Abstract
Bacterial selenocysteine incorporation occurs in response to opal stop codons and is dependent on the presence of a selenocysteine insertion sequence (SECIS) element, which recruits the selenocysteine specific elongation factor and tRNASec needed to reassign the UGA codon. The SECIS element is a stem-loop RNA structure immediately following the UGA codon and forms part of the coding sequence in bacterial selenoproteins. Although the site specific incorporation of selenocysteine is of great interest for protein engineering, the sequence constraints imposed by the adjoining SECIS element severely limit its use. We have evolved an E. coli tRNASec that is compatible with the canonical translation machinery and can suppress amber stop codons to incorporate selenocysteine with high efficiency. This evolved tRNASec allows the production of new recombinant selenoproteins containing structural motifs such as selenyl-sulfhydryl and diselenide bonds.
Incorporation of the rare amino acid selenocysteine into proteins confers unique biophysical properties and is essential for life in organisms spanning all three domains.1 Unlike the 20 canonical amino acids, selenocysteine lacks an aminoacyl-tRNA synthetase, and is instead a modification of a precharged serine and is inserted into proteins in response to opal stop codons. The overall mechanism for cotranslational incorporation at particular stop codons requires several specific cis and trans acting protein and RNA factors,2 including a dedicated selenocysteine tRNA (tRNASec), a selenophosphate synthase (SelD), and selenocysteine synthase (SelA), which are required to form Sec-tRNASec,3 a selenocysteine-specific elongation factor (SelB), and a stop codon-adjacent selenocysteine insertion sequence (SECIS) element that forms a conserved stem-loop RNA structure.4 SelB, which is structurally related to EF-Tu, is capable of discriminating between serylated and selenylated tRNASec,5,6 and the SelB:Sec-tRNASec complex is recruited by the SECIS element during translation to facilitate recoding of the UGA stop codon.5,7
Selenocysteine has a significantly lower pKa than cysteine (5.2 vs 8.5 for free amino acid) and much stronger nucleophilic properties, making it an attractive target for altering protein chemistry and function. Unfortunately, most selenoproteins have proven difficult to produce in E. coli, the standard host for recombinant protein production.2 This is due to the inherently low efficiency of selenocysteine incorporation in bacteria (4–5% vs termination of protein synthesis). In addition, the requirement that the SECIS element immediately follows the UGA codon, forming part of the coding sequence, greatly limits which proteins are amenable to selenocysteine insertion.8 Recently, Aldag et al.9 developed a hybrid tRNA (designated tRNAUTu) in which the acceptor stem of E. coli tRNASer was replaced with that of tRNASec, and the anticodon was changed to CUA to enable recognition of amber stop codons. Unlike wild type tRNASec, the hybrid tRNA was a substrate for EF-Tu (rather than SelB) and was shown to be compatible with canonical translation, greatly reducing the sequence constraints for selenocysteine incorporation.
Unfortunately, compared to wild-type tRNASec, selenylation of Ser-tRNAUTu was impaired, and in turn, serine was incorporated at a significant rate (35–45%).9 We hypothesized that the impairment was due to the loss of important contacts between SelA and the D- and T-loops of tRNASec.10 To overcome this problem, we used E. coli tRNASec as a scaffold for mutagenesis to identify tRNASec variants capable of participating in canonical translation. While the unusual 8 bp acceptor stem in tRNASec was thought to impair interactions with EF-Tu,11 Rudinger and co-workers12 showed that extended acceptor stems were compatible with EF-Tu binding and that a specific antideterminant sequence in tRNASec was instead responsible for blocking the interaction. This sequence corresponded to the final base pair of the acceptor stem (C7-G66) and to the first two base pairs of the T-arm (G49-U65 and C50-G64) (Figure 1c). Sequence changes at either location abolished antideterminant activity. Replacement of this sequence in tRNASec was shown to yield a tRNA capable of minimal interaction with EF-Tu.12 Similarly, a partially overlapping region of the T-arm covering base pairs 49–65, 50–64, and 51–63 is known to modulate the affinity of canonical tRNAs for EF-Tu.13 On the basis of this information, we opted to randomize the antideterminant region of tRNASec to identify sequences capable of interaction with EF-Tu and able to participate in canonical translation.
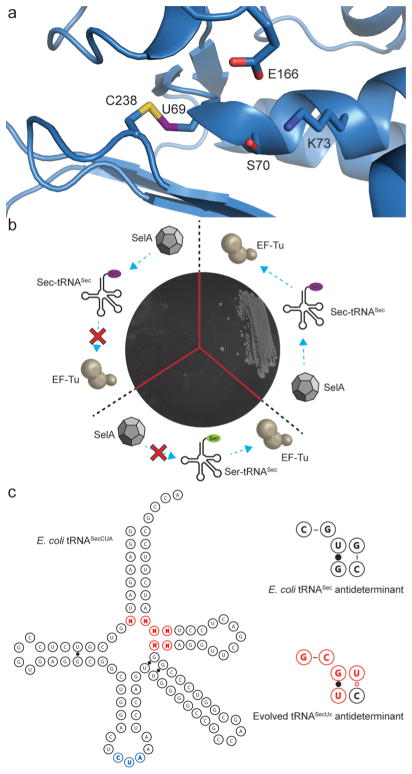
Figure 1.
Selection of tRNAs capable of canonical incorporation of selenocysteine. (a) Representation of the NMC-A β-lactamase from Enterobacter cloacae (PDB: 1BUE) showing the engineered selenyl-sulfhydryl bond between residues 69 and 238 and its proximity to the catalytic site. (b) Validation of a β-lactamase reporter capable of discriminating serine from selenocysteine. Three NMC-A variants, C69X (TAG), C69, and C69S (clockwise from left), were constructed to determine the threshold for selection (<50 μg·mL−1). These approximate the following outcomes during selection: termination at the amber stop codon due to inefficient interaction with EF-Tu, impaired interaction with SelA promoting incorporation of serine, and efficient interactions with both SelA and EF-Tu resulting in incorporation of selenocysteine. (c) Representation of E. coli tRNASecCUA and the evolved antideterminant sequence. The antideterminant library is shown in red, and the CUA anticodon is shown in blue.
While traditionally genetic code expansions have been evolved using reporter proteins containing amber stop codons,14,15 success is measured solely by the ability to make a full-length protein. Such selections are blind to the identity of the amino acid incorporated and rely on further negative selections to eliminate variants capable of nonspecific interactions. To prevent selection of tRNASec variants that interact with EF-Tu but are poor substrates for SelA, we developed a novel genetic selection capable of discriminating different levels of selenocysteine incorporation. To specifically “addict” a reporter protein to selenocysteine rather than serine we used the NMC-A β-lactamase from Enterobacter cloacae. This enzyme has high sequence similarity to the SME-1 β-lactamase from Serratia marcescens, an enzyme that has previously been shown to require a disulfide bond adjacent to the active site serine residue for activity but that confers a significant fitness cost on E. coli.16 We first constructed a C69S mutant of NMC-A, which failed to confer resistance to ampicillin (MIC < 50 μg·mL−1), indicating that the disulfide bond was essential for activity (Figure 1b). We then replaced cysteine 69 with an amber stop codon (X69; Figure 1a) for library selection, hypothesizing that the incorporation of selenocysteine and the formation of a selenyl-sulfhydryl bond would restore activity.17
To eliminate any crosstalk between the tRNASec library and the endogenous selenocysteine incorporation machinery, the selA, selB, and selC genes (encoding SelA, SelB, and tRNASec, respectively) were deleted from E. coli DH10B (designated DHΔabc). Cells containing the reporter plasmid pNMC-A C69X and the accessory plasmid pRSF-eSelA (expressing SelA) were transformed with plasmid pMB1-ZU containing the tRNASec antideterminant library. Transformants were plated on media containing a gradient of ampicillin concentrations for selection of mutants capable of selenocysteine-specific suppression. The single colonies that arose covered a range of ampicillin concentrations. Some 12 colonies from each plate were sequenced and revealed three distinct tRNASec mutants: G7-C66:U49-G65:C50-U64, C7-G66:U49-G65:C50-U64, and C7-U66:U49-A65:A50-Δ64 (where underlined, bases represent changes from the parental antideterminant sequence). Of these tRNASec variants, only G7-C66:U49-G65:C50-U64 was detected at the two highest ampicillin concentrations (200 and 250 μg·mL−1).
The tRNASec variant containing the G7-C66:U49-G65:C50-U64 antideterminant sequence was designated tRNASecUx and was compared with the previously designed chimera (tRNAUTu) and with a tRNASec derivative designed to have an antideterminant region that should tightly bind EF-Tu (tRNAUG; Figure 2a). The parental tRNASec containing a CUA anticodon and tRNAUG failed to produce active β-lactamase. Cells containing the hybrid tRNAUTu incorporated selenocysteine and could grow on 75 μg·mL−1. In contrast, expression of tRNASecUx resulted in significantly higher β-lactamase activity (up to 400 μg·mL−1), but only when coexpressed with SelA, confirming activity was selenocysteine dependent. To further confirm tRNASecUx incorporated selenocysteine in response to amber stop codons we employed a standard colorimetric assay based on the activity of the endogenous E. coli selenoprotein formate dehydrogenase H (FdhH) (Figure 2b). FdhH is expressed under anaerobic conditions and catalyzes the oxidation of formate to produce CO2 with the concomitant reduction of the electron acceptor benzyl viologen resulting in the development of a deep purple color.18 Formate oxidation by FdhH is strictly dependent on the selenocysteine residue at position 140; the mutant FdhH U140S was completely inactive. Only tRNASecUx and tRNAUTu when coexpressed with SelA produced active FdhH.
Figure 2.

Activity of tRNASec variants in canonical translation. (a) NMC-A β-lactamase reporter assay. From top row: Wild type NMC-A β-lactamase, NMC-A C69S, NMC-A C69X with tRNASecCUA, NMC-A C69X with tRNASecUx, NMC-A C69X with tRNASecUx and inactive SelA, NMC-A C69X with tRNASecUG containing a strong EF-Tu binding sequence, and NMC-A C69X with tRNAUTu. The observed dynamic range of the selection exceeds 20-fold. (b) Reduction of benzyl viologen by E. coli formate dehydrogenase H confirms selenocysteine is incorporated by the tRNASecUx. Plate layout is identical to panel a with FdhH, FdhH U140S, and FdhH U140TAG reporters replacing the NMC-A reporter.
The selected tRNA contained a nonstandard sequence in the junction that normally interacts with EF-Tu. Given that neither the base of the acceptor stem nor the adjoining T-arm base pairs are believed to play a role in the interaction between tRNASec and SelA, our results suggest that the selected C:U leads to stronger binding to EF-Tu than the wild-type tRNASec sequence.10 That said, the unusual C50-U64 base pair is not predicted to bind strongly to EF-Tu based on models developed for canonical tRNAs,13 and expression of a hybrid tRNAUG containing the strong EF-Tu binding region from the major E. coli tRNAGly did not lead to the production of active β-lactamase, suggesting that the nonstandard sequence was functionally important. Thus, it is possible that portions of the engineered tRNASec bind to EF-Tu differently than do canonical tRNAs, which would not necessarily be surprising given that tRNASec normally interacts with SelB.23
The development of engineered E. coli strains lacking the prfA gene encoding release factor 1 (RF1) has allowed efficient incorporation of a range of unnatural amino acids,19,20 and the development of the genome-engineered “Amberless” E. coli C321.ΔA20 provided an excellent opportunity to determine whether we could express proteins that efficiently incorporated selenocysteine. The selA, selB, and selC genes were deleted in C321.ΔA (designated strain RTΔA), and cells were transformed with the amber-containing NMC-A reporter and accessory plasmids (Figure S1, Supporting Information (SI)). β-lactamase activity was dramatically increased in RF1-deficient cells compared to prfA+ DHΔabc cells that still contain RF1. In addition, in a RF1-deficient background tRNASecUx could now support the formation of a functional diselenide bond (via amber-mediated incorporation of two selenocysteine residues, U69 and U238; Figure S1, SI).
To further enhance the efficiency of selenocysteine incorporation, a number of steps were taken to improve the levels of Sec-tRNASec relative to Ser-tRNASec, including increasing the level of SelA, decreasing the gene dose of tRNASecUx, and coexpressing a phosphoseryl-tRNASec kinase (Methods, SI). To monitor the efficiency of selenocysteine incorporation and demonstrate the possibilities for protein engineering, E. coli dihydrofolate reductase (DHFR) was produced containing an engineered nonessential selenyl-sulfhydryl bond.21 Top down mass spectrometry showed close to 100% selenocysteine incorporation with no detectable background corresponding to DHFR containing serine (Figure 3). The rationally designed tRNAUTu chimera was also observed to incorporate selenocysteine in DHFR containing a P39X substitution, but resulted in a much lower level of selenocysteine incorporation (38%) and significant serine incorporation (62%) (Figure S1, SI). No masses corresponding to the incorporation of other canonical amino acids were observed in the mass spectra (Figure S4, SI). In order to further validate selenocysteine incorporation, we also expressed the Pseudomonas aeruginosa metalloprotein azurin with its essential cysteine (C112) replaced by selenocysteine and the human selenoprotein cellular glutathione peroxidase (GPx-1) (Figures S3 and S5, SI). For azurin, this chemical change had previously been proven possible only through expressed protein ligation;22 the essential cysteine could now be biologically replaced with selenocysteine with good efficiency as measured by mass spectrometry of the intact protein.
Figure 3.
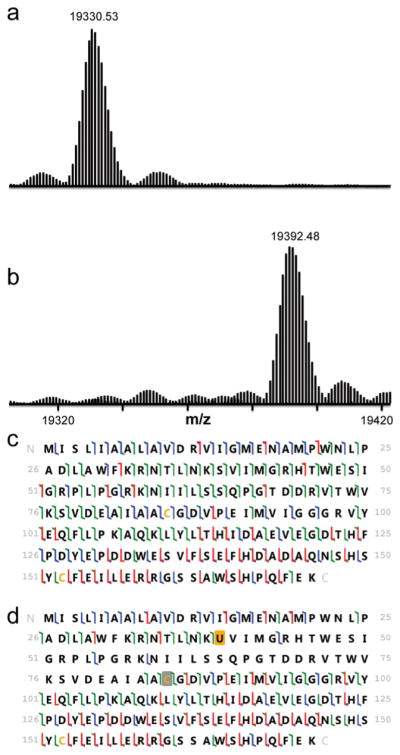
Intact mass and 193 nm UVPD results are shown for DHFR and seleno-DHFR produced using tRNASecUx. For the sequence information shown, disulfide-bound cysteines are shaded in gray, and selenocysteine is shaded in orange. When selenocysteine is located at position 39, it forms a selenyl-sulfhydryl bond with the cysteine at position 85. The different colors of slash marks represent the cleavage sites that lead to different N-terminal and C-terminal ions (green is used for a and x ions, blue is for b and y ions, and red is for c and z ions). (a) Expanded region of the deconvoluted mass spectrum of DHFR with serine at position 39 (expected average mass m/z 19331.72). (b) Expanded region of the deconvoluted mass spectrum of DHFR with selenocysteine incorporated at position 39 using tRNASecUx (expected average mass m/z 19392.66). (c) UVPD fragmentation map of DHFR P39S obtained for the 20+ charge state. (d) UVPD fragmentation map of DHFR with selenocysteine at position 39 (18+ charge state).
In this study we developed a new and robust genetic selection that relies on an engineered, essential selenyl-sulfhydryl bond in NMC-A β-lactamase. The selection system has a wide dynamic range and can function independently of the host genetic background. A new tRNASec variant with a nonstandard T-arm sequence and dramatically improved activity in canonical translation was evolved. A simple two-plasmid system carrying the new tRNA enabled highly efficient, site-specific incorporation of selenocysteine under culture conditions optimal for E. coli with no observable toxicity. Using this system, we have demonstrated the production of recombinant selenoproteins containing structural motifs such as selenyl-sulfhydryl and diselenide bonds at yields comparable to those obtained using expressed protein ligation. Such motifs have a number of protein engineering applications including bond formation in reducing intracellular environments and directing ordered folding of peptide scaffolds.24,25 Furthermore, this system will complement the development of new RF1-deficient strains derived from genetic backgrounds optimized for protein production,26 forming a powerful platform for numerous synthetic biology and biotechnology applications as well as a useful tool for potentially exploring the evolutionary expansion of the bacterial selenoproteome.
Supplementary Material
Acknowledgments
Funding from the NSF (CHE-1402753) and the Welch Foundation (F-1155) is acknowledged.
Footnotes
The authors declare no competing financial interest.
Experimental details and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Copeland PR. Genome Biol. 2005;6:27. doi: 10.1186/gb-2005-6-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arner ES, Sarioglu H, Lottspeich F, Holmgren A, Bock A. J Mol Biol. 1999;292:1003. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 3.Yuan J, O’Donoghue P, Ambrogelly A, Gundllapalli S, Sherrer RL, Palioura S, Simonovic M, Soll D. FEBS Lett. 2010;584:342. doi: 10.1016/j.febslet.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mansell JB, Guevremont D, Poole ES, Tate WP. EMBO J. 2001;20:7284. doi: 10.1093/emboj/20.24.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leibundgut M, Frick C, Thanbichler M, Bock A, Ban N. EMBO J. 2005;24:11. doi: 10.1038/sj.emboj.7600505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paleskava A, Konevega AL, Rodnina MV. J Biol Chem. 2010;285:3014. doi: 10.1074/jbc.M109.081380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshizawa S, Rasubala L, Ose T, Kohda D, Fourmy D, Maenaka K. Nat Struct Mol Biol. 2005;12:198. doi: 10.1038/nsmb890. [DOI] [PubMed] [Google Scholar]
- 8.Suppmann S, Persson BC, Bock A. EMBO J. 1999;18:2284. doi: 10.1093/emboj/18.8.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aldag C, Brocker MJ, Hohn MJ, Prat L, Hammond G, Plummer A, Soll D. Angew Chem, Int Ed. 2013;52:1441. doi: 10.1002/anie.201207567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh Y, Brocker MJ, Sekine S, Hammond G, Suetsugu S, Soll D, Yokoyama S. Science. 2013;340:75. doi: 10.1126/science.1229521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baron C, Bock A. J Biol Chem. 1991;266:20375. [PubMed] [Google Scholar]
- 12.Rudinger J, Hillenbrandt R, Sprinzl M, Giege R. EMBO J. 1996;15:650. [PMC free article] [PubMed] [Google Scholar]
- 13.Schrader JM, Uhlenbeck OC. Nucleic Acids Res. 2011;39:9746. doi: 10.1093/nar/gkr641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Schultz PG. Chem Biol. 2001;8:883. doi: 10.1016/s1074-5521(01)00063-1. [DOI] [PubMed] [Google Scholar]
- 15.Ellefson JW, Meyer AJ, Hughes RA, Cannon JR, Brodbelt JS, Ellington AD. Nat Biotechnol. 2014;32:97. doi: 10.1038/nbt.2714. [DOI] [PubMed] [Google Scholar]
- 16.Majiduddin FK, Palzkill T. Antimicrob Agents Chemother. 2003;47:1062. doi: 10.1128/AAC.47.3.1062-1067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swarén P, Maveyraud L, Raquet X, Cabantous S, Duez C, Pédelacq JD, Mariotte-Boyer S, Mourey L, Labia R, Nicolas-Chanoine MH, Nordmann P, Frère JM, Samama JP. J Biol Chem. 1998;273:26714. doi: 10.1074/jbc.273.41.26714. [DOI] [PubMed] [Google Scholar]
- 18.Zinoni F, Birkmann A, Leinfelder W, Bock A. Proc Natl Acad Sci USA. 1987;84:3156. doi: 10.1073/pnas.84.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukai T, Hayashi A, Iraha F, Sato A, Ohtake K, Yokoyama S, Sakamoto K. Nucleic Acids Res. 2010;38:8188. doi: 10.1093/nar/gkq707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, Isaacs FJ. Science. 2013;342:357. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villafranca JE, Howell EE, Oatley SJ, Xuong NH, Kraut J. Biochemistry. 1987;26:2182. doi: 10.1021/bi00382a017. [DOI] [PubMed] [Google Scholar]
- 22.Clark KM, van der Donk WA, Lu Y. Methods Enzymol. 2009;462:97. doi: 10.1016/S0076-6879(09)62005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li WQ, Yarus M. J Mol Biol. 1992;223:9. doi: 10.1016/0022-2836(92)90709-s. [DOI] [PubMed] [Google Scholar]
- 24.Shchedrina VA, Novoselov SV, Malinouski MY, Gladyshev VN. Proc Natl Acad Sci USA. 2007;104:13919. doi: 10.1073/pnas.0703448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armishaw CJ, Daly NL, Nevin ST, Adams DJ, Craik DJ, Alewood PF. J Biol Chem. 2006;281:14136. doi: 10.1074/jbc.M512419200. [DOI] [PubMed] [Google Scholar]
- 26.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, Emig CJ, Bang D, Hwang SJ, Jewett MC, Jacobson JM, Church GM. Science. 2011;333:348. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.