i.e., a c-di-GMP-degrading enzyme) with a critical role in controlling motility (46, 47), was more highly expressed in planktonic cells, and this correlated with smaller amounts of c-di-GMP. Genes encoding three diguanylate cyclases characterized as having positive effects on invasion (yeaJ) (48), motility (STM2503) (49), or motility and virulence (STM4551) (49) were more highly expressed in planktonic cells at TP2, which correlated with the predicted phenotype of these cells. nlpI, encoding a putative membrane protein with a repressive effect on CsgD synthesis (50), was also more highly expressed in planktonic cells, which matched the lower levels of CsgD in these cells. In contrast, high expression of ycgR and ydiV in planktonic cells at TP2 did not match the predicted phenotypes. YcgR contains a PilZ domain for binding c-di-GMP and represses motility by acting as a type of flagellar brake (51, 52). Based on the published function, we would have expected ycgR to be more highly expressed in multicellular aggregates and to function at higher c-di-GMP levels inside the cell. YdiV represses expression of yhjH and also negatively regulates motility (53). It is possible that the synthesis of YcgR and YdiV results in localized effects within the cell, but the overall c-di-GMP pool is predominantly influenced by the increased expression of yhjH (46, 47). In general, the RNA-seq findings were consistent with the levels of c-di-GMP detected and the predicted phenotypes of planktonic cells and multicellular aggregates.
TABLE 2.
Differentially expressed genes in the c-di-GMP signaling network
| Gene namea | Enzyme typeb | Functionc | Fold changed | Time point(s)e |
|---|---|---|---|---|
| STM14_5467 (STM4551) | DGC | Positive regulator of motility and virulence by both c-di-GMP-dependent and -independent mechanisms (49); an additional report states that the function is the exact opposite (48) | 4.05 | TP2, TP3, TP4 |
| yeaJ (STM1283) | DGC | Stimulates invasion (48) | 3.18 | TP2, TP3 |
| yhjH (STM3611) | PDE | Negative regulator of rdar morphotype; positive regulator of motility (46) | 4.47 | TP2 |
| STM14_3068 (STM2503) | DGC | Positive effect on motility (49) | 3.13 | TP2 |
| ycgR | C-di-GMP biosensor | Binds c-di-GMP through a high-affinity PilZ domain; represses motility by binding directly to FliG (51, 52) | 4.07 | TP2 |
| ydiV (STM1344) | EAL-like protein | Positive regulator of csgD transcription and negative regulator of motility, but does not have a direct role in c-di-GMP turnover; represses expression of yhjH (53) | 3.58 | TP2 |
| nlpI | Outer membrane protein | NlpI can negatively influence the expression of yjcC, which codes for a DGC affecting CsgD expression (50) | 3.89 | TP2, TP3, TP4 |
| adrA (yaiC) | DGC | Positive regulator of cellulose production (46) | 0.34 | TP2, TP3, TP4 |
| STM14_2408 (STM1987) | DGC | Positive effect on rdar morphotype, CsgD expression, and cellulose (45) | 0.54 | TP2 |
Gene names are listed as they appear in the S. Typhimurium 14028 genome (39); the S. Typhimurium LT2 gene names are listed in parentheses.
An enzyme that generates c-di-GMP is called a DGC; an enzyme that breaks down c-di-GMP is called a PDE. PDE enzymes contain EAL domains.
Functions are listed in the context of c-di-GMP regulation and turnover, CsgD expression, motility, and virulence.
Fold change represents the expression ratios of planktonic cells/aggregates at TP2; genes with values of >1 have higher expression in planktonic cells, whereas genes with values of <1 have higher expression in multicellular aggregates.
Time points where the expression difference between planktonic cells and multicellular aggregates was statistically significantly (P < 0.05) are listed.
Bistable gene expression in bacteria is usually the result of positive autoregulation of a master regulatory gene (54, 55). A feed-forward loop has been proposed for csgD in E. coli, but it has not been proven in S. Typhimurium (30). CsgD is proposed to activate the expression and synthesis of IraP, which inhibits the degradation of RpoS, thus leading to greater csgD transcription (56). RNA-seq analysis revealed that multicellular aggregates had elevated transcription of rpoS at TP2 and csgD and iraP (yaiB) at TP3 and TP4, potentially confirming this regulatory loop (see Fig. S2 in the supplemental material).
SPI-1 T3SS expression, synthesis, and motility are increased in planktonic cells.
The SPI-1 T3SS is a 3.5-MDa translocation apparatus that is required for injection of Salmonella effector proteins into the cytoplasm of host cells. These effector proteins stimulate Salmonella invasion and dissemination, as well as host inflammation (6). The current dogma is that SPI-1 T3SS synthesis is activated by the body temperatures of warm-blooded hosts (57). This is primarily based on the finding that the nucleoid-associated protein H-NS represses transcription of key SPI-1 regulators at lower growth temperatures, such as 25°C (58). Biofilm growth conditions (i.e., low osmolarity, 28°C, and stationary phase) were traditionally thought to be non-SPI-1 inducing because they are so different from classical SPI-1-inducing conditions (i.e., high osmolarity, 37°C, and late exponential phase) (57, 59, 60).
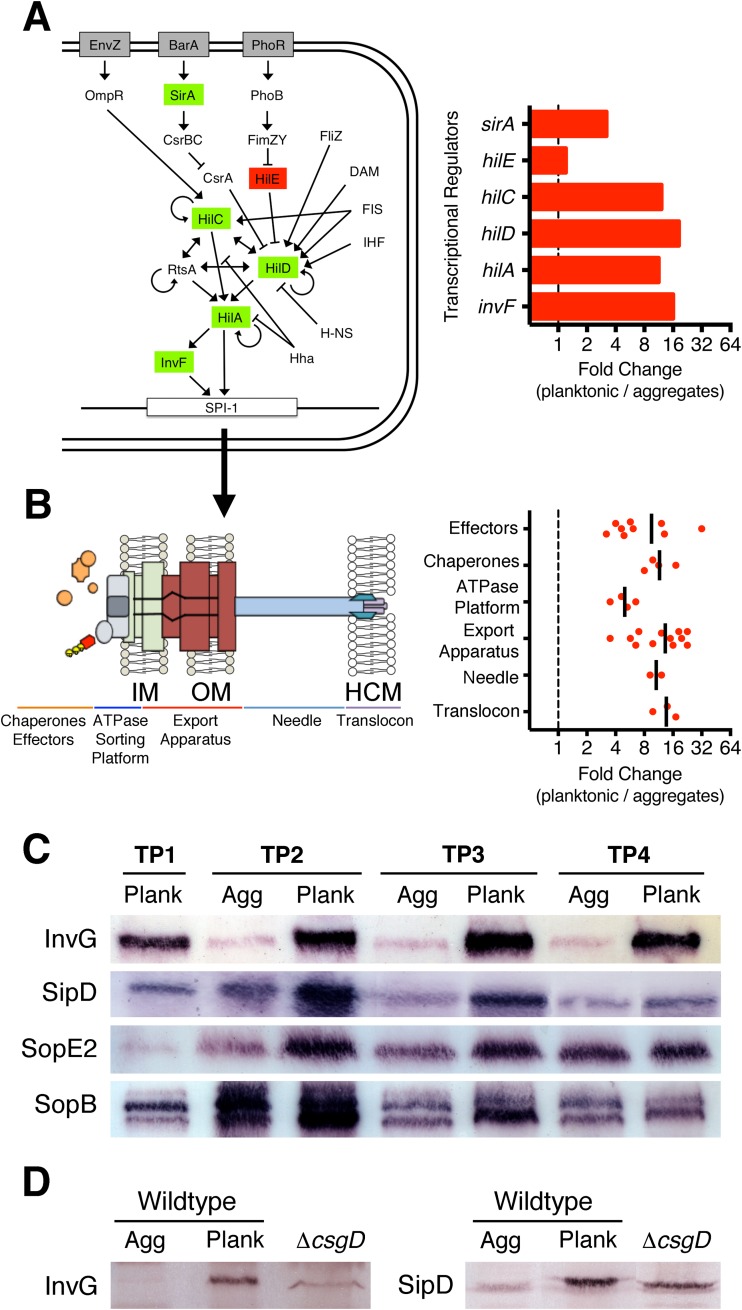
RNA-seq analysis revealed that 41 of 44 SPI-1 T3SS-related genes were more highly expressed in planktonic cells grown under biofilm-inducing conditions (see Data Set S1 in the supplemental material). These genes included pivotal transcriptional regulators (Fig. 4A), as well as apparatus, chaperone, and effector genes (Fig. 4B). HilA activates transcription of SPI-1 apparatus genes distributed in the inv-spa and prg-org operons (57). InvF is encoded as part of the inv-spa operon and, together with SicA, promotes transcription of the sic-sip operon encoding the translocon, as well as numerous effector genes. Temporal expression analysis of the SPI-1 genes followed a pattern consistent with the establishment of secretion-competent organelles in planktonic cells; apparatus genes had the highest expression at TP1, while translocon and numerous effector genes had peak expression at TP2 (see Fig. S3 in the supplemental material). In contrast, the expression levels of all SPI-1 genes were reduced in multicellular aggregates (see Fig. S3 in the supplemental material). Comparison between wild-type planktonic cells and ΔcsgD planktonic cells at TP2 revealed that sipD and sopE2 were more highly expressed in the wild-type cells (see Data Set S2 in the supplemental material). In addition, several other SPI-1 genes were more highly expressed in wild-type cells but had P values just above the cutoff for significance (data not shown). These findings suggested that there could be a potential regulatory link between CsgD and the SPI-1 T3SS.
FIG 4.
Expression and synthesis of the SPI-1 T3SS in S. Typhimurium planktonic cells. (A) Schematic of a bacterial cell showing the regulatory map and main transcriptional activators (green) and repressors (red) for SPI-1 T3SS expression (6). The fold change values for the regulators shown are displayed as red bars on the accompanying graph, calculated from RNA-seq data at TP2. (B) The SPI-1 T3SS translocation apparatus (left) (90) was divided into five component groups, including proteins embedded in the bacterial inner (IM) or outer (OM) membrane or host cell membrane (HCM). Each point on the accompanying graph represents the fold change value at TP2 for the genes for effectors (sopE2, sopB, sipA, sopD, gtgE, sopA, avrA, sptP, and gogB), chaperones (sicA, sicP, invB, and sigE [pipC]), ATPase platform (invC, spaO, orgA, and orgB), export apparatus (invG, spaR, prgH, invI, invA, invE, prgK, spaP, spaQ, invJ, spaS, and invH), needle (prgI and prgJ), and translocon (sipB, sipC, and sipD) (6). The dashed line at 1 represents no difference in gene expression; the vertical lines represent the mean fold changes for the groups. (C) Whole-cell lysates of planktonic cells and multicellular aggregates were analyzed by Western blotting to detect the SPI-1 T3SS proteins. Plank, planktonic cells; Agg, multicellular aggregates. (D) InvG and SipD were detected in ΔcsgD planktonic cells at TP2, and the relative levels were compared to those of aggregates and planktonic cells isolated from wild-type cultures.
To determine if SPI-1 transcriptional differences were reflected at the protein level, whole-cell lysates of either cell type were analyzed by Western blotting. InvG, the outer membrane (OM) secretin (61), was detected at high levels in planktonic cells at all time points and was nearly undetectable in the multicellular-aggregate samples (Fig. 4C). The on/off pattern of synthesis for this critical structural component indicated that planktonic cells had the capacity to form a greater number of SPI-1 T3SS organelles than aggregated cells. SipD, the translocon tip protein that interacts with host cell membranes (62), was also present at high levels in planktonic cells at all time points (Fig. 4C). SopB and SopE2, two of the earliest expressed effectors that stimulate host cell cytoskeleton rearrangements (63), were also synthesized at higher levels in planktonic cells at TP2 and TP3, with the difference decreasing by TP4 (Fig. 4C). SPI-1 protein levels were also analyzed in ΔcsgD cells at TP2. InvG and SipD levels were noticeably lower in ΔcsgD cells than in wild-type planktonic cells (Fig. 4D), whereas SopB and SopE2 had no detectable difference between the two strains (data not shown).
Recent evidence suggests that the SPI-1 T3SS and flagellum-mediated motility are required for targeting of permissive invasion sites on host cells (64) and that these systems are often coregulated (65). In our RNA-seq experiment, 28 genes coding for flagellar assembly and chemotaxis proteins were identified as being more highly expressed in planktonic cells (see Data Set S1 in the supplemental material). We performed a swim assay, which confirmed that planktonic cells had enhanced motility compared to multicellular aggregates but were not significantly different from ΔcsgD planktonic cells (see Fig. S4 in the supplemental material).
Transcriptional priming of the SPI-2 T3SS in planktonic cells.
SPI-2 encodes a second T3SS that has an important role once Salmonella has invaded host cells. The SPI-2 T3SS is required for modification of the phagolysosome to make it conducive for Salmonella replication, particularly inside macrophages (6). Thirty-seven of 42 genes encoding the SPI-2 T3SS were more highly expressed in planktonic cells than in the multicellular aggregates (see Data Set S1 in the supplemental material). However, unlike SPI-1, we were unable to detect SPI-2 protein synthesis, such as that of SsaC, the equivalent of InvG, in any cell fractions (data not shown). This suggested that functional SPI-2 needles were not present on the surfaces of planktonic cells and supported the possibility that transcriptional priming was occurring, as has been observed prior to host cell invasion (66, 67).
Planktonic cells display enhanced invasion of a human intestinal cell line.
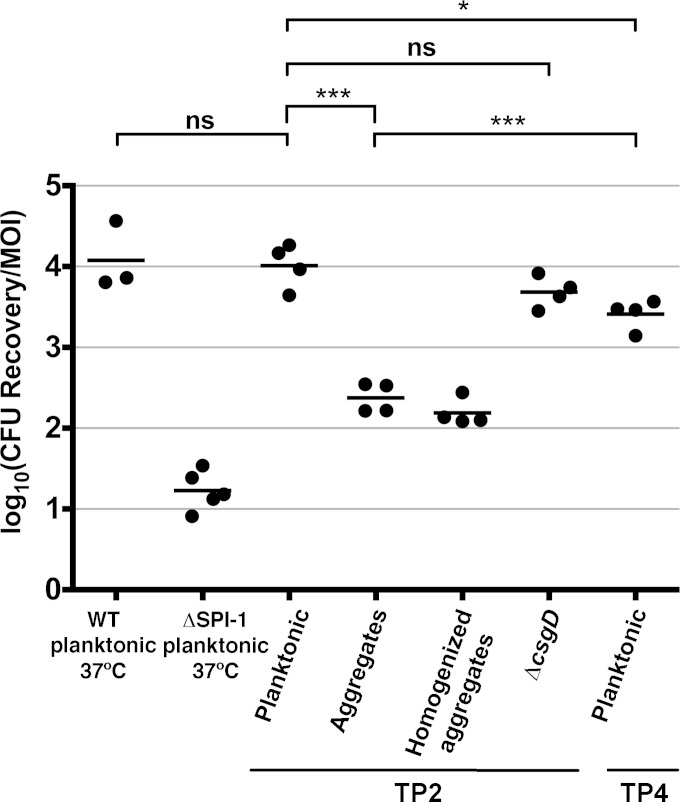
To our knowledge, SPI-1 T3SS synthesis has never been observed before under biofilm-inducing conditions but is usually correlated with increased invasion and virulence (68). Therefore, we tested the invasion by different S. Typhimurium cell types of polarized Caco-2 cells, a human intestinal cell line. Planktonic cells isolated at TP2 displayed the same invasiveness as planktonic cells grown under classical SPI-1-inducing conditions (Fig. 5), confirming that the SPI-1 organelles synthesized under biofilm-inducing conditions were functional. Planktonic cells isolated at later time points (i.e., TP4) still retained efficient invasion capability, suggesting that this phenotype was stable. In contrast, the multicellular aggregates were significantly less invasive, presumably because of reduced type three secretion synthesis. Surprisingly, ΔcsgD cells, which had reduced InvG and SipD levels, showed only a slight drop in invasion that was not statistically different than that of wild-type planktonic cells. For the aggregates, we reasoned that reduced invasion could be influenced by the presence of extracellular matrix polymers blocking access to the Caco-2 cell surface. To disrupt the matrix, we homogenized the aggregates prior to repeating the assay, but it did not lead to increased invasion (Fig. 5, homogenized aggregates). It was noted, however, that the aggregated cells invaded significantly better than a ΔSPI-1 deletion strain, suggesting that they would retain a capacity for virulence.
FIG 5.
Invasion of polarized Caco-2 cells by different S. Typhimurium subpopulations. Planktonic cells and multicellular aggregates isolated from S. Typhimurium 14028 biofilm cultures at TP2 and TP4 were used to infect polarized Caco-2 cells. Aggregates that were homogenized using a tissue grinder were also tested, along with planktonic cells prepared from ΔcsgD cultures. Wild-type (WT) and ΔSPI-1 cells grown in LB broth at 37°C were used as positive and negative invasion controls, respectively. Each point represents the average value from invasion of three replicate wells of Caco-2 cells; the horizontal bars represent the mean values. The average MOI was 17, based on an estimate of 2.5 × 105 polarized Caco-2 cells per well. Statistical significance: ***, P < 0.001; *, P < 0.05; ns, P > 0.05.
As part of the Caco-2 cell invasion assays, the aminoglycoside antibiotic gentamicin was added to kill any extracellular bacteria that had not invaded. Prior to performing the assays, we tested the susceptibilities of multicellular aggregates and planktonic cells to gentamicin. While the planktonic cells were completely killed (below the limit of detection) at standard concentrations of 200 to 400 μg ml−1 (33), the multicellular aggregates displayed enhanced resistance and were depleted only once the concentration reached 1,200 μg ml−1 (see Fig. S5 in the supplemental material). This confirmed that the multicellular aggregates represented a more resistant subpopulation of cells.
Planktonic cells display a significant virulence advantage in murine infection.
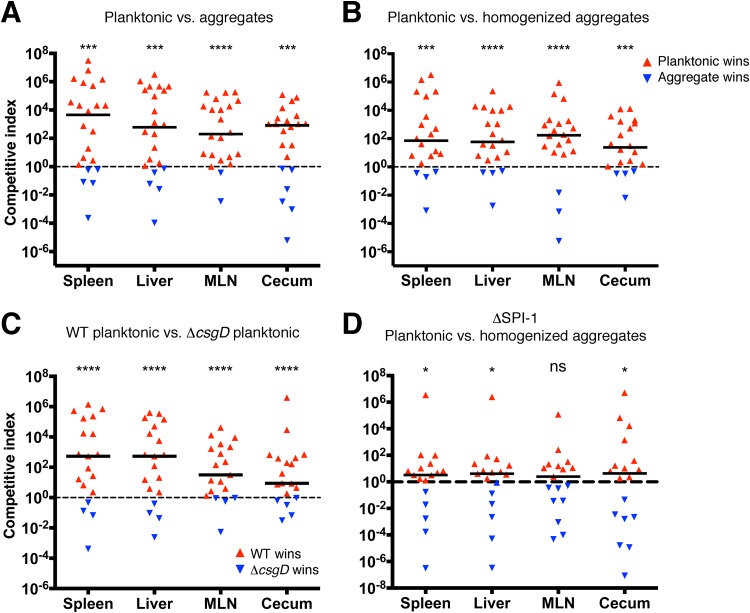
Based on the expression data and Caco-2 invasion results, we hypothesized that the planktonic cells were more virulent, whereas the multicellular aggregates were likely to have enhanced persistence. To test the colonization efficiency and virulence of each cell type, we performed formally randomized, blinded coinfections of mice with 1:1 ratios of multicellular aggregates and planktonic cells isolated from kanamycin- or chloramphenicol-resistant strains of S. Typhimurium (see Fig. S6 in the supplemental material). For the initial CI trial, we wanted to keep the cell subpopulations in their native physical states; therefore, we minimized the handling of the cells and used a wide-bore gavage needle to prevent shearing of aggregates during oral delivery. Under these conditions, the planktonic cells exhibited a significant virulence advantage, as demonstrated by their proportionally increased recovery from spleen, liver, cecum, and mesenteric lymph nodes in the majority of infected mice (Fig. 6A, red triangles). As predicted, the multicellular aggregates did retain a capacity for virulence and were recovered in larger proportions in several mice (Fig. 6A, blue triangles). We reasoned that, despite being comprised of hundreds to thousands of cells, each aggregate might function as only a single infectious unit. Therefore, we homogenized the aggregates to generate a single-cell suspension prior to repeating the CI trial. The CI values were slightly reduced, but the overall trend remained the same (Fig. 6B), indicating that the increased virulence of planktonic cells was not due to a difference in the multiplicity of infection. We also competed wild-type planktonic cells and ΔcsgD planktonic cells, and again, the wild-type planktonic cells displayed a strong virulence advantage (Fig. 6C). This was surprising, because the invasion phenotypes of the two strains were similar; however, there could be other impairments related to the ΔcsgD mutation (69). To assess the importance of SPI-1 in the colonization advantage displayed by planktonic cells, we repeated a CI trial with planktonic cells and multicellular aggregates prepared from a ΔSPI-1 mutant strain. For each organ, the median CI values dropped approximately 10-fold and were near 1 (Fig. 6D), which indicated that the SPI-1 T3SS was the primary factor responsible for increased colonization of planktonic cells. However, small differences were still detected for the spleen, liver, and cecum (Fig. 6D), indicating that the planktonic cells had other adaptations (e.g., increased motility) that provided them with a colonization advantage.
FIG 6.
Competitive infections between multicellular aggregates and planktonic cells prepared from wild-type, ΔSPI-1, and ΔcsgD cultures. The CI was determined by oral coinfection of C57BL/6 mice with a 1:1 ratio of different cell types. (A) Trial between planktonic cells and multicellular aggregates, using a wide-bore gavage needle to prevent shearing of aggregates. (B) Trial between planktonic cells and multicellular aggregates after homogenization of the aggregates to generate a single-cell population. (C) Trial between wild-type planktonic cells and ΔcsgD planktonic cells. (D) Trial between planktonic cells and multicellular aggregates isolated from a ΔSPI-1 biofilm culture. Each point represents the CI value calculated from one organ from a single mouse, with either cell type (red or blue triangles) winning the competition. The solid lines represent the median of all CI values from a particular organ. The dashed line represents a CI value of 1, which indicates no difference in virulence. Statistical significance: *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; ns, P > 0.05.
Persistence of multicellular aggregates reduces the competitive advantage of planktonic cells.
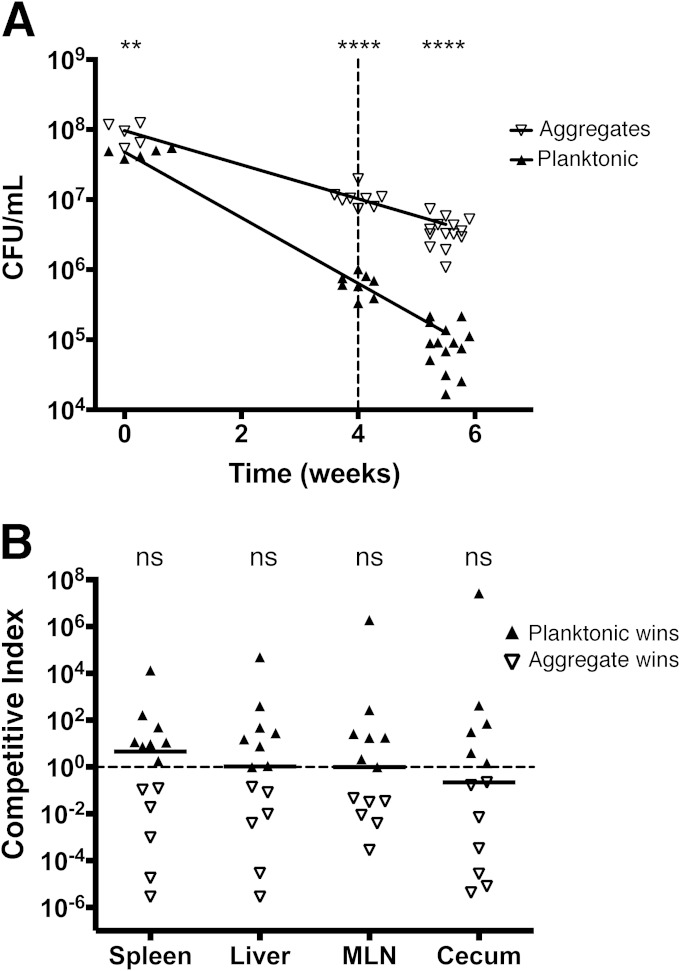
To assess how the multicellular aggregates and planktonic cells responded to a potential environmental stress, aliquots of the challenge inocula from the first CI experiment were desiccated and stored in 24-well plates at room temperature. Viability assays performed on the desiccated challenge inocula revealed that the multicellular aggregates survived significantly better than the planktonic cells (Fig. 7A). At the 4-week time point, there were approximately 10 times more aggregated cells alive (Fig. 7A, dashed line). We performed another CI trial at this time point, after rehydration and resuspension of the cells. The values for each organ indicated no significant advantage for either subpopulation, with aggregates or planktonic cells winning the competition in approximately equal numbers of mice (Fig. 7B). This indicated that upon exposure to environmental stress, the multicellular aggregates had a fitness advantage. If these results were extrapolated into the natural world, one could envision a scenario where, after extended periods of environmental exposure, only multicellular aggregates would be alive and able to infect new hosts.
FIG 7.
Enhanced persistence of S. Typhimurium multicellular aggregates. (A) Multicellular aggregates and planktonic cells used for the trial in Fig. 6A were desiccated and stored in 24-well plates. Survival of each cell type was determined after storage for the times displayed on the x axis. Statistical significance: **, P < 0.01; ****, P < 0.0001. The dashed line represents the time at which cells were resuspended and a CI trial was performed. (B) Each point represents the CI value calculated from one organ from a single mouse, and the solid lines represent the median of all CI values from a particular organ. The dashed line represents a CI value of 1, or no difference in virulence; none of the organs displayed CI values that were significantly different from 1 (ns, P > 0.05).
DISCUSSION
In this study, we isolated and analyzed two distinct subpopulations of cells within a clonal population of S. Typhimurium. Based on genetic, biochemical, and functional characterization, we hypothesize that the planktonic cells and multicellular aggregates formed under biofilm-inducing conditions represent specialized cell types that are adapted for virulence and persistence, respectively. Due to the presence of functional SPI-1 T3SS organelles, we predict that the planktonic cells are proficient at direct host-to-host transmission, whereas the resistant multicellular aggregates can survive in the environment and cause infections at later times. The connection of virulence and persistence within an individual population represents an elegantly simple strategy that would prepare S. Typhimurium for unpredictability and improve the chances for transmission each time the bacteria leave an infected host. This scenario also implies that the host immune system may respond differently to these specialized cell types. If hosts routinely encounter multicellular aggregates, it could explain why curli fimbriae are such potent stimulators of the innate immune system (29, 70, 71), even though the organelles do not appear to play a role in Salmonella infections (25).
The concept of bistability was described by Novick and Weiner over 50 years ago as the existence of different subpopulations of enzyme-producing cells within a genetically identical population of E. coli, controlled by a feed-forward loop (72). CsgD lies at the heart of a feed-forward loop in S. Typhimurium when cells are grown under biofilm-inducing conditions. CsgD is synthesized at high levels in specific cells, leading to production of extracellular matrix polymers and the formation of multicellular aggregates (25, 30). The regulation of csgD transcription and CsgD synthesis is extremely complex (reviewed in reference 31). Numerous transcription factors are involved, including OmpR, MlrA, and CpxR and the nonspecific DNA-binding proteins H-NS and IHF, in addition to the feed-forward loop involving rpoS and the factors that regulate RpoS activity, including IraP and Crl. In addition, at least five sRNAs can bind to the 5′ untranslated region of the csgD mRNA and regulate CsgD synthesis (73). Robust control of CsgD production is clearly important, given that >30% of the genes in the genome were differentially expressed between the S. Typhimurium multicellular aggregates and planktonic cells. The large number of differentially expressed genes represents a tremendous amount of nonheritable genetic change, on the order of a developmental program in bacteria (74). Since only 20 CsgD-specific targets were identified in a recent E. coli chromatin immunoprecipitation sequencing (ChIP-seq) study (75), we hypothesize that CsgD initiates the aggregation process and that a cascade of gene expression changes follows as cells produce an extracellular matrix (35). Ultimately, this cascade would be the cumulative result of numerous connected signaling pathways, including c-di-GMP (44).
Recent reports have shown that bistable gene expression can occur within bacterial virulence gene networks. In Vibrio cholerae, bimodal expression of ToxT results in virulent and nonvirulent cell subpopulations (76). Bistable production of the SPI-1 T3SS in S. Typhimurium has also recently been described (77–79). The authors observed that nearly 100% of S. Typhimurium cells interacting with intestinal tissues were positive for SPI-1 expression while only 15% of cells in the intestinal lumen were positive (77). The presence of both SPI-1+ and SPI-1− cells seems to be important for stabilizing the virulence traits of S. Typhimurium (78). At the moment, it is unclear how these findings are connected to what we have observed. Our finding that SPI-1 T3SS synthesis occurs in planktonic cells under biofilm-inducing conditions may indicate that an alternative regulatory pathway exists for SPI-1 activation. It is also possible that the planktonic population consists of both SPI-1+ and SPI-1− cells. Alternatively, the subpopulation of cells that form multicellular aggregates under biofilm-inducing conditions could be related to the SPI-1− cells described by Hardt and colleagues (e.g., similar genetic pathways activated or repressed) (77–79). One of our initial goals was to determine if the differentiation of cells was more than an in vitro phenomenon. The fact that the planktonic cells were more virulent than multicellular aggregates, even after the aggregates were homogenized into a single-cell suspension, suggested that differentiation was retained until the cells reached the murine intestine. The importance of SPI-1 in this process was confirmed, since planktonic cells and multicellular aggregates from a ΔSPI-1 mutant strain had nearly equal colonization efficiencies. Furthermore, the reduction in virulence that we observed for the ΔcsgD strain, together with recent connections between c-di-GMP, virulence (28), and motility (52), suggest that the master biofilm regulator, CsgD, can modulate the virulence capacity of S. Typhimurium.
Bet hedging is characterized as a risk-spreading strategy displayed by clonal populations, where each phenotype performs more or less well at any time, depending on the selection pressures present (80). Although we cannot be certain of the evolutionary selections that led to bistable CsgD expression, the passage of Salmonella isolates between host and nonhost environments represents a model of unpredictability (7). Aggregation is known to have many evolutionary benefits to ensure the survival of populations, but rather than being interpreted as altruistic behavior, it may be driven by harsh penalties associated with an individual not being part of the group (81). For pathogens like S. Typhimurium, the maintenance of a subset of cells primed for invasion but not ideally suited for environmental survival would impose a penalty unless a host is encountered. The energy commitment required to make SPI-1 T3SS organelles (82) or extracellular matrix polymers (83, 84) in different subpopulations indicates that it might be a strategy for S. Typhimurium to prepare for unpredictability. A proportion of cells could survive exposure to either the host or nonhost environment, with the ultimate goal being preservation of the shared genome for the next generation (54, 80). We speculate that signaling can occur between the host immune system, the microbiota, and the invading pathogen, with the distinct possibility that S. Typhimurium, and presumably other NTS isolates, can modulate its virulence (85) or persistence (7) as a result of these interactions.
As pathogens evolve, there is thought to be continual selection pressure for them to adapt to their hosts (86). Host adaptation has the benefit of reducing the variation and unpredictability that a bacterial pathogen is exposed to but makes strains more dependent on a direct means of transmission (4). Differences in transmission could potentially explain the correlation between an invasive, host-adapted way of life and loss of the rdar morphotype in Salmonella and E. coli (17, 26). In E. coli, bistable expression of CsgD does occur in specific pathogenic strains (87); however, human-adapted enteroinvasive E. coli and Shigella isolates have lost the ability to aggregate (27). Similarly, the evolution of S. Typhi has been strongly influenced by the human carrier state (5), and nearly all isolates are negative for the rdar morphotype (25, 26). We hypothesize that host-adapted or host-restricted isolates favor a single-cell mode of growth, because shedding from chronic carriers makes them less dependent on long-term survival to complete the transmission cycle. S. Typhi is capable of other forms of biofilm growth that are required for persistence inside human carriers, such as attachment to the surfaces of gallstones (88), and recent evidence indicates that S. Typhi can be transmitted indirectly via contaminated water (89). This suggests that there are other factors to consider when correlating a loss of persistence with host adaptation. Nevertheless, identifying the molecular mechanisms that connect virulence and persistence may hold the key for understanding transmission and ultimately reducing the spread of these human pathogens. We predict that the bet-hedging program described here for S. Typhimurium is a common strategy for survival, especially for other enteric pathogens that spend a significant portion of their life cycle in the environment.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by Natural Sciences and Engineering Research Council (NSERC) Discovery grants to A.P.W. (386063-2010) and A.D.S.C. (435784-2013) and through the Jarislowsky Chair in Biotechnology to A.P.W. K.D.M. was supported by a Canada Graduate Scholarship from NSERC, Y.W. by a Postdoctoral Research Award from the Saskatchewan Health Research Foundation, and C.S.W. by an Undergraduate Research Award from the University of Saskatchewan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are grateful to Don Wilson, Stew Walker, and the VIDO animal care staff for professional help with the animal experiments; to Diane Miller, Yongjun Zhao, and the research team at Canada's Michael Smith Genome Sciences Centre for RNA-seq; to Jun Han and Derek Smith at the UVic Genome BC Proteomics Centre for c-di-GMP measurements; to Alex Pasternak for assistance with confocal microscopy; to Anjuman Ara, Neil Rawlyk, and Wayne Connor for laboratory assistance; and to Mike Surette, Scott Napper, Peter Howard, and Jos Vanderleyden for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00137-15.
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winfield MD, Groisman EA. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol 69:3687–3694. doi: 10.1128/AEM.69.7.3687-3694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser MJ, Kirschner D. 2007. The equilibria that allow bacterial persistence in human hosts. Nature 449:843–849. doi: 10.1038/nature06198. [DOI] [PubMed] [Google Scholar]
- 5.Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill F-X, Goodhead I, Rance R, Baker S, Maskell DJ, Wain J, Dolecek C, Achtman M, Dougan G. 2008. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet 40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moest TP, Meresse S. 2013. Salmonella T3SSs: successful mission of the secret(ion) agents. Curr Opin Microbiol 16:38–44. doi: 10.1016/j.mib.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Monack DM. 2012. Salmonella persistence and transmission strategies. Curr Opin Microbiol 15:100–107. doi: 10.1016/j.mib.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Waldner LL, MacKenzie KD, Koster W, White AP. 2012. From exit to entry: long-term survival and transmission of Salmonella. Pathogens 1:128–155. doi: 10.3390/pathogens1020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziehm D, Dreesman J, Campe A, Kreienbrock L, Pulz M. 2013. Risk factors associated with sporadic salmonellosis in adults: a case-control study. Epidemiol Infect 141:284–292. doi: 10.1017/S0950268812000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol 173:4773–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romling U, Sierralta WD, Eriksson K, Normark S. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol 28:249–264. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- 12.Gibson DL, White AP, Snyder SD, Martin S, Heiss C, Azadi P, Surette M, Kay WW. 2006. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J Bacteriol 188:7722–7730. doi: 10.1128/JB.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 14.Doran JL, Collinson SK, Burian J, Sarlos G, Todd EC, Munro CK, Kay CM, Banser PA, Peterkin PI, Kay WW. 1993. DNA-based diagnostic tests for Salmonella species targeting agfA, the structural gene for thin, aggregative fimbriae. J Clin Microbiol 31:2263–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solano C, Garcia B, Valle J, Berasain C, Ghigo JM, Gamazo C, Lasa I. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol 43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- 16.Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol 180:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White AP, Sibley KA, Sibley CD, Wasmuth JD, Schaefer R, Surette MG, Edge TA, Neumann NF. 2011. Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Appl Environ Microbiol 77:7620–7632. doi: 10.1128/AEM.05909-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anriany YA, Weiner RM, Johnson JA, De Rezende CE, Joseph SW. 2001. Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl Environ Microbiol 67:4048–4056. doi: 10.1128/AEM.67.9.4048-4056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scher K, Romling U, Yaron S. 2005. Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl Environ Microbiol 71:1163–1168. doi: 10.1128/AEM.71.3.1163-1168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlich GA, Cooke PH, Solomon EB. 2006. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol 72:2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White AP, Gibson DL, Kim W, Kay WW, Surette MG. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J Bacteriol 188:3219–3227. doi: 10.1128/JB.188.9.3219-3227.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Austin JW, Sanders G, Kay WW, Collinson SK. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol Lett 162:295–301. doi: 10.1111/j.1574-6968.1998.tb13012.x. [DOI] [PubMed] [Google Scholar]
- 23.Barak JD, Gorski L, Naraghi-Arani P, Charkowski AO. 2005. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl Environ Microbiol 71:5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apel D, White AP, Grassl GA, Finlay BB, Surette MG. 2009. Long-term survival of Salmonella enterica serovar Typhimurium reveals an infectious state that is underrepresented on laboratory media containing bile salts. Appl Environ Microbiol 75:4923–4925. doi: 10.1128/AEM.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White AP, Gibson DL, Grassl GA, Kay WW, Finlay BB, Vallance BA, Surette MG. 2008. Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect Immun 76:1048–1058. doi: 10.1128/IAI.01383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romling U, Bokranz W, Rabsch W, Zogaj X, Nimtz M, Tschape H. 2003. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int J Med Microbiol 293:273–285. doi: 10.1078/1438-4221-00268. [DOI] [PubMed] [Google Scholar]
- 27.Sakellaris H, Hannink NK, Rajakumar K, Bulach D, Hunt M, Sasakawa C, Adler B. 2000. Curli loci of Shigella spp. Infect Immun 68:3780–3783. doi: 10.1128/IAI.68.6.3780-3783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmad I, Wigren E, Le Guyon S, Vekkeli S, Blanka A, El Mouali Y, Anwar N, Chuah ML, Lunsdorf H, Frank R, Rhen M, Liang ZX, Lindqvist Y, Romling U. 2013. The EAL-like protein STM1697 regulates virulence phenotypes, motility and biofilm formation in Salmonella Typhimurium. Mol Microbiol 90:1216–1232. doi: 10.1111/mmi.12428. [DOI] [PubMed] [Google Scholar]
- 29.Hufnagel DA, Tukel C, Chapman MR. 2013. Disease to dirt: the biology of microbial amyloids. PLoS Pathog 9:e1003740. doi: 10.1371/journal.ppat.1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grantcharova N, Peters V, Monteiro C, Zakikhany K, Romling U. 2010. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar Typhimurium. J Bacteriol 192:456–466. doi: 10.1128/JB.01826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steenackers H, Hermans K, Vanderleyden J, De Keersmaecker SCJ. 2012. Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res Int 45:502–531. doi: 10.1016/j.foodres.2011.01.038. [DOI] [Google Scholar]
- 32.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desin TS, Lam PK, Koch B, Mickael C, Berberov E, Wisner AL, Townsend HG, Potter AA, Koster W. 2009. Salmonella enterica serovar Enteritidis pathogenicity island 1 is not essential for but facilitates rapid systemic spread in chickens. Infect Immun 77:2866–2875. doi: 10.1128/IAI.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maloy SR, Stewart VJ, Taylor RK. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 35.White AP, Weljie AM, Apel D, Zhang P, Shaykhutdinov R, Vogel HJ, Surette MG. 2010. A global metabolic shift is linked to Salmonella multicellular development. PLoS One 5:e11814. doi: 10.1371/journal.pone.0011814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 37.Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, Mack D, Goldman WE, Gomelsky M, Perry RD. 2011. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol Microbiol 79:533–551. doi: 10.1111/j.1365-2958.2010.07470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J Bacteriol 192:560–567. doi: 10.1128/JB.01233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton JC. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. doi: 10.2307/2346101. [DOI] [Google Scholar]
- 43.Robinson MD, Smyth GK. 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 44.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, Lasa I. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol Microbiol 54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- 46.Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 47.Le Guyon S, Simm R, Rhen M, Romling U. 25 July 2014. Dissecting the c-di-GMP signaling network regulating motility in Salmonella enterica serovar Typhimurium. Environ Microbiol doi: 10.1111/1462-2920.12580. [DOI] [PubMed] [Google Scholar]
- 48.Ahmad I, Lamprokostopoulou A, Le Guyon S, Streck E, Barthel M, Peters V, Hardt WD, Romling U. 2011. Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS One 6:e28351. doi: 10.1371/journal.pone.0028351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solano C, Garcia B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, Casals J, Pedroso E, Lasa I. 2009. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc Natl Acad Sci U S A 106:7997–8002. doi: 10.1073/pnas.0812573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rouf SF, Ahmad I, Anwar N, Vodnala SK, Kader A, Romling U, Rhen M. 2011. Opposing contributions of polynucleotide phosphorylase and the membrane protein NlpI to biofilm formation by Salmonella enterica serovar Typhimurium. J Bacteriol 193:580–582. doi: 10.1128/JB.00905-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 52.Zorraquino V, Garcia B, Latasa C, Echeverz M, Toledo-Arana A, Valle J, Lasa I, Solano C. 2013. Coordinated cyclic-di-GMP repression of Salmonella motility through YcgR and cellulose. J Bacteriol 195:417–428. doi: 10.1128/JB.01789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simm R, Remminghorst U, Ahmad I, Zakikhany K, Romling U. 2009. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility in Salmonella enterica serovar Typhimurium. J Bacteriol 191:3928–3937. doi: 10.1128/JB.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubnau D, Losick R. 2006. Bistability in bacteria. Mol Microbiol 61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 55.Siebring J, Sorg RA, Herber M, Kuipers OP. 2012. Take it or leave it: mechanisms underlying bacterial bistable regulatory networks, p 305–332. In Filloux AAM. (ed), Bacterial regulatory networks. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 56.Gualdi L, Tagliabue L, Landini P. 2007. Biofilm formation-gene expression relay system in Escherichia coli: modulation of sigmaS-dependent gene expression by the CsgD regulatory protein via sigmaS protein stabilization. J Bacteriol 189:8034–8043. doi: 10.1128/JB.00900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190:79–90. doi: 10.1534/genetics.111.132779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ono S, Goldberg MD, Olsson T, Esposito D, Hinton JC, Ladbury JE. 2005. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem J 391:203–213. doi: 10.1042/BJ20050453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galan JE, Curtiss R III. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun 58:1879–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 61.Sanowar S, Singh P, Pfuetzner RA, Andre I, Zheng H, Spreter T, Strynadka NC, Gonen T, Baker D, Goodlett DR, Miller SI. 2010. Interactions of the transmembrane polymeric rings of the Salmonella enterica serovar Typhimurium type III secretion system. mBio 1:e00158-10. doi: 10.1128/mBio.00158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lara-Tejero M, Galan JE. 2009. Salmonella enterica serovar Typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun 77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mirold S, Ehrbar K, Weissmuller A, Prager R, Tschape H, Russmann H, Hardt WD. 2001. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J Bacteriol 183:2348–2358. doi: 10.1128/JB.183.7.2348-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Misselwitz B, Barrett N, Kreibich S, Vonaesch P, Andritschke D, Rout S, Weidner K, Sormaz M, Songhet P, Horvath P, Chabria M, Vogel V, Spori DM, Jenny P, Hardt WD. 2012. Near surface swimming of Salmonella Typhimurium explains target-site selection and cooperative invasion. PLoS Pathog 8:e1002810. doi: 10.1371/journal.ppat.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mouslim C, Hughes KT. 2014. The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog 10:e1003987. doi: 10.1371/journal.ppat.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osborne SE, Coombes BK. 2011. Transcriptional priming of Salmonella Pathogenicity Island-2 precedes cellular invasion. PLoS One 6:e21648. doi: 10.1371/journal.pone.0021648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown NF, Vallance BA, Coombes BK, Valdez Y, Coburn BA, Finlay BB. 2005. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLoS Pathog 1:e32. doi: 10.1371/journal.ppat.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galan JE, Curtiss R III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Santiviago CA, Reynolds MM, Porwollik S, Choi SH, Long F, Andrews-Polymenis HL, McClelland M. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog 5:e1000477. doi: 10.1371/journal.ppat.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tukel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, van Putten JP, Baumler AJ. 2010. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol 12:1495–1505. doi: 10.1111/j.1462-5822.2010.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rapsinski GJ, Wynosky-Dolfi MA, Oppong GO, Tursi SA, Wilson RP, Brodsky IE, Tukel C. 2015. Toll-like receptor 2 and NLRP3 cooperate to recognize a functional bacterial amyloid, curli. Infect Immun 83:693–701. doi: 10.1128/IAI.02370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Novick A, Weiner M. 1957. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci U S A 43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boehm A, Vogel J. 2012. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol Microbiol 84:1–5. doi: 10.1111/j.1365-2958.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- 74.Claessen D, Rozen DE, Kuipers OP, Sogaard-Andersen L, van Wezel GP. 2014. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol 12:115–124. doi: 10.1038/nrmicro3178. [DOI] [PubMed] [Google Scholar]
- 75.Ogasawara H, Yamamoto K, Ishihama A. 2011. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol 193:2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, Felt SA, Torreilles S, Schoolnik GK. 2010. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog 6:e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ackermann M, Stecher B, Freed NE, Songhet P, Hardt WD, Doebeli M. 2008. Self-destructive cooperation mediated by phenotypic noise. Nature 454:987–990. doi: 10.1038/nature07067. [DOI] [PubMed] [Google Scholar]
- 78.Diard M, Garcia V, Maier L, Remus-Emsermann MN, Regoes RR, Ackermann M, Hardt WD. 2013. Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494:353–356. doi: 10.1038/nature11913. [DOI] [PubMed] [Google Scholar]
- 79.Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt WD. 2011. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog 7:e1002143. doi: 10.1371/journal.ppat.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Jong IG, Haccou P, Kuipers OP. 2011. Bet hedging or not? A guide to proper classification of microbial survival strategies. Bioessays 33:215–223. doi: 10.1002/bies.201000127. [DOI] [PubMed] [Google Scholar]
- 81.Parrish JK, Edelstein-Keshet L. 1999. Complexity, pattern, and evolutionary trade-offs in animal aggregation. Science 284:99–101. doi: 10.1126/science.284.5411.99. [DOI] [PubMed] [Google Scholar]
- 82.Ali SS, Soo J, Rao C, Leung AS, Ngai DH, Ensminger AW, Navarre WW. 2014. Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathog 10:e1004500. doi: 10.1371/journal.ppat.1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davidson CJ, White AP, Surette MG. 2008. Evolutionary loss of the rdar morphotype in Salmonella as a result of high mutation rates during laboratory passage. ISME J 2:293–307. doi: 10.1038/ismej.2008.4. [DOI] [PubMed] [Google Scholar]
- 84.Smith DR, Chapman MR. 2010. Economical evolution: microbes reduce the synthetic cost of extracellular proteins. mBio 1:e00131-00110. doi: 10.1128/mBio.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mastroeni P, Morgan FJ, McKinley TJ, Shawcroft E, Clare S, Maskell DJ, Grant AJ. 2011. Enhanced virulence of Salmonella enterica serovar Typhimurium after passage through mice. Infect Immun 79:636–643. doi: 10.1128/IAI.00954-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lenski RE, May RM. 1994. The evolution of virulence in parasites and pathogens: reconciliation between two competing hypotheses. J Theor Biol 169:253–265. doi: 10.1006/jtbi.1994.1146. [DOI] [PubMed] [Google Scholar]
- 87.DePas WH, Hufnagel DA, Lee JS, Blanco LP, Bernstein HC, Fisher ST, James GA, Stewart PS, Chapman MR. 2013. Iron induces bimodal population development by Escherichia coli. Proc Natl Acad Sci U S A 110:2629–2634. doi: 10.1073/pnas.1218703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crawford RW, Rosales-Reyes R, Ramirez-Aguilar MDL, Chapa-Azuela LO, Alpuche-Aranda C, Gunn JS. 2010. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc Natl Acad Sci U S A 107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baker S, Holt KE, Clements AC, Karkey A, Arjyal A, Boni MF, Dongol S, Hammond N, Koirala S, Duy PT, Nga TV, Campbell JI, Dolecek C, Basnyat B, Dougan G, Farrar JJ. 2011. Combined high-resolution genotyping and geospatial analysis reveals modes of endemic urban typhoid fever transmission. Open Biol 1:110008. doi: 10.1098/rsob.110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kosarewicz A, Konigsmaier L, Marlovits TC. 2012. The blueprint of the type-3 injectisome. Philos Trans R Soc Lond B Biol Sci 367:1140–1154. doi: 10.1098/rstb.2011.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y, MacKenzie KD, White AP. An empirical strategy to detect bacterial transcript structure from directional RNA-seq transcriptome data. BMC Genomics, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.