Abstract
Clostridium perfringens strains produce severe diseases, including myonecrosis and enteritis necroticans, in humans and animals. Diseases are mediated by the production of potent toxins that often damage the site of infection, e.g., skin epithelium during myonecrosis. In planktonic cultures, the regulation of important toxins, such as CPA, CPB, and PFO, is controlled by the C. perfringens Agr-like (CpAL) quorum sensing (QS) system. Strains also encode a functional LuxS/AI-2 system. Although C. perfringens strains form biofilm-like structures, the regulation of biofilm formation is poorly understood. Therefore, our studies investigated the role of CpAL and LuxS/AI-2 QS systems and of QS-regulated factors in controlling the formation of biofilms. We first demonstrate that biofilm production by reference strains differs depending on the culture medium. Increased biomass correlated with the presence of extracellular DNA in the supernatant, which was released by lysis of a fraction of the biofilm population and planktonic cells. Whereas ΔagrB mutant strains were not able to produce biofilms, a ΔluxS mutant produced wild-type levels. The transcript levels of CpAL-regulated cpa and pfoA genes, but not cpb, were upregulated in biofilms compared to planktonic cultures. Accordingly, Δcpa and ΔpfoA mutants, in type A (S13) or type C (CN3685) backgrounds, were unable to produce biofilms, whereas CN3685Δcpb made wild-type levels. Biofilm formation was restored in complemented Δcpa/cpa and ΔpfoA/pfoA strains. Confocal microscopy studies further detected CPA partially colocalizing with eDNA on the biofilm structure. Thus, CpAL regulates biofilm formation in C. perfringens by increasing levels of certain toxins required to build biofilms.
INTRODUCTION
The Gram-positive, spore-forming bacterium Clostridium perfringens is the most widely distributed pathogen in nature (1–3). This anaerobe affects humans and animals, producing severe diseases, including food poisoning (4), gastrointestinal syndromes and enterotoxemias (3, 5, 6), gas gangrene, and numerous histotoxic infections (7–9). With the exception of food poisoning caused by C. perfringens enterotoxin (CPE), a common characteristic in most toxigenic C. perfringens vegetative infections is the rapid progression of the disease to a final, often fatal, outcome. The virulence of C. perfringens is directly related to its prolific production of more than 17 different potent toxins. Current schemes, however, utilize differential production of four lethal typing toxins (alpha [CPA], beta [CPB], epsilon [Etx], and/or iota [Itx]) to classify C. perfringens isolates into five pathogenic types (A to E) (2, 10–13). C. perfringens type A, for example, must produce CPA, whereas type C strains produce CPA and CPB. All toxinotypes make other biomedically important toxins such as perfringolysin O (PFO) or CPB2 (10).
C. perfringens type A and type C strains have been isolated from human cases of severe disease (2). C. perfringens type A produces nearly ∼90% of all gas gangrene cases (3, 9, 13). Gas gangrene, or myonecrosis, is considered one of the most fulminate infections caused by a Gram-positive organism in humans and animals (8, 9). Infection starts from the site of a recent surgical wound or trauma, where C. perfringens type A strains must first attach to the disrupted epidermal epithelium and proliferate while at the same time producing toxins that necrotize the tissue (8, 14–16). Tissue destruction associated with C. perfringens infection progresses rapidly to involve an entire extremity (8, 14, 17–19). Amputation remains the single best life-saving treatment, although mortality still remains high (8, 9, 15, 16).
The production of C. perfringens alpha toxin (CPA) and, to some extent, PFO has largely been implicated in clostridial myonecrosis (3, 9, 13, 20, 21). For example, a C. perfringens type A strain (cpa+ pfoA+) was highly virulent in a mouse model of clostridial myonecrosis, whereas isogenic cpa or pfoA mutants showed reduced virulence (including reduced tissue destruction) (17, 19, 22, 23). Elimination of both CPA and PFO production (a double cpa-pfoA toxin gene mutant) removed most of the histopathological features typical of clostridial myonecrosis (24).
C. perfringens type C is the etiologic agent of human enteritis necroticans (also called pigbel, darmbrand, or “gangrene of the bowel”), which originates in the intestine (2). Enteritis necroticans (EN) is particularly aggressive in diabetics and immunocompromised patients from developed countries (they survive less than 48 h after the first appearance of symptoms) (25–30). EN is characterized by its sudden onset with abdominal cramps, shock, bloody diarrhea, acute inflammation, and pronounced necrosis of intestinal mucosa (27, 31). Necrosis of the intestinal epithelium always coincides with the presence of Gram-positive rods characteristic of C. perfringens attached to the surface of the necrotic mucosa that, when visualized by microscopy, appear to form biofilm-like structures (28, 29). CPB, the most important toxin for developing EN (32–34), has been detected on the apical side of intestinal cells underneath those attached bacteria, indicating that in situ production of CPB is important for pathogenesis (29). A similar EN disease produced by type A and type C strains affects chickens and other domestic birds. Chicken EN globally costs the poultry industry $2 billion per year (35).
We recently demonstrated that C. perfringens strains encodes a functional agr-like (accessory gene regulator) quorum sensing (QS) system that controls production of some C. perfringens toxins (36, 37). The C. perfringens Agr-like (CpAL) system is related to the Agr system from Staphylococcus aureus and other Gram positives (36, 38–41). Signaling through CpAL requires a secreted pheromone encoded by the agrD gene, which is processed to a functional cyclic peptide by a transmembrane protein encoded by the agrB gene (36, 37). The CpAL system regulates in vitro production of CPA and PFO in all studied toxin types, including type A and type C strains (36, 42–44). CpAL also regulates production of C. perfringens beta toxin (CPB) in type B and type C strains (42, 43). Moreover, a functional CpAL system was required in vivo to produce necrotizing enteritis in rabbit ileal loops by regulating intestinal levels of CPB (42).
Another QS system encoded by genome-sequenced C. perfringens strains, the LuxS/AI-2 system, was implicated in transcriptional regulation of pfoA, but not cpa or other toxin genes, in mid-exponential-phase planktonic cultures of type A strain 13 (S13) (45). Although studied in vivo and in vitro, LuxS-AI-2 regulation has not been identified in pfoA-encoded C. perfringens type C strain CN385 (42, 46).
As mentioned earlier, C. perfringens strains may form biofilm-like structures on host tissues or epithelia. Biofilms are bacterial communities surrounded by a matrix providing the mechanical stability, mediating their adhesion to surfaces, and forming a cohesive, three-dimensional polymer network that protects them from the host response and antibiotics (47). Biofilm production in C. perfringens has been poorly studied. Initial attempts to characterize C. perfringens biofilms demonstrated that biofilms are produced in vitro by some strains (48, 49). The matrix has been recently studied and found to be made of some proteins, β-1,4-linked polysaccharides and extracellular DNA (eDNA) (49, 50). A type IV pilus (TFP) is also required for biofilm formation; these pili were located on the biofilm matrix (48).
More recently, Obana et al. reported in 2014 that biofilms formed at 37°C have an increased biomass and a phenotypically different biofilm matrix in comparison to those formed at 25°C. Furthermore, incubation at 37°C, but not at 25°C, upregulated levels of pilA2 mRNA, which encodes the main pilin subunit of the matrix-associated C. perfringens TFP (50). These data suggest that C. perfringens biofilms are differentially regulated. In the present study, we investigated QS regulation of C. perfringens biofilms and demonstrated that the CpAL, but not the LuxS/AI-2 system, is required for proper formation of the biofilm structure. Moreover, CpAL-regulated CPA and PFO toxins, but not CPB, were required for biofilm formation, and transcription of the genes cpa and pfoA, but not cpb, was upregulated in biofilms compared to planktonic cultures.
MATERIALS AND METHODS
Strains, culture media, and chemicals.
Wild-type strains and mutant derivatives utilized in the present study are listed in Table 1. Media for culturing C. perfringens included FTG (fluid thioglycolate medium; Difco Laboratories), TGY (3% tryptic soy broth [Becton Dickinson]; 2% glucose [Fisher Scientific]; 1% yeast extract [Becton Dickinson], 0.1% sodium thioglycolate [Sigma-Aldrich]), brain heart infusion (BHI) agar (Becton Dickinson). Sodium thioglycolate, a reducing agent, present in TGY maintains a low oxygen tension in the medium. Blood agar plates were prepared with BHI and 5% defibrinated sheep blood (Remel, Lenexa, KS); egg yolk agar plates were prepared with BHI containing 4% (vol/vol) egg yolk. For culturing Escherichia coli, Luria-Bertani (LB) broth (1% tryptone [Becton Dickinson], 0.5% yeast extract, 1% NaCl) and LB agar (1.5% agar [Becton Dickinson]) medium were used. All antibiotics used in the present study were purchased from the Sigma-Aldrich Chemical Company.
TABLE 1.
Strains and plasmids utilized in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| CN3685 | Type C strain isolated from peritoneal fluid of a sheep with struck | 33, 34 |
| JGS1495 | Type C strain isolated from a piglet with necrotic enteritis | 60 |
| Strain 13 | Type A, gangrene strain | 75 |
| JGS1075 | Type C strain isolated from porcine enteritis | 76 |
| JGS1076 | Type C strain isolated from porcine enteritis | 76 |
| JGS1659 | Type C strain isolated from porcine enteritis | 76 |
| CPJV20 | CN3685ΔluxS | This study |
| CPJV21 | CN3685ΔagrB | This study |
| CPJV22 | CN3685Δplc | This study |
| CPJV23 | CN3685ΔpfoA | This study |
| CPJV25 | CN3685Δcpb | This study |
| CPJV26 | S13ΔagrB | This study |
| CPJV27 | S13Δplc | This study |
| CPJV28 | S13ΔpfoA | This study |
| CPJV30 | CPJV27 carrying pCP13plc | This study |
| CPJV31 | CPJV28 carrying pCP13pfoA | This study |
| CPJV32 | CPJV21 carrying pCPJVagrB | This study |
| Plasmids | ||
| pJIR750 | E. coli-C. perfringens shuttle vector carrying a chloramphenicol-resistant gene, catP | 54 |
| pJIR751 | E. coli-C. perfringens shuttle vector carrying an erythromycin-resistant gene, ermB | 54 |
| pJIR750ai | pJIR750 with plc targeted intron in antisense orientation | 52 |
| pCPJVlux | pJIR750ai with luxS targeted intron in an antisense orientation | This study |
| pCPJVagrB | pJIR750ai with agrB targeted intron in a sense orientation | This study |
| pCPJVpfoA | pJIR750ai with pfoA targeted intron in an antisense orientation | This study |
| pCPJVcpb | pJIR750ai with cpb targeted intron in an antisense orientation | This study |
| pCP13plc | pJIR751 carrying the plc gene from strain 13 | This study |
| pCP13pfoA | pJIR750 carrying the pfoA gene from strain 13 | This study |
Production of C. perfringens biofilms.
Strains were inoculated into FTG and grown overnight at 37°C. To produce biofilms, these FTG cultures were inoculated (diluted 1:10) in triplicate into either 8-well glass slides (Lab-Tek), polystyrene 96-well plates (Corning), CellBIND surface polystyrene 24-well plates (Corning), or tissue culture-treated polystyrene six-well plate (Corning) containing FTG or TGY with no antibiotics, followed by incubation at 37°C for the indicated time.
Quantification of biofilm biomass.
Biofilm development was evaluated using the classic crystal violet staining method as previously reported (51). Alternatively, biofilms were stained by fluorescence with membrane-permeant fluorescent reagents SYTO9 (Molecular Probes) or DAPI (4′,6′-diamidino-2-phenylindole). These molecules stain nucleic acids. Briefly, biofilms were washed three times with sterile phosphate-buffered saline (PBS; pH 7.4) and incubated with 5 mM SYTO9 for 20 min at room temperature or with DAPI (30 nM) and incubated for 5 min. After two washes with PBS, fluorescence (arbitrary units) was quantified using a VICTOR X3 multilabel plate reader (Perkin-Elmer). Biofilms were photographed using an inverted Evos fluorescence microscope (Advanced Microscopy Group). Fluorescence arbitrary units of a chosen experimental condition (e.g., wild-type [wt] biofilm 8 h postinoculation) were set to 100% of biofilm biomass and used to calculate the percentage of biofilm biomass of all other tested conditions.
Quantification of eDNA.
Strains were inoculated into 24-well plates and incubated for the indicated time at 37°C. The supernatant (∼1 ml) was removed, centrifuged for 5 min at 12,000 × g in a refrigerated centrifuged (Eppendorf) and filter sterilized using a 0.45-μm-pore-size syringe filter (Fisher Scientific). This supernatant was mixed with 0.5 volume of ethanol and vortexed for 10 s, and then extracellular DNA (eDNA) was purified using the QIAamp eDNA minikit (Qiagen) according to the manufacturer's instructions. To quantify the amounts of DNA, a quantitative PCR (qPCR) assay targeting the cpa gene was utilized. Reactions were performed with IQ SYBR green super mix (Bio-Rad), 300 nM concentrations of each primer (JEV193L and JEV194R [Table 2]) and 2 μl of DNA template. Reactions were run in duplicate using a CFX96 real-time PCR detection system (Bio-Rad) under the following conditions: 1 cycle of 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 55°C for 1 min, and 72°C for 1 min. Melting curves were generated by a cycle of 95°C for 1 min and 65°C for 1 min, followed by 80 cycles starting at 65°C with 0.5°C increments. For quantification purposes, standards (1, 10, 100, and 1,000 pg of purified chromosomal DNA per reaction) were run in parallel to generate a standard curve. Amounts of eDNA in pg/ml were calculated by using the software Bio-Rad CFX Manager.
TABLE 2.
Primers utilized in this study
| Primera | Sequence (5′–3′) | Source or reference |
|---|---|---|
| cpb-Fwd | GCGAATATGCTGAATCATCTA | 77 |
| cpb-Rev | GCAGGAACATTAGTATATCTTC | |
| JEV135L | TAAAAACCGAAAAGAAGAAAAA | This study |
| JEV126R | TATGTAGGTTAGAGTCATACA | |
| JEV165L | TTGAATTCCCCAGTTATTCACGATTAAAG | This study |
| JEV166R | TTGGATCCTTAATTGTAAGTAATACTAGATCCA | |
| JEV167L | TTGAATTCAGGTTAAAACCTGTTTTTGATTGA | This study |
| JEV168R | TTTCTAGATTTTGTAAATACCACCAAAACCAAT | |
| JEV181L | ACCTCCACTTATGGTTTCAAATGT | This study |
| JEV182R | TTGTTATGTTCTTGTGCATCTCCT | |
| JEV193L | CTAGCATGAGTCATAGTTGGGATG | This study |
| JEV194R | ACATGTAGTCATCTGTTCCAGCAT | |
| polCJVL | AATATATGATACTGAAGAGAGAGTAA | 46 |
| polCJVR | TCTAAATTATCTAAATCTATGTCTACT | |
| luxS-L | CCTAAAGGAGATATAGTTTCAAAG | 46 |
| luxS-R | TCACTAGAGTTAGCTAAGAGTCAT | |
| KO-IBS* | AAAAAAGCTTATAATTATCCTTAATTGCCATACTTGTGCGCCCAGATAGGGTG | 34 |
| KO-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCATACTTGCTAACTTACCTTTCTTTGT | |
| KO-EBS2 | TGAACGCAAGTTTCTAATTTCGATTGCAATTCGATAGAGGAAAGTGTCT | |
| IBS-pfoA** | AAAAAAGCTTATAATTATCCTTAATTGCCATACTTGTGCGCCCAGATAGGGTG | 52 |
| EBS1d-pfoA | CAGATTGTACAAATGTGGTGATAACAGATAAGTCATACTTGCTAACTTACCTTTCTTTGT | |
| EBS2-pfoA | TGAACGCAAGTTTCTAATTTCGATTGCAATTCGATAGAGGAAAGTGTCT | |
| 566/567s-IBS† | AAAAAAGCTTATAATTATCCTTATTCCACACTGAAGTGCGCC-CAGATAGGGTG | 53 |
| 566/567s-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCACTGAATATAACTTACCTTTCTTTGT | |
| 566/567s-EBS2 | TGAACGCAAGTTTCTAATTTCGGTTTGGAATCGATAGAGGAAAGTGTCT | |
| luxS-IBS‡ | AAAAAAGCTTATAATTATCCTTATACTCCAAAGCTGTGCGCCCAGATAGGGTG | 46 |
| luxS-EBS1d | CAGATTGTACAAATGTGGTGATAACAGATAAGTCAAAGCTTCTAACTTACCTTTCTTTGT | |
| luxS-EBS2 | TGAACGCAAGTTTCTAATTTCGGTTGAGTATCGATAGAGGAAAGTGTCT |
Some primers were used to prepare a cpb mutant (*), a pfoA mutant (**), an agrB mutant (†), or an luxS mutant (‡), as indicated.
Viability of biofilms and planktonic cells using the Live/Dead assay.
C. perfringens type A strain 13 or type C strain CN3685 was inoculated in 96-well plates containing TGY, followed by incubation for 4 or 8 h at 37°C. Biofilms made 4 or 8 h postinoculation were washed three times with sterile PBS and immediately stained utilizing a Live/Dead BacLight bacterial viability kit (L7012; Invitrogen, Grand Island, NY). The assay has successfully been used to investigate biofilms made by Clostridium difficile and C. perfringens (48, 57). Planktonic cells were removed at the indicated time and stained as described above. The concentration of dyes was utilized according to the manufacturer's recommendations. Stained biofilms, or planktonic cells, were washed additionally two times with PBS, and fluorescence measurements (arbitrary units) were obtained with a Victor X3 multilabel plate reader (Perkin-Elmer). Fluorescent dyes included in the kit are incorporated, or not, into bacterial cells as a function of membrane integrity and therefore viability. Preparations were also observed and photographed utilizing an inverted Evos fluorescence microscope (Advanced Microscopy Group).
Construction of CN3685 and strain 13 null mutants using TargeTron technology.
The following genes were insertionally inactivated using a Clostridium TargeTron gene knockout system (Sigma-Aldrich): cpb (34), pfoA (34), cpa (also known as plc) (52), luxS (46), and agrB (53). The cpa mutant was prepared with plasmid pJIR750ai, included in the TargeTron gene knockout system. Derivative plasmids to insertionally inactivate all others were prepared by cloning target-specific sequences as follows: a set of primers (IBS, EBS1d, and EBS2) that had been previously published (Table 2) were utilized to generate a target-specific ∼350-bp PCR product; this target-specific fragment replaced the cpa-specific fragment on pJIR750ai. An intron plasmid which integrated an ∼900-bp intron fragment into the target gene was then electroporated into either strain 13 or CN3685. Except for the mutation into the agrB gene, in which the intron was integrated in the sense orientation, the intron fragment was inserted in the antisense orientation in all other target genes. Transformants were selected on BHI agar plates containing 15 μg of chloramphenicol (Cm)/ml and then PCR screened for an intron-disrupted gene using the following target-specific primers: cpb (cpb-Fwd and cpb-Rev), cpa (JEV193L and JEV194R), pfoA (JEV181L and JEV182R), agrB (JEV135L and 126R), and luxS (luxS-L and luxS-R). Our PCR screening was designed as to amplify an ∼200- to 300-bp product from wild-type genes but amplified a larger approximately 1.1- to 1.2-kbp product from those mutants. Mutants carrying an intron insertion were grown in FTG medium without Cm for ∼10 days, with daily subculturing, to cure the intron-carrying donor plasmid. Curing was shown by lack of growth on Cm-containing BHI plates.
Preparation of cpa and pfoA complemented strains.
Plasmids pJIR750 (ATCC 87015) and pJIR751 (ATCC 87016) (54) were purified and doubly digested with restriction enzymes, EcoRI and BamHI or EcoRI and XbaI, respectively. Digested plasmids were again purified using a QIAquick gel extraction kit (Qiagen). Simultaneously, the pfoA or plc gene, including promoter regions, was amplified by PCR using the primers JEV165L and JEV166R (pfoA) or the primers JEV167L and JEV168R (plc), purified and doubly digested with enzymes EcoRI and BamHI or EcoRI and XbaI, respectively. Digested fragments were ligated to pJIR750-pfoA (pCP13-pfoA) or pJIR751-plc (pCP13plc) using T4 DNA ligase (Promega) and transformed into competent cells of S13ΔpfoA (pJIR750-pfoA) or S13Δplc (pJIR751-plc) by electroporation as previously described (36). Transformants were plated onto BHI plates containing Cm or erythromycin, respectively, and isolated colonies were screened for the presence of both the intron-disrupted mutant gene and plasmid-encoded wt genes as mentioned above.
Preparation of an agrB complemented strain.
Taking advantage of the fact that the insertion of the intron within agrB created a conditional mutation (i.e., the intron fragment was integrated in the sense strand), complementation of the agrB-null mutant CPJV21 was achieved at the mRNA level by removing the intron from disrupted agrB mRNA, thus restoring a functional agrB transcript (55). Removal of this intron was achieved by reintroducing pCPJVagrB, which encodes the LtrA protein required to splice out the intron insertion, into CPJV21. Transformants were selected on BHI agar containing 15 μg of chloramphenicol/ml. Strain CPJV32 was grown at 30°C to allow maximum expression of the LtrA protein.
RNA extraction and analysis of RNA preparations.
Planktonic or biofilm cells were added with 1 volume of RNAprotect Bacteria (Qiagen) and immediately centrifuged for 15 min at 15,000 × g in a refrigerated centrifuge (Eppendorf). Total RNA was then extracted from the pellet by using an RNeasy minikit (Qiagen) as outlined by the manufacturer and additionally treated with 2 U of DNase I (Promega) essentially as previously described (46, 51). The integrity of our RNA preparations (i.e., the RNA integrity number [RIN]) and RNA concentration of samples were obtained by using an RNA 6000 Nano kit and a 2100 Bioanalyzer (Agilent Technologies).
RT-PCRs.
Reactions were performed with 100 ng of DNase I-treated RNA as the template. Those reaction mixtures contained 500 nM concentrations of each primer (Table 2), 1× PCR master mix (New England BioLabs), molecular-grade water, and 10 U of avian myeloblastosis virus retrotranscriptase (Promega). Control reverse transcription-PCRs (RT-PCRs) were similarly performed, except for the omission of reverse transcriptase (negative) or the addition of 100 ng of genomic DNA (positive). Reactions conditions were as follows: an initial incubation at 42°C for 30 min and denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 15 s, 55°C for 30 s, and 68°C for 1 min and then a final extension at 68°C for 10 min. RT-PCR products were run in 2% agarose gels and stained with SYBR Safe DNA gel stain (Invitrogen).
Quantitative RT-PCR (qRT-PCR).
Reactions were performed with an iScript one-step RT-PCR kit with SYBR green (Bio-Rad), 300 nM concentrations of the indicated primers (Table 2), and 10 ng of DNase I-treated RNA template. Reactions were run in duplicate using a CFX96 real-time PCR detection system (Bio-Rad) and the following conditions: 1 cycle at 50°C for 30 min, 1 cycle at 95°C for 10 min, and 40 cycles of 95°C for 15 and 55°C for 1 min. Melting curves were generated as described above. The relative quantitation of mRNA expression was normalized to the constitutive expression of the housekeeping polC gene and calculated by the comparative CT (2−ΔΔCT) method (56).
Hb release assay.
C. perfringens strains were grown in 24-well plates containing TGY for 4 h; maximal expression of PFO, and therefore hemoglobin (Hb) release, has been observed at this time point (36). Culture supernatant was obtained by centrifugation at 20,000 × g for 10 min and then filter sterilized using a 0.45-μm-pore-size filter (Fisher Scientific). The Hb release assay was then performed essentially as previously described (46). Photographs of erythrocyte pellets in ΔpfoA mutants were also obtained.
Detection of CPA by dot blot.
Strains were grown in 24-well plates containing TGY for 8 h at 37°C, after which the culture was centrifuged at 20,000 × g for 10 min. An aliquot (50 μl) of the supernatant was spotted onto a nitrocellulose membrane, air dried, and then blocked with PBS-Tween 20 (0.05% [vol/vol]) and nonfat dry milk (5% [wt/vol]) for 1 h at room temperature. Membranes were probed with a polyclonal anti-CPA antibody (Bioss Antibodies, Woburn, MA) followed by incubation with horseradish peroxidase-conjugated secondary anti-rabbit antibodies (Jackson ImmunoResearch) and the addition of SuperSignal West Pico chemiluminescent substrate (Pierce).
Confocal microscopy studies.
C. perfringens strains were inoculated in 8-well glass slides (Lab-Tek) containing TGY and incubated for 24 h at 37°C. Biofilms produced were washed twice with PBS and fixed with 2% paraformaldehyde for 15 min at room temperature. Fixed biofilms were then permeabilized with 0.5% Triton X-100 for 5 min, washed again with PBS, and blocked with 1% bovine serum albumin for 30 min at 37°C. Nucleic acids were stained with SYTO9 as described above. CPA was stained with a rabbit polyclonal anti-CPA antibody, followed by a goat anti-rabbit secondary antibody conjugated to Alexa Fluor 555 (Molecular Probes). Stained biofilms were finally washed three times with PBS, mounted with Vectashield mounting medium (Vector Laboratories), and analyzed with an Olympus FV1000 confocal microscope. Confocal images were analyzed with ImageJ (v1.49k; National Institutes of Health, Bethesda, MD).
Statistical analysis.
All statistical analyses presented in the present study were performed using the Mann-Whitney U test and the software SigmaPlot (v12.0; Systat Software, Inc.).
RESULTS
Production of C. perfringens biofilms is regulated by environmental signals.
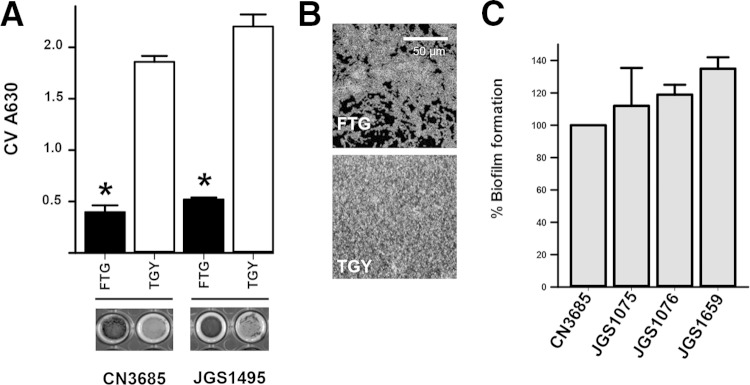
Environmental signals, such as those present in bacterial culture media, have been associated with biofilm production in some Gram-positive bacteria (51, 57, 58). We therefore investigated whether the culture medium would impact the formation of C. perfringens biofilms. To assess this, we used two classic C. perfringens broth media, FTG and TGY (59). As shown in Fig. 1A, we found that the biofilm mass produced in TGY by two different C. perfringens type C reference strains, CN3685 (33, 34) and JGS1495 (60), was significantly greater than that produced on FTG. Biofilms produced on TGY were observed in the wells as a thick white pellicle (Fig. 1A, bottom panels). Compared to biofilms made on FTG that covered ∼70% of the substrate, biofilms produced on TGY covered the entire surface (Fig. 1B). Similar results were obtained when C. perfringens type A strain 13 was inoculated in FTG and TGY (data not shown). We next investigated the biofilm phenotype of other C. perfringens type C strains isolated from porcine enteritis (Table 1). All tested strains produced similar biofilm biomass on a microtiter plate at 24 h postinoculation (Fig. 1C). Together, our results suggest that C. perfringens biofilms are regulated by environmental signals.
FIG 1.
C. perfringens biofilms are differentially produced depending on the bacterial culture medium. (A) Type C strain CN3685, or JGS1495, was inoculated into either FTG or TGY, and biofilms were incubated for 24 h. A set of wells containing biofilms were washed and photographed (bottom panel), and then the crystal violet absorbance was obtained. Asterisks indicate statistical significance (P < 0.05), as evaluated by a Mann-Whitney U test. (B) Another set of wells was stained with DAPI, and fluorescence micrographs were obtained. Experiments were repeated at least three times. (C) C. perfringens strains were inoculated in TGY medium and incubated for 24 h. Biofilms were stained with crystal violet, and the A630 was determined. Arbitrary units obtained from CN3685 were set to 100% biofilms, and those made by the other strains were calculated. Error bars represent the standard errors of the mean calculated using data from at least three independent experiments.
Development of a fluorescent assay to study C. perfringens biofilms.
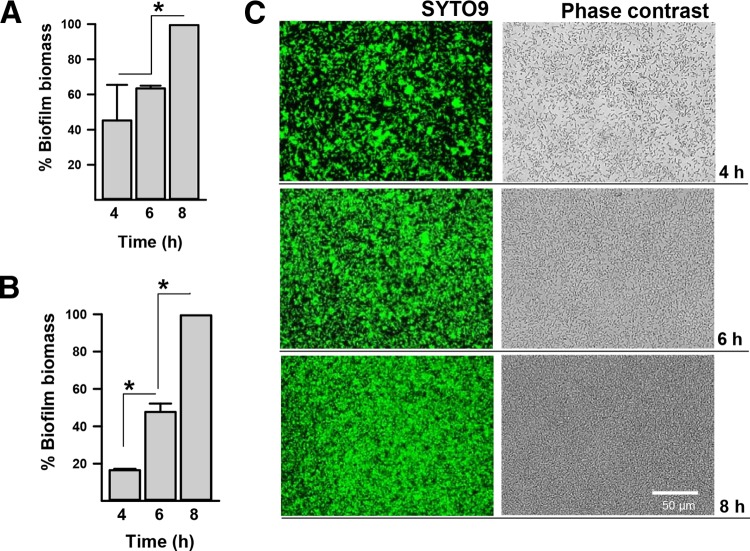
To improve current methods to study C. perfringens biofilms, we developed a fluorescence-based method using DAPI or SYTO9. These fluorophores have different excitation and emission spectra and are readily available. C. perfringens biofilms stained with these fluorescence reporters were compared to the biofilm mass obtained using the classic crystal violet procedure. Fluorescence arbitrary units of biofilms, produced 8 h postinoculation, were set to 100%, and the biomass of all other time points was calculated. The classic protocol, utilizing crystal violet, revealed similar biomass 4 and 6 h postinoculation and a statistically different mass 8 h postinoculation (Fig. 2A). The fluorescence method revealed a statistically different biomass that increased at each time point (Fig. 2B). The latter correlated with microscopic observations of C. perfringens biofilms (Fig. 2C). Similar results were obtained when biofilms were stained with DAPI (data not shown).
FIG 2.
Fluorescence-based method to quantify C. perfringens biofilms. Strain CN3685 was inoculated in 96-well plates containing THY and incubated at 37°C for the indicated time. Biofilms were washed with PBS and then stained with crystal violet, after which the absorbance (A630) was obtained (A), or fluorescently stained with SYTO9, and fluorescence arbitrary units were obtained with a fluorometer (B). The biofilm biomass (%) was calculated in panels A and B by setting 100% biomass units obtained for the 8-h time point to calculate the biomass (%) of the other two. *, Statistical significance (P < 0.05), as evaluated by the Mann-Whitney U test. Error bars represent the standard errors of the mean calculated using data from three independent experiments. (C) Biofilms stained in panel B were photographed using an inverted microscope and either the EVOS light-cube green fluorescent protein 470/510 (SYTO9) or phase contrast. The scale bar at the bottom right applies to all six panels.
C. perfringens releases eDNA into the culture medium.
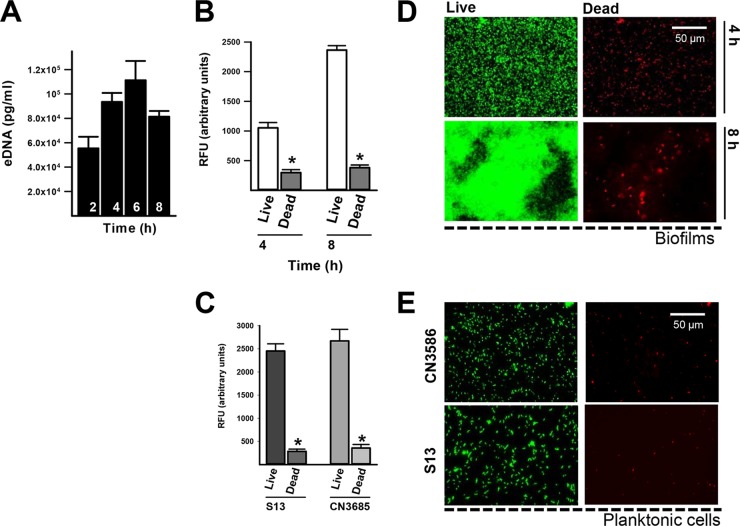
The biofilm matrix of other pathogens, such as Streptococcus pneumoniae (61, 62) or Enterococcus faecalis (63), is partially composed of DNA that has been excreted into the medium (i.e., eDNA). It has been recently reported that eDNA is present in the C. perfringens biofilm matrix (49, 50). To assess whether C. perfringens cells release DNA to the extracellular milieu, DNA was purified from supernatants of cultures of C. perfringens type C strain CN3685. Released eDNA was quantified using qPCRs targeting the chromosomally encoded cpa gene (13). The presence of eDNA was detected as early as 2 h postinoculation (5.5 × 104 pg/ml), reaching a maximum peak at 6 h postinoculation (1.1 × 105 pg/ml) with a non-statistically significant decrease in eDNA in the supernatant after 8 h of incubation (Fig. 3A). Increasing presence of eDNA in the supernatant is consistent with the increased biomass formed over time (see Fig. 2). Since the release of eDNA in other species has been linked in part to autolysis, we sought to investigate the viability of C. perfringens biofilms and planktonic cells. Our experiments showed that ∼25% of the biofilm bacteria appeared to be dead, as scored using the Live/Dead stain, at 4 h after inoculation of strain CN3685 (Fig. 3B) or strain 13 (data not shown). Likewise, ∼16% of the biofilm population at 8 h postinoculation consisted of dead bacteria (Fig. 3B and D). Similar populations of dead planktonic cells were observed in 8-h-old cultures of strain 13 or CN3685 (Fig. 3C and E). Our data suggest that eDNA is in part provided by lysis of both planktonic and biofilm bacteria.
FIG 3.
eDNA is released by bacterial lysis into the supernatant of growing C. perfringens biofilms. Strain CN3685 was inoculated in 24-well plates containing TGY, followed by incubation at 37°C for the indicated times, after which 1 ml of supernatant was removed. (A) eDNA was purified and quantified utilizing qPCR. In another set of experiments, 4- or 8-h biofilms (B) or planktonic cells (C) were stained using a Live/Dead BacLight kit, and fluorescence arbitrary units were obtained using a fluorometer. *, Statistical significance (P < 0.05), evaluated by the Mann-Whitney U test, compared to live bacteria. Biofilms (D) or 8-h planktonic cultures (E) were also photographed with an Evos inverted microscope using the light-cube green fluorescent protein 470/510 (live) or red fluorescent protein 531/593 (dead). The scale bar at the upper right applies to all panels.
The CpAL QS system, but not the LuxS/AI-2 system, regulates C. perfringens biofilms.
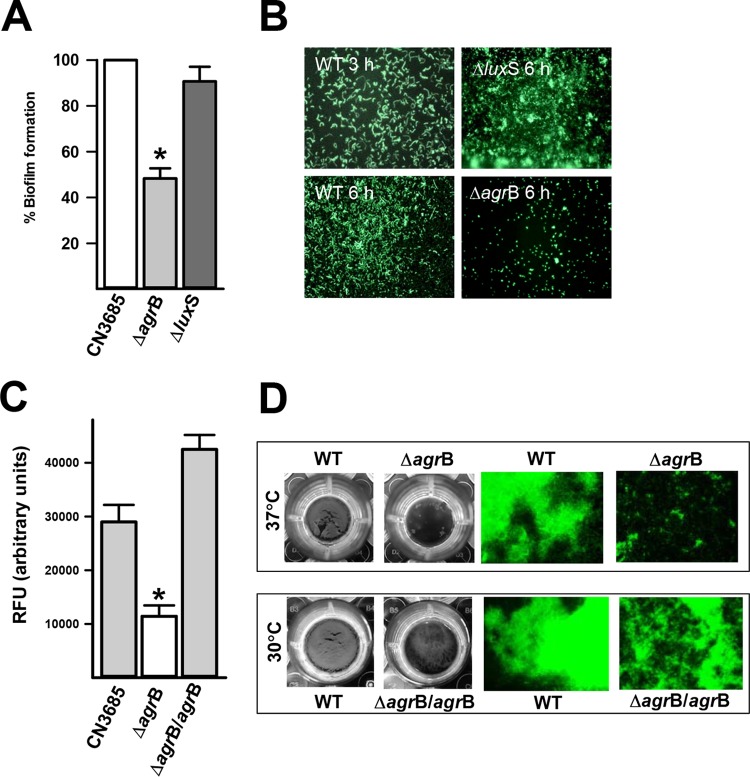
C. perfringens strains produce two different quorum sensing (QS) systems, an Agr-like QS system and the LuxS/AI-2 system (36, 37, 45). To investigate whether biofilms are regulated by QS, we prepared CN3685 derivatives CN3685ΔagrB and CN3685ΔluxS using the TargeTron technology. While the mutant CN3685ΔluxS strain produced biofilm levels similar to those of the wt strain, the mutant strain CN3685ΔagrB produced significantly less biofilm mass by 24 h (Fig. 4A) or 48 h (data not shown) postinoculation. A time course study revealed that C. perfringens wt bacteria had already attached to the substrate at 3 h postinoculation (early biofilms) and covered the entire surface by 6 h later (Fig. 4B). Similarly, the luxS mutant produced wt biofilm levels 6 h postinoculation, whereas biofilms of the ΔagrB mutant, at the same time point, were not observed (Fig. 4B). Similar results were obtained when the agrB was insertionally inactivated in the gangrene-producing strain 13 (data not shown).
FIG 4.
CpAL control of C. perfringens biofilms. Strain CN3685 or mutant derivatives were inoculated in 96-well plates containing THY, and biofilms were incubated for 3, 6, or 24 h at 37°C. (A) Biofilms (24 h) were stained with SYTO9, and their biomass was obtained using a fluorometer. (B) Micrographs of fluorescence-stained biofilms at 3 or 6 h postinoculation were obtained using an Evos inverted microscope. In another set of experiments, strain CN3685 or CPJV21 (ΔagrB) was incubated at 37°C, whereas strain CN3685 or CPJV32 (ΔagrB/agrB) was incubated at 30°C. (C) SYTO9-stained biofilms were quantified by fluorescence. *, Statistical significant (P < 0.05), as evaluated by the Mann-Whitney U test. (D) The macroscopic aspect of these strains was photographed, and micrographs were also obtained with a fluorescence microscope. Where shown, error bars indicate the standard errors of the mean calculated using data from three independent experiments.
To confirm the role of the CpAL system in controlling C. perfringens biofilm formation, the mobile group II intron that had been inserted in the sense orientation (i.e., relative to the direction of transcription) was removed by trans-splicing from the intron-disrupted agrB gene. To remove the intron, the plasmid pCPJVagrB was reintroduced into CN3685ΔagrB to create CPJV32. This strain, CPJV32, was then grown at 30°C to produce maximal expression of the temperature-sensitive LtrA protein which is required for the splicing-induced intron removal (34, 55). Incubation at 30°C allowed strain CPJV32 to produce a significant increase of the biomass, as quantified by fluorescence (Fig. 4C). CPJV32 biofilms were observed as a thick biofilm biomass on the bottom of the wells that covers most of the substrate (Fig. 4D). These experiments confirmed the CpAL system regulates biofilm formation of C. perfringens type C strain CN3685.
Upregulated expression of cpa and pfoA in C. perfringens biofilm cells.
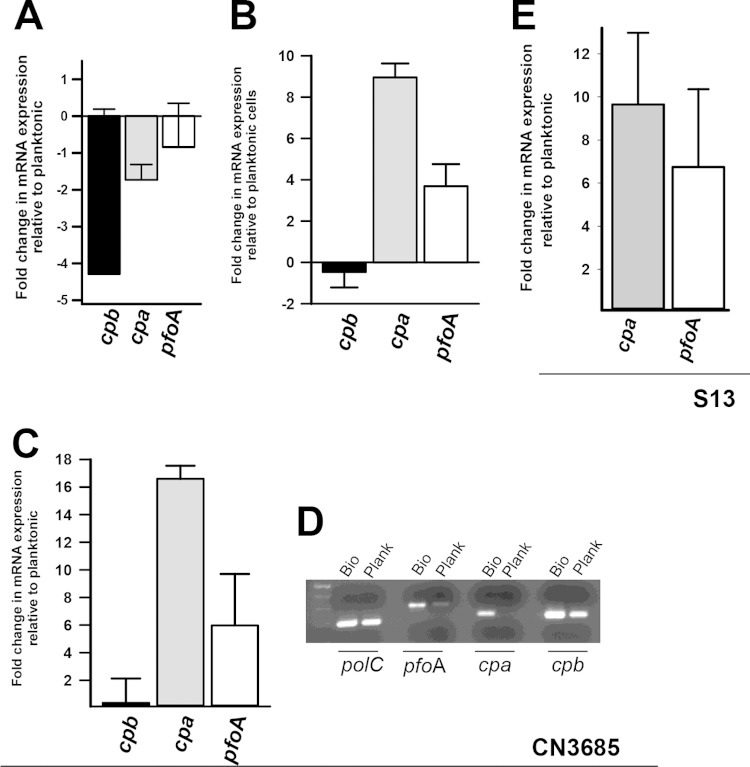
The CpAL system controls mRNA levels of genes encoding important toxins such as cpb, cpa, and pfoA in planktonic cultures of type A, type B, and type C strains (36, 42, 43). To evaluate biofilm levels of these transcripts, total RNA was extracted from CN3685 biofilms and planktonic cultures and mRNA expression was assessed by qRT-PCR. Of evaluated genes, the cbp gene, encoding C. perfringens beta toxin (61), is maximally expressed in planktonic cultures during the early-mid-log phase of growth (60). In comparison to planktonic cultures, the population of biofilm bacteria 4 h postinoculation had downregulated transcription of the cbp gene (∼4-fold decrease) but mRNA levels of cpa and pfoA remained similarly expressed (Fig. 5A). Expression of the cpa or pfoA transcript was ∼9-fold or ∼4-fold upregulated in 24-h biofilms, respectively, relative to planktonic cells grown for 4 h (Fig. 5B), whereas cpb expression did not change. Moreover, mRNA levels of cpa or pfoA in biofilm cells increased ∼17- or ∼6-fold compared to planktonic cells by 24 h postinoculation, as quantified by qRT-PCR (Fig. 5C) or detected by conventional RT-PCR (Fig. 5D).
FIG 5.
Upregulated transcription of cpa and pfoA, but not cpb, in C. perfringens biofilms versus planktonic cells. C. perfringens type C CN3685 or type A strain 13, was inoculated in TGY, followed by incubation at 37°C for 4 or 24 h. Planktonic cells and biofilms were harvested, RNA extracted, and used as the template in qRT-PCRs targeting the cpb, cpa, or pfoA gene. Average CT values were normalized to the polC gene, and the fold differences were calculated using the comparative CT method (2−ΔΔCT). Graphs show the fold changes in mRNA expression of 4-h biofilms relative to planktonic cells (A), 24-h biofilms relative to 4-h planktonic cells (B), and 24-h biofilms relative to 24-h planktonic cells (C and E). (D) RNA from 24-h biofilms (Bio) or planktonic (Plank) was utilized in conventional RT-PCRs targeting the indicated gene. Error bars represent the standard errors of the mean calculated using data from at least three independent experiments.
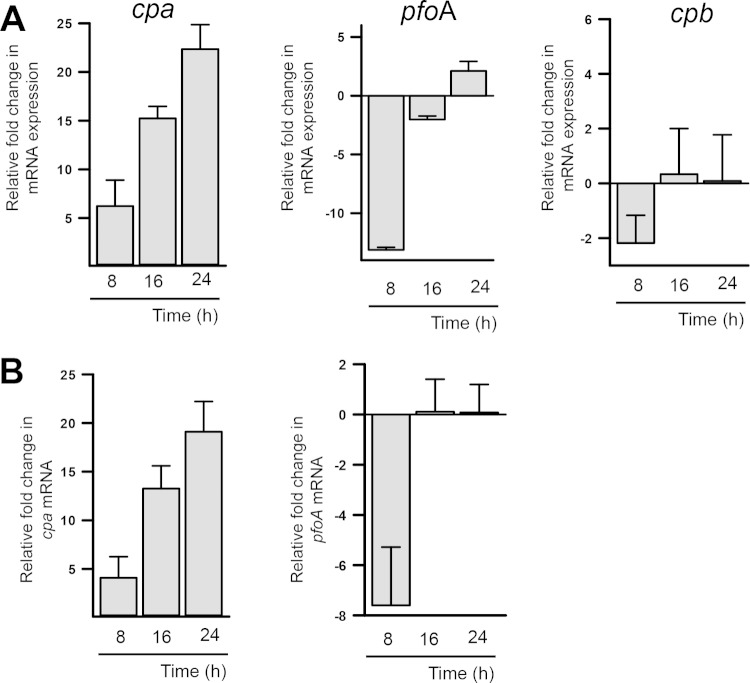
Relative to 4-h biofilms, however, the levels of cpa mRNA gradually increased 8, 16, or 24 h postinoculation, whereas pfoA mRNA levels were downregulated in 8-h biofilms, after which its expression remained the same (Fig. 6A). The levels of the cpb message did not change in biofilms incubated for 8, 16, or 24 h compared to 4-h biofilms (Fig. 6A). These results indicate that mRNA levels of both cpa and pfoA are increased in biofilms compared to planktonic cells, at the time points tested here, but that only mRNA levels of cpa are increasingly produced as the biofilm structure matures.
FIG 6.
Upregulated transcription of cpa in C. perfringens biofilms. C. perfringens type C CN3685 (A) or type A strain 13 (B) was inoculated into TGY, followed by incubation at 37°C for 4 or 24 h. Biofilms were harvested, and RNA was extracted and used as the template in qRT-PCRs targeting the cpb, cpa, or pfoA gene. Average CT values were normalized to the polC gene, and the fold differences were calculated using the comparative CT method (2−ΔΔCT). The panels show the fold changes in mRNA expression relative to biofilms at 4 h postinoculation compared to 8-, 16-, or 24-h biofilms. Error bars represent the standard errors of the mean calculated using data from at least four independent experiments.
To further confirm the upregulated transcription of cpa and pfoA in biofilm cells, the mRNA levels of those genes were evaluated in total RNA extracted from the gangrene producer, type A, strain 13. As shown in Fig. 5E, mRNA expression for cpa and pfoA increased ∼9- and ∼6-fold in 24-h biofilm cells relative to planktonic cells, respectively. Similar expression of cpa and pfoA transcripts to that presented above for strain CN3685 was observed when expression in 8-, 16-, or 24-h biofilms was compared to the expression in biofilms produced 4 h postinoculation (Fig. 6B).
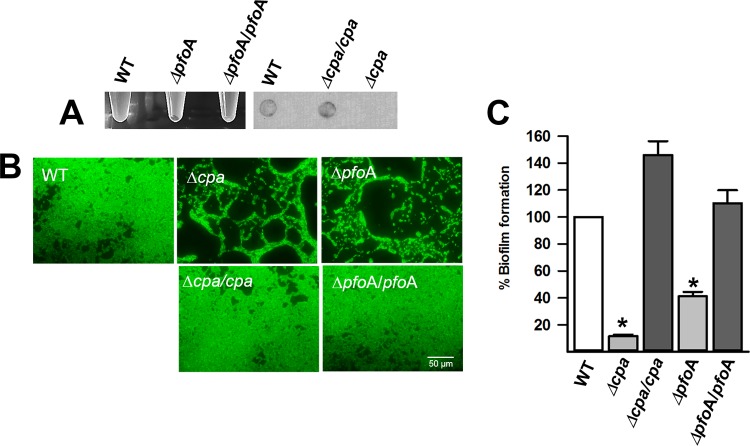
CPA and PFO are required to produce biofilms.
The upregulated production of toxin genes in biofilms and recent findings demonstrating that the CpAL system regulates production of C. perfringens alpha toxin (CPA), CPB and perfringolysin O (PFO) (36, 42, 43) prompted us to directly evaluate the role of these toxins in biofilm formation. To assess this, we inserted an intron separately into cpa, cpb, or pfoA, encoding CPA, CPB, or PFO, respectively (34, 52). The expected phenotypes for the mutation were evaluated in each mutant strain; for example, a strain with an insertionally inactivated cpa gene (CN3685Δcpa or S13Δcpa) was not able to hydrolyze phospholipids in egg yolk agar plates (not shown) and did not produce CPA in the culture supernatant (Fig. 7A). An Hb release assay confirmed that strains CN3685ΔpfoA and S13ΔpfoA did not lyse red blood cells (data not shown), whereby a compact pellet of erythrocytes was observed after 2 h of incubation (Fig. 7A).
FIG 7.
CPA and PFO required for C. perfringens biofilms. (A) The phenotypes of wild-type strain 13 (WT) and the S13Δcpa and S13ΔpfoA mutants and the corresponding complemented strains were evaluated. The left panel shows the lysis of erythrocytes (note an intact erythrocytes pellet when supernatants from the ΔpfoA mutant were used); the right panel shows CPA production as assessed by dot blotting with anti-CPA antibodies. (B and C) Strain 13 (WT), or its derivatives, was inoculated into 96-well plates containing TGY, and biofilms were incubated for 24 h at 37°C. The biofilms were stained with SYTO9, and micrographs of fluorescence-stained biofilms were obtained using an Evos inverted microscope (B), or their biomass (arbitrary units) was obtained using a fluorometer (C). Asterisks indicate statistical significance (P < 0.05) compared to the wild type (WT), as evaluated by the Mann-Whitney U test. Error bars represent the standard errors of the mean calculated using data from at least four independent experiments.
Although the gangrene-causing, genome-sequenced, strain 13 produced robust biofilms 24 h postinoculation (Fig. 7B and C), the Δplc and ΔpfoA mutants produced significantly less biofilm than did their parent strains. Similar results were obtained for CN3685 when biofilms made by Δplc and ΔpfoA mutant derivatives were compared to those produced by the parent strain. In contrast, CN3685Δcpb produced wt biofilm levels at 24 h postinoculation (not shown).
Mutant strains S13Δplc and S13ΔpfoA were each complemented with the cpa or the pfoA wt gene, respectively. As expected, the complemented strains regained the ability to produce CPA and hydrolyze phospholipids or lyse red blood cells (Fig. 7A). Biofilms produced by complemented strains, S13Δplc/plc and S13ΔpfoA/pfoA, were restored to wt levels (Fig. 7B and C), confirming the role of these hemolysins in the formation of C. perfringens biofilms.
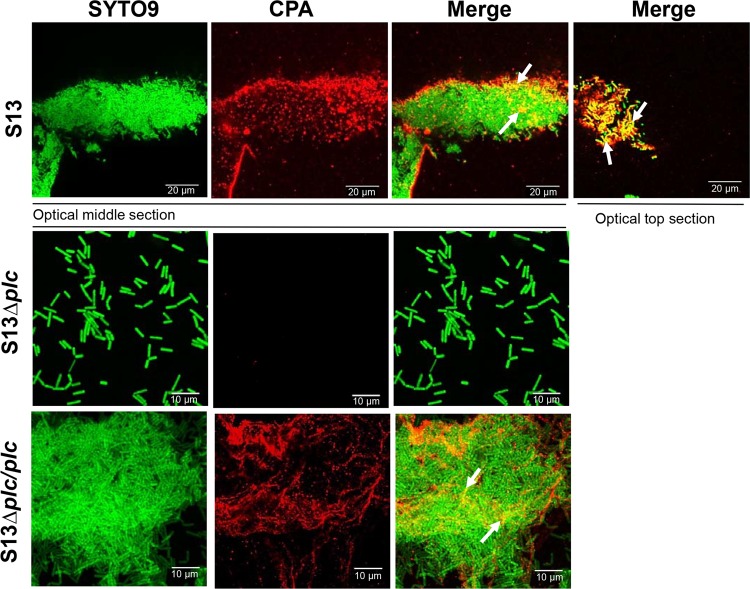
CPA is detected on C. perfringens biofilms.
Given the increasing upregulated expression of cpa in biofilms, the presence of CPA in biofilms was investigated by confocal microscopy. The signal of CPA was detected in optical middle and top sections of biofilms made by S13 and S13Δplc/plc, whereas, at 24 h postinoculation, the CPA signal was absent in the few S13Δplc bacteria that remained attached to the substrate (Fig. 8). In S13 and S13Δplc/plc biofilms, CPA colocalized in specific areas with DNA staining, suggesting that CPA may form a complex with eDNA in the biofilm matrix.
FIG 8.
Detection of CPA on C. perfringens biofilms. Strain S13Δplc or S13Δplc/plc was inoculated into a four-well chamber slide containing TGY, followed by incubation for 24 h at 37°C. Bacteria were stained with SYTO9, and CPA was detected using rabbit polyclonal anti-Clostridium perfringens alpha toxin antibodies, followed by goat anti-rabbit secondary antibodies conjugated to Alexa Fluor 555. Optical middle or top sections were obtained with a confocal microscope. Arrows point to areas of colocalization.
DISCUSSION
Clostridium perfringens infections have been associated with the production of potent and often lethal toxins (11, 64), and yet the microenvironments where strains must produce toxins are different. We have demonstrated here that CPA, a toxin produced by all toxigenic types, and PFO are necessary for C. perfringens type A strain 13 and type C strain CN3685 to produce biofilms, since their absence rendered strains unable to form wt levels of biofilms. This absence did not have an effect on growing rates of planktonic cells (data not shown). The need of CPA and PFO production for biofilm formation correlated well with the upregulated transcription of cpa and pfoA toxin genes observed in biofilms compared to planktonic cultures. The specific role that these toxins play in the building of the C. perfringens biofilm structure needs, however, further elucidation. Perhaps CPA helps to stabilize the structure by forming complexes with other molecules within the biofilm matrix, e.g., eDNA.
CPA and PFO produce lysis of red blood cells of different species; whereas CPA has phospholipase activity and produces alpha hemolysis on blood agar plates, PFO is a potent pore-forming toxin that induces beta hemolysis of red blood cells (65). Another hemolysin produced by the Gram-positive bacterium S. pneumoniae, the pneumolysin Ply, was recently found to be essential for proper assemblage of early pneumococcal biofilms in vitro and in a life-like environment (66). PFO and Ply belong to the cholesterol-dependent cytolysin family, sharing∼70% homology and ∼60% identities. Ply is located on the pneumococcal cell wall (67) and appears to link pneumococcal cells within a growing biofilm structure (66). In contrast, PFO has a signal peptide for its secretion into the supernatant. It is unclear at this point why, or how, secreted toxins such as PFO and CPA may help in the building of C. perfringens biofilms. CPA may be incorporated into the matrix, where it was seen partially colocalizing with eDNA in our confocal studies. Although not structurally related to CPA or PFO, recent discoveries showed that a hemolysin produced by Staphylococcus aureus forms a nucleoprotein complex with released DNA, leading to a solid biofilm matrix required to build staphylococcal biofilms both in vitro and in vivo (68). Other exotoxins, such as alpha-toxin (also known as HLA) secreted by S. aureus strains, have also been implicated in the formation of staphylococcal biofilms on mucosal tissues and virulence (69).
The composition of the C. perfringens biofilm matrix is under active investigation. Two different research groups recently showed that treatment of preformed biofilms with proteinase K, cellulose, or DNase I dispersed C. perfringens biofilms, suggesting the presence of proteins, β-1,4-linked polysaccharides, and eDNA (49, 50). Our studies further demonstrated the presence of eDNA in the supernatant of biofilm cultures acting as a source of DNA to be incorporated into the matrix. Although C. perfringens strain CN3685, utilized in these studies, carries plasmids ranging in size from ∼65 to ∼110 kb (70), the eDNA identified corresponded to chromosomal DNA, since our quantitative assays targeted a chromosomally encoded cpa gene (13). Chromosomal eDNA was, in part, provided by lysis of a subpopulation of biofilm bacteria and planktonic cells as quantified and visualized by using the Live/Dead cell assay. Since most C. perfringens strains carry virulence plasmids (70, 71), the possibility exists that plasmids are also released as a source of eDNA for the matrix.
Regulation of C. perfringens biofilms appears to be a complex mechanism which we demonstrated to be mediated by the CpAL QS system but not by the LuxS/AI-2 system. Whereas a mutation in the luxS gene did not affect the biomass, our experiments showed that CpAL was required for proper formation of the structure, since the absence of agrB (encoding a transmembrane protein required for processing the AgrD pheromone [36]) rendered strain CN3685, or strain 13, unable to produce mature biofilm structures. Instead, ΔagrB mutants grew mostly as planktonic bacteria. CpAL regulation of biofilms might involve only a subset of CpAL-regulated factors since the CPB toxin, which is regulated by this system, was not implicated in the formation of biofilms by CN3685. Although not addressed here, the CpAL system could activate biofilm-specific downstream regulators. For example, it has been demonstrated that the two-component regulatory system VirR/S transcriptionally regulates toxin levels of CPA, PFO, and other virulence factors in planktonic cultures (72, 73). Whereas the VirR/S system is the logical downstream candidate, since the VirS transmembrane protein appears to be the receptor for the AgrD pheromone (37), another transcriptional regulator called CtrAB has been recently described to indirectly (i.e., via an unknown factor) control the transcript levels of cpa and pfoA (74). A ctrAB-null mutant, however, produced wt biofilm biomass when incubated at 37°C but demonstrated some effects on biofilms formed at 25°C (50). Our data showing upregulated transcription in biofilm cells of CpAL-regulated cpa and pfoA genes, but not cbp, further reinforce our hypothesis that biofilms activate specific downstream signals in response to the CpAL-released AgrD pheromone. Homolog QS systems have also been described in most toxigenic clostridia (41), with most producing biofilms to survive in different hostile environments. Whether these biofilms are controlled by Agr-like systems remains to be investigated.
An additional contribution of the present study was the protocol we developed and validated to fluorescently stain C. perfringens biofilms. The fluorescence-based protocol was compared against the classic crystal violet procedure, which, although helpful, was not as sensitive as fluorescence staining of biofilms, which allowed us to detect statistically significant differences of the biofilm biomass at each time point tested. The fluorescence protocol has the additional advantage that biofilms can be stained and quantified within minutes, after which fluorescence micrographs can be obtained from the same preparations. Since there is increasing evidence that other clostridia may produce biofilms in different environments, having a simplified and more sensitive protocol will help to improve our knowledge of biofilms produced by other anaerobes.
In summary, we hypothesize that proliferation of C. perfringens strains at the site of the infection may result in activation of the CpAL system, which regulates production of components of the biofilm matrix. The biofilm structure may confer resistance to innate immune molecules and antibiotics while also inducing the degradation of epithelial tissue by the production of toxins. In cases of gangrene produced by type A strains, for example, the border between healthy and necrotic tissue often advances several inches per hour, despite appropriate antibiotic therapy. Therefore, studies of the C. perfringens biofilm structure are necessary to better understand the diseases and ultimately help improve the prognosis for these types of infections.
ACKNOWLEDGMENTS
We are grateful to Tohru Shimizu and Kaori Othani from Kanazawa University, Japan, for their gift of strain 13. We also thank Julian I. Rood from Monash University, Victoria, Australia, for providing strain CN3685 and Glenn Songer from Iowa State University for providing C. perfringens strains JGS1075, JGS1076, JGS1495, and JGS1659. We acknowledge contributions of Shazaib Jiwani from Oglethorpe University to some laboratory procedures. We also thank June R. Scott, Emory University, for thoughtful suggestions on the manuscript.
REFERENCES
- 1.Fosse J, Laroche M, Oudot N, Seegers H, Magras C. On-farm multi-contamination of pigs by food-borne bacterial zoonotic hazards: an exploratory study. Vet Microbiol 147:209–213. doi: 10.1016/j.vetmic.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 2.McClane BA, Uzal FA, Fernandez-Miyakawa M, Lyerly D, Wilkins TD. 2004. The enterotoxigenic clostridia, p 698–752. In Dworkin M, Rosenburg E, Schleifer KF, Stackebrandt E (ed), The prokaryotes, vol 4 Springer-Verlag, New York, NY. [Google Scholar]
- 3.Rood JI, Cole ST. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev 55:621–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClane BA. 2001. The complex interactions between Clostridium perfringens enterotoxin and epithelial tight junctions. Toxicon 39:1781–1791. doi: 10.1016/S0041-0101(01)00164-7. [DOI] [PubMed] [Google Scholar]
- 5.Songer JG. 1996. Clostridial enteric diseases of domestic animals. Clin Microbiol Rev 9:216–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Songer JG. 2010. Clostridia as agents of zoonotic disease. Vet Microbiol 140:399–404. doi: 10.1016/j.vetmic.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Hirn M, Niinikoski J, Lehtonen OP. 1993. Effect of hyperbaric oxygen and surgery on experimental multimicrobial gas gangrene. Eur Surg Res 25:265–269. doi: 10.1159/000129288. [DOI] [PubMed] [Google Scholar]
- 8.Finsterer J, Hess B. 2007. Neuromuscular and central nervous system manifestations of Clostridium perfringens infections. Infection 35:396–405. doi: 10.1007/s15010-007-6345-z. [DOI] [PubMed] [Google Scholar]
- 9.Bryant AE. 2003. Biology and pathogenesis of thrombosis and procoagulant activity in invasive infections caused by group A streptococci and Clostridium perfringens. Clin Microbiol Rev 16:451–462. doi: 10.1128/CMR.16.3.451-462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uzal FA, Vidal JE, McClane BA, Gurjar AA. 2010. Clostridium perfringens toxins involved in mammalian veterinary diseases. Open Toxinol J 2:24–42. doi: 10.2174/1875414701003010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smedley JG III, Fisher DJ, Sayeed S, Chakrabarti G, McClane BA. 2004. The enteric toxins of Clostridium perfringens. Rev Physiol Biochem Pharmacol 152:183–204. doi: 10.1007/s10254-004-0036-2. [DOI] [PubMed] [Google Scholar]
- 12.Popoff MR, Bouvet P. 2009. Clostridial toxins. Future Microbiol 4:1021–1064. doi: 10.2217/fmb.09.72. [DOI] [PubMed] [Google Scholar]
- 13.Rood JI. 1998. Virulence genes of Clostridium perfringens. Annu Rev Microbiol 52:333–360. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 14.Lanting B, Athwal GS, Naudie DD. 2007. Spontaneous Clostridium perfringens myonecrosis of the shoulder: a case report. Clin Orthop Relat Res 461:20–24. [DOI] [PubMed] [Google Scholar]
- 15.Schropfer E, Rauthe S, Meyer T. 2008. Diagnosis and misdiagnosis of necrotizing soft tissue infections: three case reports. Cases J 1:252. doi: 10.1186/1757-1626-1-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroda S, Okada Y, Mita M, Okamoto Y, Kato H, Ueyama S, Fujii I, Morita S, Yoshida Y. 2005. Fulminant massive gas gangrene caused by Clostridium perfringens. Intern Med 44:499–502. doi: 10.2169/internalmedicine.44.499. [DOI] [PubMed] [Google Scholar]
- 17.Stevens DL, Tweten RK, Awad MM, Rood JI, Bryant AE. 1997. Clostridial gas gangrene: evidence that alpha and theta toxins differentially modulate the immune response and induce acute tissue necrosis. J Infect Dis 176:189–195. doi: 10.1086/514022. [DOI] [PubMed] [Google Scholar]
- 18.Bryant AE, Bergstrom R, Zimmerman GA, Salyer JL, Hill HR, Tweten RK, Sato H, Stevens DL. 1993. Clostridium perfringens invasiveness is enhanced by effects of theta toxin upon PMNL structure and function: the roles of leukocytotoxicity and expression of CD11/CD18 adherence glycoprotein. FEMS Immunol Med Microbiol 7:321–336. doi: 10.1111/j.1574-695X.1993.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 19.Ellemor DM, Baird RN, Awad MM, Boyd RL, Rood JI, Emmins JJ. 1999. Use of genetically manipulated strains of Clostridium perfringens reveals that both alpha-toxin and theta-toxin are required for vascular leukostasis to occur in experimental gas gangrene. Infect Immun 67:4902–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy CL, Krejany EO, Young LF, O'Connor JR, Awad MM, Boyd RL, Emmins JJ, Lyras D, Rood JI. 2005. The alpha-toxin of Clostridium septicum is essential for virulence. Mol Microbiol 57:1357–1366. doi: 10.1111/j.1365-2958.2005.04774.x. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy CL, Lyras D, Cheung JK, Hiscox TJ, Emmins JJ, Rood JI. 2009. Cross-complementation of Clostridium perfringens PLC and Clostridium septicum alpha-toxin mutants reveals PLC is sufficient to mediate gas gangrene. Microbes Infect 11:413–418. doi: 10.1016/j.micinf.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Hickey MJ, Kwan RY, Awad MM, Kennedy CL, Young LF, Hall P, Cordner LM, Lyras D, Emmins JJ, Rood JI. 2008. Molecular and cellular basis of microvascular perfusion deficits induced by Clostridium perfringens and Clostridium septicum. PLoS Pathog 4:e1000045. doi: 10.1371/journal.ppat.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awad MM, Bryant AE, Stevens DL, Rood JI. 1995. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol Microbiol 15:191–202. doi: 10.1111/j.1365-2958.1995.tb02234.x. [DOI] [PubMed] [Google Scholar]
- 24.Awad MM, Ellemor DM, Boyd RL, Emmins JJ, Rood JI. 2001. Synergistic effects of alpha-toxin and perfringolysin O in Clostridium perfringens-mediated gas gangrene. Infect Immun 69:7904–7910. doi: 10.1128/IAI.69.12.7904-7910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gui L, Subramony C, Fratkin J, Hughson MD. 2002. Fatal enteritis necroticans (pigbel) in a diabetic adult. Mod Pathol 15:66–70. doi: 10.1038/modpathol.3880491. [DOI] [PubMed] [Google Scholar]
- 26.Tonnellier M, Maury E, Guglielminotti J, Offenstadt G. 2001. A fatal sandwich. Lancet Infect Dis 1:202. doi: 10.1016/S1473-3099(01)00097-4. [DOI] [PubMed] [Google Scholar]
- 27.Severin WP, de la Fuente AA, Stringer MF. 1984. Clostridium perfringens type C causing necrotising enteritis. J Clin Pathol 37:942–944. doi: 10.1136/jcp.37.8.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrillo TM, Beck-Sague CM, Songer JG, Abramowsky C, Fortenberry JD, Meacham L, Dean AG, Lee H, Bueschel DM, Nesheim SR. 2000. Enteritis necroticans (pigbel) in a diabetic child. N Engl J Med 342:1250–1253. doi: 10.1056/NEJM200004273421704. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda T, Okada Y, Inagi E, Tanabe Y, Shimizu Y, Nagashima K, Sakurai J, Nagahama M, Tanaka S. 2007. Enteritis necroticans ‘pigbel’ in a Japanese diabetic adult. Pathol Int 57:622–626. doi: 10.1111/j.1440-1827.2007.02149.x. [DOI] [PubMed] [Google Scholar]
- 30.Farrant JM, Traill Z, Conlon C, Warren B, Mortensen N, Gleeson FV, Jewell DP. 1996. Pigbel-like syndrome in a vegetarian in Oxford. Gut 39:336–337. doi: 10.1136/gut.39.2.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke LE, Diekmann-Guiroy B, McNamee W, Java DJ Jr, Weiss SM. 1994. Enteritis necroticans with midgut necrosis caused by Clostridium perfringens. Arch Surg 129:557–560. doi: 10.1001/archsurg.1994.01420290103015. [DOI] [PubMed] [Google Scholar]
- 32.Sakurai J, Duncan CL. 1978. Some properties of beta-toxin produced by Clostridium perfringens type C. Infect Immun 21:678–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vidal JE, McClane BA, Saputo J, Parker J, Uzal FA. 2008. Effects of Clostridium perfringens beta-toxin on the rabbit small intestine and colon. Infect Immun 76:4396–4404. doi: 10.1128/IAI.00547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayeed S, Uzal FA, Fisher DJ, Saputo J, Vidal JE, Chen Y, Gupta P, Rood JI, McClane BA. 2008. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol Microbiol 67:15–30. doi: 10.1111/j.1365-2958.2007.06007.x. [DOI] [PubMed] [Google Scholar]
- 35.Shojadoost B, Vince AR, Prescott JF. 2012. The successful experimental induction of necrotic enteritis in chickens by Clostridium perfringens: a critical review. Vet Res 43:74. doi: 10.1186/1297-9716-43-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidal JE, Chen J, Li J, McClane BA. 2009. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS One 4:e6232. doi: 10.1371/journal.pone.0006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohtani K, Yuan Y, Hassan S, Wang R, Wang Y, Shimizu T. 2009. Virulence gene regulation by the agr system in Clostridium perfringens. J Bacteriol 191:3919–3927. doi: 10.1128/JB.01455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet 248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 39.Ji G, Beavis RC, Novick RP. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci U S A 92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wuster A, Babu MM. 2008. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J Bacteriol 190:743–746. doi: 10.1128/JB.01135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooksley CM, Davis IJ, Winzer K, Chan WC, Peck MW, Minton NP. 2010. Regulation of neurotoxin production and sporulation by a putative agrBD signaling system in proteolytic Clostridium botulinum. Appl Environ Microbiol 76:4448–4460. doi: 10.1128/AEM.03038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal JE, Ma M, Saputo J, Garcia J, Uzal FA, McClane BA. 2012. Evidence that the Agr-like quorum sensing system regulates the toxin production, cytotoxicity, and pathogenicity of Clostridium perfringens type C isolate CN3685. Mol Microbiol 83:179–194. doi: 10.1111/j.1365-2958.2011.07925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, McClane BA. 2012. Role of the Agr-like quorum-sensing system in regulating toxin production by Clostridium perfringens type B strains CN1793 and CN1795. Infect Immun 80:3008–3017. doi: 10.1128/IAI.00438-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Rood JI, McClane BA. 2011. Epsilon-toxin production by Clostridium perfringens type D strain CN3718 is dependent upon the agr operon but not the VirS/VirR two-component regulatory system. mBio 2:e00275–11. doi: 10.1128/mBio.00275-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Ohtani K, Hayashi H, Shimizu T. 2002. The luxS gene is involved in cell-cell signaling for toxin production in Clostridium perfringens. Mol Microbiol 44:171–179. doi: 10.1046/j.1365-2958.2002.02863.x. [DOI] [PubMed] [Google Scholar]
- 46.Vidal JE, Ohtani K, Shimizu T, McClane BA. 2009. Contact with enterocyte-like Caco-2 cells induces rapid upregulation of toxin production by Clostridium perfringens type C isolates. Cell Microbiol 11:1306–1328. doi: 10.1111/j.1462-5822.2009.01332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 48.Varga JJ, Therit B, Melville SB. 2008. Type IV pili and the CcpA protein are needed for maximal biofilm formation by the gram-positive anaerobic pathogen Clostridium perfringens. Infect Immun 76:4944–4951. doi: 10.1128/IAI.00692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charlebois A, Jacques M, Archambault M. 2014. Biofilm formation of Clostridium perfringens and its exposure to low-dose antimicrobials. Front Microbiol 5:183. doi: 10.3389/fmicb.2014.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obana N, Nakamura K, Nomura N. 2014. A sporulation factor is involved in the morphological change of Clostridium perfringens biofilms in response to temperature. J Bacteriol 196:1540–1550. doi: 10.1128/JB.01444-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vidal JE, Ludewick HP, Kunkel RM, Zahner D, Klugman KP. 2011. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun 79:4050–4060. doi: 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, McClane BA, Fisher DJ, Rood JI, Gupta P. 2005. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl Environ Microbiol 71:7542–7547. doi: 10.1128/AEM.71.11.7542-7547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Chen J, Vidal JE, McClane BA. 2011. The Agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infect Immun 79:2451–2459. doi: 10.1128/IAI.00169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bannam TL, Rood JI. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:233–235. doi: 10.1006/plas.1993.1025. [DOI] [PubMed] [Google Scholar]
- 55.Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM. 2006. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 12:1271–1281. doi: 10.1261/rna.68706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Ethapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M. 2013. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol 195:545–555. doi: 10.1128/JB.01980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. 2004. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol 186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez-Miyakawa ME, Marcellino R, Uzal FA. 2007. Clostridium perfringens type A toxin production in 3 commonly used culture media. J Vet Diagn Invest 19:184–186. doi: 10.1177/104063870701900208. [DOI] [PubMed] [Google Scholar]
- 60.Fisher DJ, Fernandez-Miyakawa ME, Sayeed S, Poon R, Adams V, Rood JI, Uzal FA, McClane BA. 2006. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect Immun 74:5200–5210. doi: 10.1128/IAI.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vidal JE, Howery KE, Ludewick HP, Nava P, Klugman KP. 2013. Quorum-sensing systems LuxS/autoinducer 2 and Com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect Immun 81:1341–1353. doi: 10.1128/IAI.01096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moscoso M, Garcia E, Lopez R. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol 188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas VC, Thurlow LR, Boyle D, Hancock LE. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol 190:5690–5698. doi: 10.1128/JB.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petit L, Gibert M, Popoff MR. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol 7:104–110. doi: 10.1016/S0966-842X(98)01430-9. [DOI] [PubMed] [Google Scholar]
- 65.Uzal FA, Freedman JC, Shrestha A, Theoret JR, Garcia J, Awad MM, Adams V, Moore RJ, Rood JI, McClane BA. 2014. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol 9:361–377. doi: 10.2217/fmb.13.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shak JR, Ludewick HP, Howery KE, Sakai F, Yi H, Harvey RM, Paton JC, Klugman KP, Vidal JE. 2013. Novel role for the Streptococcus pneumoniae toxin pneumolysin in the assembly of biofilms. mBio 4:e00655–13. doi: 10.1128/mBio.00655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price KE, Camilli A. 2009. Pneumolysin localizes to the cell wall of Streptococcus pneumoniae. J Bacteriol 191:2163–2168. doi: 10.1128/JB.01489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huseby MJ, Kruse AC, Digre J, Kohler PL, Vocke JA, Mann EE, Bayles KW, Bohach GA, Schlievert PM, Ohlendorf DH, Earhart CA. 2010. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc Natl Acad Sci U S A 107:14407–14412. doi: 10.1073/pnas.0911032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson MJ, Lin YC, Gillman AN, Parks PJ, Schlievert PM, Peterson ML. 2012. Alpha-toxin promotes Staphylococcus aureus mucosal biofilm formation. Front Cell Infect Microbiol 2:64. doi: 10.3389/fcimb.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gurjar A, Li J, McClane BA. 2010. Characterization of toxin plasmids in Clostridium perfringens type C isolates. Infect Immun 78:4860–4869. doi: 10.1128/IAI.00715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sayeed S, Li J, McClane BA. 2007. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect Immun 75:2391–2398. doi: 10.1128/IAI.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ba-Thein W, Lyristis M, Ohtani K, Nisbet IT, Hayashi H, Rood JI, Shimizu T. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J Bacteriol 178:2514–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lyristis M, Bryant AE, Sloan J, Awad MM, Nisbet IT, Stevens DL, Rood JI. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol Microbiol 12:761–777. doi: 10.1111/j.1365-2958.1994.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 74.Obana N, Nakamura K. 2011. A novel toxin regulator, the CPE1446-CPE1447 protein heteromeric complex, controls toxin genes in Clostridium perfringens. J Bacteriol 193:4417–4424. doi: 10.1128/JB.00262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci U S A 99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sawires YS, Songer JG. 2006. Clostridium perfringens: insight into virulence evolution and population structure. Anaerobe 12:23–43. doi: 10.1016/j.anaerobe.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 77.Meer RR, Songer JG. 1997. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am J Vet Res 58:702–705. [PubMed] [Google Scholar]