Abstract
Thus far, the focus of personalized medicine has been the prevention and treatment of conditions that affect adults. Although advances in genetic technology have been applied more frequently to prenatal diagnosis than to fetal treatment, genetic and genomic information is beginning to influence pregnancy management. Recent developments in sequencing the fetal genome combined with progress in understanding fetal physiology using gene expression arrays indicate that we could have the technical capabilities to apply an individualized medicine approach to the fetus. Here I review recent advances in prenatal genetic diagnostics, the challenges associated with these new technologies and how the information derived from them can be used to advance fetal care. Historically, the goal of prenatal diagnosis has been to provide an informed choice to prospective parents. We are now at a point where that goal can and should be expanded to incorporate genetic, genomic and transcriptomic data to develop new approaches to fetal treatment.
The goal of personalized medicine is to optimize health and develop individualized therapy for disease by combining a person's genetic and genomic data with information about their lifestyle and exposures. So far, personalized medicine has focused on the prevention and treatment of conditions affecting adults, such as cancer and cardiovascular disease1. Remarkably, few studies have addressed the therapeutic implications of recent advances in genetic technologies for the fetus. This is surprising, as progress in prenatal diagnosis2 has led to widespread antenatal screening programs that have been successfully reproduced throughout the developed world. One could argue that a personalized medicine approach would have maximal benefit over the course of an individual's lifetime if it began in the womb or at birth. However, a major limitation of applying advanced genetic and genomic techniques in the prenatal setting is that genomic variation can be identified for which the clinical implications are not known. But as knowledge of the fetus and fetal development progresses by sequencing fetal DNA and RNA, new treatment opportunities will emerge. Now is the time to acknowledge the scientific progress that has taken place in the area of prenatal genomic medicine and consider the practical and ethical considerations raised by these technologies. Furthermore, the strong interest in and acceptance of direct-to-consumer genetic testing by many pregnant women3–5 effectively mandates that multidisciplinary specialists should consider and discuss these issues before a ‘parallel universe’ populated by commercial interests is fully established outside of the traditional health care system.
Importantly, advances in prenatal diagnosis that allow for a more detailed analysis of fetal genetics and genomics provide some of the data necessary for a personalized approach to fetal medicine. I first discuss these technological advances and provide examples of how they have provided a greater understanding of the genetic basis of specific fetal disorders (Table 1). In the prenatal setting, pure fetal material may be obtained invasively through amniocentesis or chorionic villus sampling or noninvasively through maternal blood. The invasive diagnostic techniques are generally safe and accurate, although they need to be performed by obstetricians with specific expertise, and they carry with them a small but measurable chance of miscarriage. There is expanding interest in noninvasive techniques of fetal assessment using cell-free DNA and RNA molecules that circulate in the maternal blood. Maternal venipuncture has no associated risk of fetal loss; however, maternal blood contains a mixture of both fetal and maternal nucleic acids, which increases the downstream analytic complexity.
Table 1.
Overview of advances in molecular testing that affect fetal diagnostics and treatment
| Genetic material analyzed | Type of testing | Consequences |
|---|---|---|
| Chromosomal DNA | Chromosome microarray analysis | Detection of submicroscopic fetal chromosome abnormalities. Detection of copy number variation in the genome. |
| Cell-free DNA in maternal blood | Real-time PCR amplification; massively parallel sequencing | Noninvasive prenatal diagnosis of fetal sex, RhD genotype, aneuploidy, other submicroscopic chromosome abnormalities and single-gene disorders. |
| Cell-free RNA in amniotic fluid | Gene expression microarray analysis; comparative transcriptomic analyses between normal and affected individuals | Improved knowledge of fetal functional development in health and in disease states. Development of new biomarkers. Identification of new therapeutic targets. |
| RNA in maternal blood | Gene expression microarray analysis; comparative transcriptomic analyses between normal and affected individuals; quantitative miRNA analysis | Improved knowledge of fetal functional development in health and in disease states. Development of new biomarkers. Identification of new therapeutic targets. |
| RNA in placenta | Gene expression microarray analysis; comparative transcriptomic analyses between normal and affected individuals | Improved knowledge of fetal functional development in health and in disease states. Development of new biomarkers. Identification of new therapeutic targets. |
| RNA in cord blood | Gene expression microarray analysis; comparative transcriptomic analyses between normal and affected individuals | Improved knowledge of fetal functional development in health and in disease states. Development of new biomarkers. Identification of new therapeutic targets. |
Transcriptomic analyses are also beginning to be applied to prenatal diagnosis; these analyses can provide a dynamic view of fetal and placental development. I discuss advances in this area and how such approaches might be used to find new biomarkers for fetal diseases or provide insights into the functional pathways involved in a particular disorder. I also discuss the practical and ethical challenges facing the field of fetal diagnostics, including how these new technologies should be incorporated into clinical practice, as well as how to move forward to translate these insights to provide new therapeutic strategies for fetal disease and achieve the goal of fetal personalized medicine.
Advances in invasive diagnosis by fetal cytogenomics
Cytogenetic diagnosis is in a transition from the microscopic analysis of chromosomes in metaphase to the analysis of DNA within chromosomes using microarrays (Fig. 1). In the postnatal setting, a chromosomal microarray analysis is now considered to be the first-tier cytogenetic diagnostic test for individuals with congenital anomalies, intellectual disability or both6. Even before the International Standard Cytogenomic Array (ISCA) Consortium (https://www.iscaconsortium.org/) published their evidence-based summary of 33 studies involving over 21,000 children and adults in 2010 (ref. 6), there was a demand for applying this technology prenatally. Therefore, in 2007 the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) funded a clinical trial comparing the accuracy of microarrays to that of standard cytogenetic analyses (NCT01279733).
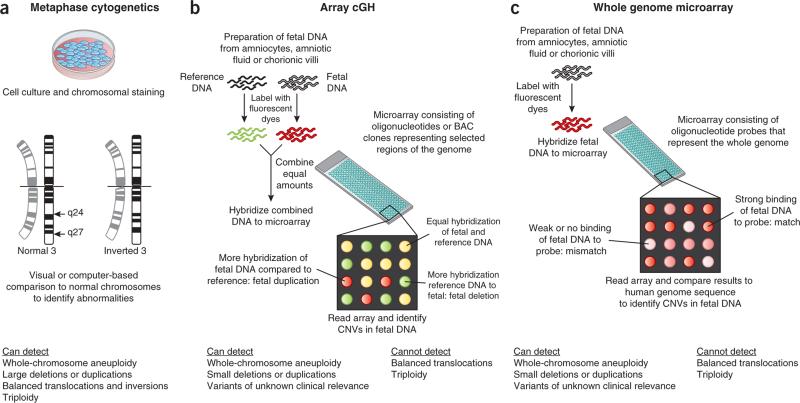
Figure 1.
Outline of the three major current techniques for analyzing fetal chromosomes. (a) Fetal cells are cultured and analyzed during cell division in metaphase. Chromosomes are analyzed under the microscope for the presence of dark and light staining bands. The staining patterns are compared with normal reference standards. Only relatively large deviations from normal (~5–10 Mb) can be detected. (b,c) The DNA within the fetal chromosomes, rather than the fetal chromosome itself, is compared to reference genomes. The DNA can be isolated from fetal cells or cell-free amniotic fluid with or without prior cell culture. In array comparative genomic hybridization (cGH) (b), patient and reference DNA samples are labeled with competing fluorescent dyes and hybridized to an array that contains DNA probes. Each probe is known to map to a specific region of the human genome. When the array is read, areas of mismatch appear as red or green. Special software converts the signal to indicate the affected area of the genome. In the method shown in c, only the patient's DNA is hybridized to an array that contains oligonucleotides (~60 bp) with coverage across the human genome. Areas of mismatch between the patient's DNA and the reference sequence are identified as CNVs. BAC, bacterial artificial chromosome.
Microarrays offer advantages over conventional karyotyping
The advantages of microarray testing over the current standard, metaphase karyotyping, include a higher sensitivity to detect chromosome deletions, duplications and unbalanced rearrangements and a shorter turnaround time. This shorter time is because the DNA isolation procedures can be automated, and there is no need to culture the fetal cells. Whereas metaphase analyses using banding techniques can identify chromosome deletions and duplications in the range of 5–10 Mb, the higher resolution provided by microarrays can detect changes as small as 50–100 kb7. The disadvantages of microarrays include their inability to detect a balanced chromosome rearrangement or a triploid karyo-type. The design of the array is crucial. Oligonucleotide and bacterial artificial chromosome arrays provide less coverage of the genome but are specifically designed to identify known chromosomal aberrations that are associated with clinically significant disease. Whole-genome arrays provide sequence information without complete knowledge of the long-term prognostic implications of all of the variants detected. Such sequence information, however, may be permanently archived and used as a reference source to advance our knowledge of the health of individuals and populations, human development and disease.
The challenge of assessing clinical meaning from microarray results
A major concern regarding the widespread application of chromosome microarray technology to low-risk pregnancies is the possibility of detecting copy number variants (CNVs) that have unknown clinical consequences. This is a problem because the phenotype of the fetus cannot be completely assessed while in the womb, so it can be difficult to determine the functional consequences of these genetic changes. When an indeterminate CNV is detected, several factors increase the probability of its pathogenicity. These include being absent in the parents (de novo), having a size of larger than 1 Mb, being a deletion rather than a duplication or involving a gene-rich area of the genome. A final consideration is whether there is a similar phenotype that involves genes within the same pathway7. When a de novo CNV is found in the fetus, publicly available references can be consulted to interpret its clinical implications, such as the database of Genotypes and Phenotypes (dbGAP) (http://www.ncbi.nlm.nih.gov/gap)8 or the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER) (http://decipher.sanger.ac.uk/)9. However, most clinicians do not have adequate training to perform such interpretations of CNVs.
Combining karyotype and microarray analyses to maximize information
In the prenatal setting, there is uncertainty as to whether it is better to use an array that only detects findings of known clinical meaning or one that has the greatest possible resolution10. Despite this uncertainty, a consensus exists with regard to the essential need for counseling before and after testing as well as the clinical scenarios in which microarrays are a useful adjunct to metaphase karyotyping. For example, in the settings of miscarriage11 and stillbirth12,13, in which tissue culture frequently fails, the quality of the chromosome preparation is suboptimal or both, microarrays have been shown to improve the detection of genomic alterations in the fetus. Similarly, the addition of a microarray analysis when there is a fetal structural anomaly and a normal metaphase karyotype results in the detection of 1–16% additional clinically relevant chromosome abnormalities14–21. The lower end of the range reflects the use of lower density arrays14–18, and the higher percentages are derived from the studies that used whole-genome platforms19–21. The use of microarrays has shown that up to 25% of apparently balanced translocations analyzed by metaphase karyotyping are in fact unbalanced and contain substantial aberrations in regions of the genome known to encode for essential genes.
A recent retrospective study performed in Israel compared antenatal karyotyping to microarrays; both options were routinely offered to couples undergoing invasive procedures22. This study illustrated the potential added value of a microarray analysis in a prenatal context. Of the 269 fetuses examined, 254 had a normal metaphase karyotype and 15 had abnormal metaphase findings of unknown clinical meaning. In the former group, 36 out of 254 fetuses had an abnormal microarray result, and 33 of these 36 fetuses were found to have benign CNVs. The remaining three fetuses had de novo duplications (two of them also had sonographic abnormalities). All three of the women carrying these fetuses opted for termination. The overall risk of having an unbalanced genomic finding after a normal karyotype was 1 in 84, or 1.1%. The microarray results were normal in 11 of the 15 fetuses (73.3%) with an abnormal metaphase finding; all of these pregnancies were continued. In four fetuses, unbalanced abnormalities in the karyotype were confirmed by a microarray analysis; the women chose to terminate their pregnancies in these instances. In the entire study, clinically relevant genetic or genomic changes that had not been previously detected were found in 18 out of 269 (6.5%, or 1 in 15) fetuses. Thus, the addition of genomic data allowed the physicians to better define the expected prognosis for the child, which substantially influenced subsequent pregnancy management and parental decisions.
Advances in noninvasive prenatal diagnosis
The clinical experience with microarrays illustrates the immediate impact that rapid advances in technology can have on clinical care, even before the completion of a prospective blinded trial. The same phenomenon occurred with the noninvasive diagnosis of fetal sex and the Rhesus D blood group (RhD) using circulating nucleic acids. In 1997, Lo and his colleagues first found that cell-free fetal DNA circulates in maternal plasma and serum by showing that gene sequences that are unique to the fetus could be amplified from maternal plasma23. Maternal blood contains a mixture of maternal and fetal (predominantly placental24–30) cell-free nucleic acids. Therefore, fetal DNA can be isolated from maternal blood and analyzed noninvasively (Fig. 2).
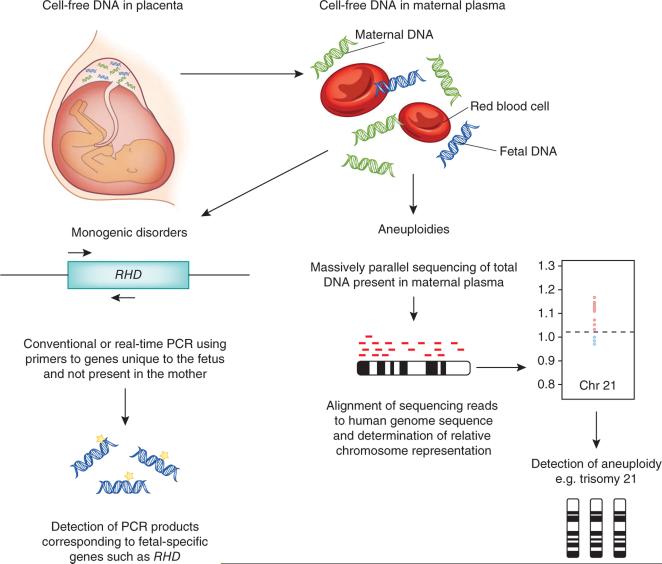
Figure 2.
Cell-free DNA analysis to diagnose fetal disorders. Cell-free DNA from maternal plasma is a mixture of maternal and fetal DNA. When testing is being performed for the diagnosis of the presence or absence of a uniquely fetal gene, the relatively low-cost method of real-time quantitative PCR can be used (left). Primers and probes that map uniquely to the fetal genome can be used to amplify the gene of interest to allow, for example, the detection of the RHD gene. For diagnosing aneuploidies such as Down's syndrome, the total cell-free DNA in maternal plasma is sequenced (right). The DNA is fragmented and analyzed in 36-bp lengths known as reads. These 36-bp reads are aligned against the human genome sequence and counted. The amount of DNA in chromosomes of interest, for example, those involved in common fetal aneuploidies such as those of chromosomes 13, 18 and 21, is normalized against the DNA from other chromosomes to determine the relative number of reads present in a given sample. The lower right image shows an increased number of sequences derived from chromosome (chr) 21 (in red) plotted against what should normally be present, indicated by the dashed line slightly above 1.0. This result is consistent with a fetus that has trisomy 21.
Noninvasive diagnosis of RhD
By the early 1990s, the gene sequence for RhD (RHD) was known, and the prenatal diagnosis of fetal Rhesus blood groups had moved from the serotyping of fetal blood to the genotyping of amniocytes or villi31. An immediate clinical application for cell-free fetal DNA in maternal blood was envisioned for the noninvasive diagnosis of fetal blood type. In an initial feasibility study, fetal RHD genotype was accurately predicted by PCR amplification of cell-free DNA isolated from peripheral blood samples in 55 of 57 RhD– pregnant women32. The definitive fetal genotype was determined by amplification of RHD in the amniotic fluid or by serology at birth. There were two false-negative calls in first-trimester samples, which were presumed to be the result of a low concentration of fetal DNA in maternal plasma early in gestation. As early as 2001, after appropriate preclinical validation studies, the International Blood Group Reference Laboratory in the UK transitioned this test to prenatal care33.
Similar large-scale clinical studies were performed in France and The Netherlands34 that identified the major challenges associated with noninvasive testing. False-negative results were generally caused by a lack of fetal DNA in the sample, either for biological (too early in gestation) or technical (poor extraction) reasons34. More recent studies have incorporated secondary PCRs using sex determining region Y (SRY), a Y chromosome sequence used as a marker of male fetal DNA, RASSF1A, a marker of differentially methylated placental DNA, or other paternal genetic markers as internal controls to verify that fetal DNA is present in the sample35. In general, instances of false positives are caused by the fact that the majority of RhD– individuals of African ancestry have one of two RHD variants, the RHD pseudo-gene or the RHD-CE-Ds hybrid sequence36. Once these variants were identified, primer and probe combinations were developed to either specifically recognize or entirely avoid the problem of false-positive amplification. Noninvasive prenatal testing (NIPT) of RHD has recently been implemented on a routine national basis in Denmark37. Prior to this policy change, all RhD– pregnant women received RhD immunoglobulin. In the Danish study, routine noninvasive prenatal testing meant that 862 women (37.2% of the total) could avoid unnecessary exposure to this blood product. NIPT has advanced antenatal care by limiting RhD prophylaxis to only those women who carry an RhD+ fetus. In addition, noninvasive fetal blood group tests are available for the blood groups C, c, E and KEL35,36.
Noninvasive diagnosis of sex-linked disorders
Similarly, PCR amplification of cell-free DNA in maternal blood has been used as a noninvasive alternative to cytogenetic diagnosis after an invasive procedure to determine fetal sex. Such diagnostic tests are recommended when knowledge of fetal sex is needed for the management of X-linked conditions or of ambiguous genitalia detected by sonogram. Furthermore, knowledge of fetal sex helps determine which women need to take steroids to prevent the masculinization of a female fetus that is at risk for congenital adrenal hyperplasia. The widespread availability of this technology over the internet and in pharmacies in the United States, however, raises substantial ethical concerns4. To determine the analytic and clinical validity of diagnosing fetal sex, a recent meta-analysis of 80 discrete datasets from 57 studies and 6,541 singleton fetuses showed that the overall diagnostic performance of noninvasive testing was high (with a 95.4% sensitivity and a 98.1% specificity) when maternal blood samples were obtained after 7 weeks of gestation38. The variables that had the biggest effect on test performance were gestational age and DNA amplification methodology. Currently in the United States, noninvasive fetal DNA testing is not available at point of care, and this testing is not approved by the Clinical Laboratory Improvement Amendments (CLIA) or reimbursed by insurers, despite the results of this meta-analysis38. In contrast, in the UK, this approach has already been incorporated into prenatal care and has led to a reduction in invasive prenatal diagnostic procedures for sex-linked disorders39.
Noninvasive prenatal testing for fetal aneuploidies
Antenatal recognition of fetuses with trisomy 21 is a major goal of all screening programs in the developed world40. Whereas NIPT of fetal sex and RHD genotype can be performed using the relatively straightforward technique of real-time PCR, identification of fetal trisomy 21 using maternal plasma is much more complicated because there are no unique fetal gene sequences to be detected. Multiple attempts over the past two decades have used intact fetal cells in maternal blood41,42, cell-free DNA43 or cell-free RNA44,45 to noninvasively diagnose Down's syndrome. The subsequent development of digital PCR techniques led to the ability to quantify the amount of nucleic acids in a maternal sample by counting amplifications from single molecules. Digital PCR has been applied to the molecular detection of aneuploidy using cell-free RNA46 or DNA47.
Although some of the early studies achieved success on a small scale, the arrival of massively parallel sequencing techniques and instrumentation substantially changed the landscape for this type of research. In 2008, two independent reports published 2 months apart each showed that fetal aneuploidy could be accurately diagnosed using DNA isolated from maternal plasma samples by mapping and aligning short sequence tags to a reference human genome, followed by counting and bioinformatics analyses48,49.
Subsequently, progress has been rapid. Since January 2011, at least ten independent large-scale clinical trials involving NIPT of trisomies 21, 18 and 13 have been published50–59. Three of these studies were prospective54–56. So far, all of them were performed with study subjects that were at a high risk for fetal aneuploidy and incorporated a case-control study design in which an independent third party matched known euploid fetuses with fetuses that had a variety of chromosomal abnormalities. The teams that performed the sample processing, sequencing and bioinformatics analyses were blinded. All of the studies achieved similar, near-perfect rates of detection of trisomy 21, including in two sets of twins (in which at least one was affected)52, three cases of mosaic trisomy 21 (ref. 56) and several cases caused by an unbalanced translocation56,60. All of the studies had very low false-positive rates (<1%). The various studies, however, differed in their bioinformatics analytical approaches. In some of them, the number of sequence tags on the chromosome of interest was normalized to the number of tags on all chromosomes in the particular sequencing run (the z-score)49–51,53–55. In other studies52,56, a normalized chromosome value (NCV) was used to calculate the ratio of the number of counts on the chromosome of interest (for example, 21) in a specific sample to the number of counts on a reference chromosome or a chromosome set derived from an unaffected group of samples. The NCV is a fixed ratio that removes variation within and between sequencing runs. A third approach incorporates information on the fraction of fetal DNA present in the sample to calculate an individual risk of trisomy59.
By late 2011, NIPT of trisomy 21 by sequencing of maternal plasma DNA began to be offered on a clinical and commercial basis in the United States and China at approximate costs ranging from $475 to $1,900, depending on the patient's insurance. This development prompted a rapid response statement to be issued by the International Society for Prenatal Diagnosis61, which cautioned that before routine population screening for fetal Down's syndrome is introduced, additional trials are needed, particularly in low-risk populations. Despite these concerns, noninvasive testing using massively parallel sequencing of maternal plasma DNA is now a clinical reality. As of April 2012, over 25,000 clinical tests have been performed in China and several thousand have been performed in the United States (http://www.genomeweb.com/mdx/non-invasive-t21-testing-space-abuzz-firms-jockey-share-1b-market-battle-over-ip).
Compared with the antenatal diagnosis of Down's syndrome, the diagnosis of other commonly occurring fetal aneuploidies, such as trisomies 13 and 18, is more challenging because the accuracy of aneuploidy detection is affected by the GC base content of an individual chromosome. Whereas chromosome 21 has a midrange percentage of GC content, chromosomes 18 and 13 have a lower percentage, which increases the coefficient of variation in the sequencing reactions of these chromosomes48,53. Specific quantitative correction of the GC content bias in the sequencing data using modified z-score equations has resulted in improved sensitivity and specificity in the detection of trisomies 13 and 18 (ref. 53). The NCV analytic method was sensitive enough to detect a case of mosaic trisomy 9, an 11q21-23 deletion, partial trisomy of 6q12-16.3 and several sex chromosome abnormalities52,56, suggesting this method may have expanded clinical utility as compared to the z-score.
Insights into fetal cell-free DNA from a noninvasive diagnosis
Crucially, the noninvasive sequencing studies have provided basic information regarding the biology of the fetal cell-free DNA in maternal plasma. The majority of the total (mainly maternal) circulating cell-free DNA derives from apoptotic hematopoietic cells62, with a peak fragment size of about 162–169 bp48,63,64. In the circulation, the DNA double helix is wound around a nucleosome, and a 20-bp fragment links the nucleosome to its core particle. Fetal DNA, which derives from the placenta24–30, is present in shorter fragments that predominantly, but not exclusively, measure around 143 bp63–65. The sizes of the maternal DNA and fetal DNA differ because in the fetal DNA, the 20-bp linker fragment has been cleaved from the nucleosome48,63. The median amount of circulating cell-free fetal DNA in maternal plasma is 10%23,63,66. Single nucleotide polymorphism (SNP) analyses reveal that the entire fetal genome is represented in maternal plasma and that the relative proportions of maternal and fetal sequence are constant63. This implies that it is theoretically possible to noninvasively screen maternal blood for both fetal DNA copy number variation and single-gene disorders. In two proof-of-concept studies of cell-free DNA analysis using blood samples from women carrying fetuses with known diagnoses, Peters et al.67 found the presence of a familial 4.2-Mb deletion in chromo-some 12p inherited by the fetus, and Lo et al.63 showed that a fetus carried the paternal (but not the maternal) mutation for β-thalassemia.
Prenatal detection of aneuploidy using maternal plasma DNA is effective because the diagnosis relies on counting sequence tags and mapping them to the clinically relevant chromosomes. NIPD of single-gene disorders, many of which are inherited as autosomal or X-linked recessive conditions, is fundamentally even more complex because of the fact that the mother and fetus share the same mutation. In preliminary studies, a digital relative mutation dosage method has been used to deduce whether the mutant or the wild-type allele is overrepresented in the maternal plasma DNA68–70. This method can be applied in autosomal or X-linked recessive conditions in which a pregnant woman carries a heterozygous gene mutation. If a woman's fetus is homozygous for either the wild-type or mutant allele, there is an underrepresentation or overrepresentation of the mutant allele in her plasma DNA. Digital PCR amplification, followed by counting and statistical analyses, determines whether an allelic imbalance is present. In small research studies, this has facilitated accurate diagnoses of hemoglobin β (HBB) mutations that cause hemoglobin E disease68, β-thalassemia68 or sickle cell anemia69, as well as of F8 and F9 mutations that cause hemophilia70. In one instance of a fetus that was at risk for β-thalassemia, a proof-of-concept whole-genome sequencing study was performed using maternal plasma DNA63, and the fetal genome was successfully inferred from SNP genotyping data of parental and chorionic villus samples. In the future, targeted sequencing approaches may be applied to noninvasively diagnose groups of single-gene disorders for which the fetus is at risk based on the ethnic background of the parents.
Advances in analyzing the fetal transcriptome
The dynamic nature of fetal developmental processes, coupled with a need to distinguish normal from abnormal physiology (especially when a fetus appears normal on sonographic examination), has led to an interest in exploring the fetal transcriptome. Unlike fetal DNA, which is released into maternal plasma in consistently increasing amounts as gestation advances71, fetal RNA levels are more variable and reflect differential expression as a function of development72. The finding that fetal Y-chromosome–specific mRNA sequences could remain intact in the maternal circulation despite the presence of circulating RNases did not occur until 2000 (ref. 73). Later work showed that fetal mRNA fragments were relatively stable in the maternal plasma74,75, probably because this mRNA circulates within apoptotic bodies that are protected from further degradation.
Interestingly, transcripts that originate from the placenta are more easily detected in maternal plasma76, whereas transcripts that originate in the fetus are more easily found in maternal whole blood77. In one study, a gene expression microarray analysis was used to compare transcripts found in maternal whole blood at term immediately before and 24–36 hours after delivery with newborn umbilical cord blood to identify circulating fetal biomarkers77. Sequences that were statistically significantly upregulated in both antepartum maternal blood and newborn blood, but not in maternal postpartum blood, were identified as possible fetal transcripts. This work showed that the majority of the circulating fetal transcripts in third-trimester maternal blood were related to visual or central nervous system development, sense of smell and the ability to mount an inflammatory response. The identification that these particular systems were actively developing at this stage made sense according to what was already known about third-trimester fetal physiology. What was new, however, was the knowledge regarding the relative proportions of the actively expressed transcripts, such as the 10% of transcripts that are devoted to development of the immune response. This study also identified specific genes that are normally expressed by the fetus before delivery at term and suggested the possibility that a multiplex, RT-PCR–based assay could be developed to track physiological gene expression78. Such an assay could then possibly be used to track abnormal patterns of gene expression and identify fetuses or infants that might be at risk for developmental delays.
Another area of current research is the investigation of placental microRNAs (miRNAs) in maternal plasma79–84. MiRNAs are small (~19–25 nt) single-stranded noncoding RNA molecules that repress protein translation by binding to the 3′ untranslated regions of their target mRNAs. Because they are remarkably stable in plasma, miRNAs are under evaluation as pregnancy-specific biomarkers in conditions such as preeclampsia80 and fetal growth restriction81. Thus far, approximately 40 placenta-specific miRNAs have been identified, although none are presently used in clinical assays82–84. MiRNAs are exported from syncytiotrophoblasts by exosomes79, which are circulating microparticles that may have a role in intercellular communication. Circulating miRNAs may therefore have a functional role in fetomaternal communication or the development of immune tolerance.
Transcriptomic analysis of fetal abnormalities
Discovery-driven fetal research using maternal blood is limited because the majority of the circulating transcripts are maternal in origin. Larrabee et al.85 hypothesized that amniotic fluid supernatant might be a useful source of pure fetal gene expression information that would provide new data on human development. In an initial proof-of-concept study, they analyzed cell-free fetal RNA in amniotic fluid samples from pregnant women in the second or third trimester undergoing amnioreduction for twin-to-twin transfusion syndrome (TTTS) or hydrops fetalis85. The results showed that fetal gene expression is dynamic and is influenced by gestational age and gender. For example, the genes encoding surfactant proteins A2, B and C, tracheobronchial, gastric and salivary mucin and statherin (a protein involved in both salivary secretion and ossification) were all upregulated as a function of gestational age. Conversely, keratin gene transcript expression decreased with gestational age, which probably reflects the decreased contact between keratin-producing cells and amniotic fluid as the fetal skin matures. Specific transcripts that were upregulated secondary to disease, such as the water transporter aquaporin 1 (AQP1) in TTTS, were also identified in this study. The authors suggested that AQP1 may have a role in TTTS by affecting water movement from the amniotic cavity across the placenta and into the fetal circulation.
The amniotic fluid transcriptome was further studied in fetuses with trisomies 21 (ref. 86M) and 18 (ref. 87) and was compared to the transcriptome of euploid fetuses that were matched for sex and gestational age. In both aneuploidies, hundreds of statistically significantly differentially regulated genes were found; however, only a handful mapped to the chromosome of interest (for example, 21 or 18). This provided strong evidence to suggest that the pathology in fetuses with aneuploidy is the result largely of complex downstream processes and not simply a gene dosage effect caused by the extra chromosome. Subsequent functional and pathway analyses suggested that each aneuploidy has a unique and characteristic transcriptome.
In the fetuses with Down's syndrome, oxidative stress, ion transport and G protein signaling were the major functional abnormalities. Whereas oxidative stress response genes have been previously examined in adults with Down's syndrome88, this study was the first to show that fetuses with Down's syndrome can also be affected by oxidative stress and its intermediate consequences, such as cell stress responses and ion transport. Furthermore, this study was the first, using the Connectivity Map database89, to identify compounds that might reverse the molecular phenotype of Down's syndrome and be considered as potential therapies that can be administered antenatally. Primary cultures of amniocytes90,91 and trophoblasts92 have also been used as a source of mRNA to further study human autosomal trisomy. Although there is always the possibility that the cell culture induces artifactual changes in gene expression, these studies concluded that the Down's syndrome phenotype derives partly from overexpressed genes on chromosome 21 and partly by secondary genome-wide transcriptional dysregulation. Interestingly, in trophoblasts, the changes in the expression of genes involved in the ubiquitin cycle were one of the greatest discriminators of trisomy 21 and euploid placentas90. This finding implies that epigenetic mechanisms that affect post- transcriptional modification by ubiquitination may also play a part in the Down's syndrome phenotype.
In the fetuses with trisomy 18, significant downregulation of genes involved in adrenal development was identified87, which could explain both the low concentrations of maternal serum estriols and the prenatal and postnatal growth restriction observed in affected fetuses and infants. A functional analysis also highlighted differential regulation of pathways related to cardiovascular disease, which is not surprising given that congenital heart disease is a major problem in affected fetuses. In particular, Rho-associated kinase 1 (ROCK1), a gene located on chromosome 18, was significantly upregulated in fetuses with trisomy 18. Prior to this study, ROCK1 was not known to be associated with trisomy 18, illustrating one of the benefits of using transcriptomic analyses to discover genes involved in fetal disease. ROCK1 has a key role in the regulation of endocardial cell differentiation and migration in early heart development. In addition, it is one of only six genes that are dysregulated in both trisomies 18 and 21 compared to euploid controls87.
In addition to the amniotic fluid transcriptome of fetuses with aneuploidies, the normal amniotic fluid transcriptome has also been analyzed in euploid fetuses93. Four hundred seventy six well-annotated genes were identified as being expressed in 12 second trimester amniotic fluid samples. Of the 23 transcripts that mapped to specific organs, six were highly expressed in fetal brain. Other transcripts originated in fetal lung, skin, thyroid, pancreas, blood, liver and placenta. A new finding from this study was the identification of the mammalian target of rapamycin (mTOR), a central regulator of cell growth, as part of a key developmental pathway in fetuses. This study showed that the amniotic fluid core transcriptome could provide information on the development of a number of different organ systems in real time from living human fetuses.
Analysis of the transcriptome in common complications of pregnancy
Subsequent translational investigations have focused on the presence of specific placental transcripts in maternal blood that can serve as biomarkers for various complications of pregnancy94–98. For example, the upregulation of the human chorionic gonadotropin β subunit, human placental lactogen and corticotrophin-releasing hormone transcripts were shown to be potential biomarkers in the blood of women who developed preeclampsia99. In a large study of placentas from 37 preeclamptic and 57 normal pregnancies, genome-wide transcriptional profiling identified 455 differentially expressed genes between preeclampsia and normal pregnancy100. New and previously described genes relating to the pathophysiology of preeclampsia were identified. The most significantly dysregulated canonical pathway identified was tryptophan metabolism. KYNU, which was upregulated in preeclampsia, encodes kynureninase, an enzyme that is key in tryptophan metabolism. This enzyme metabolizes l-kynurenine, which suppresses T cell proliferation and natural killer cells, adding to previous information that immune tolerance to foreign antigens plays a part in the pathogenesis of preeclampsia. Other cell signaling and metabolic pathways that were dysregulated in the preeclamptic placentas included linoleic, fatty acid and arachidonic metabolism, notch signaling, endoplasmic reticulum stress and oxidative stress mediated by nuclear respiratory factor 2. Notably, some differentially regulated genes are involved in the production of hydrogen peroxides and the elimination of lipid peroxidation products. These differences may be among the factors that activate the maternal endothelium and result in atherosclerotic-like lesions that trigger systemic inflammation in preeclampsia.
The placental transcriptome is also being used to understand key fetal biological processes such as intrauterine growth restriction (IUGR), in which the growth of the fetus is substantially reduced compared to normal, healthy fetuses. In one study, the analysis of differentially regulated genes in growth-restricted fetuses suggested that the affected placentas have an upregulation of inflammation that is mediated by chemokine and cytokine signaling pathways101. Importantly, none of the genes known to be imprinted in the placenta were differentially expressed between the normal placentas and those with IUGR, suggesting that epigenetic modification has a minor role in the pathogenesis of IUGR and that perhaps future therapies for this condition should be directed toward decreasing inflammation. Furthermore, hydroxysteroid (11-β) dehydrogenase 1 was upregulated in the IUGR placentas. This enzyme has a role in the regeneration of cortisol from cortisone, which enhances the effect of glucocorticoids on the production of pulmonary surfactant. This may explain why growth-restricted newborns often have substantially accelerated lung maturation for their gestational age.
In an investigation of umbilical cord blood from premature neonates with fetal inflammatory response syndrome, a gene expression analysis of leukocyte mRNA showed an enrichment of biological pathways related to antigen presentation and processing, B cell receptor and phosphatidylinositol signaling and cell adhesion and metabolism compared to neonates without evidence of inflammation102. The transcriptomic studies showed that despite age-related differences in the fetal and adult immune systems, they had many similar responses to infection and inflammation. Among the many genes that were shown to be upregulated in fetal inflammatory response syndrome were ones that are known to play a part in leukocyte adhesion, leukotriene synthesis and chemotaxis.
The All Our Babies Cohort Study103, currently enrolling subjects in Alberta, Canada, is taking a fetal personalized medicine approach to understanding preterm birth by prospectively collecting maternal blood RNA and examining environmental factors. All of these investigations use comparative microarray analyses to identify new biomarkers and potential avenues for intervention.
Advancing from diagnosis to personalized prenatal medicine Challenges for prenatal diagnostics
It should be clear from the preceding paragraphs that the technical advances in prenatal diagnosis that have occurred in the last 5 years have greatly exceeded their translation into clinical practice. The major considerations that affect their incorporation into routine obstetric care include education, cost and ethical issues (Table 2).
Table 2.
Emerging challenges for prenatal diagnosis and fetal personalized medicine
| Prenatal diagnosis | Fetal personalized medicine |
|---|---|
|
Practical issues Need for professional standards and guidelines Advanced training and education for providers Need to decide what is optimal and what is feasible Resource allocation Confidentiality of information obtained Technological issues Reduce cost and duplication of services Improve efficiency Determine clinical meaning of copy number variants detected by chromosome microarrays Ethical issues Direct-to-consumer genetic testing Possibility of detecting mistaken paternity and incest Possibility of coercion to have DNA testing Fewer opportunities for reflection and counseling Devaluation of affected individuals with trisomy Role of industry in promoting research |
Improve annotation of genes to show information about their function and expression in a fetal context Integration of the transcriptome with proteome, metabolome and epigenetic information Determine if (and what) animal models are useful in which to evaluate treatment(s) Decide whether existing drugs can be repurposed or new drugs need to be developed Need to show safety and absence of teratogenicity when developing drug trials for pregnant women and fetuses Substantial costs involved in developing large-scale clinical trials for potentially rare diseases Resource allocation Confidentiality of information obtained |
A crucial aspect of this incorporation will be education, as there is no question that improvement in the genomic literacy104 of health care providers is a fundamental requirement in all fields of medicine. This is especially true in obstetrics and gynecology because of the widespread availability of direct-to-consumer prenatal testing and intense patient interest in the well being of their fetuses. Should any of the direct-to-consumer tests reveal an abnormal fetal finding, it will ultimately be the obstetrician who will have the responsibility for follow-up management. Therefore, obstetricians in particular need frequent and comprehensive educational updates regarding the practical implications of advances in genetics and genomics. Although in many instances the new genetic tests perform better than the old ones, it is currently unknown how these new tests will be incorporated into prenatal care. With regard to cytogenetic analyses, it is cost prohibitive to offer both classical cytogenetic analyses and microarray analyses to all pregnant women undergoing invasive procedures. The average cost for a metaphase karyotype is currently $750 (ranging from $250 to $1,000), and the average cost for a microarray study is $1,500 (ranging from $750 to $3,000). The results of the prospective blinded NICHD trial of prenatal cytogenetic diagnosis by array-based copy number analysis, including data on approximately 4,400 pregnancies, were presented orally at the annual Society for Maternal Fetal Medicine meeting in February 2012 and indicated that karyotyping and microarray analysis were equally accurate in the detection of aneuploidy105. In fetuses with apparently normal metaphase karyotypes sampled for advanced maternal age or abnormal serum screens, chromosome microarray studies detected 1.7% additional clinically relevant abnormalities. In fetuses with sonographic abnormalities and normal cytogenetic studies, this figure rose to 5.8%. These data prompted the investigators to recommend that microarray investigations transition to a first-line diagnostic test in the antenatal setting.
For prenatal diagnosis of Down's syndrome, the current standard of care involves a two-tiered approach (for example, serum screening and nuchal translucency measurement, followed by an offer of invasive procedures such as amniocentesis or chorionic villus sampling to screen women who tested positive) (Fig. 3). If NIPT of Down's syndrome achieves diagnostic accuracy, the cost of testing could be offset by a reduction or elimination of these invasive procedures. Using population-based data from Victoria, Australia, Susman et al.106 investigated the impact of changing from the current screening algorithms to a one-stage noninvasive approach. Their results showed that there would be an 84% reduction in the number of invasive procedures, with an additional 7% of cases of Down's syndrome being detected, albeit with an accompanying reduction in the number of other abnormalities detected. Additional testing will require an increased commitment to pretesting counseling services that will affect the overall cost to the health care system107. Although these added costs have not yet been systematically addressed in very many studies, in one report, the costs of noninvasive prenatal diagnosis of fetal sex for X-linked conditions did not differ from invasive prenatal diagnosis108. As the number of laboratory tests increases, the cost of testing is also expected to decrease.
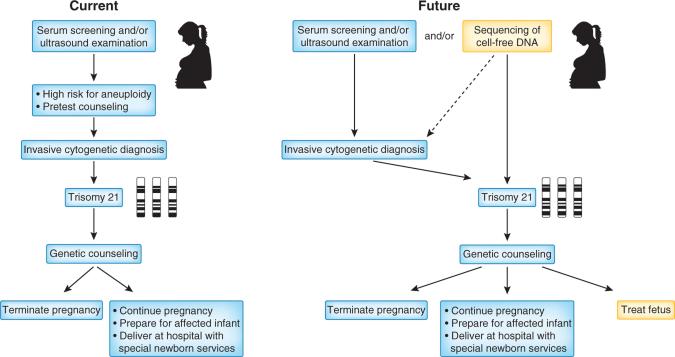
Figure 3.
A potential future diagnostic and treatment strategy for Down's syndrome. A comparison of the current two-tiered approach for the noninvasive diagnosis of fetal trisomy 21 with no fetal treatment options and a possible future approach in which sequencing of maternal plasma DNA may eliminate the need for invasive testing. Furthermore, advances in study of the fetal transcriptome may identify new treatments that could be administered to the pregnant woman as soon as the diagnosis is made.
Although NIPT of Down's syndrome has greater sensitivity and specificity compared with the serum screening algorithms that are currently used in the clinic, the expenses associated with DNA sequencing, bioinformatic analysis and data storage are considerable. Current research is therefore focused on reducing the cost and improving the efficiency of NIPT. Recent approaches have included the enrichment of fetal DNA concentrations by fragment size selection65,66, targeted sequencing of regions from specific chromosomes of clinical interest57–59,109, the possible use of antibodies to histone H1 to bind and remove circulating maternal DNA63, immunoprecipitation of methylated DNA sequences followed by real-time PCR amplification110 and the use of highly heterozygous SNPs to calculate haplotype ratios between the maternally and paternally inherited genes in maternal plasma, thereby inferring information about the fetal karyotype111.
The personalized approach to fetal diagnosis raises several ethical concerns112. With chromosome microarray studies, the detection of CNVs of unknown clinical meaning or variants that have known effects but incomplete penetrance raises parental anxiety, as well as the possibility of termination of a clinically unaffected fetus112. Furthermore, chromosome microarrays can readily detect mistaken paternity and incest, which are issues that are not usually discussed during pretesting counseling sessions113. With regard to NIPT of aneuploidy, there are multiple issues114, including how consent for the test should be obtained115. The current multistep approach to Down's syndrome screening allows several opportunities for reflection that will be lost if replaced by a single blood test116. There is also the potential for coercion to take the test by providers, peers or insurers. A proof-of-principle study63 showed that it is already feasible to noninvasively obtain information on the entire fetal genome, raising practical and ethical questions with regard to what to do with information that is not relevant in infancy, for example, predisposition to an adult-onset condition. In addition, in contrast to the prior introduction of new laboratory tests by academic laboratories, tests associated with the sequencing of fetal DNA are being developed mainly by industry112. It is unknown how intellectual property rights will affect the implementation of these tests and their costs. There are already several patent infringement lawsuits under consideration in the United States.
Challenges for developing new therapies for fetal diseases and pregnancy disorders
As presented here, the discovery-driven approach that is associated with analysis of the transcriptome has facilitated the identification of many genes that seem to have key roles in both normal and abnormal fetal and placental development. A substantial challenge that already exists is acquiring age-appropriate annotation of gene expression (Table 2). Most of the publicly available databases provide gene expression information that is annotated only for adult humans. For example, natriuretic peptide receptor A (NPR1), a gene that is significantly upregulated in all normal full-term infants, is functionally annotated as being associated with congestive heart failure. Whereas that may be true for adults, in infants, this transcript is probably upregulated as a result of the normal physiological diuresis that occurs after delivery. Another challenge will be the integration of massive amounts of data from other types of investigations, such as analyses of the fetal proteome and metabolome along with an improved understanding of epigenetic influences, with the information on the fetal genome and transcriptome presented here. There is a need to learn more about physiological fetal functional gene expression, for example, what genes must be expressed at different stages of gestation for normal fetal maturation? As key genes in fetal development are identified, more focused and cost-effective platforms can be created to measure or monitor specific fetal organ system function117. Such platforms could provide new information that would supplement fetal sonographic studies that currently detect anatomic, but not functional, abnormalities.
With regard to the discovery and evaluation of new fetal treatments, there will be a need to identify appropriate animal models with similar placentation to humans. Even if new therapeutics show promise in animal models, there will be a need to demonstrate their safety and an absence of accompanying teratogenicity in pregnant women and their fetuses. Some of the diseases discussed here are rare; it is unknown whether any organization will wish to invest the substantial costs involved in implementing large-scale clinical trials to test the therapy. The development of new treatments, whether the result of repurposing existing drugs or of developing new ones, is a logical extension of the transcriptomic studies discussed here. As an example, a potential future vision of how the transcriptome might change prenatal diagnosis and treatment of Down's syndrome is given in Figure 3.
Outlook
Prenatal genetic diagnostic technology is advancing at an astonishingly rapid pace. Of the 117 references cited in this paper, 53 (45%) of them have been published in the past 2 years. Professional education and guidelines for incorporating new genetic tests into current practice are urgently needed. High-level economic analyses are also required to assess the benefits and limitations of current compared to future approaches. Government organizations, such as the United States Food and Drug Administration, need to be more proactively involved to ensure the quality and safety of the tests. Multidisciplinary research teams consisting of basic and translational scientists, clinicians, ethicists and parents should be formed to consider many of the issues raised here. Time is of the essence because the commercial sector has already made some of these tests, such as fetal sex and determination of paternity, directly available to pregnant women.
In parallel, relatively recent developments in the ability to sequence the fetal genome, both directly from fetal tissue and indirectly from maternal blood, as well as progress in understanding normal and abnormal fetal physiology using gene expression arrays, provide evidence that we could have the technical capabilities to apply a personalized medicine approach to the fetus. Increasing amounts of fetal genetic and genomic information are now available, and that information has already influenced subsequent pregnancy management, such as decreasing the need for invasive cytogenetic procedures or the administration of steroids or blood products to pregnant women. Although we still have a way to go before new fetal therapeutics can be identified and translated to clinical care, preliminary data indicate that translational approaches based on genomic and transcriptomic information are feasible and that the fetal transcriptome contains crucial new information about fetal development and physiology that can be repeatedly mined.
For the past 30 years, the goal of prenatal diagnosis has been to provide an informed choice to prospective parents. That paradigm is now shifting. We are now at a point where that goal can and should be expanded to incorporate genetic and genomic data to pave the way for a personalized approach to fetal treatment.
ACKNOWLEDGMENTS
The author would like to thank D. Walt, E. Norwitz, J. Maron and L. Hui for their critical reading of the manuscript and their suggestions. In addition, she is grateful for the administrative support provided by R. Forman. The author's time and effort in writing this manuscript was partially supported by the US National Institutes of Health grant HD42053-09.
Footnotes
COMPETING FINANCIAL INTERESTS
The author declares competing financial interests: details are available in the online version of the paper.
References
- 1.Khoury MJ, et al. The scientific foundation for personal genomics: recommendations from an NIH-CDC multidisciplinary workshop. Genet. Med. 2009;11:559–567. doi: 10.1097/GIM.0b013e3181b13a6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferguson-Smith MA, Bianchi DW. Prenatal diagnosis: past, present, and future. Prenat. Diagn. 2010;30:601–604. doi: 10.1002/pd.2574. [DOI] [PubMed] [Google Scholar]
- 3.Wolfberg AJ. Genes on the web-direct-to-consumer marketing of genetic testing. N. Engl. J. Med. 2006;355:543–545. doi: 10.1056/NEJMp068079. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi DW. At-home fetal DNA gender testing: caveat emptor. Obstet. Gynecol. 2006;107:216–218. doi: 10.1097/01.AOG.0000199427.83503.d0. [DOI] [PubMed] [Google Scholar]
- 5.Hawkins AK, Ho A. Genetic counseling and the ethical issues around direct to consumer genetic testing. J. Genet. Counsel. 2012;21:367–373. doi: 10.1007/s10897-012-9488-8. [DOI] [PubMed] [Google Scholar]
- 6.Miller DT, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman JM. High-resolution array genomic hybridization in prenatal diagnosis. Prenat. Diagn. 2009;29:20–28. doi: 10.1002/pd.2129. [DOI] [PubMed] [Google Scholar]
- 8.Mailman MD, et al. The NCBI dbGAP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firth HV, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ACOG Committee Opinion No. 446: array comparative genomic hybridization in prenatal diagnosis. Obstet. Gynecol. 2009;114:1161–1163. doi: 10.1097/AOG.0b013e3181c33cad. Anonymous. [DOI] [PubMed] [Google Scholar]
- 11.Schaeffer AJ, et al. Comparative genomic hybridization-array analysis enhances the detection of aneuploidies and submicroscopic imbalances in spontaneous miscarriages. Am. J. Hum. Genet. 2004;74:1168–1174. doi: 10.1086/421250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raca G, et al. Array-based comparative genomic hybridization (aCGH) in the genetic evaluation of stillbirth. Am. J. Med. Genet. A. 2009;149A:2437–2443. doi: 10.1002/ajmg.a.33083. [DOI] [PubMed] [Google Scholar]
- 13.Harris RA, et al. Genome-wide array-based copy number profiling in human placentas from unexplained stillbirths. Prenat. Diagn. 2011;31:932–944. doi: 10.1002/pd.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaffer LG, et al. Comparison of microarray-based detection rates for cytogenetic abnormalities in prenatal and neonatal specimens. Prenat. Diagn. 2008;28:789–795. doi: 10.1002/pd.2053. [DOI] [PubMed] [Google Scholar]
- 15.Coppinger J, et al. Whole-genome microarray analysis in prenatal specimens identifies clinically significant chromosome alterations without increase in results of unclear significance compared to targeted microarray. Prenat. Diagn. 2009;29:1156–1166. doi: 10.1002/pd.2371. [DOI] [PubMed] [Google Scholar]
- 16.Van den Veyver IB, et al. Clinical use of array comparative genomic hybridization (aCGH) for prenatal diagnosis in 300 cases. Prenat. Diagn. 2009;29:29–39. doi: 10.1002/pd.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JH, et al. Application of a target array comparative genomic hybridization to prenatal diagnosis. BMC Med. Genet. 2010;11:102. doi: 10.1186/1471-2350-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleeman L, et al. Use of array comparative genomic hybridization for prenatal diagnosis of fetuses with sonographic anomalies and normal metaphase karyotype. Prenat. Diagn. 2009;29:1213–1217. doi: 10.1002/pd.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyreman M, et al. High resolution array analysis: diagnosing pregnancies with abnormal ultrasound findings. J. Med. Genet. 2009;46:531–541. doi: 10.1136/jmg.2008.065482. [DOI] [PubMed] [Google Scholar]
- 20.Faas BH, et al. Identification of clinically significant, submicroscopic chromosome alterations and UPD in fetuses with ultrasound anomalies using genome-wide 250k SNP array analysis. J. Med. Genet. 2010;47:586–594. doi: 10.1136/jmg.2009.075853. [DOI] [PubMed] [Google Scholar]
- 21.D'Amours G, et al. Whole-genome array CGH identifies pathogenic copy number variations in fetuses with major malformations and a normal karyotype. Clin. Genet. 2012;81:128–141. doi: 10.1111/j.1399-0004.2011.01687.x. [DOI] [PubMed] [Google Scholar]
- 22.Maya I, et al. Diagnostic utility of array-based comparative genomic hybridization (aCGH) in a prenatal setting. Prenat. Diagn. 2010;30:1131–1137. doi: 10.1002/pd.2626. [DOI] [PubMed] [Google Scholar]
- 23.Lo YM, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 24.Tjoa ML, Cindrova-Davies T, Spasic-Boskovic O, Bianchi DW, Burton GJ. Trophoblastic oxidative stress and the release of cell-free feto-placental DNA. Am. J. Pathol. 2006;169:400–404. doi: 10.2353/ajpath.2006.060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alberry M, et al. Free fetal DNA in maternal plasma in anembryonic pregnancies: confirmation that the origin is the trophoblast. Prenat. Diagn. 2007;27:415–418. doi: 10.1002/pd.1700. [DOI] [PubMed] [Google Scholar]
- 26.Bischoff FZ, Lewis DE, Simpson JL. Cell-free fetal DNA in maternal blood: kinetics, source and structure. Hum. Reprod. Update. 2005;11:59–67. doi: 10.1093/humupd/dmh053. [DOI] [PubMed] [Google Scholar]
- 27.Masuzaki H, et al. Detection of cell free placental DNA in maternal plasma: direct evidence from three cases of confined placental mosaicism. J. Med. Genet. 2004;41:289–292. doi: 10.1136/jmg.2003.015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekizawa A, et al. Evaluation of bidirectional transfer of plasma DNA through placenta. Hum. Genet. 2003;113:307–310. doi: 10.1007/s00439-003-0987-4. [DOI] [PubMed] [Google Scholar]
- 29.Chim SS, et al. Detection of the placental epigenetic signature of the maspingene in maternal plasma. Proc. Natl. Acad. Sci. USA. 2005;102:14753–14758. doi: 10.1073/pnas.0503335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan KC, et al. Hypermethylated RASSF1A in maternal plasma: a universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin. Chem. 2006;52:2211–2218. doi: 10.1373/clinchem.2006.074997. [DOI] [PubMed] [Google Scholar]
- 31.Bennett PR, et al. Prenatal determination of fetal RhD type by DNA amplification. N. Engl. J. Med. 1993;329:607–610. doi: 10.1056/NEJM199308263290903. [DOI] [PubMed] [Google Scholar]
- 32.Lo YM, et al. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N. Engl. J. Med. 1998;339:1734–1738. doi: 10.1056/NEJM199812103392402. [DOI] [PubMed] [Google Scholar]
- 33.Finning KM, Martin PG, Soothill PW, Avent ND. Prediction of fetal D status from maternal plasma: introduction of a new noninvasive fetal RHD genotyping service. Transfusion. 2002;42:1079–1085. doi: 10.1046/j.1537-2995.2002.00165.x. [DOI] [PubMed] [Google Scholar]
- 34.Bianchi DW, Avent ND, Costa JM, van der Schoot EM. Noninvasive prenatal diagnosis of fetal Rhesus D: ready for prime(r) time. Obstet. Gynecol. 2005;106:841–844. doi: 10.1097/01.AOG.0000179477.59385.93. [DOI] [PubMed] [Google Scholar]
- 35.Scheffer PG, van der Schoot CE, Page-Christiaens GC, de Haas M. Noninvasive fetal blood group genotyping of rhesus D, c, E and of K in alloimmunised pregnant women: evaluation of a 7-year clinical experience. BJOG. 2011;118:1340–1348. doi: 10.1111/j.1471-0528.2011.03028.x. [DOI] [PubMed] [Google Scholar]
- 36.Daniels G, Finning K, Martin P. Noninvasive fetal blood grouping: present and future. Clin. Lab. Med. 2010;30:431–442. doi: 10.1016/j.cll.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Clausen FB, et al. Report of the first nationally implemented clinical routine screening for fetal RHD in D-pregnant women to ascertain the requirements for antenatal D prophylaxis. Transfusion. 2012;52:752–758. doi: 10.1111/j.1537-2995.2011.03362.x. [DOI] [PubMed] [Google Scholar]
- 38.Devaney SA, Palomaki GE, Scott JA, Bianchi DW. Noninvasive fetal sex determination using cell-free fetal DNA. J. Am. Med. Assoc. 2011;306:627–636. doi: 10.1001/jama.2011.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill M, et al. Non-invasive prenatal determination of fetal sex: translating research into clinical practice. Clin. Genet. 2011;80:68–75. doi: 10.1111/j.1399-0004.2010.01533.x. [DOI] [PubMed] [Google Scholar]
- 40.ACOG Committee on Practice Bulletins ACOG practice bulletin no. 77: screening for fetal chromosomal abnormalities. Obstet. Gynecol. 2007;109:217–227. doi: 10.1097/00006250-200701000-00054. [DOI] [PubMed] [Google Scholar]
- 41.Bianchi DW, et al. Fetal gender and aneuploidy detection using fetal cells in maternal blood: analysis of NIFTY I data. National Institute of Child Health and Development Fetal Cell Isolation Study. Prenat. Diagn. 2002;22:609–615. doi: 10.1002/pd.347. [DOI] [PubMed] [Google Scholar]
- 42.Bianchi DW, Hanson J. Sharpening the tools: a summary of a National Institutes of Health workshop on new technologies for detection of fetal cells in maternal blood for early prenatal diagnosis. J. Matern. Fetal Neonatal Med. 2006;19:199–207. doi: 10.1080/14767050600676851. [DOI] [PubMed] [Google Scholar]
- 43.Lo YM. Noninvasive prenatal detection of fetal chromosomal aneuploidies by maternal plasma nucleic acid analysis: a review of the current state of the art. BJOG. 2009;116:152–157. doi: 10.1111/j.1471-0528.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- 44.Lo YM, et al. Plasma placental RNA allelic ratio permits noninvasive prenatal chromosomal aneuploidy detection. Nat. Med. 2007;13:218–223. doi: 10.1038/nm1530. [DOI] [PubMed] [Google Scholar]
- 45.Tsui NB, et al. Synergy of total PLAC4 RNA concentration and measurement of the RNA single-nucleotide polymorphism allelic ratio for the noninvasive prenatal detection of trisomy 21. Clin. Chem. 2010;56:73–81. doi: 10.1373/clinchem.2009.132662. [DOI] [PubMed] [Google Scholar]
- 46.Lo YM, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc. Natl. Acad. Sci. USA. 2007;104:13116–13121. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan HC, Quake SR. Detection of aneuploidy with digital polymerase chain reaction. Anal. Chem. 2007;79:7576–7579. doi: 10.1021/ac0709394. [DOI] [PubMed] [Google Scholar]
- 48.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiu RW, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl. Acad. Sci. USA. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiu RW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. Br. Med. J. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehrich M, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am. J. Obstet. Gynecol. 2011;204:205.e1–11. doi: 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- 52.Sehnert AJ, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel sequencing of cell-free fetal DNA from maternal blood. Clin. Chem. 2011;57:1042–1049. doi: 10.1373/clinchem.2011.165910. [DOI] [PubMed] [Google Scholar]
- 53.Chen EZ, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS ONE. 2011;6:e21791. doi: 10.1371/journal.pone.0021791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palomaki GE, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet. Med. 2011;13:913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 55.Palomaki GE, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet. Med. 2012;14:296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bianchi DW, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet. Gynecol. 2012;119:890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 57.Sparks AB, et al. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat. Diagn. 2012;32:3–9. doi: 10.1002/pd.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashoor G, Syngelaki A, Wagner M, Birdir C, Nicolaides KH. Chromosome-selective sequencing of maternal plasma cell-free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012;206:322.e1–5. doi: 10.1016/j.ajog.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 59.Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Non-invasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012;206:319.e1–9. doi: 10.1016/j.ajog.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 60.Lun FM, et al. Noninvasive prenatal diagnosis of a case of Down syndrome due to Robertsonian translocation by massively parallel sequencing of maternal plasma DNA. Clin. Chem. 2011;57:917–919. doi: 10.1373/clinchem.2011.161844. [DOI] [PubMed] [Google Scholar]
- 61.Benn P, et al. Prenatal detection of Down syndrome using massively parallel sequencing (MPS): a rapid response statement from a committee on behalf of the Board of the International Society for Prenatal Diagnosis, 24 October 2011. Prenat. Diagn. 2012;32:1–2. doi: 10.1002/pd.2919. [DOI] [PubMed] [Google Scholar]
- 62.Lui YY, et al. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 2002;48:421–427. [PubMed] [Google Scholar]
- 63.Lo YM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 64.Fan HC, et al. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin. Chem. 2010;56:1279–1286. doi: 10.1373/clinchem.2010.144188. [DOI] [PubMed] [Google Scholar]
- 65.Chan KC, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clin. Chem. 2004;50:88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 66.Lun FM, et al. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin. Chem. 2008;54:1664–1672. doi: 10.1373/clinchem.2008.111385. [DOI] [PubMed] [Google Scholar]
- 67.Peters D, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N. Engl. J. Med. 2011;365:1847–1848. doi: 10.1056/NEJMc1106975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lun FM, et al. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc. Natl. Acad. Sci. USA. 2008;105:19920–19925. doi: 10.1073/pnas.0810373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrett AN, McDonnell TC, Chan KC, Chitty LS. Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin. Chem. 2012 doi: 10.1373/clinchem.2011.178939. [DOI] [PubMed] [Google Scholar]
- 70.Tsui NB, et al. Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood. 2011;117:3684–3691. doi: 10.1182/blood-2010-10-310789. [DOI] [PubMed] [Google Scholar]
- 71.Lo YM, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am. J. Hum. Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chiu RW, et al. Time profile of appearance and disappearance of circulating placenta-derived mRNA in maternal plasma. Clin. Chem. 2006;52:313–316. doi: 10.1373/clinchem.2005.059691. [DOI] [PubMed] [Google Scholar]
- 73.Poon LL, Leung TN, Lau TK, Lo YM. Presence of fetal RNA in maternal plasma. Clin. Chem. 2000;46:1832–1834. [PubMed] [Google Scholar]
- 74.Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin. Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 75.Ng EK, et al. mRNA of placental origin is readily detectable in maternal plasma. Proc. Natl. Acad. Sci. USA. 2003;100:4748–4753. doi: 10.1073/pnas.0637450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heung MM, et al. Placenta-derived fetal specific mRNA is more readily detectable in maternal plasma than in whole blood. PLoS ONE. 2009;4:e5858. doi: 10.1371/journal.pone.0005858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maron JL, et al. Gene expression analysis in pregnant women and their infants identifies unique fetal biomarkers that circulate in maternal blood. J. Clin. Invest. 2007;117:3007–3019. doi: 10.1172/JCI29959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bianchi DW, Maron JL, Johnson KL. Insights into fetal and neonatal development through analysis of cell-free RNA in body fluids. Early Hum. Dev. 2010;86:747–752. doi: 10.1016/j.earlhumdev.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo SS, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 80.Pineles BL, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am. J. Obstet. Gynecol. 2007;196:261.e1–6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 81.Mouillet JF, et al. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta. 2010;31:781–784. doi: 10.1016/j.placenta.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chim SS, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 83.Miura K, et al. Identification of pregnancy-associated microRNAs in maternal plasma. Clin. Chem. 2010;56:1767–1771. doi: 10.1373/clinchem.2010.147660. [DOI] [PubMed] [Google Scholar]
- 84.Kotlabova K, Doucha J, Hromadnikova I. Placental-specific microRNA in maternal circulation-identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J. Reprod. Immunol. 2011;89:185–191. doi: 10.1016/j.jri.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 85.Larrabee PB, et al. Global gene expression analysis of the living human fetus using cell-free messenger RNA in amniotic fluid. J. Am. Med. Assoc. 2005;293:836–842. doi: 10.1001/jama.293.7.836. [DOI] [PubMed] [Google Scholar]
- 86.Slonim DK, et al. Functional genomic analysis of amniotic fluid cell-free mRNA suggests that oxidative stress is significant in Down syndrome fetuses. Proc. Natl. Acad. Sci. USA. 2009;106:9425–9429. doi: 10.1073/pnas.0903909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koide K, et al. Transcriptomic analysis of cell-free fetal RNA suggests a specific molecular phenotype in trisomy 18. Hum. Genet. 2011;129:295–305. doi: 10.1007/s00439-010-0923-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zana M, Janka Z, Kalman J. Oxidative stress: a bridge between Down's syndrome and Alzheimer's disease. Neurobiol. Aging. 2007;28:648–676. doi: 10.1016/j.neurobiolaging.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 89.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 90.FitzPatrick DR, et al. Transcriptome analysis of human autosomal trisomy. Hum. Mol. Genet. 2002;11:3249–3256. doi: 10.1093/hmg/11.26.3249. [DOI] [PubMed] [Google Scholar]
- 91.Chung IH, et al. Gene expression analysis of cultured amniotic fluid cell with Down syndrome by DNA microarray. J. Korean Med. Sci. 2005;20:82–87. doi: 10.3346/jkms.2005.20.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rozovski U, et al. Genome-wide expression analysis of cultured trophoblast with trisomy 21 karyotype. Hum. Reprod. 2007;22:2538–2545. doi: 10.1093/humrep/dem214. [DOI] [PubMed] [Google Scholar]
- 93.Hui L, et al. The amniotic fluid transcriptome: a source of novel information about human fetal development. Obstet. Gynecol. 2012;119:111–118. doi: 10.1097/AOG.0b013e31823d4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsui NB, et al. Systematic micro-array based identification of placental mRNA in maternal plasma: towards non-invasive prenatal gene expression profiling. J. Med. Genet. 2004;41:461–467. doi: 10.1136/jmg.2003.016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Purwosunu Y, et al. Cell-free mRNA concentrations of CRH, PLAC1, and selectin-P are increased in the plasma of pregnant women with preeclampsia. Prenat. Diagn. 2007;27:772–777. doi: 10.1002/pd.1780. [DOI] [PubMed] [Google Scholar]
- 96.Okazaki S, et al. Placenta-derived, cellular messenger RNA expression in the maternal blood of preeclamptic women. Obstet. Gynecol. 2007;110:1130–1136. doi: 10.1097/01.AOG.0000286761.11436.67. [DOI] [PubMed] [Google Scholar]
- 97.Miura K, et al. The possibility of microarray-based analysis using cell-free placental mRNA in maternal plasma. Prenat. Diagn. 2010;30:849–861. doi: 10.1002/pd.2570. [DOI] [PubMed] [Google Scholar]
- 98.Miura K, et al. Increased levels of cell-free placenta mRNA in a subgroup of placenta previa that needs hysterectomy. Prenat. Diagn. 2008;28:805–809. doi: 10.1002/pd.2056. [DOI] [PubMed] [Google Scholar]
- 99.Ng EK, et al. The concentration of circulating corticotrophin-releasing hormone mRNA in maternal plasma is increased in preeclampsia. Clin. Chem. 2003;49:727–731. doi: 10.1373/49.5.727. [DOI] [PubMed] [Google Scholar]
- 100.Løset M, et al. A transcriptional profile of the decidua in preeclampsia. Am. J. Obstet. Gynecol. 2011;204:84.e1–27. doi: 10.1016/j.ajog.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sitras V, Paulssen R, Leirvik J, Vartun A, Acharya G. Placental gene expression profile in intrauterine growth restriction due to placental insufficiency. Reprod. Sci. 2009;16:701–711. doi: 10.1177/1933719109334256. [DOI] [PubMed] [Google Scholar]
- 102.Madsen-Bouterse SA, et al. The transcriptome of the fetal inflammatory response syndrome. Am. J. Reprod. Immunol. 2010;63:73–92. doi: 10.1111/j.1600-0897.2009.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gracie SK, et al. All Our Babies Cohort Study: recruitment of a cohort to predict women at risk of preterm birth through the examination of gene expression profiles and the environment. BMC Pregnancy Childbirth. 2010;10:87. doi: 10.1186/1471-2393-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guttmacher AE, et al. Educating health-care professionals about genetics and genomics. Nat. Rev. Genet. 2007;8:151–157. doi: 10.1038/nrg2007. [DOI] [PubMed] [Google Scholar]
- 105.Wapner R. A multicenter, prospective, masked comparison of chromosomal microarray with standard karyotyping for routine and high risk prenatal diagnosis. Am. J. Obstet. Gynecol. 2012;206:S2. [Google Scholar]
- 106.Susman MR, et al. Using population-based data to predict the impact of introducing noninvasive prenatal diagnosis for Down syndrome. Genet. Med. 2010;12:298–303. doi: 10.1097/GIM.0b013e3181d5d022. [DOI] [PubMed] [Google Scholar]
- 107.Benn PA, Chapman AR. Practical and ethical considerations of noninvasive prenatal diagnosis. J. Am. Med. Assoc. 2009;301:2154–2156. doi: 10.1001/jama.2009.707. [DOI] [PubMed] [Google Scholar]
- 108.Hill M, et al. Incremental cost of non-invasive prenatal diagnosis versus invasive prenatal diagnosis of fetal sex in England. Prenat. Diagn. 2011;31:267–273. doi: 10.1002/pd.2680. [DOI] [PubMed] [Google Scholar]
- 109.Liao GJ, et al. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clin. Chem. 2011;57:92–101. doi: 10.1373/clinchem.2010.154336. [DOI] [PubMed] [Google Scholar]
- 110.Papageorgiou EA, et al. Fetal-specific DNA methylation ratio permits noninvasive prenatal diagnosis of trisomy 21. Nat. Med. 2011;17:510–513. doi: 10.1038/nm.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ghanta S, et al. Non-invasive prenatal detection of trisomy 21 using tandem single nucleotide polymorphisms. PLoS ONE. 2010;5:e13184. doi: 10.1371/journal.pone.0013184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Benn PA, Chapman AR. Ethical challenges in providing noninvasive prenatal diagnosis. Curr. Opin. Obstet. Gynecol. 2010;22:128–134. doi: 10.1097/GCO.0b013e3283372352. [DOI] [PubMed] [Google Scholar]
- 113.Beaudet AL. Ethical issues raised by common copy number variants and single nucleotide polymorphisms of certain and uncertain significance in general medical practice. Genome Med. 2010;2:42. doi: 10.1186/gm163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Greely HT. Get ready for the flood of fetal gene screening. Nature. 2011;469:289–291. doi: 10.1038/469289a. [DOI] [PubMed] [Google Scholar]
- 115.Deans Z, Newson AJ. Should non-invasiveness change informed consent procedures for prenatal diagnosis? Health Care Anal. 2011;19:122–132. doi: 10.1007/s10728-010-0146-8. [DOI] [PubMed] [Google Scholar]
- 116.Hall A, Bostanci A, Wright CF. Non-invasive prenatal diagnosis using cell-free fetal DNA technology: applications and implications. Public Health Genomics. 2010;13:246–255. doi: 10.1159/000279626. [DOI] [PubMed] [Google Scholar]
- 117.Massingham LJ, et al. Proof of concept study to assess fetal gene expression in amniotic fluid by nanoarray PCR. J. Mol. Diagn. 2011;13:565–570. doi: 10.1016/j.jmoldx.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]