Abstract
Francisella tularensis is a facultative intracellular bacterium utilizing macrophages as its primary intracellular habitat and is therefore highly capable of resisting the effects of reactive oxygen species (ROS), potent mediators of the bactericidal activity of macrophages. We investigated the roles of enzymes presumed to be important for protection against ROS. Four mutants of the highly virulent SCHU S4 strain with deletions of the genes encoding catalase (katG), glutathione peroxidase (gpx), a DyP-type peroxidase (FTT0086), or double deletion of FTT0086 and katG showed much increased susceptibility to hydrogen peroxide (H2O2) and slightly increased susceptibility to paraquat but not to peroxynitrite (ONOO−) and displayed intact intramacrophage replication. Nevertheless, mice infected with the double deletion mutant showed significantly longer survival than SCHU S4-infected mice. Unlike the aforementioned mutants, deletion of the gene coding for alkyl-hydroperoxide reductase subunit C (ahpC) generated a mutant much more susceptible to paraquat and ONOO− but not to H2O2. It showed intact replication in J774 cells but impaired replication in bone marrow-derived macrophages and in internal organs of mice. The live vaccine strain, LVS, is more susceptible than virulent strains to ROS-mediated killing and possesses a truncated form of FTT0086. Expression of the SCHU S4 FTT0086 gene rendered LVS more resistant to H2O2, which demonstrates that the SCHU S4 strain possesses additional detoxifying mechanisms. Collectively, the results demonstrate that SCHU S4 ROS-detoxifying enzymes have overlapping functions, and therefore, deletion of one or the other does not critically impair the intracellular replication or virulence, although AhpC appears to have a unique function.
INTRODUCTION
Francisella tularensis is a highly virulent facultative intracellular pathogen that causes tularemia in humans and many other mammalian species. Two F. tularensis subspecies are of clinical importance: tularensis (type A) and holarctica (type B). Strains of both subspecies are highly infectious since inoculation of as few as 10 CFU is sufficient to cause disease in humans (1); however, strains of F. tularensis subsp. tularensis show distinctly higher virulence for humans and cause high mortality if the infection is untreated. Therefore, the bacterium is classified as a tier 1 select agent (i.e., a category containing the most likely bioterrorist agents) (2). Experimentally, the live vaccine strain (LVS) of F. tularensis is widely used as a model for virulent strains of the species. It was derived from a type B strain in the former Soviet Union during the 1950s (3). Although widely used, the mechanisms behind its attenuation are still not fully understood.
The ability to survive or suppress the normally antibacterial activities of host macrophages is considered to be critical for the virulence of intracellular pathogens like F. tularensis, and both LVS and fully virulent strains proliferate effectively within resting macrophages. Gamma interferon (IFN-γ) activation of macrophages enhances their bactericidal activity, and this effectively restricts the multiplication of F. tularensis. The bactericidal ability of macrophages that control LVS is conferred by mechanisms involving both reactive oxygen and nitrogen species (ROS and RNS, respectively), whereas their roles are less evident with regard to virulent strains, such as the prototypic SCHU S4 strain of F. tularensis subsp. tularensis (4–8). In fact, the mechanisms mediating the bactericidal activity of IFN-γ-activated macrophages toward virulent F. tularensis strains are still unclear (8).
Like other intracellular pathogens of professional phagocytes, F. tularensis expresses an array of enzymes that metabolize and neutralize ROS and RNS, such as superoxide dismutases (SODs), catalase (KatG), glutathione peroxidase (Gpx), organic hydroperoxides, alkyl-hydroperoxide reductase (AhpC), and MoxR ATPase (9–13), and all of the published F. tularensis genomes contain genes encoding each of these enzymes, most of which are highly conserved. They have overlapping catalytic activities and, among other things, catalyze reduction of hydrogen peroxide (H2O2), organic hydroperoxides, and peroxynitrite (ONOO−) (14–16). Previously, we observed that LVS is more susceptible than virulent strains, such as SCHU S4, to killing mediated by H2O2 or ONOO− (17). KatG has a pivotal role for resistance to H2O2 in vitro, but in vivo, KatG was demonstrated to be essential only for the full virulence of LVS but played a less important role for SCHU S4 (17). Therefore, virulent strains of F. tularensis possess KatG-independent mechanisms contributing to oxidative stress resistance that are lacking in LVS. FTT0086 is an example of a putative KatG-independent, ROS-protective mechanism, since the encoding gene is predicted to encode a DyP-type peroxidase, but the gene is truncated in the LVS strain and, therefore, it has been postulated to lack peroxidase activity (18, 19).
In the present study, the aim was to obtain an improved understanding of ROS-detoxifying enzymes for the ability of F. tularensis to counteract the toxic effects of various types of ROS by investigating the phenotypes of relevant mutants under defined conditions in vitro. Moreover, we also wanted to compare how the in vitro phenotypes correlated to the ability of intramacrophage replication and virulence. To this end, we investigated the roles of the SCHU S4 mutants lacking expression of the FTT0086, KatG, Gpx, or AhpC enzyme for ROS detoxification and for the intracellular survival and virulence of the pathogen. We also investigated the effects of combining the gene deletions to better understand if the detoxifying functions are overlapping or complementary. In addition, it was assessed if the FTT0086 peroxidase plays a role in the differential detoxifying capabilities of SCHU S4 and LVS since it has been postulated to be enzymatically inactive in the latter strain.
MATERIALS AND METHODS
Plasmids and bacterial strains.
All strains and plasmids used in this study are described in Table 1. F. tularensis LVS was originally obtained from the American Type Culture Collection (ATCC 29684). F. tularensis strains SCHU S4 (F. tularensis subsp. tularensis) and FSC200 (F. tularensis subsp. holarctica) were obtained from the Francisella Strain Collection of the Swedish Defense Research Agency, Umeå, Sweden. In-frame deletion of the FTT0086 gene was constructed by allelic exchange using a previously described technique based on integration and excision of a suicide plasmid carrying upstream and downstream sequences of the target gene (17). Briefly, 1,500 bp of the upstream and downstream regions of each of the genes was amplified by PCR. The PCR fragments for each gene contained complementary sequences in the 3′ end of the upstream fragment and the 5′ end of the downstream fragment, which were annealed during a second round of PCR. After restriction enzyme digestion and purification, the PCR fragments were cloned into the suicide vector pDMK2, which was later transformed into Escherichia coli S17-1. Conjugation to F. tularensis SCHU S4 was carried out as described previously (20). Conjugants were selected on media containing 10 μg/ml kanamycin and 50 μg/ml of polymyxin B and confirmed by PCR. To select for a second recombination event, conjugants were plated on medium containing 5% sucrose, and the deletion of the genes was verified by sequencing and PCR (see Fig. S1 in the supplemental material).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmida | Origin, genetic feature, or reference(s) |
|---|---|
| Strains | |
| F. tularensis | |
| SCHU S4 | Human, USA, 1941 |
| ΔFTT0086 mutant | This study |
| ΔkatG mutant | 17 |
| ΔkatG ΔFTT0086 mutant | This study |
| ΔahpC mutant | 29 |
| Δgpx mutant | 29 |
| FSC200 | Human ulcer |
| LVS | ATCC 29684 |
| FSC741 | 18, 19 |
| E. coli | |
| S17-1 | 38 |
| BL21 | 23 |
| Plasmids | |
| pDMK2 | 20 |
| pBAD24 | 23 |
| pBAD24::yfeX | This study |
| pBAD24::FTT0086 | This study |
| pBAD24::FTL1773 | This study |
The mutant strains listed were derived from the SCHU S4 strain of F. tularensis subsp. tularensis.
Plasmids used for trans-complementation studies were constructed by PCR amplification of the ahpC or katG genes and introduction of them into plasmid pKK289Km, to allow constitutive expression from the groEL promoter. Plasmids were transferred into the SCHU S4 strain by electroporation. The FTT0086 gene was used for cis-complementation of LVS as previously described (19). FTT0086 was introduced into LVS by conjugation and localized in the chromosome under the control of its native promoter.
Escherichia coli strain BL21 and plasmids pBAD24 and pBAD24::yfeX were gifts from Cécile Wandersman (Institut Pasteur, Paris, France). The genes FTL1773 and FTT0086 were amplified by PCR from genomic DNA of LVS and SCHU S4, respectively. These fragments were cloned into pBAD24 using XbaI and SalI restriction sites to give pBAD24::FTL1773 and pBAD24::FTT0086.
Mice.
For virulence testing, specific-pathogen-free female BALB/c mice were purchased from Charles River Laboratories, St. Constant, Quebec, Canada. Mice were maintained and used in accordance with the recommendations of the Canadian Council on Animal Care Guide to the Care and Use of Experimental Animals in federally licensed, select agent-approved, small-animal containment level 3 facilities at the National Research Council, Ottawa, Ontario, Canada.
Bone marrow-derived macrophages (BMDM) were derived from C57BL/6 mice, which were purchased from Charles River Laboratories, Germany. Mice were maintained and used in accordance with the recommendations of the Swedish Guidelines for Animal Care.
Preparation of growth media.
Chamberlain's defined medium (CDM) with iron depleted by chelation (C-CDM) was produced as described previously (10, 21, 22). Briefly, 1% (wt/vol) Chelex-100 (Bio-Rad, Hercules, CA) was added to CDM without FeSO4, and the mixture was kept in rotation for 24 h at 4°C. The Chelex-100 was removed by filtering the medium through a 0.45-μm-pore-size Millipore filter (Biosciences, Stockholm, Sweden), and the chelation step was repeated once. The medium was thereafter supplemented with essential cations (MgSO4, 0.55 mM; ZnCl2, 1.4 μM; CuSO4, 0.2 μM; MnCl2, 1.0 μM; CaCl2, 5 μM). Sterility was ensured by a second filtration through a 0.22-μm-pore-size Millipore filter (Biosciences, Stockholm, Sweden).
Deferoxamine (DFO) is an iron chelator and was used to prepare agar plates lacking free iron in the medium (DFO plates). These plates were composed of 1 part 4% agar GC medium base (BD Diagnostic Systems, Sparks, MD), 1 part C-CDM without FeSO4, and 25 μg/ml of DFO (Sigma-Aldrich, St. Louis, MO). Modified Mueller-Hinton (MC plates) contained 1% (wt/vol) hemoglobin (Oxoid, Ltd., Hampshire, United Kingdom), 3.6% (wt/vol) GC agar base (BD Diagnostic Systems), and 1% (vol/vol) IsoVitaleX (BD Diagnostic Systems), and plates containing this medium were used to propagate strains.
Heme-dependent growth.
SCHU S4 and LVS and their respective isogenic ΔFTT0086 and ΔFTL1773 mutants were grown on C-CDM plates overnight. These iron-depleted strains were inoculated into C-CDM supplemented with heme (200 μg/ml) and the iron chelator deferoxamine (10 μg/ml). The growth of the respective strain was followed by measuring the optical density at 600 nm.
H2O2 susceptibility assay.
Bacteria cultivated overnight on MC plates were resuspended and diluted in phosphate-buffered saline (PBS) to an estimated density of 3 × 106 bacteria/ml. The bacterial suspension of each F. tularensis strain was divided into test tubes with H2O2 or control tubes that were untreated. The tubes were incubated for 2 h at room temperature (RT) without shaking. Thereafter, the samples were serially diluted and spread on MC plates that were incubated at 37°C in 5% CO2 for 3 days before enumeration of the CFU. The concentrations of H2O2 used were chosen to best reflect the differences among the strains and were not intended to mirror any physiological concentrations.
Paraquat susceptibility assay.
Bacteria cultivated overnight on MC plates were resuspended and diluted in PBS to an estimated density of 3 × 107 bacteria/ml. One hundred microliters of bacterial suspension was spread on an MC plate. Once dried, a Whatman paper disc containing 10 μl of a paraquat (Sigma) solution (H2O) was deposited on each plate and incubated overnight at 37°C in 5% CO2 before determination of the inhibition zone. The concentrations of paraquat used were chosen to best reflect the differences among the strains and were not intended to mirror any physiological concentrations.
SIN-1 susceptibility assay.
Bacteria cultivated overnight on MC plates were resuspended and diluted in PBS to an estimated density of 3.0 × 106 bacteria/ml. The bacterial suspension of each F. tularensis strain was divided into test tubes with SIN-1 or control tubes that were untreated. The tubes were incubated for 4 h at RT without shaking. Thereafter, the samples were serially diluted and spread on MC plates that were incubated at 37°C in 5% CO2 for 3 days before enumeration of the CFU. The concentrations of SIN-1 used were chosen to best reflect the differences among the strains and were not intended to mirror any physiological concentrations.
Deferrochelatase assay.
pBAD24::FTL1773 and pBAD24::FTT0086 were introduced into the E. coli C600 strain by electrotransformation. C600 strains containing the different vectors were streaked on LB agar plate supplemented with 0.2% l-arabinose. After an overnight incubation at 37°C, fluorescence was investigated by near-UV light irradiation (405 nm) (23).
Infection of macrophages.
J774A.1 macrophages or bone marrow-derived macrophages (BMDM) from C57BL/6 mice were used in the cell infection assays. J774A.1 macrophages were cultured and maintained in DMEM (GIBCO BRL, Grand Island, NY) with 10% heat-inactivated fetal bovine serum (FBS) (GIBCO). BMDM were obtained by flushing bone marrow cells from the femurs and tibias of C57BL/6 mice. These cells were cultured for 7 days in DMEM containing 10% FBS, 5 μg/ml gentamicin, and 20% conditioned medium (CM) from L929 cells (ATCC no. CCL-1), after which they were grown in medium lacking gentamicin. CM was replaced every 2 to 3 days. The day before infection, macrophages were seeded at a density of 4 × 105 in a 24-well tissue culture plates in DMEM with 10% FBS. Following incubation overnight, cells were washed, reconstituted with fresh culture medium, and allowed to recover for at least 30 min prior to infection. A multiplicity of infection (MOI) of 30 was used for nonstimulated cells and an MOI of 200 was used for IFN-γ-stimulated cells, and phagocytosis of the bacteria was allowed to proceed for 90 min. Thereafter, the monolayer was washed twice and incubated with gentamicin (5 μg/ml) to eradicate any remaining extracellular bacteria. The time point after 30 min of gentamicin treatment was defined as 0 h. For the remaining time of the experiment, the medium was supplied with 2 μg/ml gentamicin. To some wells, 1,000 U/ml of IFN-γ (Peprotech. London, United Kingdom) was supplied 12 h before bacteria were added and thereafter. This treatment elicits effective control of intracellular F. tularensis (8) and primes the cells for ROS production (24).
At 0 and 18 h, the macrophage monolayers were lysed with 200 μl of 0.1% deoxycholate, and the number of intracellular bacteria was determined by plating 10-fold serial dilutions of the lysates in PBS. The assay was performed in triplicate and repeated three times.
Mouse infections.
For virulence testing, specific-pathogen-free female BALB/c mice were injected intradermally with F. tularensis strains in a volume of 50 μl. Actual concentrations of inocula were determined by plating 10-fold serial dilutions. Mice were examined daily for signs of infection and were euthanized by CO2 asphyxiation as soon as they displayed signs of irreversible morbidity.
To determine the proliferative capacity of the strains, the bacterial numbers in liver, spleen, lungs, and blood from infected BALB/c mice were analyzed. The mice were given approximately 20 CFU intradermally of the indicated F. tularensis strain. At day 4 of infection, the mice were euthanized by CO2, and organs and blood were withdrawn. The number of bacteria was determined by plating 10-fold serial dilutions of homogenized organs in PBS.
Statistical analysis.
Independent-sample t tests (two-tailed) were used to identify differences between groups. Statistical analyses were carried out using SPSS software, version 21.0.
RESULTS
Identification of F. tularensis enzymes for resistance to H2O2-mediated killing.
The roles of the potential ROS-detoxifying enzymes AhpC, KatG, Gpx, and FTT0086 of SCHU S4 were assessed by analyzing the survival of the corresponding deletion mutants after exposure to various concentrations of H2O2. FTT0086 belongs to the superfamily of heme-containing peroxidases with structures distinct from other peroxidases that show wide substrate specificities, but their roles to protect against ROS have been little studied (25).
After 2 h of exposure to 4 mM H2O2, the viability of SCHU S4 or the ΔahpC mutant was not affected, whereas the viabilities of the Δgpx, ΔFTT0086, ΔkatG, and ΔFTT0086 ΔkatG mutants decreased significantly (Fig. 1). Of the single mutants, the viability of the Δgpx strain decreased the least (P < 0.05 versus SCHU S4), and the ΔkatG strain was the most susceptible, whereas the ΔFTT0086 strain showed an intermediate phenotype (P < 0.001 and P < 0.01, respectively). The ΔFTT0086 ΔkatG mutant was more susceptible than any of the single mutants (P < 0.001), showing a >2-log10 decrease (Fig. 1). The ΔkatG strain complemented with katG in trans exhibited essentially the same susceptibility to 4 mM H2O2 as SCHU S4 (see Fig. S2 in the supplemental material). When the ΔkatG and ΔFTT0086 strains were exposed to 8 mM H2O2, the ΔkatG mutant showed a complete loss of survival (a decrease of >6 log10 CFU), whereas the viability of the ΔFTT0086 mutant decreased 1.5 log10 CFU and that of SCHU S4 decreased 0.5 log10 CFU.
FIG 1.
Changes in bacterial numbers of indicated F. tularensis strains after exposure to 4 mM H2O2 for 2 h. The bars represent the average decrease in bacterial viability from three replicates, and the error bars represent the standard error of the mean (SEM) (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). Similar results were observed in two additional experiments. The bacterial number at the initiation of the experiment for each strain was approximately 6.0 log10 CFU.
In summary, AhpC does not contribute to the protection against H2O2, whereas FTT0086 as well as Gpx and KatG protect F. tularensis against relatively low or intermediate concentrations of H2O2. Among the mutants tested, KatG is the most important enzyme to protect SCHU S4 against high concentrations of H2O2.
Identification of F. tularensis enzymes for resistance to paraquat-mediated killing.
The SCHU S4, ΔFTT0086, ΔkatG, ΔFTT0086 ΔkatG, and Δgpx mutant strains were exposed to paraquat using a disc diffusion assay. Paraquat reacts with components of the respiratory chain in bacteria, leading to the reduction of O2 and the generation of superoxide anion O2− (26). By the action of cytoplasmic superoxide dismutases, or spontaneously, this leads to generation of H2O2. Thus, addition of paraquat leads to increased intracellular levels of H2O2. The most marked effect of paraquat was observed for ΔahpC, which was highly susceptible (P < 0.001) compared to SCHU S4 (Fig. 2). The other mutants tested, genotypes ΔkatG, ΔFTT0086, Δgpx, and ΔFTT0086 ΔkatG, were less susceptible (P < 0.01) than the ΔahpC mutant but more susceptible (P < 0.01) than SCHU S4 to the relatively low concentrations of 5 or 20 mM (see Fig. S3 in the supplemental material). At the intermediate concentration of 80 mM, the ΔahpC and ΔkatG strains were still more susceptible than SCHU S4, whereas the other mutants were not (see Fig. S3). This increased susceptibility was sustained for ΔahpC alone at concentrations up to 500 mM (see Table S1 in the supplemental material). The ΔahpC strain complemented with ahpC in trans exhibited essentially the same susceptibility to paraquat as SCHU S4 (see Fig. S3).
FIG 2.
Inhibition of the growth of F. tularensis strains after exposure to paraquat. A disk containing 10 μl of the indicated concentration of paraquat was placed on an agar plate. The bars represent the average of the growth inhibition zone (diameter) from three separate plates. The error bars represent the SEM (*, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
In summary, AhpC appears to constitute the primary detoxifying mechanism against the paraquat-induced toxicity among the mutants tested, whereas the other enzymes had less important roles, and differences were observed only at relatively low concentrations of paraquat.
Identification of F. tularensis enzymes for resistance to SIN-mediated killing.
A variant of highly reactive and bactericidal species is represented by ONOO−, resulting from the reaction between O2− and nitric oxide. SIN-1 generates O2− and nitric oxide, which spontaneously form ONOO−. The effect of this agent on the survival of SCHU S4 or the ΔahpC, ΔFTT0086, ΔkatG, ΔFTT0086 ΔkatG, or Δgpx mutant was investigated. No killing was observed when the strains were exposed to a concentration of 4.8 mM for 5 h (data not shown), whereas 8.0 mM resulted in significant killing of the ΔahpC mutant (P < 0.01 versus SCHU S4) but not of the other mutants or SCHU S4 (Fig. 3A). When the concentration was increased to 10 mM, the effect on the ΔahpC mutant was even more marked (P < 0.001), whereas SCHU S4 was not affected (Fig. 3B).
FIG 3.
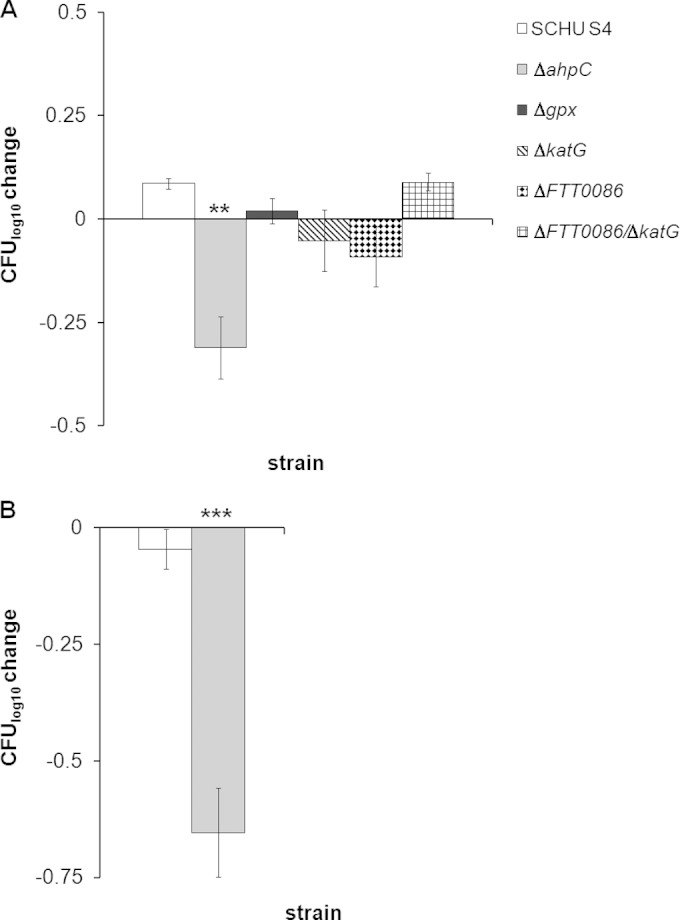
Changes in bacterial numbers of indicated F. tularensis strains after exposure to SIN-1. The bars represent the average change in bacterial viability from three replicates, and the error bars represent the SEM (**, P ≤ 0.01; ***, P ≤ 0.001). Similar results were observed in two additional experiments. (A) Exposure to 8.0 mM SIN-1 for 2 h. (B) Exposure to 10 mM SIN-1 for 2 h. The bacterial number at the initiation of the experiment for each strain was approximately 6.0 log10 CFU.
Intracellular replication and virulence of mutants defective for ROS-detoxifying enzymes.
The Δgpx, ΔahpC, ΔkatG, ΔFTT0086, and ΔFTT0086 ΔkatG mutants were tested for intracellular growth in J774 macrophage-like cells, and all of them showed the same degree of replication as SCHU S4. In BMDM, which demonstrate a higher state of activation than J774 cells (27), ΔahpC, but not the other mutants, showed compromised replication versus SCHU S4 (P < 0.01) (Fig. 4). Importantly, the ΔahpC mutant grew as well as SCHU S4 in CDM (data not shown). We hypothesized that the role of each of the enzymes would become more critical if the macrophages were endowed with enhanced bactericidal capabilities. The essential role of IFN-γ in this respect is very well documented (5, 6, 8, 28). In IFN-γ stimulated BMDM, the net growth of SCHU S4 was marginal, and there was no difference in this regard compared to the mutant strains (P > 0.05) (Fig. 4).
FIG 4.
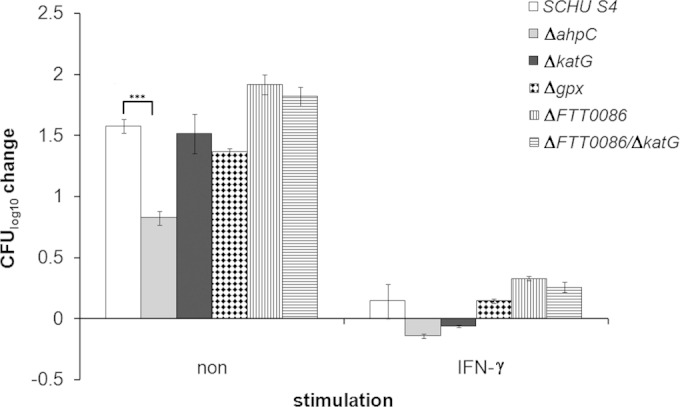
Intracellular replication of the indicated F. tularensis strains in BMDM with or without IFN-γ. Cells were infected at an MOI of 30 (without IFN-γ) or 200 (with IFN-γ), and bacterial numbers were determined after 24 h. ***, P < 0.001.
The virulence of the ΔFTT0086 and ΔFTT0086 ΔkatG mutants was tested after intradermal injection of 10 CFU into mice. The ΔFTT0086 mutant killed mice as quickly as SCHU S4, whereas the average time to death of mice infected with the ΔFTT0086 ΔkatG mutant was 6.8 ± 0.26 days, significantly delayed (P < 0.01) compared to mice infected with SCHU S4 (5.4 ± 0.32 days) (Fig. 5). Previously, we observed that the ΔahpC mutant displayed significantly delayed time to death (9.4 days versus 6.0 days for SCHU S4 [29]), and we now investigated if the replication of the ΔFTT0086 ΔkatG and ΔahpC mutants was affected in internal organs after intradermal infection. This was indeed the case, since both mutants showed lower bacterial numbers than SCHU S4; however, although there was an approximately 1-log10 difference for the ΔFTT0086 ΔkatG mutant, this was not significant, whereas the ΔahpC mutant displayed significantly compromised replication, almost 2 log10 lower CFU, in liver, spleen, and lung (P < 0.01 for spleen and liver and P < 0.001 for lung) (Table 2). No ΔahpC bacteria were recovered from blood and significantly, lower numbers were recovered from blood of ΔFTT0086 ΔkatG strain-infected mice versus SCHU S4-infected mice (P < 0.05) (Table 2).
FIG 5.
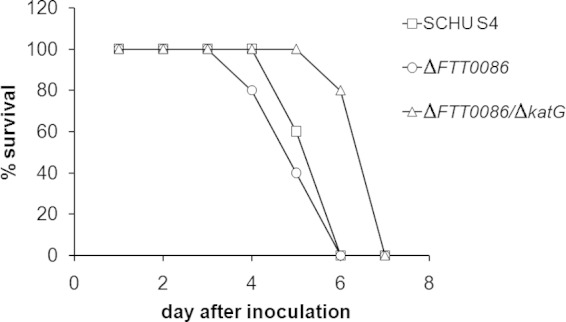
Survival of C57BL/6 mice after infection with SCHU S4 (squares), the ΔFTT0086 mutant (circles), and the ΔFTT0086 ΔkatG mutant (triangles). Mice were injected intradermally with 10 CFU of the indicated strain. Each group contained 5 mice.
TABLE 2.
Bacterial burdens in organs and blood of BALB/c mice infected with F. tularensis
| Strain | Bacterial burden ina: |
|||
|---|---|---|---|---|
| Organ (log10 CFU/organ)b |
Blood (log10 CFU/ml)b | |||
| Liver | Spleen | Lung | ||
| SCHU S4 | 6.81 ± 0.25 | 7.7 ± 0.38 | 3.84 ± 0.49 | 3.38 ± 0.44 |
| ΔkatG ΔFTT0086 mutant | 6.08 ± 0.33 | 6.95 ± 0.36 | 2.90 ± 0.55 | 1.27 ± 0.78* |
| ΔahpC mutant | 5.36 ± 0.34** | 6.00 ± 0.30** | 0.26 ± 0.26*** | 0 ± 0*** |
Organs were harvested from mice at day 4 after intradermal infection with approximately 20 CFU of the indicated F. tularensis strain per mouse. Asterisks indicate significant difference versus SCHU S4-infected mice for the indicated organ: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Values are means ± SEM from 5 mice.
Previously, we observed that the Δgpx and ΔkatG mutants showed no attenuation with regard to time to death (17, 29). Nevertheless, significantly fewer ΔkatG bacteria were recovered from skin, spleen, and liver of infected mice (17).
Thus, the data demonstrate that neither FTT0086, KatG, nor Gpx is crucial for intramacrophage growth or in vivo infection. Nevertheless, infection with the strain with combined ΔFTT0086 and ΔkatG mutations resulted in significantly delayed time to death of mice. AhpC was required for effective replication in BMDM and also demonstrated an important role in vivo, since replication of the ΔahpC mutant in internal organs was very significantly impaired.
Presence of an altered Dyp-type peroxidase FTT0086 homolog in F. tularensis LVS.
Most of the genes encoding the ROS-detoxifying enzymes of F. tularensis are highly conserved among subsp. tularensis and holarctica, with the exception of FTT0086. A BLAST analysis of the protein sequence using the PeroxiBase database (https://peroxibase.toulouse.inra.fr) confirmed that FTT0086 belongs to the DyP-type peroxidase B class and demonstrates 38% protein identity to CtesDyPrx01 of Comamonas testosteroni and 25% identity to YfeX from E. coli. FTT0086 is predicted to reside in the cytosol but lacks signal peptide and the TAT motif. The predicted primary structure reveals the presence of a heme-binding site in the C-terminal part of the protein.
The encoding gene is highly conserved in the published F. tularensis genomes, with the exception of the LVS homolog FTL1773, which contains a deletion of 93 nucleotides in the C-terminal part (18, 19). It has been previously postulated that the deletion would inactivate the peroxidase function (18, 19); however, our bioinformatic analysis demonstrated that it contains all the conserved residues required for heme-binding and peroxidase activities, as illustrated in the protein alignment (Fig. 6). The motif GXXDG corresponds to the catalytic domain of the peroxidase and is strictly conserved. Other conserved residues important for heme binding like D134, H204, and R220 are present in both proteins. The C-terminal truncation of 31 amino acids in FTL1773 may, however, affect the tertiary structure and thereby the function of the protein.
FIG 6.
Multiple sequence alignments of FTT0086, FTL1773, and homologous DyP-type peroxidases. The sequence alignments were performed using ClustalW, and ESPript was used to display the protein alignment (36, 37). Conserved residues are indicated in white on black. Similar amino acid residues are in bold. The D134, H203, and R220 residues of DyP are indicated with black stars. The GXXDG motif is boxed. The sequences of TyrA and BtDyP were retrieved from PDB under accession no. 2hag and 2gvk, respectively. BtDyp is the Dyp homologue from Bacteroides thetaiotaomicron and TyrA the homologue from Shewanella oneidensis.
In view of the uncertain function of the peroxidase in the LVS strain, we investigated the roles of the two Francisella homologues for resistance to ROS. LVS and the ΔFTL1773 mutant were exposed to a concentration of 1 mM H2O2. At this concentration, a decrease of slightly more than 1.0 log10 was observed for both strains (Fig. 7). Thus, the lack of FTL1773 did not affect the susceptibility of LVS to H2O2. In contrast, FSC741—LVS complemented with the SCHU S4 FTT0086 gene (18, 19)—showed less than a 0.5-log10 decrease in bacterial numbers when exposed to 1 mM H2O2 (Fig. 7), which was significantly less than LVS (P < 0.001). Thus, our findings suggest that FTT0086 but not FTL1773 has the capacity to degrade H2O2.
FIG 7.
Changes in bacterial numbers of the indicated F. tularensis strains after exposure to 1 mM H2O2 for 2 h. The bars represent the average decrease in bacterial viability from three separate tubes, and the error bars represent the SEM (***, P ≤ 0.001). Similar results were observed in two additional experiments. The bacterial number at the initiation of the experiment for each strain was approximately 6.0 log10 CFU.
We also investigated if FTT0086 possessed deferrochelatase activity like its homolog YfeX in E. coli (23). To this end, YfeX and the two F. tularensis homologs were expressed in a recombinant system by E. coli C600, and it was found that YfeX led to the release of iron from the heme, and this resulted in the accumulation of proporphyrin, which was detected as red fluorescence (see Fig. S4 in the supplemental material). When FTT0086 or FTL1773 was expressed under the same conditions as yfeX, there was no manifestation of red fluorescence (see Fig. S4). The expression in E. coli of the proteins encoded by FTT0086 and FTL1773, respectively, was confirmed by SDS-PAGE (see Fig. S5 in the supplemental material). Therefore, we conclude that FTT0086 is not involved in iron acquisition from heme through deferrochelation, like its homolog YfeX. In line with this result, SCHU S4 and LVS and their respective ΔFTT0086 and ΔFTL1773 isogenic mutants grew with heme as the exclusive iron source (data not shown).
DISCUSSION
F. tularensis is highly adapted to the intracellular habitat and capable of multiplying in many cell types, most notably within macrophages, a normally highly bactericidal cell type (18, 19). The intracellular growth of Francisella, which occurs in the cytosol, is considered a key factor for the virulence of the bacterium since it allows for escape from many immune mechanisms and access to nutrients and thereby enables its multiplication in the host. The prerequisite of intracellular growth for the bacterial virulence is demonstrated by the close correlation between the capability of F. tularensis mutants to replicate in macrophages and their virulence in the mouse model. To survive within the macrophage, the bacterium must first counteract the normally bactericidal environment of the phagosome, which is replete with antibacterial agents, such as ROS and RNS, which indicates that potent defenses against such reactive species are critical to successful intracellular bacteria, such as F. tularensis (30, 31). The bactericidal effects of various types of macrophages vary considerably; however, activation almost invariably endows them with highly effective capabilities to control infection and IFN-γ (8, 24).
In the present study, the aim was to investigate the roles of ROS-detoxifying enzymes for the survival of F. tularensis in vitro in the presence of specific bactericidal agents and to determine how this susceptibility is correlated to intramacrophage growth and virulence. Thereby, we hypothesized that we would be able to understand indirectly how infection is controlled. Previously, we demonstrated that KatG plays an essential role for the resistance of SCHU S4 and LVS to ROS-mediated killing in vitro and that the enzyme is essential for the virulence of LVS but not that of SCHU S4 (17). Here, we demonstrated that not only KatG but also Gpx and FTT0086 of SCHU S4 contributed significantly to the resistance to H2O2. At high concentrations, however, resistance relied exclusively on expression of KatG. As the putative Dyp peroxidase of LVS, FTL1773, is truncated in its C terminus, we wanted to know if this truncation altered the peroxidase activity. Although we observed that the known characteristic features of DyP-type peroxidases were preserved in FTL1773, the corresponding mutant, in contrast to the SCHU S4 FTT0086 mutant, showed no enhanced susceptibility to H2O2, indicating that, in fact, it is nonfunctional. Presumably, the lack of the C terminus affects the overall protein conformation, leading to impairment of its peroxidase function, or the protein is inherently unstable. In further support of a functional difference between FTT0086 and FTL1773, we observed that FSC741 (LVS complemented with the SCHU S4 FTT0086 gene) showed increased resistance to H2O2. Interestingly, as previously demonstrated, the virulence of the complemented strain is not enhanced (19). Therefore, our findings collectively suggest that the ability of LVS to degrade ROS—for example, via the function of KatG—is sufficient for its survival in vivo and that the enhanced detoxifying capability conferred by a functional DyP peroxidase does not confer a survival advantage in the mouse.
FTT0086 endows SCHU S4 with an efficient tool to handle oxidative stress that is missing from LVS; however, since the single mutants and the ΔkatG ΔFTT0086 double mutant showed significant survival at fairly high H2O2 concentrations, the bacterium must possess additional detoxifying mechanisms besides KatG and FTT0086. The most likely culprits in this regard are the peroxiredoxin, AhpC, and glutathione peroxidase, Gpx, since both enzymes have been shown to be potent detoxifiers of H2O2 in other bacterial species (32). Indeed, we observed that the susceptibility to H2O2 of the Δgpx mutant was increased to the same extent as for the ΔFTT0086 mutant. AhpC is involved in resistance to endogenous H2O2 and has been demonstrated to be essential for the virulence of the facultative intracellular bacterium Brucella abortus (33). We observed no enhanced susceptibility of the mutant to exogenous H2O2, but in view of its postulated role to protect against endogenous H2O2, we tested its sensitivity to paraquat, a molecule that via generation of O2− leads to generation of intracellular H2O2, and SIN-1, which indirectly generates ONOO− (26). In both instances, the ΔahpC mutant was found to be highly susceptible, and the ahpC deletion resulted in much diminished bacterial load and significantly prolonged time to death for infected mice.
Altogether, the data indicate that virulent F. tularensis strains rely on an intricate network of overlapping functions to protect themselves against the effects of ROS. Apparently, each of the FTT0086 and Gpx enzymes possesses a significant ability to degrade H2O2; however, these functions are not critical for the bacterium to survive in macrophages or in vivo, presumably due to their overlapping activities. As we previously demonstrated, this also holds true for the ΔkatG mutant of SCHU S4 (17). In our previous study, we observed, however, that the ΔkatG mutant showed some degree of attenuation since the bacterial numbers in liver and spleen were decreased, although this did not affect the survival of the mice. Here, we observed that SCHU S4 was dependent on KatG to degrade high concentrations of H2O2, and in fact, the mutant was eradicated by a concentration of 8 mM, but presumably, such high concentrations are not encountered in vivo, and therefore not even KatG is critically required. In contrast, the findings on the ΔahpC mutant show that the function of AhpC is distinct from the other enzymes studied, since it was not required for resistance to H2O2 but was the only enzyme that played a critical role in protection against the paraquat- and ONOO−-mediated effects. Thus, the findings imply that AhpC performs a very important role for detoxification in view of the enhanced susceptibility to the two compounds. In view of these findings, it was somewhat surprising that the mutant was capable of nearly normal intramacrophage replication; however, it showed marked attenuation in vivo as evidenced by very impaired growth in liver, spleen, and lung. Together, the findings indicate that the critical role of AhpC for virulence may not be related to the macrophage infection. Also, the ΔkatG ΔFTT0086 double mutant showed attenuation in vivo as evidenced by significantly longer time to death than SCHU S4.
Collectively, our data suggest that F. tularensis possesses complementary detoxifying systems, meaning that the absence of one or the other of these detoxifying enzymes will not be detrimental. In addition to the enzymes studied here, there are also F. tularensis enzymes that may contribute to the overall capability of virulent F. tularensis strains to withstand oxidative stress. One such example is the FTN1133 homolog that originally was identified in Francisella novicida and also in LVS was found to contribute to resistance to organic hydroperoxides but not to H2O2 (9), and also the MoxR ATPase that was important for resistance of the LVS strain to H2O2 (13). In addition, each of the LVS SodB and SodC mutants has been demonstrated to display an attenuated phenotype (9); however, we observed no attenuation of the SCHU S4 sodC mutant (29), presumably again emphasizing the more elaborate network of resistance to ROS in the highly virulent F. tularensis strains than in the LVS strain. The critical role of the antioxidative defense of F. tularensis is further emphasized by the recent demonstration that an emrA1 mutant is highly susceptible to hydrogen peroxide and attenuated for virulence by affecting secretion of SodB and KatG (34).
The correlation we found between the lack of detoxifying ability and attenuation demonstrated the relevance of the in vitro assays for the situation in vivo; however, exactly when during the life cycle the critical roles of the ROS-detoxifying enzymes are executed are unclear. Since F. tularensis replicates in the cytosol and appears to be capable of effective phagosomal escape even in activated macrophages, control of infection must be mediated in the macrophage cytosol. However, no cytosolic effector mechanisms have been directly identified, although inhibition of RNS partially reverses the effects (17). These facts, together with the present findings that the ΔahpC mutant was capable of normal intramacrophage replication, indicate that the relevance of ROS detoxification may be most important either during the extracellular phase or in cell types other than macrophages.
It may be difficult to pinpoint a definite role for many enzymes in vivo since their activities may be overlapping, as is the case for FTT0086. It has been found that F. tularensis gene expression inside macrophage was characterized by the upregulation of many genes involved in resistance to oxidative stress during the early phagosomal phase of infection (11). These findings together with our present and previous findings indicate that the redundancy and overlapping enzymatic functions that we observe with regard to detoxification of ROS reflect the very important overall roles of these functions for the survival of intracellular pathogens, such as F. tularensis. It would be possible to further delineate these overlapping and redundant functions by combining deletions of the detoxifying genes, as exemplified by the ΔkatG ΔFTT0086 double mutant. However, we have repeatedly attempted to generate other combinations, such as ΔahpC plus ΔkatG, but these attempts have been futile. One possible explanation is that such combinations severely compromise the ability of the bacterium to cope with the oxidative levels normally encountered, and, therefore, the combinations are not possible to select for during the genetic recombination.
Future studies will be aimed at identifying the regulations and roles of the enzymes implicated in oxidative stress resistance at different stages of infection with F. tularensis. Their regulation should also be analyzed in relation to iron metabolism, since we have demonstrated that the high degree of oxidative stress resistance of F. tularensis subsp. tularensis is related to low levels of intracellular free iron (35).
Supplementary Material
ACKNOWLEDGMENTS
Grant support was obtained from the Umeå Centre for Microbial Research (UCMR) Linnaeus Postdoctoral Program at Umeå University. Grant support was also obtained from the Swedish Medical Research Council (K2010-56X-09485 and K2013-56X-22356), the Medical Faculty, Umeå University, Umeå, Sweden, and a postdoctoral stipend from MIMS to J.B. The work was performed in part at the Umeå Centre for Microbial Research (UCMR).
We thank Céline Loot, Pasteur Institute, Paris, France, for help with determination of deferrochelatase activity.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02488-14.
REFERENCES
- 1.Sjöstedt A. 2007. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci 1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 2.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Tonat K. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 3.Conlan JW. 2004. Vaccines against Francisella tularensis—past, present and future. Expert Rev Vaccines 3:307–314. doi: 10.1586/14760584.3.3.307. [DOI] [PubMed] [Google Scholar]
- 4.Fortier AH, Green SJ, Polsinelli T, Jones TR, Crawford RM, Leiby DA, Elkins KL, Meltzer MS, Nacy CA. 1994. Life and death of an intracellular pathogen: Francisella tularensis and the macrophage. Immunol Ser 60:349–361. [PubMed] [Google Scholar]
- 5.Fortier AH, Polsinelli T, Green SJ, Nacy CA. 1992. Activation of macrophages for destruction of Francisella tularensis: identification of cytokines, effector cells, and effector molecules. Infect Immun 60:817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindgren H, Stenman L, Tarnvik A, Sjöstedt A. 2005. The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect 7:467–475. doi: 10.1016/j.micinf.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Polsinelli T, Meltzer MS, Fortier AH. 1994. Nitric oxide-independent killing of Francisella tularensis by IFN-gamma-stimulated murine alveolar macrophages. J Immunol 153:1238–1245. [PubMed] [Google Scholar]
- 8.Edwards JA, Rockx-Brouwer D, Nair V, Celli J. 2010. Restricted cytosolic growth of Francisella tularensis subsp. tularensis by IFN-gamma activation of macrophages Microbiology 156:327–339. doi: 10.1099/mic.0.031716-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melillo AA, Mahawar M, Sellati TJ, Malik M, Metzger DW, Melendez JA, Bakshi CS. 2009. Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J Bacteriol 191:6447–6456. doi: 10.1128/JB.00534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindgren H, Honn M, Golovlev I, Kadzhaev K, Conlan W, Sjöstedt A. 2009. The 58-kilodalton major virulence factor of Francisella tularensis is required for efficient utilization of iron. Infect Immun 77:4429–4436. doi: 10.1128/IAI.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, Brouwer D, Nair V, Fischer ER, Wicke L, Curda AJ, Kupko JJ III, Martens C, Crane DD, Bosio CM, Porcella SF, Celli J. 2009. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol 11:1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llewellyn AC, Jones CL, Napier BA, Bina JE, Weiss DS. 2011. Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS One 6:e24201. doi: 10.1371/journal.pone.0024201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieppedale J, Sobral D, Dupuis M, Dubail I, Klimentova J, Stulik J, Postic G, Frapy E, Meibom KL, Barel M, Charbit A. 2011. Identification of a putative chaperone involved in stress resistance and virulence in Francisella tularensis. Infect Immun 79:1428–1439. doi: 10.1128/IAI.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryk R, Griffin P, Nathan C. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407:211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- 15.Carmel-Harel O, Storz G. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol 54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 16.Smulevich G, Jakopitsch C, Droghetti E, Obinger C. 2006. Probing the structure and bifunctionality of catalase-peroxidase (KatG). J Inorg Biochem 100:568–585. doi: 10.1016/j.jinorgbio.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 17.Lindgren H, Shen H, Zingmark C, Golovliov I, Conlan W, Sjöstedt A. 2007. Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect Immun 75:1303–1309. doi: 10.1128/IAI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohmer L, Brittnacher M, Svensson K, Buckley D, Haugen E, Zhou Y, Chang J, Levy R, Hayden H, Forsman M, Olson M, Johansson A, Kaul R, Miller SI. 2006. Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect Immun 74:6895–6906. doi: 10.1128/IAI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salomonsson E, Kuoppa K, Forslund AL, Zingmark C, Golovliov I, Sjöstedt A, Noppa L, Forsberg A. 2009. Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infect Immun 77:3424–3431. doi: 10.1128/IAI.00196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golovliov I, Sjöstedt A, Mokrievich A, Pavlov V. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol Lett 222:273–280. doi: 10.1016/S0378-1097(03)00313-6. [DOI] [PubMed] [Google Scholar]
- 21.Cox CD. 1994. Deferration of laboratory media and assays for ferric and ferrous ions. Methods Enzymol 235:315–329. doi: 10.1016/0076-6879(94)35150-3. [DOI] [PubMed] [Google Scholar]
- 22.Chamberlain RE. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl Microbiol 13:232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letoffe S, Heuck G, Delepelaire P, Lange N, Wandersman C. 2009. Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc Natl Acad Sci U S A 106:11719–11724. doi: 10.1073/pnas.0903842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noubade R, Wong K, Ota N, Rutz S, Eidenschenk C, Valdez PA, Ding J, Peng I, Sebrell A, Caplazi P, DeVoss J, Soriano RH, Sai T, Lu R, Modrusan Z, Hackney J, Ouyang W. 2014. NRROS negatively regulates reactive oxygen species during host defence and autoimmunity. Nature 509:235–239. doi: 10.1038/nature13152. [DOI] [PubMed] [Google Scholar]
- 25.Sugano Y. 2009. DyP-type peroxidases comprise a novel heme peroxidase family. Cell Mol Life Sci 66:1387–1403. doi: 10.1007/s00018-008-8651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassan HM, Fridovich I. 1979. Paraquat and Escherichia coli. Mechanism of production of extracellular superoxide radical. J Biol Chem 254:10846–10852. [PubMed] [Google Scholar]
- 27.Gersuk GM, Razai LW, Marr KA. 2008. Methods of in vitro macrophage maturation confer variable inflammatory responses in association with altered expression of cell surface dectin-1. J Immunol Methods 329:157–166. doi: 10.1016/j.jim.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Lindgren H, Golovliov I, Baranov V, Ernst RK, Telepnev M, Sjöstedt A. 2004. Factors affecting the escape of Francisella tularensis from the phagolysosome. J Med Microbiol 53:953–958. doi: 10.1099/jmm.0.45685-0. [DOI] [PubMed] [Google Scholar]
- 29.Kadzhaev K, Zingmark C, Golovliov I, Bolanowski M, Shen H, Conlan W, Sjöstedt A. 2009. Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS One 4:e5463. doi: 10.1371/journal.pone.0005463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clemens DL, Lee BY, Horwitz MA. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun 72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santic M, Al-Khodor S, Abu Kwaik Y. 2010. Cell biology and molecular ecology of Francisella tularensis. Cell Microbiol 12:129–139. doi: 10.1111/j.1462-5822.2009.01400.x. [DOI] [PubMed] [Google Scholar]
- 32.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steele KH, Baumgartner JE, Valderas MW, Roop RM II. 2010. Comparative study of the roles of AhpC and KatE as respiratory antioxidants in Brucella abortus 2308. J Bacteriol 192:4912–4922. doi: 10.1128/JB.00231-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Z, Banik S, Rane H, Mora VT, Rabadi SM, Doyle CR, Thanassi DG, Bakshi CS, Malik M. 2014. EmrA1 membrane fusion protein of Francisella tularensis LVS is required for resistance to oxidative stress, intramacrophage survival and virulence in mice. Mol Microbiol 91:976–995. doi: 10.1111/mmi.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindgren H, Honn M, Salomonsson E, Kuoppa K, Forsberg A, Sjöstedt A. 2011. Iron content differs between Francisella tularensis subspecies tularensis and subspecies holarctica strains and correlates to their susceptibility to H2O2-induced killing. Infect Immun 79:1218–1224. doi: 10.1128/IAI.01116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:787–796. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.