Abstract
Pseudomonas aeruginosa is an important human opportunistic pathogen, accounting for a significant fraction of hospital-acquired lung infections. CD1d-restricted NKT cells comprise an unusual innate-like T cell subset that plays important roles in both bacterial and viral infections. Previous reports have differed in their conclusions regarding the role of NKT cells in clearance of P. aeruginosa from the lung. Since there is significant strain-dependent variation in NKT cell number and function among different inbred strains of mice, we investigated whether the role of NKT cells was dependent on the host genetic background. We found that NKT cells did indeed play a critical role in the clearance of P. aeruginosa from the lungs of BALB/c mice but that they played no discernible role in clearance from the lungs of C57BL/6 mice. We found that the strain-dependent role of NKT cells was associated with significant strain-dependent differences in cytokine production by lung NKT cells and that impaired clearance of P. aeruginosa in BALB/c CD1d−/− mice was associated with an increase in neutrophil influx to the lung and increased levels of proinflammatory cytokines and chemokines after infection. Finally, we found that the role of alveolar macrophages was also dependent on the genetic background. These data provide further support for a model in which the unusually high level of variability in NKT cell number and function among different genetic backgrounds may be an important contributor to infectious-disease susceptibility and pathology.
INTRODUCTION
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium that is an important human opportunistic pathogen. P. aeruginosa infection is responsible for a significant fraction of hospital-acquired infections (1). It commonly affects patients with impaired lung function due to disease (e.g., cystic fibrosis) or mechanical ventilation. While neutrophils appear to be the principal mediators of host resistance to P. aeruginosa in the lung (2), evidence suggests that other leukocyte subsets of both the innate and adaptive arms of the immune system also play significant roles in the host clearance of Pseudomonas bacteria from the lungs (1, 3).
CD1d-restricted NKT cells comprise an unusual innate-like αβ T cell subset that plays important roles in both bacterial and viral infections (4, 5). NKT cells can be activated directly, through the recognition of glycolipids presented by CD1d (6), or indirectly, through interleukin-12 (IL-12)) and IL-18 produced upon recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (7). Once activated, NKT cells can rapidly produce large amounts of a wide variety of cytokines and chemokines which have significant modulatory effects during the early stages of infection on the function of leukocytes of both the innate and adaptive arms of the immune system, including neutrophils and macrophages (8–10). Previous investigations into the role of NKT cells in the host response to P. aeruginosa infection of the lungs have yielded conflicting results, with one report demonstrating that NKT cells were critical in mediating clearance of lung P. aeruginosa (11), while another found little evidence of a role for NKT cells (12).
NKT cell number and function differ considerably among different mouse genetic backgrounds (13–18). In addition, levels of susceptibility to P. aeruginosa differ significantly among different inbred strains of mice (19). Since a key difference between these two studies was the use of different host genetic backgrounds, C57BL/6 and BALB/c, we hypothesized that the role of NKT cells may be dependent on the host genetic background. We found that NKT cells did indeed play a critical role in the clearance of Pseudomonas bacteria from the lungs of BALB/c mice but that they played no discernible role in clearance from the lungs of C57BL/6 mice. Furthermore, we found that this strain-dependent role of NKT cells was associated with significant differences in NKT cell cytokine production by lung NKT cells. These data not only reconcile disparate results in the field but also provide further support for a model in which the unusually high level of variability in NKT cell number and function among different genetic backgrounds may be an important contributor to disease susceptibility and pathology.
MATERIALS AND METHODS
Mice.
C57BL/6J, BALB/cJ, C57BL/6 CD1d−/−, C57BL/6 γδ−/−, and BALB/c CD1d−/− mice were obtained from Jackson Laboratory (Bar Harbor, ME). C57BL/6J JA18−/− mice were a gift from Mark Exley. All mice were housed in the specific-pathogen-free facility at the University of Vermont. All experiments were approved by the University of Vermont Institutional Animal Care and Use Committee.
P. aeruginosa preparation and infection.
Preparation of Pseudomonas aeruginosa (PAO1 strain) for infection was performed using Lennox broth cultures as previously described (20). Mice were infected by anesthetization using isoflurane and administration of the P. aeruginosa inoculum (∼2 × 107 CFU in a 40-μl volume) oropharyngeally (o.p.). At various times after infection, mice were euthanized, and lungs were obtained for P. aeruginosa enumeration. Lungs were homogenized in Dulbecco's phosphate-buffered saline (DPBS) as previously described (21), and viable bacterial counts were determined by serial dilution plating onto Pseudomonas isolation agar (PIA) (BD-Difco) followed by incubation at 37°C for 24 h.
BAL fluid analysis.
Mice were euthanized using carbon dioxide inhalation. Bronchoalveolar lavage (BAL) fluid was collected by cannulating the trachea and flushing the lungs with 0.9% saline solution. BAL fluid cell counts were obtained using an Advia 120 hematology system (Bayer HealthCare, Morristown, NJ). Differentials were obtained using cytospin, followed by staining with a modified Wright-Giemsa stain (Hema3; Biochemical Sciences). Cytokine, chemokine, and growth factor levels were quantitated using Bioplex (Bioplex Pro 23-plex) or an enzyme-linked immunosorbent assay (ELISA) (BD Biosciences).
Lung leukocyte isolation.
Mice were euthanized using carbon dioxide inhalation. Lungs were inflated with an enzymatic digestion buffer (Dubecco's modified Eagle's medium [DMEM], 1 mg/ml collagenase type IV [Invitrogen], 0.2 mg/ml DNase [Sigma]), after which they were dissected away from the trachea and then finely minced with scissors. Lung tissue was resuspended in additional enzymatic digestion buffer and shaken at 200 rpm at 37°C for 20 min. The samples were triturated with a 16-gauge needle and allowed to digest for an additional 20 min, after which they were triturated one last time. The digested tissue was passed through a 70-μm-pore-size filter and washed in PBS. Red blood cells were lysed using Gey's solution and washed in PBS–2% fetal calf serum [FCS], after which cells were counted and resuspended either in cell culture medium or in flow cytometry staining buffer.
Liver intrahepatic leukocytes (IHLs) were isolated as previously described (16). Briefly, anesthetized mice were perfused with PBS, after which the liver was removed, minced, and gently pressed through nylon mesh. The resulting cell suspension was washed twice and then resuspended in isotonic 33.8% Percoll (GE Healthcare). After centrifugation, the IHL cell pellet was resuspended and washed in PBS. Red blood cells were lysed using Gey's solution.
Flow cytometry.
Cells were stained at 4°C in PBS–2% FCS containing 0.1% sodium azide. TruStain FcX (Biolegend) was used in all samples prior to the addition of antibodies (Abs) to block nonspecific antibody binding. Antibodies used in these experiments were as follows: anti-T cell receptor (TCR)-β (H57-597; BD Biosciences) and anti-CD45 (30-F11; Life Technologies). CD1d tetramer (CD1dtet) loaded with PBS57 was obtained from the NIH tetramer facility (Emory University Vaccine Center, Atlanta, GA). Data were collected on an LSRII system (BD Biosciences) and analyzed using FlowJo (TreeStar, Ashland, OR) software.
For cell sorting, lung NKT cells, enzymatically digested lung samples from 5 C57BL/6 mice and 5 BALB/c mice, were first enriched for CD45+ leukocytes using a MACS column (Miltenyi Biotec) and CD45-enriched cells from each strain were pooled. CD45-enriched samples were stained at 4°C, in PBS–2% FCS with anti-TCR and CD1d tetramer. Cell sorting was conducted using a FACSAria instrument (BD Biosciences). Postsort analysis indicated that the sorted NKT cells were 86% to 93% pure.
Cell culture.
Lung leukocytes (100,000 cells) or liver intrahepatic leukocytes (50,000 cells) were resuspended in complete cell culture medium (RPMI 1640 supplemented with 10% FCS [Atlanta Biologicals], penicillin-streptomycin, nonessential amino acids, β-mercaptoethanol, and sodium pyruvate) at 37°C in 5% CO2 in the presence of various amounts of the CD1d ligand alpha-galactosylceramide (αGalCer; Axxora Pharmaceuticals, San Diego, CA). Leukocytes were incubated with αGalCer for 96 h (lung leukocytes) or 48 h (liver IHLs), after which cytokine levels were assessed using ELISA. Prior to cell culture, aliquots of each sample were taken to assess NKT cell frequencies using flow cytometry. These data were used to normalize NKT cell cytokine levels as measured by ELISA.
Sorted lung NKT cells (CD45+ TCR-β+ CD1dtet/PBS57+) were resuspended in complete cell culture medium and cultured in 96-well plates previously coated with various amounts of anti-CD3 monoclonal antibody (MAb) (145-2C11). NKT cells were incubated at 37°C in 5% CO2 for 72 h, after which cytokine levels were assessed using Bioplex (Bioplex Pro 23-plex) or ELISA (BD Biosciences).
Alveolar macrophage depletion.
Clodronate-loaded liposomes (a gift from Nico van Rooijen) were used to deplete lung phagocytic cells, the vast majority of which are alveolar macrophages in the naive mouse lung. Clodronate-loaded and control liposomes were administered oropharyngeally to anesthetized mice. The efficiency of alveolar macrophage depletion was assessed by assessing cytospin samples of BAL fluid 24 h after clodronate-loaded liposome administration. Control experiments yielded a depletion of 85% to 90% at 24 h.
Statistics.
Statistical analysis was conducted using Prism 6.04 (GraphPad Software). Analysis of strain-dependent differences in CFU and of cytokine and chemokine production by sorted NKT cells was conducted using a 1-way analysis of variance (ANOVA) followed by a Bonferroni post hoc comparison. Data were log transformed prior to analysis. Analysis of strain-dependent differences in BAL fluid differentials was conducted using 2-way ANOVA followed by a Bonferroni post hoc comparison. Analysis of dose-response curves of lung and liver leukocytes or of sorted NKT cells was conducted by fitting the data using nonlinear regression to a 3-parameter dose-response curve and then testing whether the resulting curve fits differed significantly between the strains tested. P values below 0.05 were deemed significant.
RESULTS
Strain-dependent role of CD1d-restricted T cells in the clearance of P. aeruginosa from the lung.
Previous studies examining whether CD1d-restricted NKT cells played a role in the clearance of P. aeruginosa from the lungs produced conflicting results (11, 12). One of the major differences between these reports was the use of different inbred strains of mice (C57BL/6 in one and BALB/cJ in the other). Since we previously demonstrated that these two strains differ both in the numbers of NKT cells and in their responsiveness to αGalCer (16), we hypothesized that strain-dependent differences in NKT cell function could explain the discrepant results. Therefore, we directly compared levels of P. aeruginosa PAO1 clearance from the lungs of wild-type C57BL/6J (B6), B6 CD1d−/−, B6 JA18−/−, BALB/cJ, and BALB/c CD1d−/− mice. CD1d-deficient mice lack the CD1d protein and lack all CD1d-restricted T cells (22). JA18−/− mice possess CD1d but lack the type I NKT cell population (23).
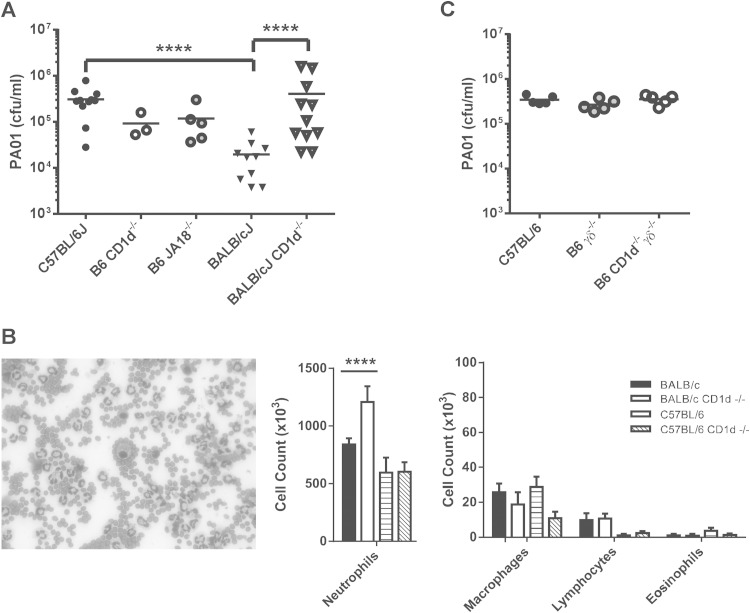
This comparison revealed that the BALB/cJ mice exhibited increased clearance of PAO1 from the lung compared to their B6 counterparts at 24 h (Fig. 1A), consistent with previous reports demonstrating that BALB/c mice are more resistant to PAO1 infection (24–26). Interestingly, a comparison of BALB/c and BALB/c CD1d−/− mice revealed that BALB/c CD1d−/− mice were impaired in their ability to clear PAO1 from the lungs; a comparison of B6 to B6 CD1d−/− and B6 JA18−/− mice revealed no such impairment (Fig. 1A). Examination of the bronchoalveolar lavage (BAL) fluid revealed predominant neutrophil infiltration in all strains (Fig. 1B). Examination of BAL fluid cell differentials revealed that BAL fluid from BALB/c CD1d−/− mice contained significantly more neutrophils 24 h after infection than that from their wild-type counterparts, whereas no difference was observed between B6 CD1d−/− mice and their wild-type counterparts (Fig. 1B). The increased number of neutrophils in the BALB/c CD1d−/− mice was an unexpected result, given the demonstrated role of neutrophils in PAO1 clearance (2) coupled with our observation that these mice were impaired in their ability to clear PAO1. No significant differences among macrophage, lymphocyte, or eosinophil populations were observed (Fig. 1B). Although B6 mice, in general, appeared to possess fewer BAL fluid lymphocytes than their BALB/c counterparts, this difference was not significant after correction for multiple comparisons.
FIG 1.
Strain-dependent role of NKT cells in clearance of Pseudomonas aeruginosa. Wild-type (WT), B6 CD1d−/−, B6 Jα18−/−, B6 γδ−/−, or B6 CD1d−/−γδ−/− mice were infected oropharyngeally with 2 × 107 CFU per mouse. (A) The number of live colonies in lungs 24 h after infection. Each symbol represents a single mouse. Lines denote the mean CFU. (B) Strain-dependent difference in BAL fluid leukocytic infiltrate 24 h after infection. (Left) Representative Giemsa staining of BAL fluid leukocytic infiltrate that is predominantly neutrophils. (Right) Cell counts of BAL fluid leukocytic infiltrate. Data represent means ± standard errors of the means (SEM); n = 5 to 9 mice/strain. Results are representative of three separate experiments. ****, P < 0.0001. (C) No effect of γδ T cells in Pseudomonas clearance on the B6 background. Data represent numbers of live colonies in lungs 24 h after infection. Each symbol represents a single mouse. Lines denote the mean CFU.
To investigate the possibility that γδ T cells, another innate-like T cell subset, were serving a function similar to that of the NKT cells in BALB/c mice, we examined PAO1 clearance from the lung in B6 γδ−/− and B6 γδ−/−CD1d−/− mice. No effect on PAO1 clearance was observed in mice lacking γδ T cells and in mice lacking both γδ T and NKT cells (Fig. 1C). Taken together, these data indicated that NKT cells did indeed play a role in PAO1 clearance from the lungs but that the critical nature of NKT cells depended on the genetic background of the host.
Strain-dependent resistance to P. aeruginosa is associated with enhanced NKT cell cytokine production.
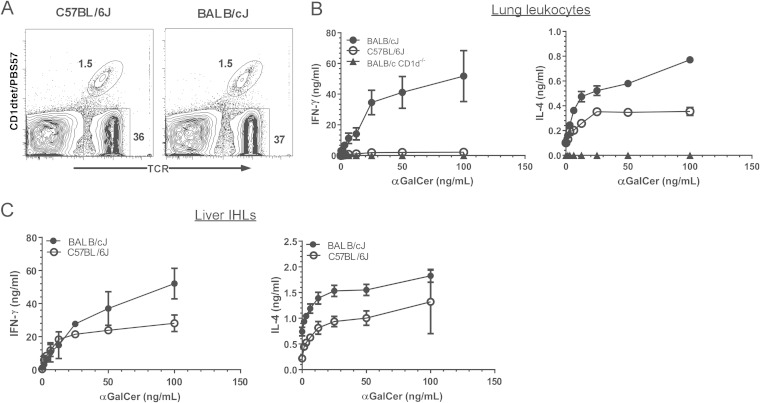
Since our observations suggested that NKT cells were important in PAO1 lung clearance in BALB/c mice but not in B6 mice, we compared the lung NKT cell functions of these two strains. Lung leukocytes were isolated from BALB/c, B6, and BALB/c CD1d−/− mice. Analysis of the lung NKT cell population using flow cytometry revealed there was no difference between the B6 and BALB/c strains in the frequency of lung NKT cells (Fig. 2A). To determine whether B6 and BALB/c lung NKT cells exhibited functional differences, we assessed cytokine production after incubation of lung leukocytes with various concentrations of the NKT cell-specific agonist αGalCer. Interestingly, we found that BALB/c lung leukocytes produced significantly more gamma interferon (IFN-γ) and IL-4 than their B6 counterparts (Fig. 2B). Indeed, the level of IFN-γ produced from the B6 lung leukocytes was only slightly higher than that produced by CD1d−/− mice. To investigate whether this difference was specific to lung NKT cells, we also compared the levels of liver NKT cytokine production by the two strains. Because there are more significantly more liver NKT cells in B6 mice than in BALB/c mice (16), we normalized the cytokine production in our analysis of both the lung and liver NKT cells based on the percentage of NKT cells in each culture, as described in Materials and Methods (B6 mice possessed an average of 28% ± 2.8% NKT cells in the liver; BALB/c mice possessed an average of 8.6% ± 1.7% NKT cells in the liver). This analysis suggested that BALB/c liver NKT cells produced more IFN-γ and IL-4 than their B6 counterparts (Fig. 2C). Therefore, lung leukocyte populations from the resistant BALB/c strain were significantly more responsive to the NKT cell agonist αGalCer than their B6 counterparts, and this strain-dependent difference was not confined to the lung NKT cell population.
FIG 2.
Strain-dependent response to αGalCer by lung leukocytes. Lung and liver leukocytes were prepared from 8-week-old C57BL/6J and BALB/cJ mice. (A) NKT and conventional T cells as a percentage of CD45+ lung leukocytes. A representative contour plot is shown. (B) Cytokine production by CD45+ lung leukocytes stimulated in vitro with αGalCer. Data represent means ± SEM; n = 6 BALB/c, 8 C57BL/6J mice, and 2 BALB/c CD1d−/− mice. (C) Cytokine production by CD45+ liver leukocytes stimulated in vitro with αGalCer. Data were normalized based on the frequency of TCR+CD1dtet/PBS57+ NKT cells, as assessed by flow cytometry. Data represent means ± SEM; n = 4 mice. The data represent the combined results of two separate experiments.
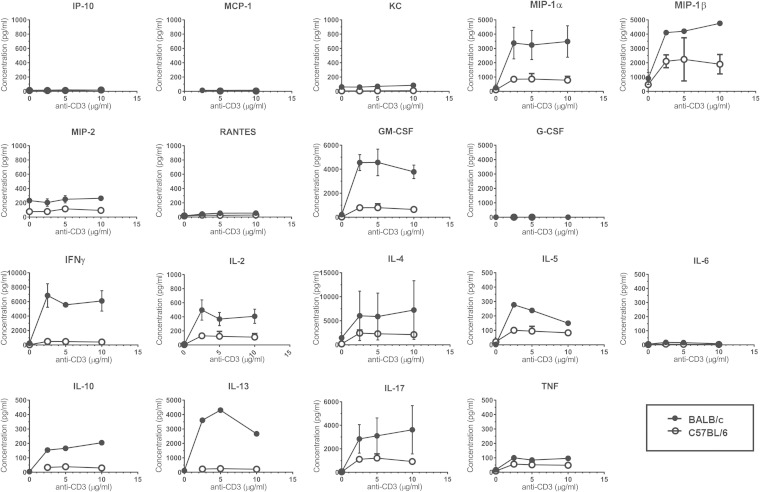
We next considered whether these strain-dependent differences in cytokine production were intrinsic to NKT cells or whether they were due to some difference in the quality of antigen presentation of αGalCer. To directly compare NKT cell function characteristics, we used fluorescence-activated cell sorter (FACS) analysis to sort NKT cells from CD45-enriched B6 and BALB/c lung leukocytes and assessed cytokine production after stimulation in anti-CD3-coated plates. Of the cytokines measured, lung NKT cells produced IFN-γ, IL-2, IL-4, IL-5, IL-10, IL-13, IL-17, and tumor necrosis factor (TNF) but no IL-6 (Fig. 3). Of the growth factors measured, lung NKT cells produced granulocyte-macrophage colony-stimulating factor (GM-CSF). Of the chemokines measured, lung NKT cells produced monocyte chemoattractant protein-1α (MIP-1α), MIP-1β, and some MIP-2 and KC but no RANTES, 10-kDa interferon-inducible protein (IP-10), or MCP-1.
FIG 3.
The strain-dependent difference in lung NKT cell function is NKT cell intrinsic. NKT cells from 8-week-old C57BL/6J and BALB/cJ mice were subjected to FACS analysis using CD45-enriched lung leukocytes based on TCR and CD1dtet/PBS57 staining. Equal numbers of NKT cells were incubated in anti-CD3 coated plates for 96 h, after which cytokine and chemokine production was assessed using BioPlex. Data represent means ± standard deviations (SD) of cytokine and chemokine production from duplicate wells.
Interestingly, we found that in nearly all cases, BALB/c NKT cells produced significantly larger amounts of cytokines, growth factors, and chemokines than did their B6 counterparts (Fig. 3). The exception was TNF, which was produced at similar levels by NKT cells from the two strains. Taken together, these data indicated that the enhanced cytokine production by BALB/c lung leukocytes after αGalCer challenge was due to an NKT-intrinsic strain-dependent difference in the ability of these cells to produce cytokines, chemokines, and growth factors upon stimulation.
Increased inflammatory response in CD1d−/− mice after PAO1 infection.
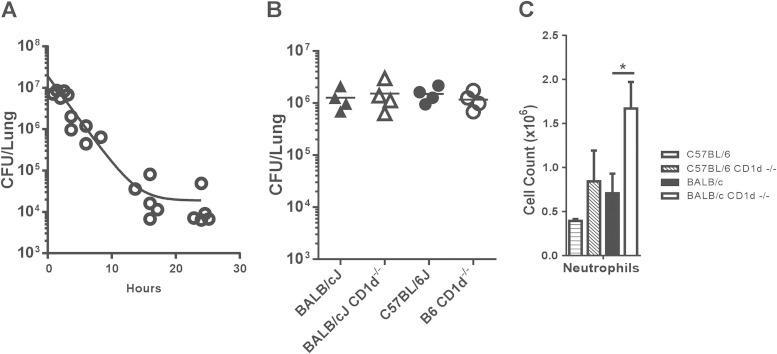
Next, we investigated more closely the relationship between enhanced neutrophil influx into the lungs of BALB/c mice and PAO1 clearance. We reasoned that the increased number of neutrophils in the BALB/c CD1d−/− mice 24 h after infection could have been due to the persistence of PAO1 bacteria in the lungs of these mice. Therefore, we investigated the effect that deletion of CD1d and CD1d-restricted T cells had on BAL fluid neutrophil numbers at an early time point after PAO1 infection, when lung PAO1 numbers of BALB/c and BALB/c CD1d−/− mice were similar. After conducting a time course of PAO1 clearance from BALB/c lungs (Fig. 4A), we chose to examine the host inflammatory response at 6 h postinfection. At this time point, we found that there was no difference in the amounts of PAO1 we could recover from the lungs of wild-type B6 and BALB/c mice and their CD1d−/− counterparts (Fig. 4B). This observation suggested that both the strain-dependent differences in lung PAO1 clearance between wild-type B6 and BALB/c mice and the effect of CD1d and NKT cells on PAO1 clearance became visible between 6 and 24 h after infection. However, despite the fact that we recovered equivalent numbers of PAO1 from the lungs of all four strains, we again found that the BALB/c CD1d−/− mice had significantly higher numbers of polymorphonuclear leukocytes (PMNs) in the BAL fluid than did BALB/c mice (Fig. 4C). This trend was also observed in the comparison of B6 and B6 CD1d−/− mice, though these results were not statistically significant. These data suggested that the presence of CD1d and/or CD1d-restricted T cells was influencing neutrophil influx into the airway irrespective of the number of P. aeruginosa bacteria in the lung.
FIG 4.
Role of CD1d and NKT cells in neutrophil influx to the airways early in infection. (A) Time course of PAO1 clearance. BALB/cJ mice were infected with 2 × 107 CFU, and live colonies in lungs were counted 24 h later. Each symbol represents the CFU from a single mouse. Lines denote the mean CFU. (B) Comparison of levels of PAO1 clearance in the lungs of different strains 6 h after infection. Lines denote the mean CFU. WT and CD1d-deficient mice were infected with 2 × 107 PAO1 o.p., and live colonies in lungs were counted 6 h later. (C) Increased neutrophil numbers in the BAL fluid of BALB/c CD1d−/− mice 6 h after infection. Data represent means ± SEM; n = 4 mice per strain. *, P < 0.05.
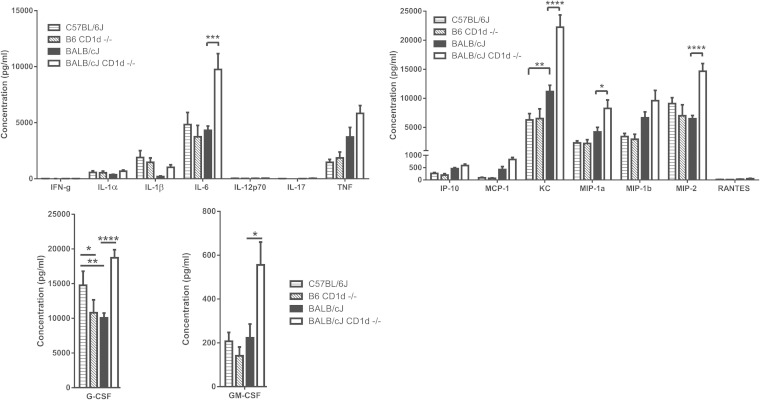
To examine the mechanisms underlying the increased neutrophil influx into the lung, we examined the cytokine and chemokine profiles in the BAL fluid of mice 6 h after infection with PAO1. These data indicated that, in comparison to that from their wild-type BALB/c counterparts, BAL fluid from the BALB/c CD1d−/− mice contained significantly larger amounts of the proinflammatory cytokine IL-6 and the chemokines KC, MIP-1α, and MIP-2 as well as of granulocyte colony-stimulating factor (G-CSF) and GM-CSF (Fig. 5). In contrast, the comparison of the B6 CD1d−/− mice with the B6 mice revealed that only the G-CSF levels were significantly different between the two strains, with the B6 CD1d−/− mice having lower levels than the wild-type B6 mice.
FIG 5.
Enhanced inflammatory response in the absence of CD1d. Results represent cytokine and chemokine profiles of BAL fluid 6 h after infection with PAO1. Data represent means ± SEM; n = 4 mice per strain. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We also noted differences in both IL-1β and TNF levels in the BAL fluid, although these differences did not reach a level of statistical significance after correction for multiple comparisons. Smaller amounts of IL-1β were detected in resistant BALB/c than in susceptible B6 mice (Fig. 5). While CD1d deficiency had no effect on BAL fluid IL-1β levels on the B6 background, loss of CD1d resulted in increased BAL fluid IL-1β levels in BALB/c CD1d−/− mice. In contrast, larger amounts of TNF were detected in the BAL fluid of resistant BALB/c mice than in that from their susceptible B6 counterparts. While CD1d deficiency had no effect in the B6 background, loss of CD1d resulted in the presence of even more TNF in the BAL fluid of BALB/c CD1d−/− mice. Taken together, the cytokine and chemokine data from the BAL fluid analyses suggested that enhanced recruitment of neutrophils to the lungs of BALB/c CD1d−/− mice was due to the enhanced production of chemokines (KC, MIP-1α, and MIP-2) and growth factors (GM-CSF) associated with neutrophil generation and recruitment. The identity of the relevant cell sources of these cytokines and chemokines is still unclear. Importantly, these differences were observed at a time when the numbers of viable PAO1 in the lungs of mice from all four strains were approximately similar.
Strain-dependent requirement for alveolar macrophages.
A previous report examining the role of NKT cells in Pseudomonas clearance concluded that NKT cells influenced the uptake of Pseudomonas by alveolar macrophages (11). Given the strain-dependent role for NKT cells reported here, we reasoned that there may be a similar strain-dependent role for macrophages in Pseudomonas clearance. To explore this issue, we used clodronate-loaded liposomes to deplete lung phagocytic cells, the majority of which are alveolar macrophages in the uninfected mouse. Examination of BAL fluid 24 h after a single administration of clodronate-loaded liposomes revealed an ∼87% decrease in the number of alveolar macrophages (data not shown).
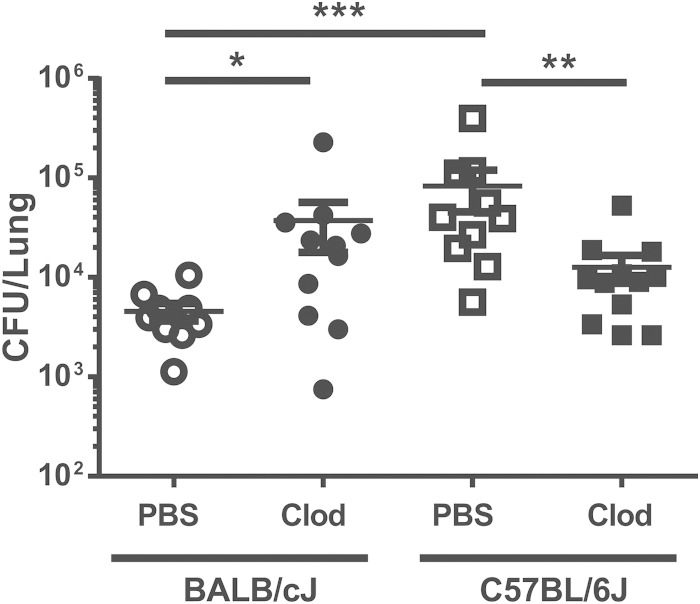
Next, we administered PBS-loaded or clodronate-loaded liposomes to either BALB/cJ or C57BL/6J mice 24 h prior to infection with P. aeruginosa PAO1 strain, and we assessed the number of PAO1 bacteria in the lungs 24 h after infection. These data demonstrated that depletion of alveolar macrophages significantly impaired PAO1 clearance in the lungs of BALB/cJ mice almost to the level seen with wild-type B6 mice (Fig. 6). In contrast, we found that depletion of alveolar macrophages significantly enhanced the rate of PAO1 clearance in the lungs of B6 mice in the same time frame (Fig. 6). These data indicated that alveolar macrophages played a role in PAO1 clearance in the BALB/c but not in the B6 mouse. If anything, the alveolar macrophages in the B6 mouse impaired the ability of B6 mice to clear P. aeruginosa from the lungs 24 h after infection.
FIG 6.
Strain-dependent effect of alveolar macrophage depletion. Mice were depleted of alveolar macrophages using clodronate (Clod)-loaded liposomes and infected with P. aeruginosa 24 h later. The numbers of live colonies in lungs 24 h after infection are shown. Each symbol represents a single mouse. Lines denote the mean CFU. The combined results of two separate depletions and infections are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
CD1d and the innate-like NKT cell subset have been demonstrated to play roles in a number of bacterial infections, including infections by P. aeruginosa and Streptococcus pneumoniae (11, 12, 27, 28). While variability in NKT cell number and function as a consequence of the host genetic background is now well appreciated (13–16, 18, 29), less attention has been paid to how this variability might affect the contribution of NKT cells in infectious-disease models. The data presented here reconcile two previous studies that reached seemingly opposite conclusions regarding the role of NKT cells in P. aeruginosa clearance by demonstrating that NKT cells make significant contributions to P. aeruginosa clearance in BALB/cJ mice but are completely dispensable in C57BL/6J mice. Moreover, by demonstrating a difference in the intrinsic cytokine-producing capability of C57BL/6 and BALB/c NKT cells, these data suggest the existence of a causative link between the disparate NKT cell functions and levels of clearance of P. aeruginosa from the lung in the two genetic backgrounds.
Strain-dependent differences in P. aeruginosa infection models are well documented (19, 25, 26, 30). Our data demonstrating that BALB/c mice are more resistant to Pseudomonas bacteria than C57BL/6/J mice in an acute model of infection corroborate previous reports of studies performed using a model of chronic Pseudomonas infection (19, 25, 26). Interestingly, in that model, proliferation of lung T cells from infected C57BL/6 mice was significantly lower than proliferation of T cells from infected BALB/c mice (19), which was similar to our observation of suppressed cytokine production by C57BL/6 NKT cells compared to BALB/c NKT cells. Taken together, these observations suggest that the NKT-intrinsic difference in cytokine production may not be unique to NKT cells but may be reflective of a genetically controlled difference common to all αβ T cells.
Previous evidence that host genetic backgrounds may dictate the role of NKT cells in infection can be seen in the differing pathologies that have resulted from infection of NKT cell-deficient C57BL/6 and BALB/c mice with Plasmodium berghei (31) or with Borrelia burgdorferi (32, 33). Examination of cerebral inflammation following infection with Plasmodium parasites revealed that a deficiency of CD1d resulted in more-severe disease in the disease-resistant BALB/c background, while a deficiency of CD1d in the disease-susceptible background appeared to afford a small measure of protection (31). In a model of Borrelia-induced Lyme disease, a deficiency of NKT cells in the normally disease-resistant C57BL/6 background resulted in more-severe carditis. In contrast, a deficiency in NKT cells in BALB/c mice manifested as a more severe arthritis. These observations suggest a complex interplay between the roles played by NKT cells in a given model of infectious disease and the complement of genetic loci governing disease susceptibility and pathogenesis. Even so, given the striking difference in NKT cell function between C57BL/6 and BALB/c mice reported here and the observation that elimination of CD1d-restricted T cells in the resistant BALB/c strain yielded lung P. aeruginosa CFU levels similar to that of the susceptible B6 strain, it is tempting to speculate that the strain-dependent differences in NKT cell function contribute to the differences in Pseudomonas clearance from the lungs in these strains. Additional investigation needs to be done, however, to determine whether this is the case.
The mechanism through which NKT cells contribute to clearance of P. aeruginosa in the BALB/c lung remains unclear. Nieuwenhuis et al. concluded that NKT cells contribute through enhancement of alveolar macrophage function, which could then influence neutrophil recruitment (11). This is consistent with data indicating that lung NKT cell activation can result in neutrophil recruitment to the lung (8) and that neutrophils are the dominant effector cells in the host response to P. aeruginosa (2). Yet researchers who have performed several studies that have directly examined the contribution of macrophages to lung Pseudomonas clearance have come to differing conclusions on their importance (2, 34, 35). Our data demonstrating that both CD1d-restricted NKT cells and alveolar macrophages contribute to Pseudomonas clearance in resistant BALB/c mice but that neither appears to contribute to Pseudomonas clearance in susceptible C57BL/6 mice support a model in which the functions of these leukocyte subsets are linked.
Our data differed from those Nieuwenhuis et al. in that we found no evidence of a reduction in the number of neutrophils in BAL fluid from BALB/c CD1d-deficient mice, despite the fact that αGalCer administration to lungs of BALB/c mice promotes efficient neutrophil recruitment to the lung airway (data not shown). Rather, we observed increased numbers of BAL fluid neutrophils in BALB/c mice that lacked CD1d and NKT cells. The reason for the differences between the two studies in the effect of NKT cells on neutrophil recruitment is unclear but may represent a difference between our study and that of Niewenhuis with respect to the strain of Pseudomonas that was used. Whereas we used the standard PAO1 laboratory strain, Niewenhuis et al. used the D4 strain, and it is possible that different P. aeruginosa strains may elicit slightly different host immune responses (36).
Interestingly, we observed increased neutrophil numbers 6 h after infection, when there was no difference in bacterial burden among the strains examined. Therefore, the increased number of neutrophils at this time point did not appear to be simply due to a higher P. aeruginosa burden in the lungs. In other words, although BALB/c CD1d−/− mice recruited more neutrophils to the lungs early in infection than their wild-type counterparts, these mice appeared impaired in their ability to clear P. aeruginosa from the lungs. The reason for this unusual observation is still unclear and requires additional investigation. It is, however, reminiscent of a previous report demonstrating that strain-dependent susceptibility to P. aeruginosa was associated with an exaggerated inflammatory response in the lungs (25). Moreover, in that study, the authors concluded that the response in susceptible C57BL/6J mice is dominated by neutrophils whereas the response in resistant BALB/c mice is dominated by macrophages (25), highlighting the strain-dependent variability in the innate immune response in these strains. Since NKT cell function impinges on each of these innate immune subsets, these observations raise the possibility that strain-dependent variability in NKT cell function could regulate these strain-dependent differences in neutrophil and macrophage function.
The results reported here reconcile disparate reports regarding the role of innate-like CD1d-restricted T cells in Pseudomonas clearance from the lung, and they highlight the importance of the consideration of host genetic background in evaluating the host response to P. aeruginosa infection. Future research will be needed to determine whether such a genetic predisposition also occurs in patients infected with P. aeruginosa. While clearance of Pseudomonas from the lungs is primarily dependent on the innate arm of the immune system, our data confirm the involvement of the adaptive arm of the immune system. Recognition of such a role could open up new avenues of treatment via manipulation of NKT cells using synthetic glycolipid agonists.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI067897 from the NIAID (J.E.B.), GM103496 (M.J.W.) from NIGMS, AI103003 (M.J.W.) from NIAID, and HL107291 (M.E.P.) from NHLBI.
We thank Julia Macauley for technical assistance and thank Collette Charland in the University of Vermont Flow Cytometry and Cell Sorting Facility for help in flow cytometry.
REFERENCES
- 1.Williams BJ, Dehnbostel J, Blackwell TS. 2010. Pseudomonas aeruginosa: host defence in lung diseases. Respirology 15:1037–1056. doi: 10.1111/j.1440-1843.2010.01819.x. [DOI] [PubMed] [Google Scholar]
- 2.Koh AY, Priebe GP, Ray C, Van Rooijen N, Pier GB. 2009. Inescapable need for neutrophils as mediators of cellular innate immunity to acute Pseudomonas aeruginosa pneumonia. Infect Immun 77:5300–5310. doi: 10.1128/IAI.00501-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavoie EG, Wangdi T, Kazmierczak BI. 2011. Innate immune responses to Pseudomonas aeruginosa infection. Microbes Infect 13:1133–1145. doi: 10.1016/j.micinf.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kronenberg M, Kinjo Y. 2009. Innate-like recognition of microbes by invariant natural killer T cells. Curr Opin Immunol 21:391–396. doi: 10.1016/j.coi.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juno JA, Keynan Y, Fowke KR. 2012. Invariant NKT cells: regulation and function during viral infection. PLoS Pathog 8:e1002838. doi: 10.1371/journal.ppat.1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. 1997. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science 278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 7.Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. 26 October 2003, posting date. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat Immunol doi: 10.1038/ni1002. [DOI] [PubMed] [Google Scholar]
- 8.Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, Wong CH, Schneider E, Dy M, Leite-de-Moraes MC. 2007. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med 204:995–1001. doi: 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoyagi T, Yamamoto N, Hatta M, Tanno D, Miyazato A, Ishii K, Suzuki K, Nakayama T, Taniguchi M, Kunishima H, Hirakata Y, Kaku M, Kawakami K. 2011. Activation of pulmonary invariant NKT cells leads to exacerbation of acute lung injury caused by LPS through local production of IFN-gamma and TNF-alpha by Gr-1+ monocytes. Int Immunol 23:97–108. doi: 10.1093/intimm/dxq460. [DOI] [PubMed] [Google Scholar]
- 10.Wintermeyer P, Cheng CW, Gehring S, Hoffman BL, Holub M, Brossay L, Gregory SH. 2009. Invariant natural killer T cells suppress the neutrophil inflammatory response in a mouse model of cholestatic liver damage. Gastroenterology 136:1048–1059. doi: 10.1053/j.gastro.2008.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieuwenhuis EE, Matsumoto T, Exley M, Schleipman RA, Glickman J, Bailey DT, Corazza N, Colgan SP, Onderdonk AB, Blumberg RS. 2002. CD1d-dependent macrophage-mediated clearance of Pseudomonas aeruginosa from lung. Nat Med 8:588–593. doi: 10.1038/nm0602-588. [DOI] [PubMed] [Google Scholar]
- 12.Kinjo T, Nakamatsu M, Nakasone C, Yamamoto N, Kinjo Y, Miyagi K, Uezu K, Nakamura K, Higa F, Tateyama M, Takeda K, Nakayama T, Taniguchi M, Kaku M, Fujita J, Kawakami K. 2006. NKT cells play a limited role in the neutrophilic inflammatory responses and host defense to pulmonary infection with Pseudomonas aeruginosa. Microbes Infect 8:2679–2685. doi: 10.1016/j.micinf.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 13.Aktan I, Chant A, Borg ZD, Damby DE, Leenstra PC, Lilley GW, Petty J, Suratt BT, Teuscher C, Wakeland EK, Poynter ME, Boyson JE. 2010. Slam haplotypes modulate the response to lipopolysaccharide in vivo through control of NKT cell number and function. J Immunol 185:144–156. doi: 10.4049/jimmunol.0902658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borg ZD, Benoit PJ, Lilley GW, Aktan I, Chant A, DeVault VL, Rincon M, Boyson JE. 2014. Polymorphisms in the CD1d promoter that regulate CD1d gene expression are associated with impaired NKT cell development. J Immunol 192:189–199. doi: 10.4049/jimmunol.1301451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyson JE, Nagarkatti N, Nizam L, Exley MA, Strominger JL. 2006. Gestation stage-dependent mechanisms of invariant natural killer T cell-mediated pregnancy loss. Proc Natl Acad Sci U S A 103:4580–4585. doi: 10.1073/pnas.0511025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rymarchyk SL, Lowenstein H, Mayette J, Foster SR, Damby DE, Howe IW, Aktan I, Meyer RE, Poynter ME, Boyson JE. 2008. Widespread natural variation in murine natural killer T-cell number and function. Immunology 125:331–343. doi: 10.1111/j.1365-2567.2008.02846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan MA, Fletcher JM, Pellicci D, Baxter AG. 2007. Slamf1, the NKT cell control gene Nkt1. J Immunol 178:1618–1627. doi: 10.4049/jimmunol.178.3.1618. [DOI] [PubMed] [Google Scholar]
- 18.Esteban LM TT, Jordan MA, Roach D, Poulton LD, Brooks A, Naidenko OV, Sidobre S, Godfrey DI, Baxter AG. 2003. Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol 171:2873–2878. doi: 10.4049/jimmunol.171.6.2873. [DOI] [PubMed] [Google Scholar]
- 19.Stevenson MM, Kondratieva TK, Apt AS, Tam MF, Skamene E. 1995. In vitro and in vivo T cell responses in mice during bronchopulmonary infection with mucoid Pseudomonas aeruginosa. Clin Exp Immunol 99:98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaBauve AE, Wargo MJ. 2014. Detection of host-derived sphingosine by Pseudomonas aeruginosa is important for survival in the murine lung. PLoS Pathog 10:e1003889. doi: 10.1371/journal.ppat.1003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wargo MJ, Gross MJ, Rajamani S, Allard JL, Lundblad LK, Allen GB, Vasil ML, Leclair LW, Hogan DA. 2011. Hemolytic phospholipase C inhibition protects lung function during Pseudomonas aeruginosa infection. Am J Respir Crit Care Med 184:345–354. doi: 10.1164/rccm.201103-0374OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smiley ST, Kaplan MH, Grusby MJ. 1997. Immunoglobulin E production in the absence of interleukin-4-secreting CD1-dependent cells. Science 275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 23.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. 1997. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science 278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 24.McClellan SA, Huang X, Barrett RP, van Rooijen N, Hazlett LD. 2003. Macrophages restrict Pseudomonas aeruginosa growth, regulate polymorphonuclear neutrophil influx, and balance pro- and anti-inflammatory cytokines in BALB/c mice. J Immunol 170:5219–5227. doi: 10.4049/jimmunol.170.10.5219. [DOI] [PubMed] [Google Scholar]
- 25.Sapru K, Stotland PK, Stevenson MM. 1999. Quantitative and qualitative differences in bronchoalveolar inflammatory cells in Pseudomonas aeruginosa-resistant and -susceptible mice. Clin Exp Immunol 115:103–109. doi: 10.1046/j.1365-2249.1999.00762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam M, Snipes GJ, Stevenson MM. 1999. Characterization of chronic bronchopulmonary Pseudomonas aeruginosa infection in resistant and susceptible inbred mouse strains. Am J Respir Cell Mol Biol 20:710–719. doi: 10.1165/ajrcmb.20.4.3223. [DOI] [PubMed] [Google Scholar]
- 27.Cohen NR, Garg S, Brenner MB. 2009. Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv Immunol 102:1–94. doi: 10.1016/S0065-2776(09)01201-2. [DOI] [PubMed] [Google Scholar]
- 28.Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, Li Y, Imamura M, Kaneko Y, Okawara A, Miyazaki Y, Gomez-Velasco A, Rogers P, Dahesh S, Uchiyama S, Khurana A, Kawahara K, Yesilkaya H, Andrew PW, Wong CH, Kawakami K, Nizet V, Besra GS, Tsuji M, Zajonc DM, Kronenberg M. 2011. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol 12:966–974. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan MA, Fletcher J, Baxter AG. 2004. Genetic control of NKT cell numbers. Immunol Cell Biol 82:276–284. doi: 10.1111/j.0818-9641.2004.01264.x. [DOI] [PubMed] [Google Scholar]
- 30.Jensen PO, Moser C, Kobayashi O, Hougen HP, Kharazmi A, Hoiby N. 2004. Faster activation of polymorphonuclear neutrophils in resistant mice during early innate response to Pseudomonas aeruginosa lung infection. Clin Exp Immunol 137:478–485. doi: 10.1111/j.1365-2249.2004.02554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen DS, Siomos MA, Buckingham L, Scalzo AA, Schofield L. 2003. Regulation of murine cerebral malaria pathogenesis by CD1d-restricted NKT cells and the natural killer complex. Immunity 18:391–402. doi: 10.1016/S1074-7613(03)00052-9. [DOI] [PubMed] [Google Scholar]
- 32.Tupin E, Benhnia MR, Kinjo Y, Patsey R, Lena CJ, Haller MC, Caimano MJ, Imamura M, Wong CH, Crotty S, Radolf JD, Sellati TJ, Kronenberg M. 2008. NKT cells prevent chronic joint inflammation after infection with Borrelia burgdorferi. Proc Natl Acad Sci U S A 105:19863–19868. doi: 10.1073/pnas.0810519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson CM Jr, Bates TC, Izadi H, Radolf JD, Huber SA, Boyson JE, Anguita J. 2009. Local production of IFN-gamma by invariant NKT cells modulates acute Lyme carditis. J Immunol 182:3728–3734. doi: 10.4049/jimmunol.0804111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kooguchi K, Hashimoto S, Kobayashi A, Kitamura Y, Kudoh I, Wiener-Kronish J, Sawa T. 1998. Role of alveolar macrophages in initiation and regulation of inflammation in Pseudomonas aeruginosa pneumonia. Infect Immun 66:3164–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung DO, Halsey K, Speert DP. 2000. Role of pulmonary alveolar macrophages in defense of the lung against Pseudomonas aeruginosa. Infect Immun 68:4585–4592. doi: 10.1128/IAI.68.8.4585-4592.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, Zhang H, Song Y, Lynch SV, Lowell CA, Wiener-Kronish JP, Caughey GH. 2012. Strain-dependent induction of neutrophil histamine production and cell death by Pseudomonas aeruginosa. J Leukoc Biol 91:275–284. doi: 10.1189/jlb.0711356. [DOI] [PMC free article] [PubMed] [Google Scholar]