Abstract
Erythrocytes infected with malaria parasites have increased permeability to ions and nutrients, as mediated by the plasmodial surface anion channel (PSAC) and recently linked to parasite clag3 genes. Although the encoded protein is integral to the host membrane, its precise contribution to solute transport remains unclear because it lacks conventional transmembrane domains and does not have homology to ion channel proteins in other organisms. Here, we identified a probable CLAG3 transmembrane domain adjacent to a variant extracellular motif. Helical-wheel analysis revealed strict segregation of polar and hydrophobic residues to opposite faces of a predicted α-helical transmembrane domain, suggesting that the domain lines a water-filled pore. A single CLAG3 mutation (A1210T) in a leupeptin-resistant PSAC mutant falls within this transmembrane domain and may affect pore structure. Allelic-exchange transfection and site-directed mutagenesis revealed that this mutation alters solute selectivity in the channel. The A1210T mutation also reduces the blocking affinity of PSAC inhibitors that bind on opposite channel faces, consistent with global changes in channel structure. Transfected parasites carrying this mutation survived a leupeptin challenge significantly better than a transfection control did. Thus, the A1210T mutation contributes directly to both altered PSAC activity and leupeptin resistance. These findings reveal the molecular basis of a novel antimalarial drug resistance mechanism, provide a framework for determining the channel's composition and structure, and should guide the development of therapies targeting the PSAC.
INTRODUCTION
The human malaria parasite Plasmodium falciparum remodels its host erythrocyte by exporting many proteins, generating membranous structures in the host cytosol, and increasing erythrocyte permeability to many solutes. Studies by multiple groups have determined that anions, sugars, purines, organic cations, and some vitamins have increased permeability after infection (1–4). The increase in permeability is primarily mediated by a parasite-derived ion and nutrient channel known as the plasmodial surface anion channel (PSAC) (5). Importantly, both PSAC single-channel properties and the relative increases in solute permeabilities are conserved in divergent malaria parasites (6). Because Babesia parasites do not induce PSAC-like activity in erythrocytes that they invade (7), this channel is thought to be restricted to the genus Plasmodium.
The clag multigene family, also conserved in and restricted to malaria parasites (8), has recently been linked to PSAC activity (9–11). Two paralogs on parasite chromosome 3, known as clag3.1 and clag3.2, appear to play a critical role, as identified by genetic mapping experiments with ISPA-28, an isolate-specific PSAC antagonist that blocks channels on the Dd2 laboratory clone but is ineffective against channels from other lines. These genes undergo epigenetic switching to encode a single protein that is initially packaged in specialized organelles known as rhoptries (12, 13). The protein is trafficked to the host erythrocyte membrane by poorly understood mechanisms (9, 14–17), consistent with a role in PSAC activity. However, because the protein lacks homology to known ion channels in other systems, it is not clear how the CLAG3 protein contributes to host cell permeability changes (18). It may contribute directly to pore formation, either in isolation or by interacting with as-yet-unknown channel subunits (18). Alternatively, CLAG3 may have only a modulatory role, such as through enzymatic activation of quiescent channels in the host membrane (19). Distinguishing between these models is an important step in understanding the structural basis of solute and nutrient transport at the host membrane and in determining whether the channel can be targeted for antimalarial drug development (10, 20).
One way to address this uncertainty involves molecular studies with available PSAC mutants that have been generated through in vitro selection with toxins that require channel-mediated uptake (11, 21–24). A leupeptin-resistant clone, HB3-leuR1, carries a nonsynonymous mutation in the clag3.2 gene. Because this parasite preferentially expresses this mutant allele, this parasite line expresses a modified CLAG protein with a single A1210T mutation. However, because in vitro selection with leupeptin may lead to multiple genome level changes, this mutation may be only coincidental with altered erythrocyte permeability. Here, we have examined the A1210T mutation to gain insights into the roles played by CLAG3. Our computational analyses suggest that the A1210 residue is located at a critical site within an amphipathic transmembrane domain capable of lining a water-filled pore. DNA transfection experiments to introduce the A1210T mutation provide experimental evidence supporting a direct contribution of CLAG3 to the formation of water-filled pores at the host membrane.
MATERIALS AND METHODS
Parasite cultivation and growth inhibition.
The P. falciparum HB3 clone and transfectant lines were cultured under standard conditions with O+ human erythrocytes (Interstate Blood Bank) and RPMI 1640 medium supplemented with 0.5% NZ microbiological bovine serum albumin (BSA; MP Biomedicals). Cultures were harvested at the trophozoite stage and enriched to >95% parasitemia by the Percoll-sorbitol method.
In vitro parasite growth with leupeptin was assessed by SYBR green I detection of parasite DNA as described previously (20), with modifications. Briefly, synchronized ring-stage parasite cultures were dispensed into 96-well microplates at 0.5% parasitemia and 2.5% hematocrit in the above-described medium with the concentrations of leupeptin indicated (see Fig. 5). After cultivation at 37°C for 5 days with a single medium change after 3 days, the cultures were lysed by the addition of an equal volume of 20 mM Tris–10 mM EDTA–1.6% Triton X-100–0.016% saponin, pH 7.5, with SYBR green I nucleic acid gel stain (Invitrogen) at a 2,500-fold dilution. After a 45-min incubation, DNA content was quantified by fluorescence measurements (excitation wavelength of 485 nm, emission wavelength of 528 nm). Normalized percent growth was determined by using matched controls seeded with no inhibitor and with 20 μM chloroquine. Similar results were obtained in growth inhibition studies that used microscopic examination of Giemsa-stained smears.
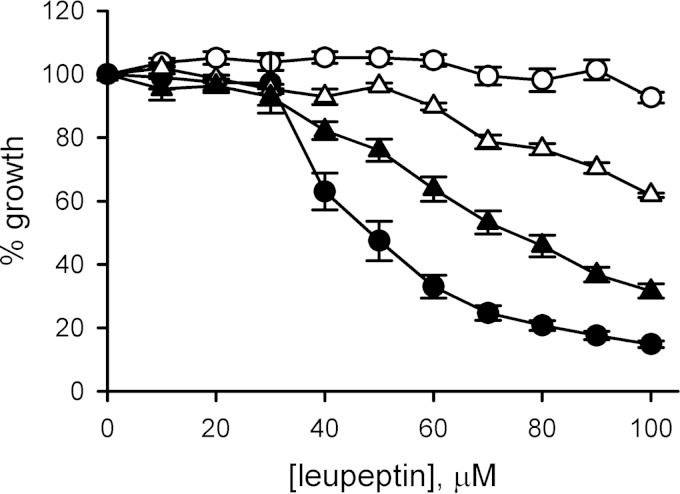
FIG 5.
The A1210T mutation contributes to leupeptin resistance. Leupeptin dose responses for in vitro growth over 5 days. Symbols represent the mean ± the standard error of the mean of replicates from four independent trials for HB3 (black circles), HB3-c3A1210T (white triangles), HB3-3rec (black triangles), and HB3-leuR1 (white circles) parasites.
Site-directed mutagenesis and DNA transfection.
Previously described plasmid pHD22Y-120w-flag-PG1 (9) was used as the template for site-directed mutagenesis with complementary primers (5′-ATGGTTTCATGTATACTTTTTGTTTTTTTGC-3′ and 5′-GCAAAAAAACAAAAAGTATACATGAAACCAT-3′). These primers carry a single desired mutation (underlined) that changes a conserved alanine at residue 1210 to threonine. Whole-plasmid PCR was performed with PfuUltra Hotstart DNA polymerase (Stratagene); following DpnI digestion, the product was used to transform chemically competent TOP10 Escherichia coli (Invitrogen). DNA sequencing confirmed the successful introduction of the mutation. The resulting plasmid was electroporated into uninfected erythrocytes and used for allelic-exchange transfection of HB3 parasites. The transfected culture was selected with 2.5 nM WR99210 and screened for homologous recombination by PCR. Integration into the clag3.1 gene was detected after 6 months; the HB3-c3A1210T clone was generated by limiting dilution (25).
Southern blot assay.
Parasite genomic DNA was digested with the enzyme HindIII, separated on a 0.8% agarose gel at 40 V, and transferred to a nylon membrane (GE Healthcare). Integration was detected with a DNA probe against the hdhfr gene with primers 5′-ATTTCCAGAGAATGACCACAAC-3′ and 5′-TTAAGATGGCCTGGGTGATTC-3′. The probe was labeled with digoxigenin-dUTP according to the manufacturer's instructions (DIG DNA Labeling and Detection kit; Roche), hybridized to the blot overnight at 39°C, and detected with allophycocyanin-conjugated anti-digoxigenin antibody and CDP-Star substrate (Roche).
Quantitative RT-PCR.
mRNA was isolated with TRIzol reagent (Invitrogen), treated with DNase, and used for reverse transcription (RT) to cDNA by using oligo(dT) priming and SuperScript III (Invitrogen). The resulting cDNA was used for quantitative real-time PCR to determine clag3 expression levels with the iCycler iQ multicolor real-time PCR system (Bio-Rad) as described previously (24). Omission of reverse transcriptase or the cDNA template was performed with each experiment to exclude contamination by genomic DNA. The results for each gene were normalized to the constitutively expressed parasite gene PF07_0073. A conserved forward primer that recognizes all clag3 genes (5′-ACCCATAACTACATATTTTCTAGTAATG-3′ [p1; see Fig. 2A]) was used with specific primers for each paralog (HB3 clag3.2, 5′-TTATAACCATTAGGAGCACTACTTTC-3′ [p2]; Dd2 clag3.1, 5′-GACAAGTTCCAGAAGCATCCT-3′ [p3]; truncated HB3 clag3.1, 5′-AGATTTAGTTACACTTGAAGAATTA-3′ [p4]). All reactions were done in triplicate; the average cycle threshold (CT) was used to quantify expression by the 2−ΔΔCT method (26).
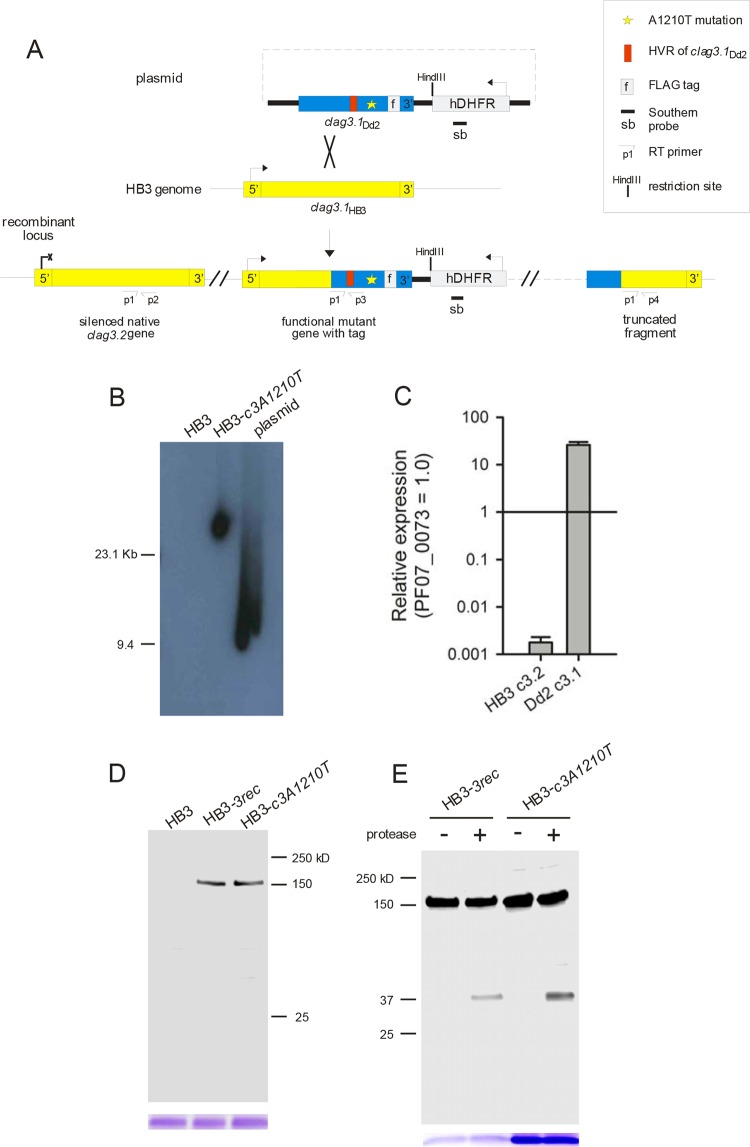
FIG 2.
Allelic-exchange transfection and expression of CLAG3 carrying the A1210T mutation. (A) Schematic showing the integration plasmid (top) and the result of allelic-exchange transfection to replace the wild-type HB3 clag3.1 gene with Dd2 clag3.1 carrying a nonsynonymous mutation encoding threonine at residue 1210. The positions of the primers (see Materials and Methods) and the probe used for Southern blotting are shown. (B) Southern blot assay with genomic DNA from the clones indicated or the integration plasmid after HindIII digestion. Plasmid size, 9.4 kb. While the hdhfr-specific probe does not recognize DNA from HB3, a single integration site is apparent in HB3-c3A1210T. (C) Mean ± the standard error of the mean transcript abundance of the genes indicated in HB3-c3A1210T from RT-PCR experiments with normalization to the PF07_0073 control. (D) Immunoblot assay of whole-cell lysates of cells infected with the parasites indicated probed with anti-FLAG antibody. A single band of the expected size (∼150 kDa) is apparent in HB3-c3A1210T and HB3-3rec. Coomassie blue staining of hemoglobin (used as a loading control) is shown at the bottom. (E) Immunoblot assay of the membrane fraction harvested from cells with or without extracellular pronase E treatment. Anti-FLAG antibody recognizes a C-terminal ∼37-kDa cleavage product in both transfectants after pronase E treatment. Loading control, Coomassie blue staining of hemoglobin.
Immunoblot assay.
Enriched trophozoite-stage-infected erythrocytes were lysed in 7.5 mM Na2HPO4–1 mM EDTA, pH 7.5, and dissolved in Laemmli sample buffer with 6% SDS. Proteins were separated on 4 to 15% Mini-PROTEAN TGX gels and transferred to nitrocellulose membrane (Bio-Rad). After blocking (Tris-buffered saline with 3% skim milk and 0.1% Tween 20), the blot was probed with anti-Flag antibody before detection with a horseradish peroxidase-conjugated anti-mouse secondary antibody (Santa Cruz) and an enhanced-chemiluminescence substrate. Matched loading was confirmed by Coomassie blue staining of gels for hemoglobin.
Where used, protease treatment was performed with enriched trophozoite-infected cells suspended at 2.5% hematocrit in 150 mM NaCl–20 mM Na2HPO4–0.6 mM CaCl2–1 mM MgCl2, pH 7.4. Freshly dissolved pronase E was added at 0.5 mg/ml; after incubation for 1 h at 37°C, protease activity was terminated by washes with ice-cold buffer and 1 mM phenylmethylsulfonyl fluoride. Hypotonic lysis (7.5 mM Na2HPO4, 1 mM EDTA, pH 7.5) and ultracentrifugation (100,000 × g, 1 h) were used to harvest the membrane fraction for SDS-PAGE and immunoblotting. A no-protease control was processed identically.
PSAC transport assays.
Solute uptake by infected cells was measured by continuous tracking of infected-cell lysis as described previously (20). Trophozoite-stage-infected cells were enriched with the Percoll-sorbitol method, washed, and resuspended in 150 mM NaCl–20 mM Na-HEPES–0.1 mg/ml BSA, pH 7.4. Solute uptake was initiated by adding 20 volumes of buffered osmotic-lysis solution (280 mosM permeant solute, 20 mM Na-HEPES, 0.1 mg/ml BSA, pH 7.4); PSAC inhibitors, where present, were added to this solution. The transmittance of 700-nm light through the cell suspension was continuously recorded to monitor the kinetics of infected-cell lysis and PSAC-mediated uptake. The solutes examined here do not produce osmotic lysis of uninfected erythrocytes (27). The time to a threshold level of osmotic lysis was determined by linear interpolation of recordings and used to estimate transport rates; untransfected cells and no-inhibitor controls were evaluated identically in each experiment.
RESULTS
Computational analysis.
The A1210T mutation in HB3-leuR1 CLAG3 is near the protein's C terminus, distal to a 10- to 30-residue motif that varies between P. falciparum lines (28, 29). Functional proteolysis studies found that this variable motif is exposed at the erythrocyte surface and that the CLAG3 C terminus is intracellular (16), implicating one or more transmembrane domains within this segment (Fig. 1A). Consistent with this prediction, this 35-kDa segment cannot be liberated from membranes by NaCO3 extraction (9). Although TMHMM 2.0 failed to identify a transmembrane domain here or at any other sites within the CLAG3 protein (30), TopPred II and Polyphobius detected a single possible membrane-spanning domain at residues 1203 to 1223 (31, 32), encompassing the A1210T mutation in HB3-leuR1 parasites. This putative transmembrane domain is conserved in P. falciparum lines and scores poorly in transmembrane prediction algorithms because there are multiple polar residues interspersed among a local enrichment of hydrophobic residues (Fig. 1B, yellow highlighting); a similar amphipathic arrangement is also apparent within this region of CLAG3 orthologs from distantly related malaria parasites.
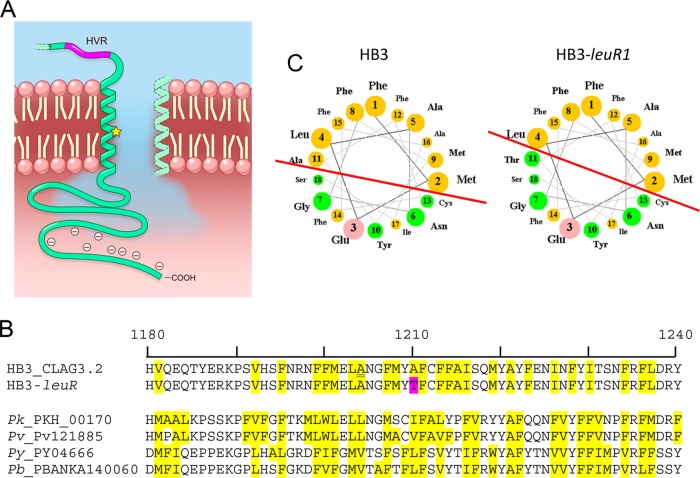
FIG 1.
A predicted amphipathic transmembrane domain near the CLAG3 C terminus. (A) Schematic showing an α-helical transmembrane domain distal to a hypervariable motif (HVR). This domain may line a water-filled pore. The A1210T mutation is indicated by a yellow star. The C-terminal cytoplasmic tail of CLAG3 is enriched with negatively charged residues (49). (B) Alignment of partial CLAG3.2 sequences from HB3 and the HB3-leuR1 PSAC mutant with the A1210T mutation highlighted in pink. The region shown is strictly conserved in P. falciparum lines on the basis of available CLAG3.1 and CLAG3.2 sequences from the 3D7, 7G8, Dd2, FVO, and HB3 lines, except for a single conservative polymorphism (A1204V, underlined). Hydrophobic residues (yellow highlighting) alternate with polar residues; this amphipathic pattern is conserved in CLAG3 orthologs in P. knowlesi, P. vivax, P. yoelii, and P. berghei, as shown in the alignment at the bottom. (C) Helical-wheel projections of the predicted transmembrane domain in HB3 and HB3-leuR1. The selected helix start site (F1190) yields strict segregation of hydrophilic residues to one face, with green and pink circles representing polar uncharged and charged residues, respectively. The other helix face has only hydrophobic residues (orange circles). Note that the red line marking the interface between the two faces has a steeper slope in HB3-leuR1 because of the A1210T mutation. This change in angle may alter the transmembrane domain orientation in the erythrocyte membrane.
To examine this putative transmembrane domain, we obtained helical-wheel projections (Fig. 1C). This approach, which assumes an α-helical domain with 3.6 residues/turn (33), revealed that all of the polar residues lie on one face of the helix (HB3 CLAG3.2, Fig. 1C); a glutamate, the single charged residue within the predicted transmembrane domain, is centered on this face of the helix. The other face of the helix consists exclusively of hydrophobic residues. Complete segregation of these residues suggests a polar helix face that lines a water-filled pore and a hydrophobic face embedded in a lipid membrane, as described in many well-characterized ion channels (34, 35). Notably, hydrophobic A1210 sits at the interface between the polar and hydrophobic faces; mutation to a polar threonine would alter the boundary between these faces and may change the tilt of the helix within the lipid bilayer (Fig. 1 C, red lines). If this predicted transmembrane domain lines the PSAC pore, this mutation may also affect channel function. Consistent with this possibility, functional studies have determined that the HB3-leuR1 mutant produces defective channels with altered solute selectivity, gating, and transport rates (22, 23).
Allelic exchange.
To directly explore the role of the A1210T mutation in channel function and leupeptin resistance, we designed an allelic-exchange strategy to replace A1210 in the wild-type HB3 parasite. We used site-directed mutagenesis to introduce the A1210T mutation on the pHD22Y-120w-flag-PG1 plasmid, which carries a 3.2-kb coding fragment from the 3′ end of the clag3.1 gene from the Dd2 parasite (Fig. 2A, where the mutant site is indicated by a yellow star). A C-terminal FLAG epitope tag is present on this plasmid in frame with the clag3.1 gene; after a stop codon, the gene is terminated with the 3′ untranslated region of clag3.1. Allelic exchange with this plasmid without the introduction of mutations into HB3 has previously generated the HB3-3rec parasite clone; this replacement conferred channel inhibition by ISPA-28, a PSAC blocker effective against Dd2 but not HB3 channels (9). ISPA-28 is thought to bind and block the channel at a small hypervariable motif on a CLAG3 extracellular loop (red shading, Fig. 1A and 2A); Dd2 CLAG3.1 carries a distinct sequence at this motif (16).
We transfected HB3 with this plasmid after introducing the A1210T mutation and selected for resistance to WR99210, as encoded by hdhfr on the plasmid. After several months, PCR screening revealed homologous recombination and integration into the HB3 clag3.1 gene. Limiting-dilution cloning was then used to obtain the HB3-c3A1210T clone, which carries a single integration event without residual episomes (Fig. 2B). Consistent with our transfection strategy, PCR and DNA sequencing indicated that this integration occurred by homologous recombination into HB3 clag3.1. DNA sequencing also implicated a recombination event between single-nucleotide polymorphisms at 3,140 and 3,577 bp from the HB3 clag3.1 start codon, conferring the entire hypervariable motif from the Dd2 clag3.1 gene and the engineered A1210T mutation.
As with most P. falciparum lines (28, 36), the transfectant parasite carries two clag3 genes. Individual parasites in a culture express only one of these two genes and silence the other through epigenetic mechanisms (13, 37). The HB3-c3A1210T clone was selected for our studies in part because it preferentially expresses the transfected clag3.1 gene and silences its native clag3.2 gene, as determined by quantitative RT-PCR (Fig. 2C). Preferential expression of one clag3 gene has been observed in a number of parasite lines (37), but the reasons for this bias remain unclear. In the present studies, it facilitated direct examination of the A1210T mutation and its effects on PSAC function.
To further examine clag3 expression in the transfectant, we performed immunoblotting with antibodies against the C-terminal FLAG epitope tag on the mutant allele. While this antibody did not recognize lysates from untransfected parasites, it detected a single ∼150-kDa band in HB3-c3A1210T and the HB3-3rec transfection control (Fig. 2D), as expected for the full-length CLAG3 protein. Treatment of these cells with extracellular pronase E confirmed cleavage within the exposed hypervariable domain, as indicated by an ∼37-kDa fragment that includes the C-terminal FLAG tag (Fig. 2E). The comparable fractional hydrolysis of the two transfectant lines suggests that the A1210T mutation does not alter the efficiency of CLAG3 trafficking to the host membrane.
Changes in PSAC activity directly associated with the A1210T mutation.
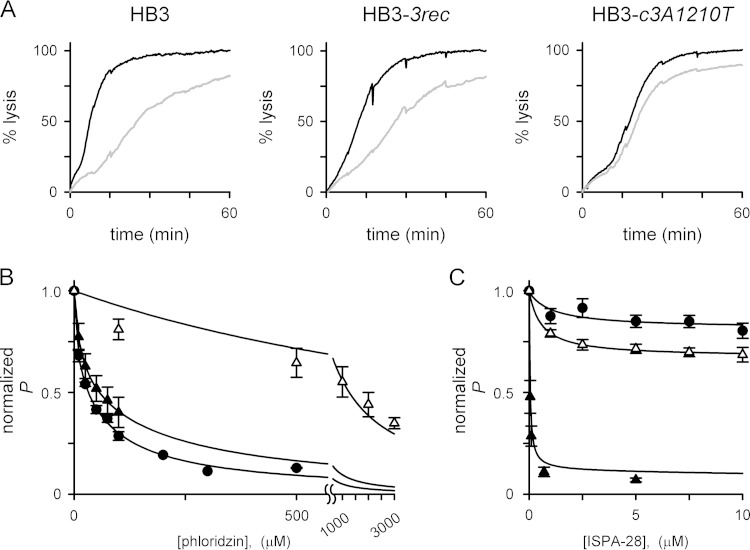
We next characterized the HB3-c3A1210T transfectant with transport studies and first measured osmotic-lysis kinetics in permeant organic solutes, an approach that correlates quantitatively with tracer flux and patch-clamp studies of this channel (20, 38). The HB3-c3A1210T parasite exhibited slower uptake of sorbitol than HB3 or the HB3-3rec control transfectant line without the A1210T mutation (Fig. 3A). The osmotic-lysis half time, which is inversely proportional to PSAC sorbitol permeability (38), was significantly increased in HB3-c3A1210T (15.7 ± 1.2 min versus 7.1 ± 0.5 and 9.3 ± 1.3 min for HB3 and HB3-3rec, respectively; P < 10−3 for HB3-c3A1210T comparison to HB3-3rec and no significant difference between HB3 and HB3-3rec [Student's t test, n = 6 to 14 trials each]). This reduction in sorbitol permeability matches that originally described for HB3-leuR1, which carries the same mutation as a result of in vitro selection with leupeptin (22). Thus, the A1210T mutation adequately accounts for reduced sorbitol permeability in leupeptin-resistant mutant parasites.
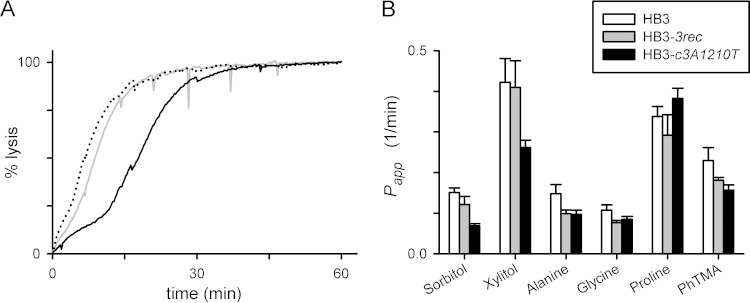
FIG 3.
The A1210T mutation alters PSAC solute selectivity. (A) Osmotic-lysis kinetics in sorbitol of HB3, HB3-3rec, and HB3-c3A1210T (dotted, gray, and black traces, respectively). Note that lysis of HB3-c3A1210T is slower, indicating reduced sorbitol permeability relative to that of wild-type HB3 or the HB3-3rec transfection control. (B) Mean ± the standard error of the mean apparent permeabilities (Papp) to the solutes indicated, determined as the reciprocal of lysis half times from kinetic experiments as in panel A (n = 4 to 14 trials for each solute and parasite). The permeability of HB3-c3A1210T to sorbitol and xylitol is significantly lower than that of the HB3-3rec transfection control, but other solutes did not produce significant differences between these parasites.
We also examined permeability to other solutes to explore possible changes in solute selectivity (Fig. 3B). Xylitol, a five-carbon sugar alcohol to which the PSAC is highly permeable, also exhibited reduced uptake with the A1210T mutation (P < 10−3 in comparison to HB3-3rec). Interestingly, however, permeability of HB3-c3A1210T to alanine, glycine, proline, and the organic cation phenytrimethylammonium, solutes with known uptake via the PSAC (3, 27, 39), did not differ from that of the HB3-3rec transfection control carrying the wild-type A1210 sequence (P > 0.05). Similar changes in solute permeability in these two transfectant lines relative to the wild-type HB3 parental line presumably reflect the effect of CLAG3 polymorphisms carried by the Dd2 clag3.1 gene used for these allelic-exchange experiments. The A1210T mutation's effect on the uptake of sugar alcohols but not of other solutes suggests that this residue is a determinant of PSAC selectivity. This finding is consistent with the residue's location within a putative pore-lining domain (Fig. 1).
We next examined the effects of the A1210T mutation on PSAC inhibition by known inhibitors because leupeptin resistance is associated with reduced PSAC blocking by phloridzin (22), a nonspecific transport inhibitor (40). Phloridzin blocking in HB3-3rec was indistinguishable from that measured in the untransfected HB3 parent (Fig. 4A and B), consistent with conserved affinity for this inhibitor in diverse parasite lines (41, 42). In contrast, the A1210T transfection mutant yielded channels with significantly reduced half-maximal affinity for phloridzin (K0.5 = 35, 47, and 1,260 μM for HB3, HB3-3rec, and HB3-c3A1210T, respectively; P ≤ 0.001 for comparison of HB3-c3A1210T to both other lines at 100 μM inhibitor and no significant difference between HB3-3rec and HB3 [n = 4 to 8 each]). Thus, the A1210T mutation adequately accounts for reduced phloridzin affinity in the mutant generated by in vitro selection with leupeptin (22).
FIG 4.
Altered PSAC pharmacology in the A1210T site-directed mutant. (A) Osmotic-lysis kinetics of the parasite lines indicated with or without 100 μM phloridzin (gray and black traces, respectively). Note the reduced inhibition in HB3-c3A1210T. (B) Phloridzin dose responses for PSAC inhibition in cells infected with HB3, HB3-3rec, and HB3-c3A1210T (circles, black triangles, and white triangles, respectively). Symbols represent the mean ± the standard error of the mean permeability (P) determined in up to eight trials at each concentration and normalized to 1.0 with no inhibitor. Allelic exchange with wild-type Dd2 clag3.1 does not affect phlordizin blocking, but addition of the A1210T mutation yields reduced inhibitor affinity. (C) Mean ± the standard error of the mean ISPA-28 inhibition dose responses (n = 5 trials for each concentration and parasite). Dd2 clag3.1 confers potent ISPA-28 blocking on HB3-3rec (black triangles); the HB3-c3A1210T parasite expresses the same clag3 chimeric gene through allelic exchange but has lost the potent ISPA-28 blocking because of the A1210T mutation (white triangles). HB3 channels are not blocked (circles). The solid lines in panels B and C represent best fits to the equation y = {a/[1 + (x/c)]} + {[1 − a]/[1 + (x/b)]}.
We also examined ISPA-28, a specific PSAC inhibitor that blocks channels on erythrocytes infected with Dd2 but is inactive against those from other lines, including HB3 (9). ISPA-28 blocking is conferred by the Dd2 clag3.1 product, which carries a distinct variant sequence at residues 1113 to 1143. Because our allelic-exchange transfection produced a chimeric clag3 gene product with this motif, we predicted that both transfectants would yield channels blocked by ISPA-28. HB3-3rec parasites indeed exhibit ISPA-28 affinity that quantitatively matches the blocking in Dd2 parasites (Fig. 4C, black triangles) (9). In contrast, HB3-c3A1210T channels were poorly blocked by ISPA-28, even though they carry the same variant motif on their extracellular face (P < 0.01 in comparisons to either HB3 or HB3-3rec at 5 μM ISPA-28 [n = 5 trials each]). Thus, the A1210T mutation leads to global changes in channel structure that affect both solute selectivity and blocking by PSAC inhibitors with distinct binding sites on the channel.
The A1210T mutation contributes directly to leupeptin resistance.
To evaluate the effect of A1210T on parasite leupeptin resistance, we performed growth inhibition studies with wild-type HB3, the transfectant HB3-c3A1210T, the HB3-3rec transfection control, and the resistant HB3-leuR1 clone generated by in vitro selection. Because HB3 parasites can become resistant within 2 months of continuous leupeptin pressure, it was important to evaluate survival and growth in short-term cultures and to use transfectant cultures with no prior exposure to this protease inhibitor. In a 5-day leupeptin challenge reported previously (22), dose-response studies revealed that HB3-c3A1210T parasites tolerated significantly higher concentrations of leupeptin than HB3-3rec (Fig. 5, P < 10−6 based on comparison at 100 μM leupeptin [n = 12 replicates from four independent trials each]). At the same time, HB3-c3A1210T was more sensitive to leupeptin than was the selected HB3-leuR1 mutant (P < 10−6), implying that additional genetic or epigenetic changes are required for greater resistance to leupeptin. These additional changes could be in either unidentified PSAC subunits or modulatory components; alternatively, they may be in one or more parasite targets of leupeptin.
Interestingly, these experiments also revealed that the HB3-3rec transfection control tolerated leupeptin better than the wild-type HB3 parasite did (50% inhibitory concentrations of 72 ± 6 and 49 ± 5 μM, respectively; P = 0.04). This might reflect an effect of selection for the hdhfr selectable marker with WR99210; it might also be due to altered leupeptin uptake in this transfection control, which expresses a chimeric CLAG3 protein that combines polymorphisms from HB3 and Dd2 parasites.
DISCUSSION
CLAG3 proteins have recently been implicated in channel-mediated ion and nutrient transport at the host membrane of infected erythrocytes. Evidence has included genetic mapping with the small-molecule PSAC inhibitor ISPA-28 and quantitative effects of externally applied proteases (9, 16), selection of recombinant parasites cultivated in nutrient-restricted media (10), and molecular studies to characterize blasticidin S-resistant parasites (11, 24). Although these multiple lines of evidence implicate this gene family in nutrient uptake, the precise structural contribution of the encoded CLAG3 protein is uncertain. A clear understanding of the role of CLAG3 has been limited by the lack of homology to known ion channels and a relative paucity of hydrophobic regions for the formation of transmembrane domains. Additional complexities include the presence of multiple CLAG paralogs in P. falciparum (12) and the possibility that parasite proteins indirectly activate host erythrocyte channels (43).
Here, we identify a conserved amphipathic domain within CLAG3 that appears to form a transmembrane domain. The position of this predicted transmembrane domain (residues 1200 to 1217 of the HB3 CLAG3.2 protein) is consistent with the prior observation that an upstream hypervariable region is extracellular while the protein's C terminus is intracellular (9, 16). Interestingly, helical-wheel analysis reveals the strict segregation of hydrophilic residues to one face of this putative α-helical transmembrane domain (Fig. 1C), suggesting that it may line a water-filled pore. Adding to this model, the HB3-leuR1 PSAC mutant generated through in vitro selection carries a critical A1210T mutation within this domain. Because this mutation lies at the interface between polar and hydrophobic faces of the helix, a modification from alanine to threonine not only increases helix polarity but may alter the helix tilt and significantly alter the local structure. To test this model, we have used DNA transfection to introduce the A1210T mutation into the HB3 wild-type parasite. This transfectant clone exhibited altered solute selectivity and pharmacology in comparison to either the untransfected parent or a transfectant that lacks this mutation but is otherwise identical. Two unrelated PSAC inhibitors, phloridzin and ISPA-28, were significantly less effective as a direct consequence of the A1210T mutation. Because these inhibitors have distinct binding sites on opposite faces of the channel (2, 9, 42), these findings imply global effects of A1210T on channel structure and support the proposed model of an amphipathic transmembrane domain.
While these studies suggest a direct and stable involvement of CLAG3 in the PSAC, the composition of the functional channel remains to be determined. Because most ion channels consist of multiple membrane-spanning domains, it seems unlikely that a CLAG3 monomer would be sufficient to form a pore capable of transporting solutes across the host membrane. One possibility is that several CLAG3 subunits associate to produce a homo-oligomeric channel. Another possibility is that one or more CLAG3 subunits interact with unrelated proteins to form the functional unit; these other proteins might be encoded by either the parasite or the host. Addressing the molecular and structural composition of the active PSAC at the host membrane is an important goal of malaria research. The present identification of a possible CLAG3 pore-lining domain should facilitate the achievement of this goal.
These findings also have implications for antimalarial drug resistance. We demonstrate that changes in the parasite clag genes are sufficient to reduce parasite susceptibility to certain water-soluble antimalarial compounds. These changes may entail either point mutations such as the A1210T mutation reported here or epigenetic silencing of one or more clag genes, as described in blasticidin S-resistant parasites (11, 24). Because leupeptin and blasticidin S act against distinct targets and have only limited structural similarity (23), reduced infected-cell uptake due to altered PSAC activity should be considered by all antimalarial drug discovery and development programs. Fortunately, the potential for this resistance mechanism can be readily evaluated. One approach involves the measurement of permeability to drugs with a radiolabeled analog. Increased uptake into infected erythrocytes combined with inhibition by specific PSAC antagonists would represent strong evidence of drug uptake by this parasite-induced channel. In cases where a radiolabeled drug analog is unavailable, in vitro parasite growth inhibition studies can also be used (23). If potent PSAC inhibitors reduce the efficacy of the drug lead in growth inhibition experiments, then uptake via this channel would be implied. Because PSAC inhibitors also interfere with parasite growth (10, 20), the relative contribution of each agent should be examined by isobologram analysis (44). A positive result of either of these tests should alert the discovery program to the risk of this additional resistance mechanism. While older antimalarial drugs tended to be hydrophobic and not require channel-mediated uptake, newer antimalarial candidates often require PSAC-mediated uptake (45–47), possibly because of evolving guidelines on desirable chemical features of drug leads (48). Because the parasite may reduce uptake through either mutations or gene silencing, early screening of candidate antimalarials is warranted; compounds that access their intracellular target after uptake through this channel now have an additional liability and should be pursued with caution.
ACKNOWLEDGMENTS
We thank Ryan Kissinger and Anita Mora for help with illustrations.
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
REFERENCES
- 1.Ginsburg H, Kutner S, Zangwil M, Cabantchik ZI. 1986. Selectivity properties of pores induced in host erythrocyte membrane by Plasmodium falciparum. Effect of parasite maturation. Biochim Biophys Acta 861:194–196. [DOI] [PubMed] [Google Scholar]
- 2.Desai SA, Bezrukov SM, Zimmerberg J. 2000. A voltage-dependent channel involved in nutrient uptake by red blood cells infected with the malaria parasite. Nature 406:1001–1005. doi: 10.1038/35023000. [DOI] [PubMed] [Google Scholar]
- 3.Staines HM, Rae C, Kirk K. 2000. Increased permeability of the malaria-infected erythrocyte to organic cations. Biochim Biophys Acta 1463:88–98. doi: 10.1016/S0005-2736(99)00187-X. [DOI] [PubMed] [Google Scholar]
- 4.Desai SA. 2014. Why do malaria parasites increase host erythrocyte permeability? Trends Parasitol 30:151–159. doi: 10.1016/j.pt.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alkhalil A, Cohn JV, Wagner MA, Cabrera JS, Rajapandi T, Desai SA. 2004. Plasmodium falciparum likely encodes the principal anion channel on infected human erythrocytes. Blood 104:4279–4286. doi: 10.1182/blood-2004-05-2047. [DOI] [PubMed] [Google Scholar]
- 6.Lisk G, Desai SA. 2005. The plasmodial surface anion channel is functionally conserved in divergent malaria parasites. Eukaryot Cell 4:2153–2159. doi: 10.1128/EC.4.12.2153-2159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkhalil A, Hill DA, Desai SA. 2007. Babesia and plasmodia increase host erythrocyte permeability through distinct mechanisms. Cell Microbiol 9:851–860. doi: 10.1111/j.1462-5822.2006.00834.x. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko O. 2007. Erythrocyte invasion: vocabulary and grammar of the Plasmodium rhoptry. Parasitol Int 56:255–262. doi: 10.1016/j.parint.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Nguitragool W, Bokhari AA, Pillai AD, Rayavara K, Sharma P, Turpin B, Aravind L, Desai SA. 2011. Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell 145:665–677. doi: 10.1016/j.cell.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillai AD, Nguitragool W, Lyko B, Dolinta K, Butler MM, Nguyen ST, Peet NP, Bowlin TL, Desai SA. 2012. Solute restriction reveals an essential role for clag3-associated channels in malaria parasite nutrient acquisition. Mol Pharmacol 82:1104–1114. doi: 10.1124/mol.112.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mira-Martínez S, Rovira-Graells N, Crowley VM, Altenhofen LM, Llinás M, Cortés A. 2013. Epigenetic switches in clag3 genes mediate blasticidin S resistance in malaria parasites. Cell Microbiol 15:1913–1923. doi: 10.1111/cmi.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko O, Yim Lim BY, Iriko H, Ling IT, Otsuki H, Grainger M, Tsuboi T, Adams JH, Mattei D, Holder AA, Torii M. 2005. Apical expression of three RhopH1/Clag proteins as components of the Plasmodium falciparum RhopH complex. Mol Biochem Parasitol 143:20–28. doi: 10.1016/j.molbiopara.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Cortés A, Carret C, Kaneko O, Yim Lim BY, Ivens A, Holder AA. 2007. Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog 3:e107. doi: 10.1371/journal.ppat.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vincensini L, Fall G, Berry L, Blisnick T, Braun BC. 2008. The RhopH complex is transferred to the host cell cytoplasm following red blood cell invasion by Plasmodium falciparum. Mol Biochem Parasitol 160:81–89. doi: 10.1016/j.molbiopara.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Alexandre JS, Xangsayarath P, Kaewthamasorn M, Yahata K, Sattabongkot J, Udomsangpetch R, Kaneko O. 2012. Stable allele frequency distribution of the Plasmodium falciparum clag genes encoding components of the high molecular weight rhoptry protein complex. Trop Med Health 40:71–77. doi: 10.2149/tmh.2012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguitragool W, Rayavara K, Desai SA. 2014. Proteolysis at a specific extracellular residue implicates integral membrane CLAG3 in malaria parasite nutrient channels. PLoS One 9:e93759. doi: 10.1371/journal.pone.0093759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck JR, Muralidharan V, Oksman A, Goldberg DE. 2014. PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature 511:592–595. doi: 10.1038/nature13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai SA. 2012. Ion and nutrient uptake by malaria parasite-infected erythrocytes. Cell Microbiol 14:1003–1009. doi: 10.1111/j.1462-5822.2012.01790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merckx A, Bouyer G, Thomas SL, Langsley G, Egee S. 2009. Anion channels in Plasmodium-falciparum-infected erythrocytes and protein kinase A. Trends Parasitol 25:139–144. doi: 10.1016/j.pt.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Pillai AD, Pain M, Solomon T, Bokhari AA, Desai SA. 2010. A cell-based high-throughput screen validates the plasmodial surface anion channel as an antimalarial target. Mol Pharmacol 77:724–733. doi: 10.1124/mol.109.062711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill DA, Pillai AD, Nawaz F, Hayton K, Doan L, Lisk G, Desai SA. 2007. A blasticidin S-resistant Plasmodium falciparum mutant with a defective plasmodial surface anion channel. Proc Natl Acad Sci U S A 104:1063–1068. doi: 10.1073/pnas.0610353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisk G, Pain M, Gluzman IY, Kambhampati S, Furuya T, Su XZ, Fay MP, Goldberg DE, Desai SA. 2008. Changes in the plasmodial surface anion channel reduce leupeptin uptake and can confer drug resistance in Plasmodium falciparum-infected erythrocytes. Antimicrob Agents Chemother 52:2346–2354. doi: 10.1128/AAC.00057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisk G, Pain M, Sellers M, Gurnev PA, Pillai AD, Bezrukov SM, Desai SA. 2010. Altered plasmodial surface anion channel activity and in vitro resistance to permeating antimalarial compounds. Biochim Biophys Acta 1798:1679–1688. doi: 10.1016/j.bbamem.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma P, Wollenberg K, Sellers M, Zainabadi K, Galinsky K, Moss E, Nguitragool W, Neafsey D, Desai SA. 2013. An epigenetic antimalarial resistance mechanism involving parasite genes linked to nutrient uptake. J Biol Chem 288:19429–19440. doi: 10.1074/jbc.M113.468371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyko B, Hammershaimb EA, Nguitragool W, Wellems TE, Desai SA. 2012. A high-throughput method to detect Plasmodium falciparum clones in limiting dilution microplates. Malar J 11:124. doi: 10.1186/1475-2875-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Bokhari AA, Solomon T, Desai SA. 2008. Two distinct mechanisms of transport through the plasmodial surface anion channel. J Membr Biol 226:27–34. doi: 10.1007/s00232-008-9136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iriko H, Kaneko O, Otsuki H, Tsuboi T, Su XZ, Tanabe K, Torii M. 2008. Diversity and evolution of the rhoph1/clag multigene family of Plasmodium falciparum. Mol Biochem Parasitol 158:11–21. doi: 10.1016/j.molbiopara.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexandre JS, Kaewthamasorn M, Yahata K, Nakazawa S, Kaneko O. 2011. Positive selection on the Plasmodium falciparum clag2 gene encoding a component of the erythrocyte-binding rhoptry protein complex. Trop Med Health 39:77–82. doi: 10.2149/tmh.2011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 31.Claros MG, von Heijne G. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10:685–686. [DOI] [PubMed] [Google Scholar]
- 32.Käll L, Krogh A, Sonnhammer EL. 2005. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21(Suppl 1):i251–i257. doi: 10.1093/bioinformatics/bti1014. [DOI] [PubMed] [Google Scholar]
- 33.Eisenberg D, Weiss RM, Terwilliger TC. 1982. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 34.Monks SA, Needleman DJ, Miller C. 1999. Helical structure and packing orientation of the S2 segment in the Shaker K+ channel. J Gen Physiol 113:415–423. doi: 10.1085/jgp.113.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu M, Akabas MH. 1996. Identification of channel-lining residues in the M2 membrane-spanning segment of the GABA(A) receptor alpha1 subunit. J Gen Physiol 107:195–205. doi: 10.1085/jgp.107.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung WY, Gardiner DL, Anderson KA, Hyland CA, Kemp DJ, Trenholme KR. 2007. The CLAG/RhopH1 locus on chromosome 3 of Plasmodium falciparum: two genes or two alleles of the same gene? Mol Biochem Parasitol 151:229–232. doi: 10.1016/j.molbiopara.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Crowley VM, Rovira-Graells N, de Pouplana LR, Cortés A. 2011. Heterochromatin formation in bistable chromatin domains controls the epigenetic repression of clonally variant Plasmodium falciparum genes linked to erythrocyte invasion. Mol Microbiol 80:391–406. doi: 10.1111/j.1365-2958.2011.07574.x. [DOI] [PubMed] [Google Scholar]
- 38.Wagner MA, Andemariam B, Desai SA. 2003. A two-compartment model of osmotic lysis in Plasmodium falciparum-infected erythrocytes. Biophys J 84:116–123. doi: 10.1016/S0006-3495(03)74836-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ginsburg H, Kutner S, Krugliak M, Cabantchik ZI. 1985. Characterization of permeation pathways appearing in the host membrane of Plasmodium falciparum infected red blood cells. Mol Biochem Parasitol 14:313–322. doi: 10.1016/0166-6851(85)90059-3. [DOI] [PubMed] [Google Scholar]
- 40.Ehrenkranz JR, Lewis NG, Kahn CR, Roth J. 2005. Phlorizin: a review. Diabetes Metab Res Rev 21:31–38. doi: 10.1002/dmrr.532. [DOI] [PubMed] [Google Scholar]
- 41.Kutner S, Breuer WV, Ginsburg H, Cabantchik ZI. 1987. On the mode of action of phlorizin as an antimalarial agent in in vitro cultures of Plasmodium falciparum. Biochem Pharmacol 36:123–129. doi: 10.1016/0006-2952(87)90389-3. [DOI] [PubMed] [Google Scholar]
- 42.Desai SA, Alkhalil A, Kang M, Ashfaq U, Nguyen ML. 2005. PSAC-independent phloridzin resistance in Plasmodium falciparum. J Biol Chem 280:16861–16867. doi: 10.1074/jbc.M414629200. [DOI] [PubMed] [Google Scholar]
- 43.Staines HM, Alkhalil A, Allen RJ, De Jonge HR, Derbyshire E, Egee S, Ginsburg H, Hill DA, Huber SM, Kirk K, Lang F, Lisk G, Oteng E, Pillai AD, Rayavara K, Rouhani S, Saliba KJ, Shen C, Solomon T, Thomas SL, Verloo P, Desai SA. 2007. Electrophysiological studies of malaria parasite-infected erythrocytes: current status. Int J Parasitol 37:475–482. doi: 10.1016/j.ijpara.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snyder C, Chollet J, Santo-Tomas J, Scheurer C, Wittlin S. 2007. In vitro and in vivo interaction of synthetic peroxide RBx11160 (OZ277) with piperaquine in Plasmodium models. Exp Parasitol 115:296–300. doi: 10.1016/j.exppara.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 45.Bray PG, Barrett MP, Ward SA, De Koning HP. 2003. Pentamidine uptake and resistance in pathogenic protozoa: past, present and future. Trends Parasitol 19:232–239. doi: 10.1016/S1471-4922(03)00069-2. [DOI] [PubMed] [Google Scholar]
- 46.Baumeister S, Wiesner J, Reichenberg A, Hintz M, Bietz S, Harb OS, Roos DS, Kordes M, Friesen J, Matuschewski K, Lingelbach K, Jomaa H, Seeber F. 2011. Fosmidomycin uptake into Plasmodium and Babesia-infected erythrocytes is facilitated by parasite-induced new permeability pathways. PLoS One 6:e19334. doi: 10.1371/journal.pone.0019334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dana S, Prusty D, Dhayal D, Gupta MK, Dar A, Sen S, Mukhopadhyay P, Adak T, Dhar SK. 2014. Potent antimalarial activity of acriflavine in vitro and in vivo. ACS Chem Biol. 9:2366–2373. doi: 10.1021/cb500476q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burrows JN, van Huijsduijnen RH, Mohrle JJ, Oeuvray C, Wells TN. 2013. Designing the next generation of medicines for malaria control and eradication. Malar J 12:187. doi: 10.1186/1475-2875-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alkhalil A, Hong L, Nguitragool W, Desai SA. 2012. Voltage-dependent inactivation of the plasmodial surface anion channel via a cleavable cytoplasmic component. Biochim Biophys Acta 1818:367–374. doi: 10.1016/j.bbamem.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]