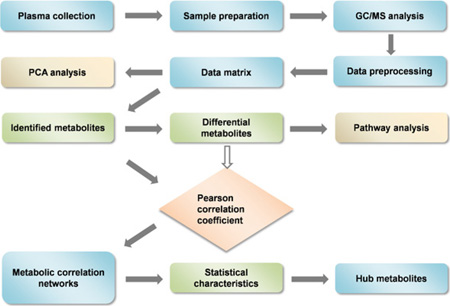
Abstract
The awareness, treatment, and control rates of hypertension for young adults are much lower than average. It is urgently needed to explore the variances of metabolic profiles for early diagnosis and treatment of hypertension. In current study, we applied a GC–MS based metabolomics platform coupled with a network approach to analyze plasma samples from young hypertensive men and age-matched healthy controls. Our findings confirmed distinct metabolic footprints of young hypertensive men. The significantly altered metabolites between two groups were enriched for the biological module of amino acids biosynthesis. The correlations of GC–MS metabolomics data were then visualized as networks based on Pearson correlation coefficient (threshold = 0.6). The plasma metabolites identified by GC–MS and the significantly altered metabolites (P < 0.05) between patients and controls were respectively included as nodes of a network. Statistical and topological characteristics of the networks were studied in detail. A few amino acids, glycine, lysine, and cystine, were screened as hub metabolites with higher values of degree (k), and also obtained highest scores of three centrality indices. The short average path lengths and high clustering coefficients of the networks revealed a small-world property, indicating that variances of these amino acids have a major impact on the metabolic change in young hypertensive men. These results suggested that disorders of amino acid metabolism might play an important role in predisposing young men to developing hypertension. The combination of metabolomics and network methods would provide another perspective on expounding the molecular mechanism underlying complex diseases.
Keywords: Metabolomics, GC–MS, Correlation network, Centrality indices, Hypertension, Hub metabolites
GRAPHICAL ABSTRACT
1. Introduction
Hypertension is a leading risk factor for heart disease, stroke, and kidney failure [1]. There are approximately one billion hypertensive individuals worldwide, and seven million deaths per year may be attributed to hypertension [2]. Although the pathogenesis of hypertension is greatly influenced by genetic, lifestyle, and environmental factors, the underlying molecular mechanisms remain to be fully elucidated.
High blood pressure has been identified as an important component of metabolic syndrome [3,4]. Recently, a growing body of exciting evidences suggested that abnormalities in cellular metabolism could be linked to hypertension [5]. For example, impaired glucose tolerance proved to be of high prevalence in patients with essential hypertension [6]. Sugar, especially fructose was reported to have a potential role in the development of hypertension, which might be caused by an increase in uric acid [7]. An association between plasma lipoprotein abnormalities and raised blood pressure was observed in young subjects with borderline hypertension [8]. It was also reported that metabolites associated with gut microbial activity might be related to blood pressure regulation [9]. Moreover, Litwin et al. reported hypertension in children and adolescents is an immuno-metabolic disease with hemodynamic consequences [10]. These studies provide further support for the increasing recognition that hypertension is a metabolic disease. However, much more work needs to be done to prove this hypothesis.
Metabolomics is a valuable emerging tool to study changes in phenotype caused by disease [11]. It focuses on low molecular weight compounds, which are the final downstream products of gene expression [12]. Mass spectrometry (MS) has grown rapidly these years, and that several new MS methods have been developed for various uses [13–17]. As MS could provide a blend of rapid, sensitive, and selective analyses for the identification and quantification of metabolites [18], it is widely applied in metabolomics studies. Recently, metabolomics has been increasingly used to evaluate metabolic disorders associated with hypertension [19,20]. AGC–MS based metabolomics study showed significantly elevated free fatty acids (heptanoic, oleic, and hexanoic acids) levels in serum of hypertensive patients compared to normotensive controls [21]. Altered carbohydrate metabolism was also found in these patients, including high levels of glucose and galactose, coupled with a decreased level of fructose [21]. An analysis using 1H NMR reported a relationship between serum metabolic profiles and blood pressure, which was partly due to lipoprotein particle composition differences [22]. Another large-scale metabolomics study revealed an inverse association between urinary excretion of formate and blood pressure [9]. These studies demonstrate that characteristic changes in metabolic profiles can be indicative of hypertensive phenotype.
The biological system is a complex physicochemical system consisting of numerous dynamic networks of biochemical reactions and signaling interactions between cellular components [23]. The variety of organic metabolites and the wide range of their concentrations always puzzled researchers in understanding metabolomics experimental results [24]. Several methods have been optimized to find valuable information hidden in metabolic data sets. Network theory, which extracts elements of systems into nodes and reduces interactions between elements to edges, has proven to be particularly suitable to explore relationships among large amount of biological data [25–28]. Various biomolecular networks have been built and studied, such as gene regulation network and protein interaction network [29]. As one of complex biological networks, metabolic correlation network has comparative advantages in interpreting the results of metabolomics. Many studies about correlation analysis in metabolomics have been reported. For instance, Chen et al. performed a LC–MS-based metabolomics study combined with metabolic correlation network to explore potential biomarkers for breast cancer [30]. Allen et al. reported a sequential reorganization of metabolic and transcriptional states in developing seedlings of Arabidopsis, using NMR-based metabolomics and metabolic correlation networks [31]. Another study revealed metabolomic correlation-network modules in Arabidopsis based on a graph-clustering approach [32]. Castro et al. used metabolomics and differential correlation networks to study Caenorhabditis elegans DAF-2 mutants [33]. As correlation network could obtain additional information about the physiological state of the system [34], it is expected that this approach would offer a novel perspective for the research of hypertension.
Aging exerts a marked effect on the cardiovascular system [35]. The number of patients suffering from hypertension increases with age [36]. Despite the importance of early diagnosis and treatment of hypertension, the awareness, treatment and controls rates of hypertension for young adults are much lower than average [37,38]. Data from National Health and Nutrition Examination Surveys (NHANES) showed that when compared to older adults, young adults ages 18–39 years in the United States are less likely to be aware they are hypertensive (59% vs. 84%), and are less likely to be receiving treatment for their hypertension (40% vs. 77%) [39]. Another study on adults with regular primary care use reported that those ages 18–24 years were 28% less likely to have their hypertension diagnosed compared to those at least 60 years of age [40]. Therefore, improved access and quality improvement efforts might need to be particularly focused on young adults. In this study, we used a GC–MS-based platform to explore the association between alterations in cellular intermediary metabolism and predisposition to hypertension in young men. Multivariate data analysis enabled to distinguish plasma metabolic profiles between young hypertensive men and age-matched healthy controls. Moreover, correlation network technology was applied to visualize the relationships between identified plasma metabolites. The statistical and topological characteristics of the network were studied in detail to characterize hub metabolites which might be responsible for hypertension in young men.
2. Materials and methods
2.1. Subjects
We recruited patients diagnosed as hypertensive by the Xijing Hospital (Xi’an, China) according to 2010 Chinese guidelines for the management of hypertension [41]. Blood pressure (BP) values of patients were measured using mercury sphygmomanometer just before sample collection. Mean systolic blood pressure (SBP) and mean diastolic blood pressure (DBP) were calculated by averaging 3–4 BP measurements. Patients were eligible if they met the hypertension diagnostic criteria (SBP≥140mmHg and DBP ≥ 90 mmHg). Twenty patients (male, 18–35 years of age) at stage 1 hypertension were enrolled in this research, classified according to the Seventh Report of the Joint National Committee on prevention, detection, evaluation, and treatment of high blood pressure (JNC VII report) [42]. All enrolled patients were not on any medication in at least 1 month. Healthy volunteers were selected by a routine physical examination in the same hospital and their blood pressure was measured using the same protocol as patients. Twenty age-matched controls (male, 20–35 years of age) were enrolled in this research. Body weights and heights of all participants were measured in unclothed subjects in the morning to assess body mass index (BMI). Any subjects with heart failure, renal dysfunction, diabetic mellitus, liver disease, or other metabolism disorders were excluded. All samples were collected following the guidelines of the local ethics committee and that written informed consents were obtained from all participants.
2.2. Sample preparation
Venous blood was collected in polypropylene tubes with EDTA in the early morning after overnight fasting, then immediately centrifuged at 3000 × g for 10 min. The resulting plasma was transferred into a clean tube and stored at −80°C. Prior to GC–MS analysis, plasma was thawed at 4°C. Plasma metabolites were derivatized and analyzed following our previously published procedures with minor modifications [43]. A measure of 50 µL of plasma was vortexed after being spiked with internal standard solution (5 µL L-2-chlorophenylalanine in water, 1 mmol L−1). A 150 µL aliquot of methanol-chloroform (3:1, v/v) was added in to the mixed solution and vortexed for 30 s. After storing at −20 °C for 10 min, the samples were centrifuged at 12,000 × g for 10 min. A 150 µL aliquot of the supernatant was transferred to a clean tube and lyophilized. Then, 80 µL of methoxyamine hydrochloride (15 mgm L−1 in pyridine) was used to dissolve dried residue, and stayed at 37 °C for 90 min. Subsequently, 80 µL of N,O-bis (trimethylsilyl)-trifluoroacetamide (BSTFA) with 1% TMCS (Sigma–Aldrich) were added to interchange the acidic protons at 70°C for 60 min.
2.3. GC-MS spectral acquisition
A measure of 1 µL aliquot of the derivatized solution was injected into a 7890A GC and coupled with a 5975C MS (Agilent Technologies, Wilmington, DE). Chromatographic separation was accomplished on a DB-5 column (30 m × 0.25 mm i.d.; film thickness: 0.5 µm; Agilent J&W Scientific, USA). The GC temperature programming was set to 2 min isothermal heating at 80 °C, followed by 10°C/min oven temperature ramps to 120 °C, 4 °C min−1 oven temperature ramps to 260 °C, and then increased at a rate of 10 °C min−1 oven temperature ramps to 300 °C, where it was held for 3 min. The inlet temperature was kept at 260 °C and ion source temperature was 200 °C. Helium was used as carrier gas at a constant flow rate of 1 mL min−1. Electron impact ionization source (70 eV) was used at full scan mode (m/z 50– 550), with a scan rate of 2.91 scans per second. The typical analysis order was composed of quality control (QC) samples and subject samples [44]. A pooled QC sample was prepared by taken 5 µL plasma from each sample and thoroughly mixed. Then, the mixed sample (QC) was extracted and derivatized together with other samples, using the same methods. Five QC samples were injected at the start of each analytical batch and then one QC sample was analyzed at every tenth injection. The subject samples were randomly analyzed between each QC sample.
2.4. Data pretreatment and chemometrics analyses
Raw data acquired from GC–MS were converted to Masshunter B.06.00 software (Agilent Technologies, Santa Clara, USA) for peak deconvolution and detection, with the “Find by Chromatographic Deconvolution” algorithm. Detected components with abundance less than 5000 counts and relative abundance less than 1% were removed. Subsequently, the deconvoluted data were imported into Mass Profile Professional software (Agilent Technologies, Santa Clara, USA) for peak alignment, filtering, baseline correction, and area calculation. The molecular features existed in at least 80% of the plasma samples in either group were retained. Abundance values were normalized to that of the internal standard and log transformed. Finally, a spreadsheet containing relative peak intensities, compound name, and sample information was exported for the next analyses.
Compounds were identified by comparison of mass spectra with NIST 11 (National Institute of Standards and Technology, Gaithersburg, MD, USA), with a similarity of more than 70%. A set of available authentic standards were analyzed under identical experimental conditions for further identification. In addition, a saturated n-alkane mixture from C7 to C30 (Sigma Chemical, St. Louis, MO, USA) was run at the beginning of the experimental worklist to determine retention indexes (RI). Automated mass spectrometry deconvolution and identification system (AMDIS) was configured to build a retention index calibration file and calculate RI for each metabolic features. Mass spectrum and RI of individual peaks in samples were compared with that of the standard references to verify the results.
For multivariate statistical analysis, the pre-processed data were imported to MATLAB 2009a (The MathWorks Inc., Natick, MA, USA). Principal component analysis (PCA) was carried out for group discrimination. Altered metabolites between groups were identified using the non-parametric Mann–Whitney–Wilcoxon test with the critical P-value set at 0.05. Raw P-values were adjusted using Benjamini and Hochberg procedure (BH method) [45]. A false discovery rate (FDR) control was executed to correct for multiple comparisons. The adjusted P-values less than a desired FDR (5%) were considered as significant. Non-parametric test and FDR control were carried out on the freely available software R (http://www.r-project.org/) with corresponding packages.
2.5. Functional enrichment analysis and pathway topology analysis
Pathway analyses were performed on significantly altered metabolites for further biological interpretation. Over representation analysis (ORA) was applied for functional enrichment analysis [46]. ORA was implemented using the hypergeometric test to evaluate whether a particular metabolite set is represented more than expected by chance within the metabolites sets. Relative-betweenness centrality was calculated for pathway topology analysis. Potential targets were selected either by P-values from pathway enrichment analysis, or by impact values from pathway topology analysis [47]. The impact value threshold was set to 0.10 and negative log P-value threshold was set to 10. Identification of pathways was carried out utilizing data sources such as Reactome (http://www.reactome.org), KEGG (http://www.genome.jp/kegg/), and MetaboAnalyst 2.0 (http://www.metaboanalyst.ca/).
2.6. Reconstruction and analysis of metabolic correlation networks
The correlations of GC–MS metabolomics data were visualized as networks based on Pearson correlation coefficient (ρ). Nodes correspond to the identified plasma metabolites and that edges indicate significant correlation between nodes [48]. Symbols, equations, and descriptions of all the parameters used for network analysis were listed in Table 1 [49–54]. For construction of the correlation network, an appropriate threshold (T) was first selected by evaluating the average degree (<k>), diameter (D) and the size of the largest connected components (Nmax). A connection between two nodes was determined by the comparison between the absolute value of ρ and T. Subsequently, a metabolic correlation network containing plenty of nodes and connections was constructed, which was visualized using Pajek (http://vlado.fmf.uni-lj.si/pub/networks/pajek). Finally, a series of statistical characteristics of the network were calculated and analyzed to extract hub metabolites, including degree (k), average path length (L), diameter (D), and clustering coefficient (C). Three centrality indices of the network, degree centrality (Cd), betweenness centrality (Cb), and closeness centrality (Cc), were also comprehensively analyzed to assess the impact of a node on the overall network.
Table 1.
Definitions of the statistical characteristics used for network analysis in this study.
| Statistical characteristic | Symbol | Equationa,b,c,d | Description | |
|---|---|---|---|---|
| Degree | k |
|
The total number of edges for a node i | |
| Average degree | <k> |
|
The average degree of the network | |
| Average path length | L |
|
The mean distance between two nodes, averaged over all pairs of nodes | |
| Diameter | D | D = max{dij} | The maximum distance between any pair of nodes | |
| Clustering coefficient | C |
|
The probability that two nodes are linked to each other given that they are both connected to node i | |
| Degree centrality | Cd |
|
The proportion of other nodes that are adjacent to node i | |
| Betweenness centrality | Cb |
|
The proportion of all geodesics between pairs of other nodes that include this node i | |
| Closeness centrality | Cc |
|
The number of other nodes divided by the sum of the distances between the node i and all the other nodes |
eij is the numbers of edges from node i to j.
dij is the shortest path length from node i to j.
N is the total number of nodes in the network.
gjk is the numbers of geodesics connecting nodes j and k.
3. Results and discussion
3.1. Sample characteristics
A total of 20 young hypertensive men (26.7 ± 8.6 years, SBP = 148.75 ± 6.6 mm Hg, DBP = 98.1 ± 6.7 mm Hg) and 20 healthy controls (27.4 ± 7.3 years, SBP = 119.6 ± 9.4 mm Hg, DBP = 72.4 ± 7.0 mm Hg) were allocated to GC–MS analysis. Table 2 presented demographic characteristics of the participants. There were no significant differences in age and body mass index (BMI) between patients and the healthy controls (t-test), suggesting a similar energy intake and expenditure between the two groups.
Table 2.
Demographic details of patients and controls involved in the study.a
| Group | Age (Years) | SBP (mm Hg)b | DBP (mm Hg)c | Height (cm) | Weight (Kg) | BMI (kgm−2)d |
|---|---|---|---|---|---|---|
| Young hypertensive men (n = 20) | 26.7 ±8.6 | 148.8 ± 6.6 | 98.1 ± 6.7 | 171.9 ± 6.8 | 68.4 ± 13.1 | 24.16 ± 3.16 |
| Healthy controls (n = 20) | 27.4 ± 7.3 | 119.6 ± 9.4 | 72.4 ± 7.0 | 174.9 ± 7.9 | 67.4 ± 13.2 | 23.92 ± 2.28 |
Values are means ± SD.
SBP, systolic blood pressure.
DBP, diastolic blood pressure.
BMI, body mass index.
3.2. Different plasma metabolic profiles between young hypertensive men and healthy controls
A GC–MS based analytical platform was applied to obtain metabolic profiles of young hypertensive men and healthy controls. After excluding internal standard, a total of 70 metabolites were consistently identified in at least 80% of the plasma samples in either group, and their relative peak intensities were used for the next analyses. Among them, 27 metabolites were identified to metabolomics standards initiative (MSI) level 1 [55], with a comparison of retention index and mass spectrum with authentic reference standards. All the identified metabolites were listed in Supplementary Table S1.
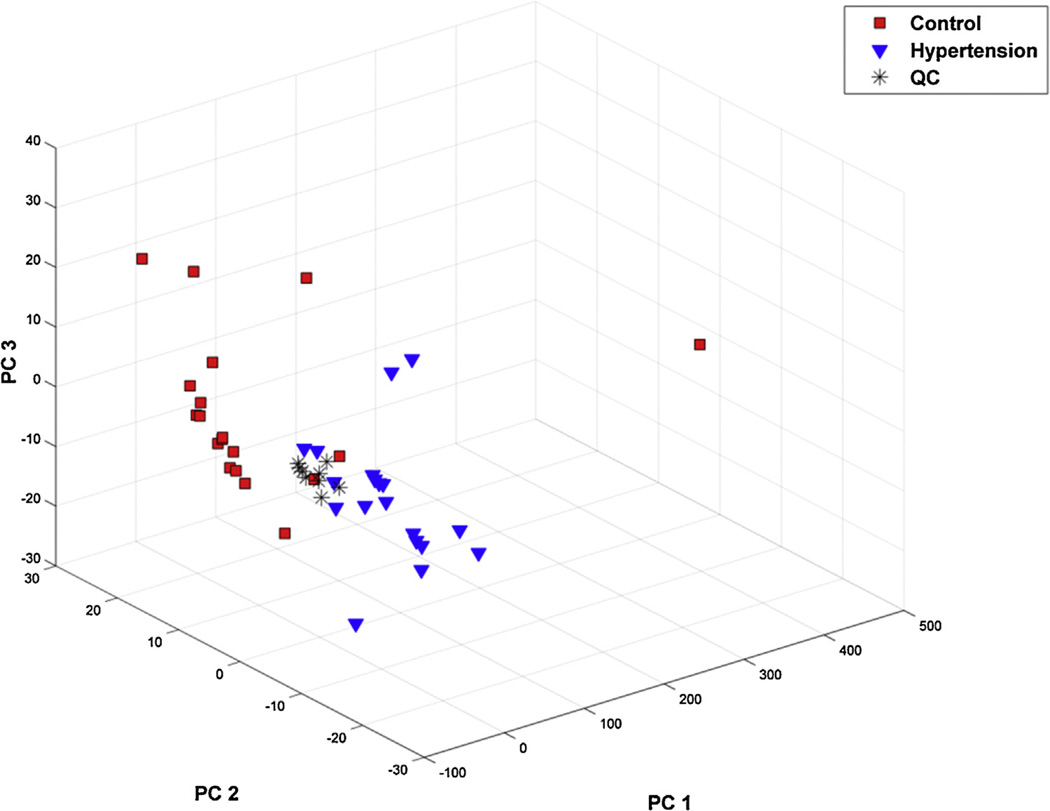
Principal component analysis (PCA) was applied to generate an unbiased overview of the metabolic data sets. The PCA scoring plot (Fig. 1) showed a clear clustering trend of the metabolic data sets of different classes. A point far from others was observed, which reflected unknown disorder in the subject. This outlier had been excluded to avoid affecting further analysis. Using the first three principal components (R2 (cum) = 72.0%), this model exhibited an acceptable discriminating ability. Moreover, the loading plot (Supplementary Fig. S1) illustrated the contributions of variables to the formation of PCA matrix. A few data points appeared to play an important role in the metabolic discrimination, including uric acid, capric acid, cystine, beta-alanine, lysine, and 5-hydroxy caproic acid, etc. The results indicated a unique metabolic signature in young hypertensive men and that some metabolites might account for the variation, which need further validation. Nine QC data were also included into pattern recognition, which showed an acceptable variance of the GC–MS analysis [56].
Fig. 1.
Young hypertensive men and healthy controls show different metabolic profiles. PCA scores plot of the metabolic profiling data obtained from plasma of young hypertensive men and healthy controls. Blue triangle, young hypertensive men (n = 20), red square, healthy controls (n = 20), black asterisk, QC samples (n = 9).(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The significance of the change in relative concentrations for each metabolite was determined by a non-parametric Mann– Whitney–Wilcoxon test. P-values were adjusted using Benjamini and Hochberg procedure (BH method) with an FDR control (<5%) to correct multiple comparisons. Adjusted P-values and fold change of all the 70 identified metabolites between groups were listed in Supplementary Table S1. A set of 20 variables were screened as significantly changed, including organic acids, amino acids, carbohydrates, and lipids (Table 3). A total of 13 amino acids were observed to be significantly decreased in the patient group. Among these compounds, isoleucine, threonine, methionine, valine, lysine, and tryptophan belong to essential amino acids, which cannot be synthesized in human. As the levels of metabolites reflect most closely the operation of biological system [57], these altered metabolites might be responsible for the high blood pressure in young men.
Table 3.
Altered plasma metabolites between young hypertensive men and healthy controls.
| Group | Compound | Fold changeb,c | Regulationd | Adjusted P-valuee | Retention time (min) | Retention indexf |
|---|---|---|---|---|---|---|
| Organic acids | Oxalic acid | 4.47 | Up | 1.05E-05 | 6.8 | 1119.6 |
| Fumaric acid | 2.8 | Up | 6.20E-05 | 14.18 | 1339.2 | |
| L-valinea | −1.17 | Down | 4.17E-02 | 10.63 | 1204.6 | |
| L-isoleucinea | −1.22 | Down | 1.99E-02 | 12.59 | 1281.3 | |
| Glycinea | −2.98 | Down | 4.78E-06 | 12.99 | 1296.7 | |
| L-threoninea | −1.23 | Down | 1.98E-03 | 15.05 | 1370.5 | |
| L-methioninea | −1.47 | Down | 1.76E-02 | 18.97 | 1506.8 | |
| Ornithinea | −3.42 | Down | 1.64E-03 | 21.63 | 1601.8 | |
| Amino acids | L-asparaginea | −1.42 | Down | 1.89E-03 | 23.14 | 1656.9 |
| L-glutaminea | −1.67 | Down | 3.90E-03 | 25.98 | 1762.2 | |
| Citrullinea | −1.85 | Down | 1.70E-04 | 27.33 | 1804.2 | |
| L-lysinea | −1.9 | Down | 1.22E-05 | 29.81 | 1908.1 | |
| L-tyrosinea | −2.8 | Down | 3.90E-03 | 30.24 | 1924.9 | |
| L-tryptophana | −1.42 | Down | 3.98E-04 | 36.72 | 2200.1 | |
| L-cystinea | −1.95 | Down | 5.30E-05 | 38.42 | 2277 | |
| Carbohydrates | Glycerol | 1.26 | Up | 4.32E-02 | 11.98 | 1257.4 |
| Lipids | Capric acid | −1.58 | Down | 6.51E-03 | 17.49 | 1448.2 |
| Adenine | 3.4 | Up | 4.47E-06 | 20.35 | 1555.9 | |
| Others | Pyrophosphate | 2.38 | Up | 1.69E-02 | 22.81 | 1645.5 |
| Uric acid | 1.28 | Up | 1.37E-02 | 34.06 | 2083.2 |
Metabolites confirmed by measuring reference substances.
Fold change was calculated as a relatively ratio of the relative peak intensities between different groups.
Positive value, patients vs. controls; negative value, controls vs. patients.
“Up” or “Down” means the trend of regulation for these metabolites in young hypertensive patients versus controls.
P-values were adjusted using Benjamini and Hochberg procedure (BH method) with an FDR of less than 5%.
Retention index was calculated relative to n-alkane standards.
Uric acid was found to be significantly elevated in plasma of young hypertensive men. Uric acid is a product of purine metabolism that circulates in the blood, and plays a role in initiating the development of hypertension [58]. It has been known that elevations of uric acid in blood are common in subjects with hypertension [59]. An elevated plasma uric acid level in childhood also predicts hypertension as an adult [60].
An increased concentration of fumarate was observed in plasma of young hypertensive men. Fumarate is a key intermediate of citric acid cycle and present at micromolar concentration in blood, which is regulated by many factors [61]. TCA cycle is central to the regulation of energy homeostasis and cell metabolism [62]. The accumulation of fumarate in young hypertensive men suggested an intrinsic disorder of the critical pathway. Our previous study also revealed a relationship between fumarate excess and salt-induced hypertension [63,64].
Adenine, glycerol, and pyrophosphate were remarkably increased in young patients, whereas capric acid significantly decreased compared with normotensive controls. These compounds were involved in various pathways, such as purine metabolism, oxidative phosphorylation, glycerolipid metabolism, and fatty acid biosynthesis. Alteration of these metabolites reflected a disorder of systemic metabolism, which might contribute to the hypertensive phenotype in young men.
3.3. Pathway analyses uncover disorders of amino acid metabolism in young hypertensive men
Global metabolomics strategies hold a significant potential for translating discriminating molecules into functional modules. In order to identify metabolic pathways associated with hypertension, pathway analyses were performed on the altered metabolites between young hypertensive men and healthy, including functional enrichment analysis and pathway topology analysis.
For enrichment analysis, over representation analysis (ORA) was used to identify biologically meaningful patterns in a list of the 20 altered metabolites. This method allows one to extract relevant information from the metabolite sets. Hypergeometric test was selected to evaluate whether a particular metabolite set was represented more than expected by chance [65]. One-tailed P-values are calculated after adjusting for multiple testing (Benjamini and Hochberg procedure). Supplementary Fig. S2 exhibited an enrichment overview of the altered metabolite sets. The findings indicated that the clusters of metabolites were involved in multiple pathways, and enriched for the pathway class of protein biosynthesis (P = 2.94 × 10−11).
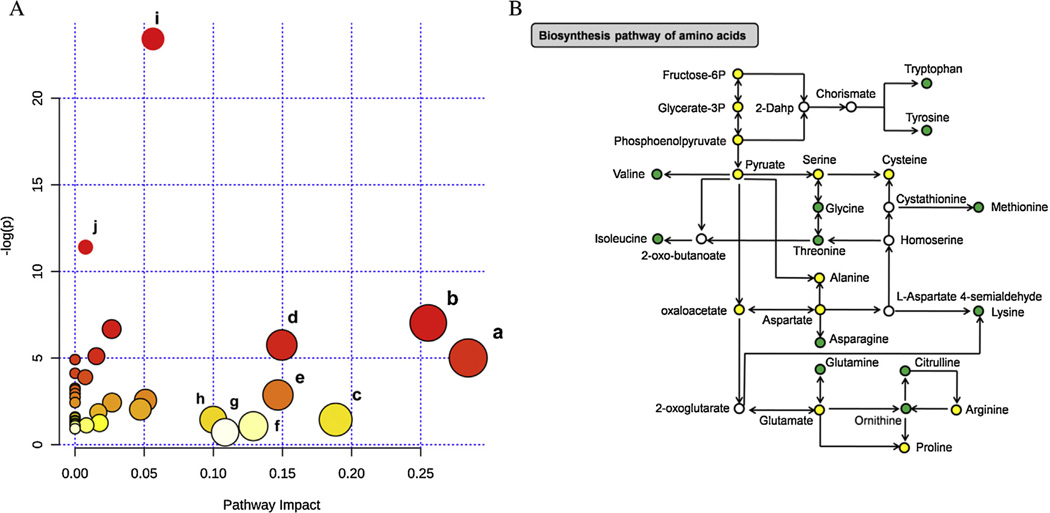
Relative-betweenness centrality algorithm was applied for pathway topology analysis [66], measuring numbers of shortest paths going through a node in the network. It takes into consideration of global network structure, not only immediate neighbor of the current node. The acquired impact value is cumulative percentage of importance for the matched metabolite nodes involved in a pathway. Relevant pathways for the 20 altered metabolites were visualized via an interactive visualization framework (Fig. 2A). We selected potential target pathways either by P-values from pathway enrichment analysis, or by impact values from pathway topology analysis. Supplementary Table S2 listed the result of pathway topology analysis. A set of 8 pathways were selected at an impact value threshold of 0.10. Among these pathways, 6 biological modules were involved in amino acid metabolism, such as glycine, serine, and threonine metabolism (impact = 0.284), alanine, aspartate, and glutamate metabolism (impact = 0.255), etc. In addition, aminoacyl-tRNA biosynthesis and nitrogen metabolism were considered as potential targets with the negative log P-values more than 10. These data demonstrated possible relationship between disorders of amino acid metabolism and hypertension pathogenesis. It has been reported that decreased levels of threonine, valine, tryptophan, and histidine existed in pulmonary arterial hypertension (PAH) patients [67]. Another research showed that a higher intake of tyrosine (0.3% of protein) was significantly related to a 2.4 mm Hg lower SBP [68].
Fig. 2.
Pathway analyses reveal disorders of amino acid metabolism in young hypertensive men. (A) Summary of pathway topology analysis of altered metabolites between young hypertensive men and controls. Potential target pathways were selected either by negative log P-values from pathway enrichment analysis, or by impact values from pathway topology analysis. (a) Glycine, serine and threonine metabolism; (b) alanine, aspartate and glutamate metabolism; (c) glycerolipid metabolism; (d) arginine and proline metabolism; (e) lysine degradation; (f) glyoxylate and dicarboxylate metabolism; (g) tryptophan metabolism; (h) lysine biosynthesis; (i) aminoacyl-tRNA biosynthesis; (j) nitrogen metabolism. (B) Changes associated with biosynthesis pathway of amino acids in young hypertensive men. The metabolites are shown in color: green represents decreased metabolites, yellow represents no change, and the open circles represent no detected metabolites. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Among the 20 significantly altered metabolites,12 amino acids belonged to a modular architecture of the biosynthesis pathway of amino acids, including lysine, glycine, tryptophan, etc. Fig. 2B shows a schematic overview of amino acids biosynthesis pathway in young hypertensive men compared with healthy controls. Obviously, a considerable reduction of levels of all the 12 intermediates was exhibited. Biosynthesis pathway of amino acids is the most conserved one in almost all the completely sequenced genomes, containing biosynthesis of basic amino acids, aromatic amino acids, and branched-chain amino acids (BCAA). Observational studies have reported an inverse association between BCAA intake and blood pressure [69]. Moreover, BCAA have a potential to improve glucose metabolism through the accelerated utility of glucose and glucose-6-phosphate in the liver [70], and were negatively correlated with prevalence of overweight status/obesity, which were major risk factors for hypertension [71]. Consequently, deficiency in amino acid synthesis might be related to high blood pressure.
3.4. Construction and analyses of metabolic correlation network for young hypertensive men
Typical metabolomics data show a few but significant correlations among metabolite levels [32]. Although the relationships among metabolites are complex, correlation network has been developed as a powerful tool for integrating information from the high throughput experiments. In this part of the study, a correlation network was constructed and analyzed based on 70 identified metabolites. Metabolic data of both the patient and control groups were used for the network construction to avoid being affected by secondary effects of high blood pressure. Pearson correlation coefficient (ρ) was used for correlation analysis.
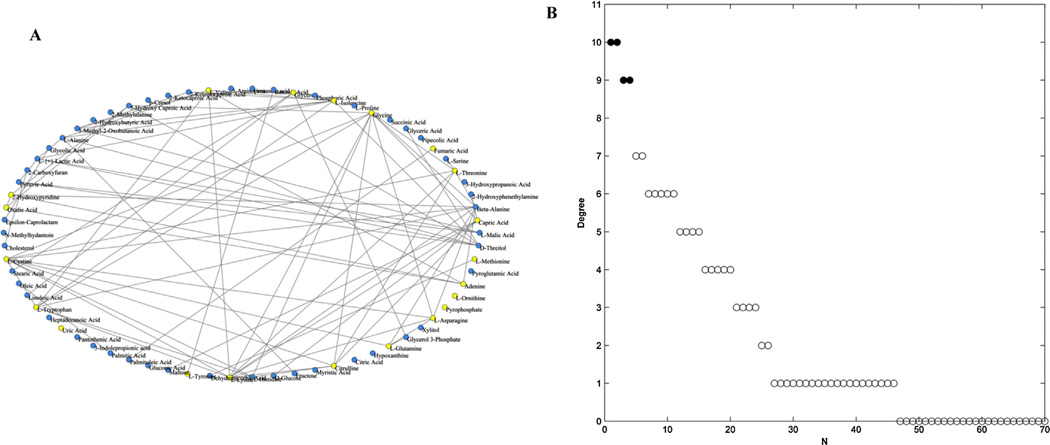
The selection of an appropriate correlation threshold (T) is of critical importance in the network construction. The network would be sparse with a high value of T, or has larger size of connected components with a relatively low one [32]. To evaluate threshold-dependent changes in the network topology, the average degree (<k>), diameter (D), and the size of the largest connected components (Nmax) of the network were assessed from T = 0.4 to T = 1 (Supplementary Table S3). <k> represents the mean value of degree (k) over all nodes, and D is the maximum distance in the network. As shown in Supplementary Fig. S3, the three topological parameters of the network exhibited various change trends at different values of T, and showed a transition at T = 0.6. Therefore, a correlation threshold of 0.6 (|ρ|> 0.6) was selected to visualize strong correlation between metabolites. Interactions between pair-wise metabolites were then reduced to edges and visualized with a heat map (Supplementary Fig. S4). The abbreviations of metabolites in Supplementary Fig. S4 were listed in Supplementary Table 1. Positive correlation could be interpreted as the result of two metabolites occurring adjacently in a metabolic pathway or derived from a common precursor. A negative correlation could indicate one metabolite is used to directly or indirectly generate the other. The final metabolic correlation network was showed in Fig. 3A, containing 70 nodes and 79 edges.
Fig. 3.
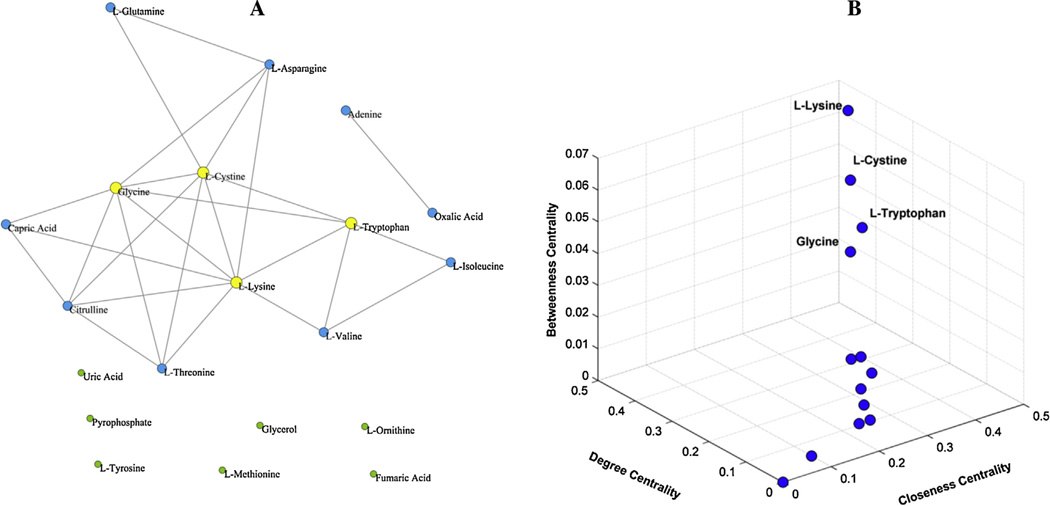
Analysis of the metabolic correlation network based on the identified plasma metabolites. (A) Metabolic correlation network based on 70 identified plasma metabolites, containing 79 edges (T = 0.6). Yellow nodes represented the significantly altered metabolites between patients and controls (P < 0.05). (B) The values of degree of 70 nodes in descending order. Filled circles represent the top highly connected metabolites in the network. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In order to confirm the most important metabolites of the correlation network, degree (k) of each node was calculated and ranked in a descending order (Fig. 3B, Supplementary Table S4). Obviously, a few nodes had most connections with others, whereas most nodes only had few links. It is well-known that changes in important positions of a network will trigger a more severe impact than changes occurring in marginal or relatively isolated positions [72]. Therefore, the top four highly connected compounds, glycine (k = 10), lysine (k = 10), cystine (k = 9), and beta-alanine (k = 9), were screened as hubs of the network, which appeared as filled circles in Fig. 3B. These data suggested a possible relation between amino acid metabolism and hypertension.
Subsequently, a set of statistical characteristics were assessed for further understanding of the metabolic correlation network (Table 4), including the average path length (L), clustering coefficient (C), average degree (<k>), diameter (D), the size of the largest cluster (Nmax), and the percentage of Nmax in all nodes (Nmax%). L determines the effective size and offers a measure of the overall navigability of the network [73]. The clustering coefficient is the ratio of all existing edges between the neighbors and the maximum number of edges possible between these neighbors, ranging from 0 to 1 [74]. The parameter describes the cohesiveness of the neighborhood of a node. The clustering coefficients of all the nodes were listed in Supplementary Table S4.
Table 4.
Statistical characteristics of metabolic correlation networks.
| Number of nodes | <k>c | Ld | Ce | Df | Nmaxg | Nmax%h |
|---|---|---|---|---|---|---|
| 70a | 2.26 | 3.43 | 0.50 | 8 | 34 | 48.57 |
| 20b | 2.60 | 1. 7 3 | 0.60 | 3 | 11 | 55.00 |
Metabolic correlation network based on 70 identified plasma metabolites.
Critical metabolic correlation network based on 20 altered metabolites between young hypertensive men and healthy controls.
<k>the average degree.
L, the average path length.
C, the clustering coefficient.
D, the diameter.
Nmax, the size of the largest cluster.
Nmax%, the percentage of Nmax in all nodes of the network.
In summary, the metabolic correlation network for young hypertensive men showed a relatively short average path length (L = 3.43) and high clustering coefficient (C = 0.5), suggesting a small-world property of the network. The small-world effect consists of two properties, short average path length and relatively high clustering coefficient [52,75]. As properties of a network often depend on a small number of hub nodes, these findings indicated that the metabolic network was organized as many small, but highly connected modules that combine in a hierarchical manner to larger, less cohesive units [76]. Metabolomic correlations complement information would help to elucidate the organization of metabolic functional modules associated with hypertension in young men.
3.5. Network analyses of altered metabolites between young hypertensive men and healthy controls
As shown in the previous section, three amino acids, glycine, lysine, and cystine were selected as hub metabolites of the correlation network. Interestingly, they were also significantly altered between young hypertensive men and the healthy. It is well known that altered metabolites are potentially more significant as they directly contribute to the metabolic discrimination. Hence, a separate network analysis of these metabolites would provide further interpretation for metabolic characteristics of young hypertensive men. As shown in Fig. 4A, a network was built based on Pearson correlation coefficients of 20 significantly altered metabolites. This network contained 20 nodes and 26 edges (T = 0.6). The statistical characteristics were calculated as mentioned above, including, L,C, D, Nmax, and Nmax% (Table 4). Similarly, a small-world property of the network was observed with a small average path length (L = 1.73) and large clustering coefficient (C = 0.6). This property demonstrated that alterations of a few metabolites would be responsible for the difference of metabolic profiles.
Fig. 4.
Analysis of the metabolic correlation network based on significantly altered metabolites between young hypertensive men and healthy controls. (A) Metabolic correlation network based on 20 significantly altered metabolites (P < 0.05), containing 26 edges (T = 0.6). Yellow nodes represented hub metabolites of the network. (B) Three-dimensional diagram of three centrality indices for the 20 nodes of the network, including betweenness centrality, closeness centrality and degree centrality. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Three important centrality indices of the 20 nodes [53,54], degree centrality (Cd), betweenness centrality (Cb), and closeness centrality (Cc) were comprehensively analyzed to evaluate the contribution of each altered metabolite to the network (Supplementary Table S5). Cd of a node was the proportion of other nodes adjacent to it, indicating the immediate influence. Cb was measured by the total number of shortest paths going through a node, and that Cc depicted how close a node was to others, reflecting the influence of a node on others through the network [77]. Fig. 4B provides an integrated display of these parameters with a three-dimensional diagram. Obviously, four metabolites were outliers with high scores of the centrality indices. Lysine (Cd = 0.4211, Cb = 0.0676, Cc = 0.4583), cystine (Cd = 0.3684, Cb = 0.0508, Cc = 0.4231), glycine (Cd = 0.3684, Cb = 0.0282, Cc = 0.4231), and tryptophan (Cd = 0.2632, Cb = 0.0453, Cc = 0.3667) were then screened as hubs of this metabolic correlation network. All these hub metabolites are amino acids, and exhibited a significantly down-regulation in plasma of young hypertensive men. More importantly, lysine, cystine, glycine were hubs of both the large metabolic correlation network (70 nodes) and the critical metabolic correlation network (20 nodes), which complemented each other. These data provided further support for the major impact of a disordered amino acid metabolism on the development of hypertension.
Lysine, an essential amino acid, proved to be hub of the metabolic correlation networks for young hypertensive men. Previous studies has revealed a significantly lower level of lysine in plasma of patients with inflammation [78]. Furthermore, plasma concentration of lysine was negatively correlated with serum concentrations of C-reactive protein (CRP), neopterin, and fibrinogen, whereas positively correlated with serum albumin concentration [78]. These data suggested a potential role of decreased lysine in cardiovascular disease (CVD) such as hypertension.
Another hub metabolite, cystine, is a conjugation of two cysteines. Cysteine is generally assumed to be a rate-limiting substrate for the synthesis of antioxidant glutathione (GSH) [79]. GSH is essential for the cellular detoxification of reactive oxygen species (ROS), the main cardiovascular risk factor for hypertension [80]. Maintenance of intracellular GSH could drive cystine transport across plasma membrane under oxidative stress [81]. Decreased plasma cystine observed in young hypertensive men might be indicative of a compromised GSH system under increased oxidative stress.
Glycine, the simplest non-essential amino acid, was also screened as a key node. It is involved in the synthesis of the structural proteins such as elastin, and has been found to protect against oxidative stress in several pathological situations [82]. It is well known that hypertension is a disease in which free radicals and large vessel elasticity are involved [83]. The lower level of glycine in young hypertensive men implied an impaired elastin formation in the aorta and increased generation of free radicals. Moreover, the addition of glycine to the diet has proven to reduce hypertension in a rat model of the metabolic syndrome [84].
In summary, a few amino acids play an important role in the metabolic correlation networks for young hypertensive men. Cellular demand for these key intermediates might contribute to a raise in blood pressure of young men. However, the possibility needs to be tested in future experiments.
4. Conclusions
This study presents one of the first integrated analyses of metabolomics and correlation network on hypertension in young adults. Our data show that metabolic variations take place at an early stage of hypertension, which is well in line with previous demonstration that hypertension is a metabolic disease [5]. Furthermore, a few amino acids were screened as hub metabolites in the correlation network for young hypertensive men. These findings suggested an association between disorders of amino acid metabolism and hypertension, and that the hypothesis was strongly supported by earlier reports on dietary amino acids and the risk of hypertension [68,69,85,86].
In the majority of hypertensive cases, the underlying cause cannot be easily identified because of the heterogeneous, polygenic, and multi-factorial nature of hypertension [87]. It is especially difficult to explore metabolic change responsible for high blood pressure when patients suffer from other major illnesses such as myocardial infarction, stroke, or kidney disease, which is common in the elderly. Therefore, studies on young hypertensive adults were more likely to uncover the molecular mechanism predisposing human to developing hypertension. In addition, we analyzed strong correlations between metabolites in both normal and pathological conditions, thus the detected hub metabolites were unlikely to be secondary effects of high blood pressure, which improved the reliability of the results.
In the next study, we will apply the methods to a larger number of subjects, and try to construct a comparison of two networks for hypertensive and healthy separately would complement the results of this study. We would also analyze the thermodynamics of these networks for a whole understanding. In all, combination of metabolomics and network would enable to generate rich biochemical insight into possible biological modules related to hypertension.
Supplementary Material
HIGHLIGHTS.
Metabolomics uncovers distinct metabolic profiles of young hypertensive men.
Several amino acids are screened as hub metabolites of the correlation networks.
Metabolic correlation networks for young hypertensive men show small-world property.
Altered metabolites are enriched for the amino acids biosynthesis pathway.
Disorders of amino acid metabolism might predispose hypertension in young men.
Acknowledgments
We are very grateful to Pu Jia-Man Wu and Shifeng Ni, from Northwest University (Xi’an 710069, China) for technical support. We also thank Shifeng Yu, Zhenhua Zhang, and Hongyan Sun from Xi’an Analytical and Monitoring Centre for Agri-Food Quality Safety (Xi’an 710077, China), for the optimization of gas chromatography/mass spectrometry.
This study was supported by the National Natural Science Foundation of China (NSFC) (Grant No. 31071029, 81270767, 81228002) and the US National Institutes of Health (Grant No. HL116264).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.aca.2014.11.009.
Footnotes
Competing interest
The authors declare no competing financial interest.
References
- 1.Vikrant S, Tiwari S. Essential hypertension – pathogenesis and pathophysiology. J. Indian Acad. Clin. Med. 2001;2:140–161. [Google Scholar]
- 2.Whitworth JA. 2003 World Health Organization (WHO)/International Society of Hypertension (ISH) statement on management of hypertension. J. Hypertens. 2003;21:1983–1992. doi: 10.1097/00004872-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Grundy SM, Brewer HB, Jr, Cleeman JI, Smith SC, Jr, Lenfant C, National Heart L, Blood I. A. American Heart, Definition of metabolic syndrome: report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Arterioscler. Thrombo. Vasc. Biol. 2004;24:e13–e18. doi: 10.1161/01.ATV.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 4.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 5.Liang MY. Hypertension as a mitochondrial and metabolic disease. Kidney Int. 2011;80:15–16. doi: 10.1038/ki.2011.84. [DOI] [PubMed] [Google Scholar]
- 6.Tugrul A, Guldiken S, Ugur-Altun B, Arikan E. An evaluation of glucose tolerance in essential hypertension. Yonsei Med. J. 2009;50:195–199. doi: 10.3349/ymj.2009.50.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson RJ, Segal MS, Sautin Y, Nakagawa T, Feig DI, Kang DH, Gersch MS, Benner S, Sanchez-Lozada LG. Potential role of sugar (fructose) in the epidemic of hypertension, obesity and the metabolic syndrome, diabetes, kidney disease, and cardiovascular disease. Am. J. Clin. Nutr. 2007;86:899–906. doi: 10.1093/ajcn/86.4.899. [DOI] [PubMed] [Google Scholar]
- 8.Pannier B, Cambillau M, Vellaud V, Atger V, Moatti N, Safar M. Abnormalities of lipid metabolism and arterial rigidity in young subjects with borderline hypertension: clinical and investigative medicine. Med. Clinique Et Experimentale. 1994;17:42–51. [PubMed] [Google Scholar]
- 9.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litwin M, Michalkiewicz J, Gackowska L. Primary hypertension in children and adolescents is an immuno-metabolic disease with hemodynamic consequences. Curr. Hypertens. Rep. 2013;15:331–339. doi: 10.1007/s11906-013-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schomburg D. A metabolic network described in absolute terms. Nat. Chem. Biol. 2009;5:535–536. doi: 10.1038/nchembio0809-535. [DOI] [PubMed] [Google Scholar]
- 13.Li A, Wang H, Ouyang Z, Cooks RG. Paper spray ionization of polar analytes using non-polar solvents. Chem. Commun. 2011;47:2811–2813. doi: 10.1039/c0cc05513a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, McLuckey SA. Electron transfer followed by collision-induced dissociation (NET-CID) for generating sequence information from backbone-modified oligonucleotide anions. Rapid Commun. Mass Spectrom. 2013;27:249–257. doi: 10.1002/rcm.6428. [DOI] [PubMed] [Google Scholar]
- 15.Li A, Wei P, Hsu HC, Cooks RG. Direct analysis of 4-methylimidazole in foods using paper spray mass spectrometry. Analyst. 2013;138:4624–4630. doi: 10.1039/c3an00888f. [DOI] [PubMed] [Google Scholar]
- 16.Li S, Limbach PA. Mass spectrometry sequencing of transfer ribonucleic acids by the comparative analysis of RNA digests (CARD) approach. Analyst. 2013;138:1386–1394. doi: 10.1039/c2an36515d. [DOI] [PubMed] [Google Scholar]
- 17.Wetzel C, Li S, Limbach PA. Metabolic de-isotoping for improved LC-MS characterization of modified RNAs. J. Am. Soc. Mass Spectrom. 2014;25:1114–1123. doi: 10.1007/s13361-014-0889-9. [DOI] [PubMed] [Google Scholar]
- 18.Dunn WB, Ellis DI. Metabolomics: current analytical platforms and methodologies. TrAC Trends Anal. Chem. 2005;24:285–294. [Google Scholar]
- 19.Rhee EP, Gerszten RE. Metabolomics and cardiovascular biomarker discovery. Clin. Chem. 2012;58:139–147. doi: 10.1373/clinchem.2011.169573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayr M. Recent highlights of metabolomics in cardiovascular research. Circulation: Cardiovasc. Genet. 2011;4:463–464. doi: 10.1161/CIRCGENETICS.111.961003. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Chen T, Qiu Y, Cheng Y, Cao Y, Zhao A, Jia W. An ultrasonication-assisted extraction and derivatization protocol for GC/TOFMS-based metabolite profiling. Anal. Bioanal. Chem. 2011;400:1405–1417. doi: 10.1007/s00216-011-4880-z. [DOI] [PubMed] [Google Scholar]
- 22.Brindle JT, Nicholson JK, Schofield PM, Grainger DJ, Holmes E. Application of chemometrics to 1H NMR spectroscopic data to investigate a relationship between human serum metabolic profiles and hypertension. Analyst. 2003;128:32–36. doi: 10.1039/b209155k. [DOI] [PubMed] [Google Scholar]
- 23.Kwoh CK, Ng PY. Network analysis approach for biology. Cell Mol. Life Sci. 2007;64:1739–1751. doi: 10.1007/s00018-007-7053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simón-Manso Y, Lowenthal MS, Kilpatrick LE, Sampson ML, Telu KH, Rudnick PA, Mallard WG, Bearden DW, Schock TB. Tchekhovskoi, metabolite profiling of a NIST standard reference material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses libraries, and web-based resources. Anal. Chem. 2013;85:11725–11731. doi: 10.1021/ac402503m. [DOI] [PubMed] [Google Scholar]
- 25.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 26.Albert R. Scale-free networks in cell biology. J. Cell. Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 27.Fadhal E, Gamieldien J, Mwambene EC. Protein interaction networks as metric spaces:a novel perspective on distribution of hubs. BMC Systems Biol. 2014;8:6. doi: 10.1186/1752-0509-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitra K, Carvunis A-R, Ramesh SK, Ideker T. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet. 2013;14:719–732. doi: 10.1038/nrg3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solé RV, Pastor-Satorras R. Complex networks in genomics and proteomics, Handbook of graphs and networks: From the genome to the internet. 2006 [Google Scholar]
- 30.Chen Y, Zhang R, Song Y, He J, Sun J, Bai J, An Z, Dong L, Zhan Q, Abliz Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: finding potential biomarkers for breast cancer. Analyst. 2009;134:2003–2011. doi: 10.1039/b907243h. [DOI] [PubMed] [Google Scholar]
- 31.Allen E, Moing A, Ebbels TM, Maucourt M, Tomos AD, Rolin D, Hooks MA. Correlation network analysis reveals a sequential reorganization of metabolic and transcriptional states during germination and gene-metabolite relationships in developing seedlings of Arabidopsis. BMC Syst. Biol. 2010;4:62. doi: 10.1186/1752-0509-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukushima A, Kusano M, Redestig H, Arita M, Saito K. Metabolomic correlation-network modules in Arabidopsis based on a graph-clustering approach. BMC Syst. Biol. 2011;5:1. doi: 10.1186/1752-0509-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castro C, Krumsiek J, Lehrbach NJ, Murfitt SA, Miska EA, Griffin JL. A study of Caenorhabditis elegans DAF-2 mutants by metabolomics and differential correlation networks. Mol. Biosyst. 2013;9:1632–1642. doi: 10.1039/c3mb25539e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steuer R. Review: on the analysis and interpretation of correlations in metabolomic data. Brief Bioinform. 2006;7:151–158. doi: 10.1093/bib/bbl009. [DOI] [PubMed] [Google Scholar]
- 35.McEniery CM, Wilkinson IB, Avolio AP. Age hypertension and arterial function. Clin. Exp. Pharmacol. Physiol. 2007;34:665–671. doi: 10.1111/j.1440-1681.2007.04657.x. [DOI] [PubMed] [Google Scholar]
- 36.Seong K, Hong J-H, Hur M-H, Lee MS. Two-week aroma inhalation effects on blood pressure in young men with essential hypertension. Euro. J. Integr. Med. 2013;5:254–260. [Google Scholar]
- 37.Gooding HC, McGinty S, Richmond TK, Gillman MW, Field AE. Hypertension awareness and control among young adults in the national longitudinal study of adolescent health. J. Gen. Intern. Med. 2014;29:1098–1104. doi: 10.1007/s11606-014-2809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.C.f.D. Control, Prevention, Vital signs: prevalence, treatment, and control of hypertension – United States, 1999–2002 and 2005–2008, Morbidity and Mortality Weekly Report. MMWR. 2011;60:10. [PubMed] [Google Scholar]
- 39.Yoon SS, Burt V, Louis T, Carroll M. Hypertension among adults in the United States, 2009–2010. NCHS data brief. 2012:1–8. [PubMed] [Google Scholar]
- 40.Johnson HM, Thorpe CT, Bartels CM, Schumacher JR, Palta M, Pandhi N, Sheehy AM, Smith MA. Undiagnosed hypertension among young adults with regular primary care use. J. Hypertens. 2014;32:65–74. doi: 10.1097/HJH.0000000000000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L. [2010 Chinese guidelines for the management of hypertension] Zhonghua Xin Xue Guan Bing Za Zhi. 2011;39:579–615. [PubMed] [Google Scholar]
- 42.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jones DW, Materson BJ, Oparil S, Wright JT. Seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Hou E, Wang Z, Sun N, He L, Chen L, Liang M, Tian Z. Analysis of metabolites in plasma reveals distinct metabolic features between Dahl salt-sensitive rats and consomic SS.13BN rats. Biochem. Biophys. Res. Commun. 2014;450:863–869. doi: 10.1016/j.bbrc.2014.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Methodol. 1995;57:289–300. [Google Scholar]
- 46.Kankainen M, Gopalacharyulu P, Holm L, Oreši9č M. MPEA—metabolite pathway enrichment analysis. Bioinformatics. 2011;27:1878–1879. doi: 10.1093/bioinformatics/btr278. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Yang B, Sun H, Zhang A. Pattern recognition approaches and computational systems tools for ultra performance liquid chromatography-mass spectrometry-based comprehensive metabolomic profiling and pathways analysis of biological data sets. Anal. Chem. 2012;84:428–439. doi: 10.1021/ac202828r. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Yu H, Luo J, Cao Z, Li Y. Complex networks theory for analyzing metabolic networks. Chin. Sci. Bull. 2006;51:1529–1537. [Google Scholar]
- 49.Newman ME. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 50.Dorogovtsev SN, Mendes JF. Evolution of networks. Adv. Phys. 2002;51:1079–1187. [Google Scholar]
- 51.Albert R, Barabási A-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47. [Google Scholar]
- 52.Watts DJ, Strogatz SH. Collective dynamics of small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 53.Freeman LC. Centrality in social networks conceptual clarification. Soc. Networks. 1979;1:215–239. [Google Scholar]
- 54.Beauchamp MA. An improved index of centrality. Behav. Sci. 1965;10:161–163. doi: 10.1002/bs.3830100205. [DOI] [PubMed] [Google Scholar]
- 55.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sangster T, Major H, Plumb R, Wilson AJ, Wilson ID. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst. 2006;131:1075–1078. doi: 10.1039/b604498k. [DOI] [PubMed] [Google Scholar]
- 57.Dunn WB, Broadhurst D, Brown M, Baker PN, Redman CWG, Kenny LC, Kell DB. Metabolic profiling of serum using Ultra Performance Liquid Chromatography and the LTQ-Orbitrap mass spectrometry system. J. Chromatogr. B-Anal. Technol. Biomed. Life Sci. 2008;871:288–298. doi: 10.1016/j.jchromb.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 58.Feig DI, Madero M, Jalal DI, Sanchez-Lozada LG, Johnson RJ. Uric acid and the origins of hypertension. J. Pediatr. 2013;162:896–902. doi: 10.1016/j.jpeds.2012.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feig DI, Kang DH, Johnson RJ. Uric acid and cardiovascular risk. N. Engl. J. Med. 2008;359:1811–1821. doi: 10.1056/NEJMra0800885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang WL, Sun K, Yang Y, Zhang HY, Hu FB, Hui RT. Plasma uric acid and hypertension in a Chinese community: prospective study and metaanalysis. Clin. Chem. 2009;55:2026–2034. doi: 10.1373/clinchem.2009.124891. [DOI] [PubMed] [Google Scholar]
- 61.He WH, Miao FJP, Lin DCH, Schwandner RT, Wang ZL, Gao JH, Chen JL, Tian H, Ling L. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature. 2004;429:188–193. doi: 10.1038/nature02488. [DOI] [PubMed] [Google Scholar]
- 62.Krebs HA. The history of the tricarboxylic acid cycle. Perspect Biol. Med. 1970;14:154–170. doi: 10.1353/pbm.1970.0001. [DOI] [PubMed] [Google Scholar]
- 63.Tian Z, Liu Y, Usa K, Mladinov D, Fang Y, Ding X, Greene AS, Cowley AW, Liang M. Novel role of fumarate metabolism in dahl-salt sensitive hypertension. Hypertension. 2009;54:255–260. doi: 10.1161/HYPERTENSIONAHA.109.129528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian Z, Greene AS, Usa K, Matus IR, Bauwens J, Pietrusz JL, Cowley AW, Liang M. Renal regional proteomes in young dahl salt-sensitive rats. Hypertension. 2008;51:899–904. doi: 10.1161/HYPERTENSIONAHA.107.109173. [DOI] [PubMed] [Google Scholar]
- 65.Brunelli L, Ristagno G, Bagnati R, Fumagalli F, Latini R, Fanelli R, Pastorelli R. A combination of untargeted and targeted metabolomics approaches unveils changes in the kynurenine pathway following cardiopulmonary resuscitation. Metabolomics. 2013;9:839–852. [Google Scholar]
- 66.Mangalam A, Poisson L, Nemutlu E, Datta I, Denic A, Dzeja P, Rodriguez M, Rattan R, Giri S. Profile of circulatory metabolites in a relapsing-remitting animal model of multiple sclerosis using global metabolomics. J. Clin. Cell. Immunol. 2013;4:e12102. doi: 10.4172/2155-9899.1000150. [PMID: 20711407] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fessel J, Fox K, Cunningham G, Summar M, Loyd J, Austin E, Hysinger E. Amino acid and urea cycle metabolism is altered in patients with pulmonary arterial hypertension. Am. J. Res. Crit. Care Med. 2013;187:A5744. [Google Scholar]
- 68.Altorf-van der Kuil W, Engberink MF, De Neve M, van Rooij FJ, Hofman A, van't Veer P, Witteman JC, Franco OH, Geleijnse JM. Dietary amino acids and the risk of hypertension in a Dutch older population: the Rotterdam Study. Am. J. Clin. Nutr. 2013;97:403–410. doi: 10.3945/ajcn.112.038737. [DOI] [PubMed] [Google Scholar]
- 69.Altorf-van der Kuil W, Engberink MF, Brink EJ, van Baak MA, Bakker SJ, Navis G, van't Veer P, Geleijnse JM. Dietary protein and blood pressure: a systematic review. PLoS ONE. 2010;5:e12102. doi: 10.1371/journal.pone.0012102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Higuchi N, Kato M, Miyazaki M, Tanaka M, Kohjima M, Ito T, Nakamuta M, Enjoji M, Kotoh K, Takayanagi R. Potential role of branched-chain amino acids in glucose metabolism through the accelerated induction of the glucose-sensing apparatus in the liver. J. Cell. Biochem. 2011;112:30–38. doi: 10.1002/jcb.22688. [DOI] [PubMed] [Google Scholar]
- 71.Qin LQ, Xun PC, Bujnowski D, Daviglus ML, Van Horn L, Stamler J, He K, Grp ICR. Higher Branched-Chain Amino Acid Intake Is Associated with a Lower Prevalence of Being Overweight or Obese in Middle-Aged East Asian and Western Adults. J. Nutr. 2011;141:249–254. doi: 10.3945/jn.110.128520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao W, Tan G, Zhu Z, Chen Q, Lou Z, Dong X, Zhang W, Pan W, Chai Y. Combined metabonomic and quantitative real-time PCR analyses reveal systems metabolic changes in Jurkat T-cells treated with HIV-1 Tat protein. J. Proteome Res. 2012;11:5109–5123. doi: 10.1021/pr300173c. [DOI] [PubMed] [Google Scholar]
- 73.Xu J. Theory and Application of Graphs. Springer; 2003. [Google Scholar]
- 74.Newman MEJ. The structure and function of complex networks. Siam Rev. 2003;45:167–256. [Google Scholar]
- 75.Amaral LA, Scala A, Barthelemy M, Stanley HE. Classes of small-world networks. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 77.Wang H, Xu CY, Hu JB, Cao KF. A complex network analysis of hypertension-related genes. Phys. Stat. Mech. Appl. 2014;394:166–176. [Google Scholar]
- 78.Suliman ME, Qureshi AR, Stenvinkel P, Pecoits-Filho R, Bárány P, Heimbürger O, Anderstam B, Ayala ER, Divino Filho JC, Alvestrand A. Inflammation contributes to low plasma amino acid concentrations in patients with chronic kidney disease. Am. J. Clin. Nutr. 2005;82:342–349. doi: 10.1093/ajcn.82.2.342. [DOI] [PubMed] [Google Scholar]
- 79.Yang J, Chen T, Sun L, Zhao Z, Qi X, Zhou K, Cao Y, Wang X, Qiu Y, Su M, Zhao A, Wang P, Yang P, Wu J, Feng G, He L, Jia W, Wan C. Potential metabolite markers of schizophrenia. Mol. Psychiatry. 2011;18:67–78. doi: 10.1038/mp.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li HG, Horke S, Forstermann U. Oxidative stress in vascular disease and its pharmacological prevention. Trends Pharmacol. Sci. 2013;34:313–319. doi: 10.1016/j.tips.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 81.Schmidt MM, Dringen R. Neural Metabolism In Vivo. Springer; 2012. Glutathione (GSH) synthesis and metabolism; pp. 1029–1050. [Google Scholar]
- 82.Zhong Z, Wheeler MD, Li X, Froh M, Schemmer P, Yin M, Bunzendaul H, Bradford B, Lemasters JJ. L-Glycine: a novel antiinflammatory, immunomodulatory, and cytoprotective agent. Curr. Opin. Clin. Nutr. Metabol. Care. 2003;6:229–240. doi: 10.1097/00075197-200303000-00013. [DOI] [PubMed] [Google Scholar]
- 83.El Hafidi M, Perez I, Banos G. Is glycine effective against elevated blood pressure? Curr. Opin. Clin. Nutr. Metabol. Care. 2006;9:26–31. doi: 10.1097/01.mco.0000196143.72985.9a. [DOI] [PubMed] [Google Scholar]
- 84.El Hafidi M, Perez I, Zamora J, Soto V, Carvajal-Sandoval G, Banos G. Glycine intake decreases plasma free fatty acids, adipose cell size, and blood pressure in sucrose-fed rats. Am. J. Physiol.-Regulatory, Integr. Comp. Physiol. 2004;287:R1387–R1393. doi: 10.1152/ajpregu.00159.2004. [DOI] [PubMed] [Google Scholar]
- 85.Tuttle KR, Milton JE, Packard DP, Shuler LA, Short RA. Dietary amino acids and blood pressure: a cohort study of patients with cardiovascular disease. Am. J. Kidney Dis. 2012;59:803–809. doi: 10.1053/j.ajkd.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 86.Elliott P, Stamler J, Dyer AR, Appel L, Dennis B, Kesteloot H, Ueshima H, Okayama A, Chan Q, Garside DB. Association between protein intake and blood pressure: the INTERMAP Study. Arch. Int. Med. 2006;166:79–87. doi: 10.1001/archinte.166.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nikolic SB, Sharman JE, Adams MJ, Edwards LM. Metabolomics in hypertension. J. Hypertens. 2014;32:1159–1169. doi: 10.1097/HJH.0000000000000168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.