Abstract
Mutations in the parkin gene cause autosomal recessive, juvenile-onset parkinsonism. Parkin is an E3 ubiquitin ligase that mediates the ubiquitination of protein substrates. Disease-associated mutations cause a loss-of-function of parkin which may compromise the poly-ubiquitination and proteasomal degradation of specific protein substrates, potentially leading to their deleterious accumulation. Here, we identify the molecular chaperones, Hsp70 and Hsc70, as substrates for parkin. Parkin mediates the ubiquitination of Hsp70 both in vitro and in cultured cells. Parkin interacts with Hsp70 via its second RING finger domain and mutations in/near this domain compromise Hsp70 ubiquitination. Ubiquitination of Hsp70 fails to alter its steady-state levels or turnover, nor does it promote its proteasomal degradation. Consistent with this observation, Hsp70 levels remain unaltered in brains from parkin-deficient autosomal recessive, juvenile-onset parkinsonism subjects, whereas alternatively, Hsp70 levels are elevated in the detergent-insoluble fraction of sporadic Parkinson’s disease/dementia with Lewy bodies brains. Parkin mediates the multiple mono-ubiquitination of Hsp70/Hsc70 consistent with a degradation-independent role for this ubiquitin modification. Our observations support a novel functional relationship between parkin and Hsc/Hsp70 and support the notion that parkin is a multi-purpose E3 ubiquitin ligase capable of modifying proteins either via attachment of alternatively linked poly-ubiquitin chains or through multiple mono-ubiquitination to achieve alternate biological outcomes.
Keywords: autosomal recessive juvenile-onset parkinsonism, PARK2, parkin, Parkinson’s disease, parkinsonism, ubiquitination
Parkinson’s disease (PD) is a chronic neurodegenerative movement disorder characterized by the progressive loss of nigrostriatal dopaminergic neurons together with other neuronal subgroups in addition to the appearance of intracy-toplasmic proteinaceous inclusions termed Lewy bodies (Lang and Lozano 1998a,b; Braak et al. 2003). The majority of PD cases appear sporadic in origin although a small proportion of cases manifest in a familial manner (Dawson and Dawson 2003; Hardy et al. 2003, 2006; Moore et al 2005a). Mutations in the parkin gene (PARK2; OMIM 600116) cause autosomal recessive juvenile-onset parkinsonism (AR-JP) (West and Maidment 2004). Parkin mutations are the most common identified cause of early-onset familial PD compatible with recessive inheritance accounting for up to 50% of all cases, and account for up to 10% of all early-onset PD cases (Lucking et al. 2000; Scott et al. 2001; West et al. 2002; West and Maidment 2004). These observations suggest a unique role for the parkin gene in the development of early-onset PD.
The parkin gene encodes a multi-domain protein containing an N-terminal ubiquitin-like (Ubl) domain and a C-terminal really interesting new gene (RING) box domain consisting of two RING finger motifs separated by an in-between-RING finger (IBR) motif. Similar to other RING finger-containing proteins, parkin can function as an E3 ubiquitin protein ligase that participates in the covalent attachment of ubiquitin to specific cellular protein substrates (Imai et al. 2000; Shimura et al. 2000; Zhang et al. 2000). Ubiquitination of proteins proceeds through the successive and highly coordinated actions of ubiquitin-activating (E1), conjugating (E2) and ligase (E3) enzymes, respectively (Pickart 2001). Ubiquitination can have a number of physiological consequences depending upon the manner in which ubiquitin moieties are conjugated to a given protein substrate, the most notable of which being the targeted degradation of certain poly-ubiquitinated protein substrates by the 26S proteasomal complex (Pickart 2001; Glickman and Ciechanover 2002). Consistent with a role as an E3 ligase, parkin can selectively interact with and utilize the E2 enzymes UbcH7, UbcH8 and the dimeric complex UbcH13/Uev1, as well as the endoplasmic reticulum-associated E2s, Ubc6 and Ubc7 (Shimura et al. 2000; Zhang et al. 2000; Imai et al. 2001; Doss-Pepe et al. 2005; Matsuda et al. 2006). Parkin may also function as part of larger protein complexes, including a Skp1-Cullin-F-box-like complex or a protein complex containing Hsp70 and CHIP (Imai et al. 2002; Staropoli et al. 2003). The majority of disease-associated mutations in parkin are considered to be loss-of-function in that they either impair the interaction of parkin with E2s, protein substrates, cofactors or other critical protein interactors, alter the biochemical solubility or cellular localization of parkin, or they reduce or abolish the catalytic activity or expression of parkin (Doss-Pepe et al. 2005; Sriram et al. 2005; Hampe et al. 2006; Matsuda et al. 2006).
A number of putative protein substrates for parkin have been identified, including CDCrel-1 and 2a (Zhang et al. 2000; Choi et al. 2003), synphilin-1 (Chung et al. 2001; Lim et al. 2005), Pael-R (Imai et al. 2001), synaptotagmin XI (Huynh et al. 2003), α- and β-tubulin (Ren et al. 2003), RanBP2 (Um et al. 2006), cyclin E (Staropoli et al. 2003), the aminoacyl-tRNA synthetase cofactor p38/AIMP2 (Corti et al. 2003; Ko et al. 2005), far upstream sequence element [FUSE]-binding protein 1 (FBP1) (Ko et al. 2006), Eps15 (Fallon et al. 2006) and protein interacting with C-kinase 1 (PICK1) (Joch et al. 2007). Parkin has been shown to mediate the ubiquitination of particular substrates which may accelerate their turnover, often through proteasomal-mediated degradation (Moore 2006). Consistent with such a mechanism, non-ubiquitinated forms of Pael-R, cyclin E, CDCrel-1 and 2a, p38/AIMP2 and FBP1 have been shown to accumulate in brain tissue from parkin-deficient AR-JP patients suggesting that parkin may normally contribute to their turnover in vivo (Zhang et al. 2000; Imai et al. 2001; Choi et al. 2003; Staropoli et al. 2003; Ko et al. 2005, 2006). Of these, only p38/AIMP2 and FBP1 additionally accumulate in brain tissue from parkin null mice, MPTP-treated mice and sporadic PD subjects suggesting that both proteins are authentic substrates of parkin that are targeted for proteasomal degradation through poly-ubiquitination (Ko et al. 2005, 2006). However, a number of previous studies with parkin have failed to convincingly demonstrate that ubiquitination of particular protein substrates (i.e. CDCrel-2a, synphilin-1) promotes their degradation by the proteasome or other degradative pathways (Chung et al. 2001; Choi et al. 2003; Lim et al. 2005; Moore 2006). Thus, parkin-mediated ubiquitination of particular protein substrates may also serve alternative degradation-independent roles in addition to proteasomal degradation. Recent examples of such a degradation-independent function include the observations that i) parkin can mediate the poly-ubiquitination of synphilin-1 predominantly through lysine-63-linked chains which fails to promote its turnover (Lim et al. 2005), and ii) parkin can catalyze the multiple mono-ubiquitination of itself in vitro (Hampe et al. 2006; Matsuda et al. 2006).
In the present study, we identify the molecular chaperone, Hsp70, as a novel substrate for the ubiquitin ligase activity of parkin. Multiple lines of evidence demonstrate that parkin mediates the multiple mono-ubiquitination of Hsp70 both in vitro and in cultured cells which fails to affect the steady-state levels, turnover or degradation of this protein. This study therefore provides additional support for an alternative, degradation-independent biological role for parkin-mediated protein ubiquitination, in addition to a novel functional relationship between parkin and Hsp70.
Materials and methods
Expression plasmids, antibodies and recombinant proteins
Mammalian expression plasmids for full-length human HA-tagged parkin, myc-tagged parkin and HA-tagged ubiquitin have been described (Zhang et al. 2000; Chung et al. 2001, 2004; Moore et al. 2005b; Sriram et al. 2005). To generate HA-tagged mutant ubiquitin cDNAs, PCR-mediated site-directed mutagenesis was performed using the QuickChange XL kit (Stratagene) to introduce lysine (K) to arginine (R) substitutions at position 48 (K48R) or all lysine residues at positions 6, 11, 27, 29, 33, 48 and 63 (K0) through subsequent rounds of mutagenesis, as previously described (Lim et al. 2005). Myc-tagged parkin constructs harboring disease-associated mutations (R42P, T240R, R256C, G328E, C431F, P437L, W453X and Q311X) and HA-tagged parkin deletion mutants (ΔUbl, R1-IBR IBR-R2, R1 and R2) have been described (Chung et al. 2001; Sriram et al. 2005). Human full-length FLAG-tagged parkin, V5-tagged Hsp70 and Hsc70 were kindly provided by Dr. Ryosuke Takahashi (RIKEN Brain Science Institute, Japan). Full-length myc-tagged human tau (shortest four-repeat isoform, 4R0N) constructs (wild-type or P301L) and myc-tagged human CHIP were kindly provided by Dr. Leonard Petrucelli (Mayo Clinic, Jacksonville).
The following antibodies were employed: mouse monoclonal antic-myc (clone 9E10), anti-c-myc-peroxidase, anti-HA (clone 12CA5) and anti-HA-peroxidase antibodies (Roche); mouse monoclonal anti-FLAG-(M2), anti-FLAG-(M2)-peroxidase, anti-β-tubulin and rabbit polyclonal anti-actin antibodies (Sigma); rabbit polyclonal anti-Hsp70 and mouse monoclonal anti-Hsp70 (clone SPA-810) antibodies (Stressgen); rabbit polyclonal anti-ubiquitin antibody (Dako); mouse monoclonal anti-V5 and anti-V5-peroxidase antibodies (Invitrogen); mouse monoclonal anti-tau (clone TAU-5) antibody (BD Biosciences); mouse monoclonal anti-polyubiquitinated conjugates (clone FK1) and anti-mono/polyubiquitinated conjugates (clone FK2) antibodies (Affiniti Research); mouse monoclonal anti-parkin (clone PRK8) antibody has been described (Pawlyk et al. 2003); rabbit polyclonal anti-superoxide dismutase 1 (SOD1) antibody was kindly provided by Dr. David Borchelt (University of Florida, Gainesville); peroxidase-coupled anti-mouse and anti-rabbit IgG secondary antibodies (Pierce Biotechnology).
Recombinant human ubiquitin-carrier protein 7 (UbcH7) protein was obtained from Sigma; rabbit ubiquitin-activating enzyme E1 protein was obtained from Calbiochem; human Hsp70 protein was from Stressgen; human ubiquitin, K48R-ubiquitin and methylated-ubiquitin proteins were obtained from Affiniti Research; baculovirus-derived His6-tagged human parkin protein has been described (Chung et al. 2004).
Cell culture and transient tranfection
Human SH-SY5Y neuroblastoma cells and HEK293 cells were maintained in DMEM media supplemented with 10% FBS and penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. For transient transfection, cells were seeded into 10-cm dishes and transfected with plasmid DNA using Lipofectamine Plus reagent (Invitrogen) accordingly to manufacturer’s recommendations. Cells were routinely harvested at 36–48 h post-transfection.
Co-immunoprecipitation and western blotting
For co-immunoprecipitation (IP) assays, SH-SY5Y or HEK293 cells were transiently transfected with 2 µg of each plasmid. After 36–48 h, cells were harvested in IP buffer [1X phosphate buffered saline (PBS), 0.5% Triton-X-100, Complete Mini protease inhibitor cocktail (Roche)]. Cell lysates were rotated at 4°C for 1 h and soluble supernatant fractions were obtained by centrifugation at 17 500 g for 15 min at 4°C. Supernatant fractions were combined with 50 µL protein G sepharose 4 fast flow (50% slurry; Amersham Biosciences) pre-incubated with mouse monoclonal anti-myc (5 µg), anti-FLAG (10 µg), anti-V5 (1 µg), or rabbit polyclonal anti-Hsp70 (1 µg) antibodies followed by overnight incubation by rotation at 4°C. Sepharose complexes were pelleted by centrifugation and washed sequentially with IP buffer supplemented with 500 mM NaCl (1×), IP buffer alone (2×) and PBS (3×). Immuno-precipitates were eluted by heating at 95°C for 5 min in 2X Laemmli sample buffer (Bio-Rad) containing 2-mercaptoethanol (5% v/v), and immunoprecipitates or inputs (1% soluble lysate) were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose (0.2 µm; Invitrogen), and subjected to Western blot analysis. Proteins were visualized by enhanced chemiluminescence (ECL; Amersham Biosciences). Quantitation of protein levels was performed using densitometry analysis software (Alphalmager, Alpha Innotech Corp.) and data were analyzed by two-tailed unpaired Student’s t-test to determine differences between the means.
Where indicated, transfected SH-SY5Y cells were treated with MG132 (5 µM; Affiniti Research), clasto-Lactacystin β-lactone (10 µM; Affiniti Research) or DMSO as a control for 24 h prior to harvesting. For cycloheximide (CHX) chase assays, CHX (100 µg/mL; Sigma) was added to transfected SH-SY5Y cells at 48 h post-transfection and cells were chased and harvested at 0, 1, 3, 6 and 24 h thereafter.
In vivo ublqultlnatlon assays
SH-SY5Y or HEK293 cells transiently transfected with HA-tagged ubiquitin and V5-tagged Hsp70 with or without FLAG- or myc-tagged parkin, were harvested at 36–48 h post-transfection in IP buffer, and IP was conducted with anti-V5 or anti-Hsp70 antibodies. IPs were washed stringently five times in IP buffer supplemented with 500 mM NaCl and once with PBS, heated at 95°C for 5 min and eluted proteins were subjected to Western blot analysis with anti-HA, anti-V5 or anti-Hsp70 antibodies to detect Hsp70-ubiquitin conjugates.
In vitro ublqultlnatlon assays
To examine parkin-mediated ubiquitination of Hsp70 in vitro, various combinations of recombinant Hsp70 (1 µg; Stressgen), ubiquitin (10 µg; Affiniti Research), E1 (100 ng; Calbiochem), UbcH7 (200 ng; Sigma) and baculovirus-derived (Bv) parkin (5 µg) proteins were incubated together in assay buffer (50 mM Tris-HCl pH 7.5, 2.5 mM MgCl2, 2 mM DTT, 2 mM ATP) at 37°C for 2 h with shaking. Reactions were terminated by addition of an equal volume of 2X Laemmli sample buffer and boiling for 5 min. In separate reactions, Bv-parkin was replaced with IP FLAG-parkin (10 µL suspension), obtained by IP with anti-FLAG antibody (10 µg) from transfected SH-SY5Y cells (10-cm dish; 2 µg DNA), washed stringently and resuspended in 50 µL 50 mM Tris-HCl pH 7.5. Ubiquitination reactions were resolved by 7.5–10% SDS-PAGE and subjected to Western blot analysis with anti-Hsp70, antiubiquitin and anti-parkin antibodies.
Human brain tissue
Human brain tissue was obtained through the brain donation program of the Morris K. Udall Parkinson’s Disease Research Center of Excellence at Johns Hopkins Medical Institutions in compliance with HIPAA regulations. Fresh-frozen cingulate gyrus tissue from two normal control brains, two Alzheimer’s disease (AD) brains and five Parkinson’s disease (PD)/dementia with Lewy bodies (DLB) brains (age: 76.7 ± 2.7 years; post-mortem delay: 11.1 ± 1.8 h) was kindly provided by Dr. Juan Troncoso (Department of Pathology, Johns Hopkins University School of Medicine) and was utilized for Western blot analysis as previously described (Moore et al. 2005b). Fresh-frozen frontal cortex tissue from four normal control brains and four parkin-linked AR-JP brains (age: 61.5 ± 2.6 years; postmortem delay: 11.6 ± 1.7 h) was collected at the Departments of Neurology and Pathology, Juntendo University School of Medicine, Japan, and was kindly provided by Drs. Nobutaka Hattori and Yoshikuni Mizuno. AR-JP cases 1 and 3 harbor a homozygous deletion of parkin exon 4, whereas cases 2 and 4 carry a homozygous deletion of parkin exon 3, as previously described (Hattori et al. 1998; Shimura et al. 1999; Moore et al. 2005b).
Fractionation of human brain tissue
Detergent-soluble and -insoluble protein fractions were prepared from human brain tissue by homogenization of samples in TNE buffer (10 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 X Complete protease inhibitor cocktail [Roche]). The homogenate was centrifuged at 100 000 g for 20 min at 4°C, and the resulting pellet (P1) and supernatant (S1, detergent-soluble) fractions were collected. The P1 fraction was washed once in TNE buffer, and the resulting pellet (P2, detergent-insoluble) was homogenized and further solubilized by sonication and boiling in TNE buffer containing 1% SDS and 0.5% sodium deoxycholate. Protein fractions were quantitated using the BCA kit (Pierce Biotech) with BSA standards. Proteins (20–30 µg/lane) were resolved by SDS-PAGE and analyzed by Western blotting with anti-Hsp70, anti-SOD1 and anti-β-tubulin antibodies.
Results
Identification of Hsp70 as a novel substrate of parkin
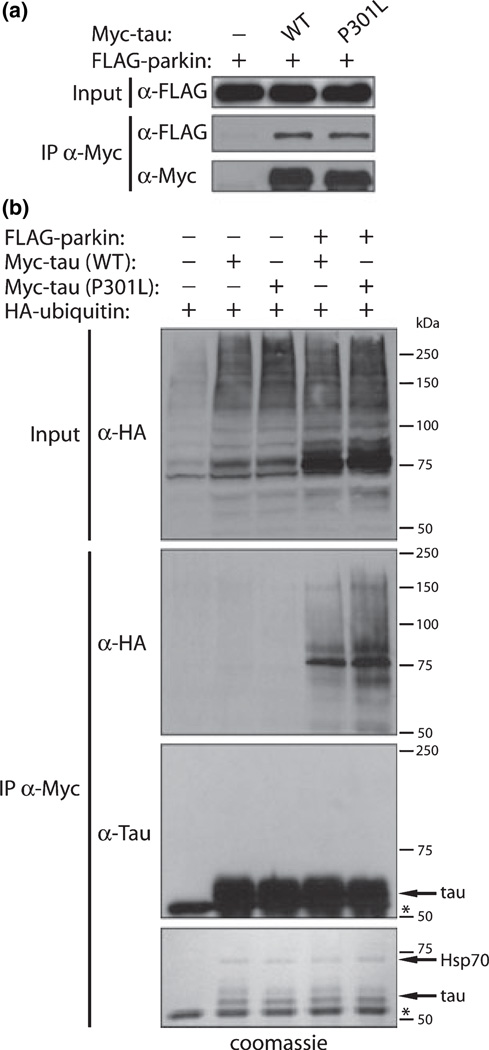
A novel interaction between parkin and the microtubule-associated protein tau has previously been reported (Petrucelli et al. 2004). The relationship between parkin and tau is of potential interest for a number of reasons, including 1) mutations in tau (i.e. P301L) are associated with the neurodegenerative tauopathy, frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) (Hutton et al. 1998), 2) neuronal tau-positive pathology is observed in brain tissue from parkin-linked AR-JP subjects (Mori et al. 1998), and 3) tau-positive inclusions associated with an array of neurodegenerative tauopathies are often also ubiquitin-positive (Lee et al. 2001; Moore et al. 2003). While tau may potentially be a substrate of parkin-mediated ubiquitination, CHIP (carboxyl terminus of Hsc70-interacting protein) has instead been identified as the major E3 ubiquitin ligase important for tau ubiquitination and metabolism (Hatakeyama et al. 2004; Petrucelli et al. 2004; Shimura et al. 2004; Sahara et al. 2005; Dickey et al. 2006). However, it is unclear whether tau can also be ubiquitinated by parkin. Therefore, the ability of parkin to ubiquitinate tau was examined. First, the robust interaction between parkin and four-repeat tau was confirmed by co-immunoprecipitation assay in human SH-SY5Y cells (Fig. 1a). To determine whether parkin can ubiquitinate tau, SH-SY5Y cells co-expressing myc-tagged tau (WT or the FTDP-17-linked mutant P301L) and HA-tagged ubiquitin with or without FLAG-tagged parkin were subjected to immunoprecipitation (IP) with anti-myc antibody and ubiquitination was monitored by probing with anti-HA antibody whereas the formation of tau-ubiquitin conjugates was examined with anti-tau antibody. In the presence of parkin, tau associates with ubiquitin but is not covalently modified by ubiquitin as revealed by the distinct absence of tau-ubiquitin conjugates of increased molecular mass (Fig. 1b). Furthermore, no evidence was found to support the ubiquitination of tau by parkin using similar ubiquitination assays in HEK293 cells or by using in vitro ubiquitination assays with recombinant components, nor were the levels of tau isoforms altered in whole brain lysates derived from parkin-deficient mice relative to their wild-type littermates (data not shown). Thus, tau is not a substrate of parkin-mediated ubiquitination despite the interaction of these two proteins. These data suggest instead that an unknown endogenous protein expressed in SH-SY5Y cells that strongly associates with tau may instead be a target of parkin-mediated ubiquitination. To identify this putative parkin substrate, anti-myc IPs from the ubiquitination assay above were resolved by SDS-PAGE and stained with coomassie colloidal blue to identify potential tau-interacting proteins (Fig. 1b). An approximate 70 kDa protein associates with tau that was subsequently identified by MALDI-TOF mass spectrometry of tryptic peptides as non-ubiquitinated Hsp70 (data not shown). Hsp70 represents a good candidate for parkin-mediated ubiquitination since the major ubiquitin-protein conjugate observed in our ubiquitination assay (Fig. 1b) is ∼78 kDa, consistent with the predicted mass of Hsp70 (∼70 kDa) covalently linked to a single ubiquitin moiety (∼8 kDa).
Fig. 1. Microtubule-associated protein tau is not a parkin substrate.
(a) Co-immunoprecipitation of parkin and four-repeat tau. SH-SY5Y cells expressing FLAG-tagged parkin and myc-tagged tau (WT or FTDP-17-linked P301L mutant) were subjected to IP with anti-myc antibody and IPs and inputs were probed with anti-FLAG or anti-myc antibodies, (b) Parkin fails to ubiquitinate tau. SH-SY5Y cells expressing myc-tagged tau (WT or P301L mutant) and HA-tagged ubiquitin with or without FLAG-tagged parkin were subjected to IP with anti-myc antibody and IPs and inputs were probed with anti-HA or anti-tau antibodies. Notice the distinct lack of tau-ubiquitin conjugates revealed by probing with anti-tau antibody. Separate SDS-PAGE gels containing identical anti-myc IPs were stained with coomassie colloidal blue (lower panel). Arrows indicate the presence of tau or associated endogenous Hsp70 that was subsequently identified by MALDI-TOF mass spectrometry. *denotes IgG heavy chain. Molecular mass markers are indicated in kilodaltons (kDa). Blots are representative of at least two independent experiments.
Hsp70 is directly ubiquitinated by parkin
To determine whether parkin can ubiquitinate endogenous Hsp70, and to ascertain whether tau is required for this modification, in vivo ubiquitination assays were performed. SH-SY5Y cells co-expressing combinations of FLAG-tagged parkin and HA-tagged ubiquitin, with or without myc-tagged tau (WT or P301L) were subjected to IP with anti-Hsp70 antibody and probed with anti-HA and anti-Hsp70 antibodies. Parkin can directly ubiquitinate endogenous Hsp70 in the absence or presence of tau as revealed by anti-HA immunoreactivity and by a corresponding shift in the molecular mass of Hsp70 (Fig. 2a). Parkin-mediated ubiquitination of Hsp70 is consistent with the covalent attachment of one or two ubiquitin monomers in this assay. Since parkin has been reported to form a protein complex with Hsp70 and the ubiquitin ligase CHIP (Imai et al. 2002), the contribution of CHIP to parkin-mediated Hsp70 ubiquitination was examined in cultured cells. Soluble lysates derived from HEK293 cells co-expressing V5-tagged Hsp70 and HA-tagged ubiquitin together with either FLAG-tagged parkin, myc-tagged CHIP or both proteins, were probed with anti-V5 antibody to monitor Hsp70-ubiquitin conjugate formation (Fig. 2b). While parkin readily ubiquitinates Hsp70, CHIP alone is unable to promote Hsp70 ubiquitination nor does it influence parkin-mediated Hsp70 ubiquitination in these cells (Fig. 2b). This finding suggests that the ubiquitin ligase activity of CHIP does not additionally contribute to the parkin-mediated ubiquitination of Hsp70. To further confirm the direct parkin-mediated ubiquitination of Hsp70 and to ascertain the requirement for potential co-factors, in vitro ubiquitination assays were developed with recombinant components. Ubiquitin, E1, UbcH7 (E2), Hsp70 and baculovirus-derived human parkin were incubated together with ATP at 37°C for 2 h and subsequently subjected to Western blotting with anti-Hsp70 antibody to monitor the formation of Hsp70-ubiquitin conjugates, or with antibodies to ubiquitin and parkin (Fig. 2c). In the presence of all components, Hsp70 is covalently modified with up to four ubiquitin monomers corresponding to an appropriate shift in the molecular mass of Hsp70. Ubiquitination of Hsp70 is prevented if parkin, UbcH7 or E1 enzymes are excluded from the assay thus highlighting the essential requirement for all three components for efficient ubiquitin conjugation to Hsp70 (Fig. 2c). Ubiquitin-protein conjugates are still produced in the absence of Hsp70 which most likely reflect the auto-ubiquitination of parkin (Fig. 2c). Parkin-mediated ubiquitination of Hsp70 could also be demonstrated in vitro using an alternate source of recombinant parkin obtained by IP from SH-SY5Y cells expressing FLAG-tagged parkin (Fig. 2d). Thus, Hsp70 is a direct target of parkin-mediated ubiquitination. Hsp70 is modified in vitro predominantly by one or two ubiquitin monomers whereas the addition of three or four monomers is less abundant. This observation suggests that Hsp70 is modified either by short poly-ubiquitin chains that are probably not sufficient in length for targeting proteasomal degradation or by multiple mono-ubiquitination. Collectively, these findings suggest that Hsp70 is a direct substrate for the E3 ubiquitin ligase activity of parkin.
Fig. 2. Hsp70 is ubiquitinated by parkin.
(a) In vivo ubiquitination assay. SH-SY5Y cells expressing combinations of HA-tagged ubiquitin, FLAG-tagged parkin and myc-tagged tau (WT or P301L mutant) were subjected to IP with anti-Hsp70 antibody to isolate endogenous Hsp70 and IPs and inputs were probed with anti-HA and anti-Hsp70 antibodies. In the presence of ubiquitin, parkin ubiquitinates endogenous Hsp70 with up to two ubiquitin (Ub) moieties. Tau is not required for parkin-mediated Hsp70 ubiquitination. (b) CHIP fails to ubiquitinate Hsp70. HEK293 cells co-expressing V5-tagged Hsp70 and HA-tagged ubiquitin together with either FLAG-tagged parkin, myc-tagged CHIP or both proteins were harvested and equivalent detergent-soluble protein fractions were probed with anti-V5 antibody to monitor Hsp70-ubiquitin conjugate formation, or with anti-FLAG and anti-myc antibodies. CHIP alone fails to appreciably ubiquitinate Hsp70 and does not influence parkin-mediated Hsp70 ubiquitination. (c) In vitro ubiquitination assay. Recombinant rabbit E1, human UbcH7 (E2), baculovirus (Bv)-derived human parkin, human Hsp70 and human ubiquitin proteins were incubated together in assay buffer containing ATP for 2 h at 37°C. Reactions were probed with anti-Hsp70, anti-parkin and anti-ubiquitin antibodies. In the presence of all components, Hsp70 is directly modified by parkin with up to four ubiquitin moieties reflected by a corresponding shift in the molecular mass of Hsp70. Parkin auto-ubiquitination is revealed by probing with anti-ubiquitin in the absence of Hsp70. (d) In vitro ubiquitination assay with FLAG-tagged parkin derived by IP from transfected SH-SY5Y cells. Reactions were probed with anti-Hsp70, anti-FLAG and anti-ubiquitin antibodies. Parkin catalyzes the ubiquitination of hsp70 and removal of UbcH7 or parkin impairs Hsp70 ubiquitination. Unmodified Hsp70 is indicated by arrows and ubiquitinated forms of Hsp70 [Hsp70-(Ubn)] or ubiquitin-protein conjugates [(Ub)n] are also indicated. Molecular mass markers are indicated in kilodaltons (kDa). Blots are representative of at least two independent experiments.
Disease-associated mutations impair parkin-mediated ubiquitination of Hsp70
Parkin has previously been shown to interact with a protein complex containing Hsp70 and CHIP in brain tissue and cultured cells (Imai et al. 2002). To further explore the interaction between parkin and Hsp70, and to determine which protein domains of parkin are responsible for this interaction, co-immunoprecipitation experiments were conducted. SH-SY5Y cells co-expressing V5-tagged Hsp70 and HA-tagged parkin deletion mutants were subjected to IP with anti-V5 antibody. Hsp70 interacts robustly with full-length parkin and more moderately with parkin deletion mutants minimally containing the RING2 (R2) domain (ΔUbl, IBR-R2 and R2 domains) but fails to interact with deletion mutants lacking the RING2 domain (R1-IBR and R1 domains) (Fig. 3a). These data suggest that the RING2 domain of parkin is both necessary and sufficient for the interaction with Hsp70.
Fig. 3. Disease-associated RING2 domain mutations impair parkin-mediated Hsp70 ubiquitination.
(a) Hsp70 specifically interacts with the RING2 domain of parkin. SH-SY5Y cells expressing V5-tagged Hsp70 and HA-tagged parkin deletion mutants were subjected to IP with anti-V5 antibody and probed with anti-HA and anti-V5 antibodies, whereas inputs were probed with anti-HA antibody. Hsp70 interacts with full-length parkin and deletion mutants (ΔUbl, IBR-R2 and R2) minimally harboring the RING2 (R2) domain. Image highlighting the domain structure and amino acid position of parkin deletion mutants, the location of disease-associated mutations, and the capacity to interact with Hsp70 (+ or −). (b) In vivo ubiquitination assay with disease-associated parkin variants. SH-SY5Y cells expressing V5-tagged Hsp70, HA-tagged ubiquitin and myc-tagged parkin harboring various disease-associated mutations, were subjected to IP with anti-V5 antibody and probed with anti-HA or anti-V5 antibodies to reveal Hsp70-ubiquitin conjugates, and inputs were probed with anti-HA and anti-myc antibodies. Mutations in the RING2 domain (C431F and P437L), the RING1 domain (T240R) or truncating mutations (Q311X and W453X) markedly impair parkin-mediated Hsp70 ubiquitination. ‘X’ represents the introduction of a premature stop codon for some mutations leading to a truncated parkin species. (c) Interaction of Hsp70 with disease-associated parkin variants. HEK293 cells co-expressing myc-tagged parkin variants (WT, T240R, C431F or P437L) with or without V5-tagged Hsp70, were subjected to IP with anti-V5 antibody and probed with anti-myc or anti-V5 antibodies to reveal interactions, and inputs were probed with anti-myc antibody to control for equivalent loading of parkin variants. The T240R and C431F mutations moderately impair the normal interaction of parkin with Hsp70. Ubiquitinated forms of Hsp70 [Hsp70-(Ub)n] and ubiquitin-protein conjugates [(Ub)n] are indicated. Molecular mass markers are indicated in kilodaltons (kDa). Blots are representative of at least two independent experiments.
The effects of disease-associated mutations on the parkin-mediated ubiquitination of Hsp70 were examined by performing in vivo ubiquitination assays. SH-SY5Y cells co-expressing V5-tagged Hsp70 and HA-tagged ubiquitin together with myc-tagged parkin harboring various disease-associated missense or truncating mutations were subjected to IP with anti-V5 antibody and probed with anti-HA and anti-V5 antibodies to monitor Hsp70 ubiquitination. Mutations in the RING2 domain of parkin (C431F and P437L), or truncating mutations that are predicted to disrupt the RING2 domain (Q311X and W453X), selectively impair the ubiquitination of Hsp70 as compared to WT parkin (Fig. 3a). The T240R mutation located in the RING1 domain of parkin appears to confer reduced activity towards Hsp70, whereas mutations in the Ubl (R42P), RING1 (R256C) or IBR (G328E) domains of parkin fail to alter the normal ubiquitination of Hsp70 (Fig. 3b). The impairment in parkin-mediated Hsp70 ubiquitination conferred by certain disease-associated mutations could be due to alterations in the interaction between these parkin variants and Hsp70. To explore this possibility, HEK293 cells co-expressing myc-tagged parkin variants (WT, T240R, C431F and P437L) with or without V5-tagged Hsp70, were subjected to IP with anti-V5 antibody and probed with anti-myc and anti-V5 antibodies to examine the interaction of parkin with Hsp70 (Fig. 3c). While WT and mutant parkin could all interact with Hsp70, the T240R and C431F mutations but not the P437L mutation moderately impairs the interaction of parkin with Hsp70, relative to WT parkin, despite equivalent expression between parkin variants (Fig. 3c). Therefore, for certain disease-associated parkin mutants (i.e. T240R and C431F), a combination of impaired ubiquitin ligase activity and a reduced interaction with Hsp70 most likely account for their impaired ability to ubiquitinate Hsp70. Collectively, these data demonstrate that parkin interacts with Hsp70 selectively via its RING2 domain and mutations in or near this domain compromise the parkin-mediated ubiquitination of Hsp70.
Parkin-mediated ubiquitination of Hsp70 fails to promote its degradation
Parkin-mediated poly-ubiquitination of its protein substrates has previously been implicated in promoting their targeted degradation by the 26S proteasomal complex (Zhang et al. 2000; Imai et al. 2001; Staropoli et al. 2003; Ko et al. 2005, 2006; Moore 2006). To determine whether ubiquitinated Hsp70 is targeted for degradation by the proteasome, in vivo ubiquitination assays were performed in the presence or absence of proteasomal inhibitors. SH-SY5Y cells expressing HA-tagged ubiquitin with or without FLAG-tagged parkin were treated with the general proteasome inhibitor MG132 (5 µM) for 24 h and subjected to IP with anti-Hsp70 antibody. Proteasomal inhibition fails to alter the pattern of parkin-mediated ubiquitination of endogenous Hsp70 as determined by anti-HA and anti-Hsp70 immunoreactivity to monitor the formation of Hsp70-ubiquitin conjugates (Fig. 4a). Similar results were obtained with the specific proteasomal inhibitor, clasto-lactacystin β-lactone (data not shown). This data suggests that ubiquitinated Hsp70 is not subjected to degradation by the proteasome.
Fig. 4. Ubiquitination of Hsp70 by parkin is degradation-independent.
(a) Ubiquitinated Hsp70 is not degraded by the proteasome. SH-SY5Y cells expressing HA-tagged ubiquitin with or without FLAG-tagged parkin were treated with or without the proteasomal inhibitor MG132 (5 µM) for 24 h, subjected to IP with anti-Hsp70 antibody, and IPs and inputs were probed with anti-HA and anti-Hsp70 antibodies to monitor Hsp70-ubiquitin conjugate formation. Proteasomal inhibition fails to affect the levels of ubiquitinated Hsp70. (b) Parkin-mediated ubiquitination does not influence the steady-state levels of Hsp70. SH-SY5Y cells expressing HA-tagged ubiquitin and increasing quantities of FLAG-tagged parkin (0.05–1 µg plasmid DNA) were harvested at 48 h post-transfection and equivalent detergent-soluble protein fractions were probed with anti-Hsp70 or anti-FLAG antibodies, or with anti-actin antibody to demonstrate equal protein loading. Over-exp; overexposed blot to highlight Hsp70-ubiquitin conjugates. (c) Parkin-mediated ubiquitination does not alter the turnover of Hsp70. SH-SY5Y cells expressing HA-tagged ubiquitin and V5-tagged Hsp70 with or without FLAG-tagged parkin were treated with cycloheximide (CHX, 100 µg/mL) at 48 h post-transfection, and cells were harvested at 0, 1, 3, 6 and 24 h thereafter. Equivalent detergent-soluble protein fractions were probed with anti-V5 and anti-FLAG antibodies to monitor Hsp70 turnover, or with anti-actin antibody to demonstrate equal protein loading. Unmodified Hsp70 is indicated by arrows and ubiquitinated forms of Hsp70 [Hsp70-(Ubn)] are also indicated. Molecular mass markers are indicated in kilodaltons (kDa). Blots are representative of at least two independent experiments.
To further clarify the consequences of parkin-mediated ubiquitination of Hsp70, experiments were performed to assess the effects of parkin on the steady-state levels and turnover of Hsp70. Soluble cell lysates from SH-SY5Y cells co-expressing HA-tagged ubiquitin and increasing quantities of FLAG-tagged parkin were probed with anti-Hsp70 antibody to assess the steady-state levels of endogenous Hsp70. Increasing concentrations of parkin fail to appreciably alter the steady-state levels of Hsp70 as compared to the absence of parkin (Fig. 4b). Parkin does, however, lead to a dose-dependent increase in the ubiquitination of Hsp70 (Fig. 4b), consistent with the notion that ubiquitinated forms of Hsp70 are relatively stable. Next, the effect of parkin on the turnover of Hsp70 was examined by performing cycloheximide chase assays. SH-SY5Y cells co-expressing V5-tagged Hsp70 and HA-tagged ubiquitin with or without FLAG-tagged parkin were treated with cycloheximide at 48 h post-transfection to inhibit new protein synthesis and Hsp70 levels were chased at different time points over the following 24 h by probing resulting soluble cell lysates with anti-V5 antibody. Parkin fails to alter the turnover of Hsp70 over the 24 h time course examined but retains the ability to promote Hsp70 ubiquitination (Fig. 4c). Together, these data suggest that parkin does not influence the steady-state levels or turnover of Hsp70 thus supporting an insignificant role for parkin-mediated ubiquitination in promoting the degradation of Hsp70.
Hsp70 levels are not altered in parkin-linked AR-JP brains
To examine the effects of parkin on the steady-state levels of Hsp70 in vivo, experiments were performed with control and parkin-deficient brain tissue. The steady-state levels and detergent solubility of Hsp70 were assessed in frontal cortex tissue from the brains of aged-matched normal control subjects and from subjects with AR-JP owing to homozygous exonic deletions within the parkin gene. The absence of parkin in AR-JP brain tissue fails to significantly alter the steady-state levels or detergent solubility of Hsp70, as compared to control brain tissue (Fig. 5a). Similar results were obtained using wild-type or parkin null mouse brain tissue (data not shown). This in vivo data further supports the contention that parkin-mediated ubiquitination of Hsp70 does not accelerate its turnover or reduce its stability.
Fig. 5. Steady-state levels and detergent-solubility of Hsp70 in parkin-linked AR-JP and sporadic PD brains.
(a) Hsp70 levels are not altered in parkin-deficient AR-JP brains. Equivalent detergent-soluble and -insoluble protein fractions extracted from frontal cortex tissue of control (1–4) and parkin-deficient AR-JP brains (1–4) were analyzed by Western blot with anti-Hsp70 antibody, or anti-actin antibody as a protein loading control. Hsp70 levels were quantified by densitometry and normalized to β-actin levels in the detergent-soluble and -insoluble fraction of control (n = 4) and AR-JP (n = 4) brains. Data are expressed as a percentage (%) of control levels as the ratio of Hsp70 to β-actin, and bars represent the mean ± SEM. (b) Hsp70 displays reduced detergent-solubility in sporadic PD/DLB brains. Equivalent detergent-soluble and -insoluble protein fractions extracted from cin-gulate cortex tissue of control (5–6), Alzheimer’s disease (AD; 1–2) and PD/DLB (1–5) brains, were analyzed by Western blot with anti-Hsp70 antibody, or with anti-SOD1 and anti-β-tubulin antibodies as protein loading controls. Hsp70 levels were quantified by densitometry and normalized in the detergent-soluble and -insoluble fraction of control (control + AD; n = 4) and PD/DLB (n = 5) brains. Data are expressed as a percentage (%) of control levels as the ratio of Hsp70 to SOD1 (soluble) or β-tubulin (insoluble), and bars represent the mean ± SEM. Differences between means were analyzed by two-tailed unpaired Student’s t-test, *p < 0.05 when compared with control, ns, non-significant.
The failure of parkin to accelerate the degradation of Hsp70 by ubiquitination perhaps suggests that the ubiquitination of Hsp70 serves an alternative degradation-independent biological role that probably acts in concert with the function of parkin. To explore whether Hsp70 may play a role in sporadic PD, the steady-state levels and detergent solubility of Hsp70 were monitored in cingulate cortex tissue from aged-matched normal control, Alzheimer’s disease (AD) and PD/dementia with Lewy bodies (DLB) subjects. The levels of Hsp70 are significantly reduced in the detergent-soluble fraction of PD/DLB brains compared to control or AD brains but are significantly elevated in the detergent-insoluble fraction of PD/DLB brains relative to control/AD brains (Fig. 5b). This finding indicates that Hsp70 is a less soluble protein species in PD/DLB brains and suggests that Hsp70 may be selectively up-regulated or stabilized in the insoluble fraction in PD/DLB brains but down-regulated or destabilized in the soluble fraction. This data supports a role for Hsp70 in the pathogenesis of sporadic PD/DLB consistent with the reported localization of Hsp70 to Lewy bodies and Lewy neurites in sporadic PD brains (Auluck et al. 2002).
Parkin mediates the multiple mono-ubiquitination of Hsp70 and Hsc70
Hsp70 can be covalently modified with one to four ubiquitin monomers by parkin which fails to facilitate its degradation suggesting that ubiquitination of Hsp70 serves an alternative biological function. To further explore the consequences of Hsp70 ubiquitination, experiments were conducted to examine the nature of parkin-mediated ubiquitin attachment to Hsp70. Ubiquitin monomers are first covalently attached to proteins through an isopeptide linkage between the extreme C-terminal glycine residue of ubiquitin (G76) and the ε-amino groups of lysine (K) side chains within the protein substrate through the cooperation of E1, E2 and E3 enzymes (Pickart 2001; Glickman and Ciechanover 2002). Thereafter, the sequential conjugation of an internal lysine residue of ubiquitin (i.e. K6, 11, 27, 29, 33, 48 or 63) to the terminal G76 residue of a new ubiquitin monomer leads to the formation of a poly-ubiquitin chain. The length and lysine linkage of the poly-ubiquitin chain dictates the fate of the target protein (Pickart 2001; Glickman and Ciechanover 2002; Pickart and Fushman 2004). Poly-ubiquitin chains linked through K48, or in some cases K29, target proteins for degradation by the 26S proteasomal complex (Pickart 2001; Glickman and Ciechanover 2002). Alternatively, poly-ubiquitin chains linked through other lysine residues, particularly K63, as well as single or multiple mono-ubiquitination have been associated with a number of diverse degradation-independent functions, including DNA repair, endocytosis, protein trafficking and transcriptional regulation (Pickart 2001; Glickman and Ciechanover 2002). There is some suggestion that poly-ubiquitin chains consisting of at least four ubiquitin monomers is the minimum length necessary to target the recipient protein substrate for proteasomal degradation (Thrower et al. 2000).
To determine the exact nature of parkin-mediated ubiquitin chain linkage to Hsp70, in vivo ubiquitination assays were conducted with various lysine point mutants of ubiquitin. Ubiquitin harboring a lysine (K) to arginine (R) point mutation at position 48 (K48R) is incapable of forming K48-linked poly-ubiquitin chains, whereas a lysine-less ubiquitin mutant (designated K0) with all seven lysine residues mutated to arginine is incapable of poly-ubiquitin chain formation but can still be singly or multiply attached to protein substrates as monomeric ubiquitin. To this end, SH-SY5Y cells co-expressing FLAG-tagged parkin and HA-tagged ubiquitin mutants (WT, K48R or K0) were subjected to IP with anti-Hsp70 antibody, and probed with anti-HA or anti-Hsp70 antibodies to monitor Hsp70 ubiquitination. Parkin-mediated ubiquitination of endogenous Hsp70 occurs using K48R- or K0-ubiquitin mutants, in a manner comparable to WT-ubiquitin, as suggested by similar patterns of anti-HA and anti-Hsp70 immunoreactivity which mainly reveal a mono- and di-ubiquitinated Hsp70 species (Fig. 6a). A smaller level of Hsp70 ubiquitination observed with K0-ubiquitin may relate to less efficient ubiquitin conjugation perhaps due to an altered protein conformation of this ubiquitin mutant owing to seven point mutations. The protein levels of each ubiquitin mutant in this experiment were comparable (Fig. 6a). The ability of parkin to mediate the conjugation of multiple K0-ubiquitin monomers to Hsp70 suggests that Hsp70 undergoes multiple mono-ubiquitination and not poly-ubiquitination.
Fig. 6. Hsp70 and Hsc70 are multiply mono-ubiquitinated by parkin.
(a) Hsp70 is mono-ubiquitinated by parkin in cultured cells. SH-SY5Y cells expressing HA-tagged ubiquitin mutants (WT, K48R or K0) and FLAG-tagged parkin were subjected to IP with anti-Hsp70 antibody, and IPs and inputs were probed with anti-HA and anti-Hsp70 antibodies to monitor Hsp70-ubiquitin conjugate formation. K48R- and K0-ubiquitin mutants fail to impair parkin-mediated Hsp70 ubiquitination. (b) Hsp70 is mono-ubiquitinated and not poly-ubiquitinated by parkin. SH-SY5Y cells expressing HA-tagged ubiquitin and V5-tagged Hsp70 with or without FLAG-tagged parkin were subjected to IP with anti-V5 antibody, and IPs and inputs were probed with ubiquitin-protein conjugate-specific antibodies that specifically recognize either poly-ubiquitinated proteins (FK1) or mono- and poly-ubiquitinated proteins (FK2). Notice only FK2 but not FK1 antibody detects ubiquitinated Hsp70. *denotes IgG heavy chain. (c) In vitro ubiquitination assay with ubiquitin mutants. Recombinant rabbit E1, human UbcH7 (E2), ba-culovirus (Bv)-derived human parkin, human Hsp70 and human ubiquitin (WT, K48R or methylated) proteins were incubated together in assay buffer containing ATP for 2 h at 37°C. Reactions were probed with anti-Hsp70 antibody to monitor Hsp70-ubiquitin conjugate formation. K48R- and methyl-ubiquitin fail to impair parkin-mediated Hsp70 ubiquitination relative to WT-ubiquitin. The absence of ubiquitin impairs Hsp70 ubiquitination. (d) Multiple mono-ubiquitination of Hsc70 by parkin. SH-SY5Y cells expressing combinations of V5-tagged Hsc70, HA-tagged ubiquitin (WT, K48R or K0) and FLAG-tagged parkin were harvested and equivalent detergent-soluble protein fractions were probed with anti-HA or anti-V5 antibodies to monitor Hsc70-ubiquitin conjugate formation. Parkin mediates Hsc70 ubiquitination that is not impaired when K48R- or K0-ubiquitin is substituted for WT-ubiquitin. Unmodified Hsp/Hsc70 is indicated by arrows and ubiquitinated forms of Hsp/Hsc70 [Hsp/Hsc70-(Ub)n] and ubiquitin-protein conjugates [(Ub)n] are indicated. Molecular mass markers are indicated in kilodaltons (kDa). Blots are representative of at least two independent experiments.
To confirm the multiple mono-ubiquitination of Hsp70 by parkin, monoclonal antibodies were employed that specifically detect ubiquitin-protein conjugates but not free monomeric ubiquitin. Antibody FK1 specifically recognizes only poly-ubiquitinated proteins whereas antibody FK2 can recognize both mono- and poly-ubiquitinated proteins (Haglund et al. 2003; Hampe et al. 2006). SH-SY5Y cells co-expressing V5-tagged Hsp70 and HA-tagged ubiquitin with or without FLAG-tagged parkin were subjected to IP with anti-V5 antibody and probed with FK1, FK2 and anti-V5 antibodies to monitor Hsp70 ubiquitination. Parkin promotes the ubiquitination of Hsp70 above background levels as determined by anti-V5 immunoreactivity which reveals Hsp70-ubiquitin conjugates consistent with the attachment of at least two ubiquitin monomers (Fig. 6b). Antibody FK2 detects Hsp70-ubiquitin conjugates consistent with mono-and di-ubiquitinated Hsp70 species, whereas antibody FK1 fails to detect any Hsp70-ubiquitin conjugates (Fig. 6b). However, both FK1 and FK2 antibodies are capable of detecting multiple ubiquitin-protein conjugates in soluble cell lysates to similar degrees (Fig. 6b). The failure of the poly-ubiquitin chain-specific FK1 antibody to detect Hsp70-ubiquitin conjugates further suggests that Hsp70 is mono-ubiquitinated and not poly-ubiquitinated by parkin.
To further confirm these observations in vitro, ubiquitination assays were performed with recombinant components as before but instead using WT- and K48R-ubiquitin, or methylubiquitin which is terminally methylated on all seven lysine residues rendering it incapable of poly-ubiquitin chain formation but still capable of single or multiple mono-ubiquitination. In the presence of E1, UbcH7, parkin and ubiquitin, Hsp70 is covalently modified with one to four ubiquitin monomers corresponding to an appropriate shift in molecular mass, as before, irrespective of the type of modified ubiquitin utilized (Fig. 6c). Parkin-mediated ubiquitination of Hsp70 is not observed in the absence of ubiquitin, as expected. A small increase in the molecular mass of each Hsp70-ubiquitin conjugate in the presence of methyl-ubiquitin is due to the addition of seven methyl groups to ubiquitin which tends to retard its electrophoretic mobility (Fig. 6c). The ability of parkin to utilize methyl-ubiquitin for Hsp70 ubiquitination in a manner comparable to WT ubiquitin further suggests that parkin mediates the multiple mono-ubiquitination of Hsp70 which varies from the conjugation of one to four ubiquitin monomers in this assay. Collectively, these data provide strong support for parkin-mediated multiple mono-ubiquitination of Hsp70 which entirely accounts for the inability of parkin to influence the steady-state levels, turnover or proteasomal degradation of Hsp70.
To determine whether parkin can also ubiquitinate other molecular chaperones closely related to Hsp70, and also to examine the type of ubiquitin attachment, in vivo ubiquitination assays were conducted with Hsc70. Soluble cell lysates derived from SH-SY5Y cells co-expressing combinations of V5-tagged Hsc70, HA-tagged ubiquitin (WT, K48R or K0) and FLAG-tagged parkin were probed with anti-V5 antibody to monitor the formation of Hsc70-ubiquitin conjugates. Parkin promotes the ubiquitination of Hsc70 consistent with the covalent attachment of up to two WT-ubiquitin monomers, as revealed by an appropriate shift in the molecular mass of Hsc70 (Fig. 6d). Parkin also mediates the attachment of K48R- and K0-ubiquitin to Hsc70 in a manner similar to WT-ubiquitin (Fig. 6d). In the absence of parkin, Hsc70 is only minimally ubiquitinated. This data suggests that Hsc70 is also multiply mono-ubiquitinated by parkin. Furthermore, these findings suggest that parkin-mediated multiple mono-ubiquitination of molecular chaperones could be a common function of this E3 ubiquitin ligase and might extend to other members of the chaperone family in addition to Hsp70 and Hsc70.
Discussion
The major finding of this study is that Hsp70 is a novel substrate for the E3 ubiquitin ligase activity of parkin. Parkin interacts with Hsp70 specifically through its second RING finger motif and disease-associated mutations in or near this RING finger selectively impair the ubiquitination of Hsp70. The ubiquitination of Hsp70 by parkin fails to alter its steady-state levels or turnover, nor does it promote its degradation by the proteasome. Consistent with these findings, the levels of Hsp70 are not altered in brain tissue from parkin-deficient AR-JP subjects. Alternatively, Hsp70 may be implicated in the sporadic form of PD since the levels of this protein are selectively increased in the detergent-insoluble fraction but are decreased in the detergent-soluble fraction of brain tissue from sporadic PD/DLB subjects. Parkin mediates the multiple mono-ubiquitination of Hsp70 consistent with a degradation-independent role for this type of ubiquitin modification. The activity of parkin also extends to Hsc70 which can similarly be multiply mono-ubiquitinated. Collectively, these data suggest that parkin-mediated ubiquitination of its protein substrates may, in some instances, serve a previously unappreciated degradation-independent biological role. Our findings contribute to the notion that parkin is a multi-purpose E3 ubiquitin ligase that is capable of modifying proteins either via attachment of alternatively linked poly-ubiquitin chains (K48 or K63) or through single or multiple mono-ubiquitination to achieve alternate biological outcomes (Moore 2006). These findings may have important implications for understanding the precise role of parkin in the molecular pathogenesis of familial and sporadic forms of PD.
The significance of parkin-mediated ubiquitination of Hsp70 is not known at present but this post-translational modification fails to promote the degradation of Hsp70. The ubiquitination of Hsp70 and Hsc70 is not unprecedented. The related U-box-containing E3 ubiquitin ligase, CHIP, has been shown to ubiquitinate both Hsc70 and Hsp70 (Jiang et al. 2001; Qian et al. 2006). However, unlike parkin-mediated Hsp70 ubiquitination, CHIP ubiquitinates Hsc70 via non-canonical ubiquitin chains that utilize either lysine 29 or 63 of ubiquitin (Jiang et al. 2001). This ubiquitin modification similarly fails to promote the proteasome-mediated degradation of Hsc70. It is conceivable that mono-ubiquitination of Hsp70 or Hsc70 could serve to mediate a docking interaction with the 19S cap of the 26S proteasomal complex similar to the proposed function of the ubiquitin-like domain of parkin (Sakata et al. 2003). The 19S cap might recognize and bind to mono-ubiquitinated Hsp70 in a degradation-independent manner thus allowing the direct delivery of unfolded protein substrates to the proteasome for subsequent degradation. A similar mechanism has recently been demonstrated for the Hsp/Hsc70 cofactor BAG-1, which acts as a coupling factor between molecular chaperones and the proteasomal complex (Luders et al. 2000). CHIP-mediated poly-ubiquitination of BAG-1 stimulates its binding to the proteasome in a degradation-independent fashion which serves as a regulatory step in the sorting of chaperone substrates to the proteasome (Alberti et al. 2002). CHIP-mediated poly-ubiquitination of Hsc70 has also been suggested to facilitate the degradation of ALS-linked mutant SOD1 through direct interaction of ubiquitinated Hsc70 with the S5a subunit of the 26S proteasomal complex and subsequent substrate delivery (Urushitani et al. 2004). A comparable role for parkin-mediated Hsp/Hsc70 mono-ubiquitination in facilitating the proteasomal degradation of chaperone substrates remains to be demonstrated. Alternatively, it is possible that ubiquitination of Hsp70 regulates its catalytic activity i.e. ATPase or refolding activities. However, since only a small proportion of total Hsp70 is modified by ubiquitin (< 5%), such experimental assays may prove difficult. Our initial attempts to artificially generate a mono-ubiquitinated form of Hsp70 through tagging ubiquitin directly to the N-terminus of Hsp70 were unsuccessful due to ubiquitin-specific proteolytic cleavage of this fusion protein (data not shown). Further studies are therefore warranted on the exact consequences of parkin-mediated Hsp/Hsc70 mono-ubiquitination, and the relationship between parkin and Hsp/Hsc70 in the protein triage decision of damaged or misfolded cellular substrates (Marques et al. 2006).
A large number of putative protein substrates have so far been identified for the E3 ubiquitin ligase activity of parkin (Moore et al. 2005a; Moore 2006). Of these, Hsp70 would appear to be one of the most robustly ubiquitinated substrates, which may, in part, be a consequence of the degradation-independent nature of this parkin-mediated modification. While a number of substrates are reported to be poly-ubiquitinated by parkin and subsequently degraded by the 26S proteasomal complex (Zhang et al. 2000; Imai et al. 2001; Huynh et al. 2003; Ren et al. 2003; Staropoli et al. 2003; Ko et al. 2005, 2006; Moore 2006; Um et al. 2006), data obtained in parkin-deficient brain tissue from AR-JP subjects and mice confirm p38/AIMP2 and FBP1 as the only parkin substrates targeted for proteasomal degradation in vivo (Ko et al. 2005, 2006). Alternatively, degradation-independent parkin-mediated ubiquitination has recently gained attention. In vitro, parkin preferentially catalyzes its own multiple mono-ubiquitination utilizing the E2s UbcH13/Uev1, Ubc7 or UbcH7 (Hampe et al. 2006; Matsuda et al. 2006), and can also partly modify p38/AIMP2 through multiple mono-ubiquitination in cultured cells (Hampe et al. 2006). Parkin also preferentially mediates the degradation-independent poly-ubiquitination of synphilin-1 via lysine-63-linked chains which has been shown to enhance the formation of Lewy body-like inclusions (Lim et al. 2005, 2006). More recently, parkin-mediated ubiquitination of Eps15, an adaptor protein involved in EGF receptor endocytosis and trafficking, failed to promote Eps15 degradation but instead interfered with Eps15-mediated EGF receptor internalization and degradation thereby leading to enhanced EGF receptor-mediated phosphoinositide 3-kinase/Akt pathway signaling (Fallon et al. 2006). Finally, parkin primarily mono-ubiquitinates the PDZ-containing protein PICK1 in a degradation-independent manner to abolish the PKC-induced, PICK1-dependent potentiation of acid-sensing ion channel 2a currents leading to reduced acid-sensing ion channel activity (Joch et al. 2007). Thus, parkin-mediated ubiquitination can clearly modify certain protein substrates in a degradation-independent manner (Moore 2006). At least some of these proteasomal-independent parkin substrates, and their ubiquitination-mediated regulation, may be of importance to the molecular pathogenesis of parkin-linked parkinsonism and potentially to sporadic PD. The identification of Hsp70 and Hsc70 as novel substrates of parkin-mediated multiple mono-ubiquitination may provide important insight into the normal biological function of parkin and the potential role of degradation-independent parkin substrates in parkin-linked parkinsonism.
Acknowledgements
The authors would like to thank Drs. Kenny Chung and Olga Pletnikova (Johns Hopkins) for provision and preparation of human brain fractions, and Dr. Robert Cole, Director of the Mass Spectrometry and Proteomics Facility at Johns Hopkins, for conducting MALDI-TOF mass spectrometry analysis. The authors are grateful for support from the NIH/NINDS P50 NS38377 and R01 NS048206, the Lee Martin Trust, the Sylvia Nachlas Trust, the American Parkinson Disease Association, the National Parkinson Foundation, the Michael J. Fox Foundation for Parkinson’s Research, and the Parkinson’s Disease Foundation. A.B.W. is supported by NIH/NINDS K99/R00 NS058111. T.M.D. is the Leonard and Madlyn Abramson Professor of Neurodegenerative Disease at Johns Hopkins University.
Abbreviations used
- AD
Alzheimer’s disease
- AR-JP
autosomal recessive juvenile-onset parkinsonism
- DLB
dementia with Lewy bodies
- FBP1
[FUSE]-binding protein 1
- IP
immunoprecipitation
- PD
Parkinson’s disease
- PBS
phosphate buffered saline
- PICK1
protein interacting with C-kinase 1
- RING
really interesting new gene
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
References
- Alberti S, Demand J, Esser C, Emmerich N, Schild H, Hohfeld J. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 2002;277:45920–45927. doi: 10.1074/jbc.M204196200. [DOI] [PubMed] [Google Scholar]
- Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Choi P, Snyder H, Petrucelli L, et al. SEPT5_v2 is a parkin-binding protein. Brain Res. Mol. Brain Res. 2003;117:179–189. doi: 10.1016/s0169-328x(03)00318-8. [DOI] [PubMed] [Google Scholar]
- Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J, Ross CA, Dawson VL, Dawson TM. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat. Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Corti O, Hampe C, Koutnikova H, et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum. Mol. Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- Dickey CA, Yue M, Lin WL, et al. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J. Neurosci. 2006;26:6985–6996. doi: 10.1523/JNEUROSCI.0746-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss-Pepe EW, Chen L, Madura K. Alpha-synuclein and parkin contribute to the assembly of ubiquitin lysine 63-linked multiubiquitin chains. J. Biol. Chem. 2005;280:16619–16624. doi: 10.1074/jbc.M413591200. [DOI] [PubMed] [Google Scholar]
- Fallon L, Belanger CM, Corera AT, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Hampe C, Ardila-Osorio H, Fournier M, Brice A, Corti O. Biochemical analysis of Parkinson’s disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum. Mol. Genet. 2006;15:2059–2075. doi: 10.1093/hmg/ddl131. [DOI] [PubMed] [Google Scholar]
- Hardy J, Cookson MR, Singleton A. Genes and parkinsonism. Lancet Neurol. 2003;2:221–228. doi: 10.1016/s1474-4422(03)00350-8. [DOI] [PubMed] [Google Scholar]
- Hardy J, Cai H, Cookson MR, Gwinn-Hardy K, Singleton A. Genetics of Parkinson’s disease and parkinsonism. Ann. Neurol. 2006;60:389–398. doi: 10.1002/ana.21022. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Matsumoto M, Kamura T, Murayama M, Chui DH, Planel E, Takahashi R, Nakayama KI, Takashima A. U-box protein carboxyl terminus of Hsc70-interacting protein (CHIP) mediates poly-ubiquitylation preferentially on four-repeat Tau and is involved in neurodegeneration of tauopathy. J. Neurochem. 2004;91:299–307. doi: 10.1111/j.1471-4159.2004.02713.x. [DOI] [PubMed] [Google Scholar]
- Hattori N, Kitada T, Matsumine H, et al. Molecular genetic analysis of a novel Parkin gene in Japanese families with auto-somal recessive juvenile parkinsonism: evidence for variable homozygous deletions in the Parkin gene in affected individuals. Ann. Neurol. 1998;44:935–941. doi: 10.1002/ana.410440612. [DOI] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Huynh DP, Scoles DR, Nguyen D, Pulst SM. The autosomal recessive juvenile Parkinson disease gene product, parkin, interacts with and ubiquitinates synaptotagmin XI. Hum. Mol. Genet. 2003;12:2587–2597. doi: 10.1093/hmg/ddg269. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 2000;275:35661–35664. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R. CHIP is associated with Parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol. Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- Joch M, Ase AR, Chen CX, Macdonald PA, Kontogiannea M, Corera AT, Brice A, Seguela P, Fon EA. Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol. Biol. Cell. 2007;18:3105–3118. doi: 10.1091/mbc.E05-11-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HS, von Coelln R, Sriram SR, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J. Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HS, Kim SW, Sriram SR, Dawson VL, Dawson TM. Identification of far upstream element-binding protein-1 as an authentic Parkin substrate. J. Biol. Chem. 2006;281:16193–16196. doi: 10.1074/jbc.C600041200. [DOI] [PubMed] [Google Scholar]
- Lang AE, Lozano AM. Parkinson’s disease. First of two parts. N. Engl. J. Med. 1998a;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- Lang AE, Lozano AM. Parkinson’s disease. Second of two parts. N. Engl. J. Med. 1998b;339:1130–1143. doi: 10.1056/NEJM199810153391607. [DOI] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Lim KL, Chew KC, Tan JM, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KL, Dawson VL, Dawson TM. Parkin-mediated lysine 63-linked polyubiquitination: a link to protein inclusions formation in Parkinson’s and other conformational diseases? Neurobiol. Aging. 2006;27:524–529. doi: 10.1016/j.neurobiolaging.2005.07.023. [DOI] [PubMed] [Google Scholar]
- Lucking CB, Durr A, Bonifati V, et al. Association between early-onset Parkinson’s disease and mutations in the parkin gene. N. Engl. J. Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 2000;275:4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- Marques C, Guo W, Pereira P, Taylor A, Patterson C, Evans PC, Shang F. The triage of damaged proteins: degradation by the ubiquitin-proteasome pathway or repair by molecular chaperones. FASEB J. 2006;20:741–743. doi: 10.1096/fj.05-5080fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Kitami T, Suzuki T, Mizuno Y, Hattori N, Tanaka K. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro . J. Biol. Chem. 2006;281:3204–3209. doi: 10.1074/jbc.M510393200. [DOI] [PubMed] [Google Scholar]
- Moore DJ. Parkin: a multifaceted ubiquitin ligase. Biochem. Soc. Trans. 2006;34:749–753. doi: 10.1042/BST0340749. [DOI] [PubMed] [Google Scholar]
- Moore DJ, Dawson VL, Dawson TM. Role for the ubiquitin-proteasome system in Parkinson’s disease and other neurodegenerative brain amyloidoses. Neuromolecular Med. 2003;4:95–108. doi: 10.1385/NMM:4:1-2:95. [DOI] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson’s disease. Annu. Rev. Neurosci. 2005a;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- Moore DJ, Zhang L, Troncoso J, Lee MK, Hattori N, Mizuno Y, Dawson TM, Dawson VL. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum. Mol. Genet. 2005b;14:71–84. doi: 10.1093/hmg/ddi007. [DOI] [PubMed] [Google Scholar]
- Mori H, Kondo T, Yokochi M, Matsumine H, Nakagawa-Hattori Y, Miyake T, Suda K, Mizuno Y. Pathologic and biochemical studies of juvenile parkinsonism linked to chromosome 6q. Neurology. 1998;51:890–892. doi: 10.1212/wnl.51.3.890. [DOI] [PubMed] [Google Scholar]
- Pawlyk AC, Giasson BI, Sampathu DM, Perez FA, Lim KL, Dawson VL, Dawson TM, Palmiter RD, Trojanowski JQ, Lee VM. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J. Biol. Chem. 2003;278:48120–48128. doi: 10.1074/jbc.M306889200. [DOI] [PubMed] [Google Scholar]
- Petrucelli L, Dickson D, Kehoe K, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum. Mol. Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Zhao J, Feng J. Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J. Neurosci. 2003;23:3316–3324. doi: 10.1523/JNEUROSCI.23-08-03316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara N, Murayama M, Mizoroki T, Urushitani M, Imai Y, Takahashi R, Murata S, Tanaka K, Takashima A. In vivo evidence of CHIP up-regulation attenuating tau aggregation. J. Neurochem. 2005;94:1254–1263. doi: 10.1111/j.1471-4159.2005.03272.x. [DOI] [PubMed] [Google Scholar]
- Sakata E, Yamaguchi Y, Kurimoto E, et al. Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep. 2003;4:301–306. doi: 10.1038/sj.embor.embor764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, Nance MA, Watts RL, et al. Complete genomic screen in Parkinson disease: evidence for multiple genes. Jama. 2001;286:2239–2244. doi: 10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, et al. Immunohistochemical and subcellular localization of Parkin protein: absence of protein in autosomal recessive juvenile parkinsonism patients. Ann. Neurol. 1999;45:668–672. doi: 10.1002/1531-8249(199905)45:5<668::aid-ana19>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- Shimura H, Schwartz D, Gygi SP, Kosik KS. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- Sriram SR, Li X, Ko HS, Chung KK, Wong E, Lim KL, Dawson VL, Dawson TM. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum. Mol. Genet. 2005;14:2571–2586. doi: 10.1093/hmg/ddi292. [DOI] [PubMed] [Google Scholar]
- Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JW, Min do S, Rhim H, Kim J, Paik SR, Chung KC. Parkin ubiquitinates and promotes the degradation of RanBP2. J. Biol. Chem. 2006;281:3595–3603. doi: 10.1074/jbc.M504994200. [DOI] [PubMed] [Google Scholar]
- Urushitani M, Kurisu J, Tateno M, Hatakeyama S, Nakayama K, Kato S, Takahashi R. CHIP promotes proteasomal degradation of familial ALS-linked mutant SOD1 by ubiquitinating Hsp/Hsc70. J. Neurochem. 2004;90:231–244. doi: 10.1111/j.1471-4159.2004.02486.x. [DOI] [PubMed] [Google Scholar]
- West AB, Maidment NT. Genetics of parkin-linked disease. Hum. Genet. 2004;114:327–336. doi: 10.1007/s00439-003-1074-6. [DOI] [PubMed] [Google Scholar]
- West A, Periquet M, Lincoln S, et al. Complex relationship between Parkin mutations and Parkinson disease. Am. J. Med. Genet. 2002;114:584–591. doi: 10.1002/ajmg.10525. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl Acad. Sci. USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]