Abstract
The adsorption behavior of hard and soft proteins under the effect of an external electric field was investigated by a combination of spectroscopic ellipsometry and molecular dynamics (MD) simulations. Optically transparent carbon electrodes (OTCE) were used as conductive, sorbent substrates. Lysozyme (LSZ) and ribonuclease A (RNase A) were selected as representative hard proteins whereas myoglobin (Mb), α-lactalbumin (α-LAC), bovine serum albumin (BSA), glucose oxidase (GOx), and immunoglobulin G (IgG) were selected to represent soft proteins. In line with recent publications from our group, the experimental results revealed that while the adsorption of all investigated proteins can be enhanced by the potential applied to the electrode, the effect is more pronounced for hard proteins. In contrast with the incomplete monolayers formed at open-circuit potential, the application of +800mV to the sorbent surface induced the formation of multiple layers of protein. These results also suggest that this effect can be related to the intrinsic polarizability of the protein (induction of dipoles), the resulting surface accessible solvent area (SASA), and structural rearrangements induced upon the incorporation on the protein layer. The described experiments are critical to understand the relationship between the structure of proteins and their tendency to form (under electric stimulation) layers with thicknesses that greatly surpass those obtained at open-circuit conditions.
1. Introduction
Protein adsorption is a spontaneous phenomenon that occurs when a solid surface is placed in contact with a protein solution.1 In general, the amount adsorbed, the adsorption rate, and the stability of the formed layer depend on the physicochemical characteristics of the protein (size, flexibility, and charge), the surface of the selected substrate (surface energy, charge, and morphology), and the environment for the interaction.2,3 Different steps (occurring at different time-scales) are involved in the attachment of protein molecules onto solid surfaces.4 At the beginning of the process, the protein molecules diffuse from the bulk of the solution to the interface. Then, the proteins are attached to the solid surface by a combination of (mostly) electrostatic and hydrophobic forces. Finally, and depending on the structure, protein molecules can relax and spread to maximize the number of interaction points with the surface. In some rare cases, proteins can also desorb from the interface and subsequently return to the bulk in either a native or structurally modified state.
Although these interactions (and the corresponding kinetics) can be controlled by the experimental conditions of the adsorption (pH, ionic strength, temperature, etc)5-7 or by chemical modifications of the protein and/or the substrate, the application of an external signal to the sorbent surface can result in a more versatile approach.8-13 In this regard, our group recently demonstrated that increases in the interfacial potential could yield to significant enhancements in the adsorption of bovine serum albumin (BSA) onto optically transparent carbon electrodes (OTCE).14 This phenomenon, that was attributed to the induction of dipoles (polarization) within the structure of the incoming protein molecules when placed in the vicinity of the electrode surface, showed a significant dependence on the potential applied, the solution pH, the concentrations of protein, and the ionic strength.14 Furthermore, subsequent studies demonstrated that the adsorption of glucose oxidase (GOx, a model protein for the development of biosensors) was also susceptible to the potential applied to the sorbent surface and that significant increases in the catalytic activity of the substrates can be achieved by this method.15 While very interesting from the catalytic point of view, the use of proteins like BSA and GOx does not provide enough information to identify the key variables involved in the effect of electric potential on the adsorption of proteins with different properties.
Aiming to address this gap in knowledge, a series of proteins with critical roles in biological purposes (ranging from molecular recognition to catalysis and cellular adhesion) and different physicochemical characteristics were selected. As proposed by W. Norde,5, 16 proteins were classified as hard or soft considering their structural stability. In one extreme are the so-called hard proteins, which present a high internal stability (given by a molecular structure with high conformational entropy) and consequently undergo limited structural changes upon the adsorption to a solid surface. On the other side, soft proteins (with a low conformational entropy) tend to adsorb to surfaces under a wider variety of conditions by permitting rearrangements of the molecular structure upon the adsorption. As a consequence, it is generally accepted that while soft proteins can adsorb to a variety of surfaces (even under unfavorable electrostatic conditions), the interaction of hard proteins require a more careful selection of the experimental conditions (surface charge, solution pH, ionic strength, etc).
Therefore, this article presents results related to adsorption of a series of proteins (with a range of properties) assisted by potential (+800 mV) onto OTCEs. Dynamic adsorption experiments were followed by spectroscopic ellipsometry and complemented by Molecular Dynamics (MD) simulations.
2. Experimental Design
Selected proteins
Ribonuclease A (RNase A, from bovine pancreas), α-Lactalbumin (α-LAC, from bovine milk), Lysozyme (LSZ, from chicken egg white), Myoglobin (Mb, from equine skeletal muscle), Glucose oxidase (GOx, Type II, from Aspergillus niger), and Immunoglobulin G (IgG, from human serum) were obtained from Sigma-Aldrich (St. Louis, MO). Bovine serum albumin (BSA) was purchased from Fisher Scientific (Fair Lawn, NJ). Table 1 provides a summary of the most relevant physicochemical properties of the selected proteins.
Table 1.
Physicochemical characteristics of hard and soft proteins selected for the experiments described in the present article. Ribonuclease A (RNase A), α-Lactalbumin (α-LAC), Lysozyme (LSZ), Myoglobin (Mb), Bovine Serum Albumin (BSA), Glucose Oxidase (GOx), and Immunoglobulin G (IgG).
| Proteins | MW (kDa) | Size (nm) | Isoelectric Point (pH units) | Ref. |
|---|---|---|---|---|
| RNase A | 13.7 | 3.8 × 2.8 × 2.2 | 9.20 | 17,18 |
| α-LAC | 14.2 | 3.7 × 3.2 × 2.5 | 4.30 | 1, 18 |
| LSZ | 14.5 | 4.5 × 3.0 × 3.0 | 11.00 | 1, 18 |
| Mb | 17.2 | 4.3 × 3.5 × 2.3 | 7.05 | 2,19-21 |
| BSA | 66.5 | 11.6 × 2.7 × 2.7 | 4.70 | 17,22 |
| GOx | 160 | 8.0 × 7.0 × 5.5 | 4.20 | 23-26 |
| IgG | 160 | 14.2 × 8.5 × 3.8 | 7.00 | 27-29 |
Stock solutions of these proteins (0.10 mg·mL−1) were prepared by dissolving a known amount of each protein in 10 mmol·L−1 buffer solution, matching the isoelectric point (IEP) of each protein (citrate was used for α-LAC, BSA, and GOx whereas phosphate was used for RNAse, LSZ, Mb, and IgG). Before the adsorption, all protein solutions were filtered through 0.2 μm poly(tetrafluoroethylene) membrane (PTFE, VWR International; Radnor, PA) to remove any aggregates.
Reagents and Solutions
All aqueous solutions were prepared using 18 MΩ·cm water (NANOpure Diamond, Barnstead; Dubuque, IA) and analytical reagent grade chemicals. Citric acid was acquired from Aldrich Chemical Company (Milwaukee, WI). N,N-Dimethylformamide (DMF, 99%) was purchased from Alfa Aesar (Ward Hill, MA). Suberic acid bis(N-hydroxysuccinimide ester) was obtained from Sigma-Aldrich (St. Louis, MO). Sodium hydroxide, and sodium phosphate monobasic anhydrous were obtained from Fisher Scientific (Fair Lawn, NJ). The pH of different solutions was adjusted using 1 mol·L−1 NaOH and measured using a glass electrode and a digital pH meter (Orion 420A+, Thermo; Waltham, MA).
Substrates
Silica wafers coated with thin optically transparent carbon films (Si/SiO2/OTCE) were used as conductive platforms to adsorb the selected proteins and investigate the effect of the potential applied. Following the procedure described in a previous paper,13 OTCE were prepared by pyrolysis of AZ P4330-RS Photoresist obtained from AZ Electronic Materials USA Corp. (Somerville, NJ). The commercial photoresist was diluted to 60% v/v of the as-received material with propylene glycol monomethyl ether acetate (PGMEA 99%, Alfa Aesar; Ward Hill, MA).
Briefly, standard <111> silicon wafers (Si/SiO2, Sumco; Phoenix, AZ) were first scored using a computer-controlled engraver (Gravograph IS400, Gravotech; Duluth, GA). The process defined pieces of 1 cm in width and 3 cm in length that were then manually cut and cleaned in piranha solution (30% hydrogen peroxide and 70% sulfuric acid) at 90 °C for 30 min. After thorough rinsing with water, the substrates were immersed and stored in ultrapure water until use. Subsequently the clean wafers were dried at 80 °C for 30 min; a thin layer of photoresist was deposited onto the silicon wafers using a spin coater (Laurell, Model WS-400-6NPP; North Wales, PA). Next, the photoresist-coated substrates were heated at 110 °C for 60 s in a convection oven to evaporate the solvent and then transferred to a tube furnace (Thermolyne F21135, Barnstead International; Dubuque, IA) for pyrolysis. The carbonization step began by flushing the system at 1 L·min−1 with forming gas (95% Ar + 5% H2, v/v) for 5 min. Next, the temperature was increased to 1000 °C at 20 °C·min−1. After pyrolysis during 1 h, the system was allowed to cool to room temperature. Finally, the samples were stored in a Petri dish for a minimum of 3 days to complete the spontaneous surface oxidation. Information related to the morphology, composition, optical properties, and conductivity of these substrates can be found elsewhere.13
Spectroscopic Ellipsometry
Adsorption experiments were performed using a variable angle spectroscopic ellipsometer (WVASE, J.A. Woollam Co.; Lincoln, NE) following previously reported procedures30-32 and summarized in the Supplementary Information. Briefly, the basis of SE is the measurement of change in the reflectance and phase difference between the parallel (RP) and perpendicular (RS) components of a polarized light beam upon reflection from a surface, which can be related to the amplitude ratio (Ψ) and phase difference (Δ). Data collected as function of wavelength or time were modeled using the WVASE software package (J.A. Woollam Co.; Lincoln, NE) and the mean square error (MSE, calculated by a built-in function in WVASE) was used to quantify the difference between the experimental and model-generated data. In agreement with previous reports, MSE < 15 were considered acceptable.30,31 The ellipsometric measurements were interpreted using a previously developed optical model13,14 allowing the description of the substrates in terms of the refractive index (n), extinction coefficient (k), and thickness (d). Consequently, five uniaxial layers with optical axes parallel to the surface substrate were considered in this optical model. Because the experiments were performed in aqueous media, the optical properties of water were also contemplated. First, the dielectric function of Si (bulk, d = 1 mm) and SiO2 (d = 2.1 ± 0.5 nm) were used to describe the optical behavior of silica wafer. Next, the optical constants of carbon13 were used to define the ellipsometric response of the OTCE (d = 19.6 ± 0.7 nm). Then, a void layer considering nano-bubbles33-36 formed on the hydrophobic and rough surface of the OTCE was also incorporated to improve the optical model. Lastly, the protein layer adsorbed on the OTCE was described using a Cauchy parametrization model, where A=1.465, B=0.01, and C=0 are computer-calculated fitting parameters describing the relationship between the refractive index and the wavelength of the incident beam.
Dynamic adsorption experiments were performed in a modified electrochemical cell30 (J.A. Woollam Co.; Lincoln, NE) mounted directly on the vertical base of the ellipsometer, with an incident angle of 70° (see Supplementary Information for more details). Before the adsorption of selected protein on the substrate, the thickness of the OTCE was measured by placing the substrate in the ellipsometry cell and performing a spectroscopic scan from 300 to 1000 nm (with 10 nm step) using 10 mmol·L−1 buffer solution as the ambient medium. Then, the adsorption experiment was started recording a baseline of the bare OTCE at open circuit potential (OCP, the potential at which no current flows through the cell) while buffer solution was pumped inside the cell at a rate of 1 mL·min−1. After 20 min of baseline, the protein solution was injected to adsorb a monolayer on the OTCE at OCP. As a result, an initial fast process followed by a slower one was always observed. When the signal reached a plateau, a potential of +800 mV was applied and kept until the end of the experiment. Previous experiments have shown that this potential is high enough to induce the accumulation of proteins on the electrode and does not produce electrochemical oxidation of the protein.14,15 The external potential was applied using a CHI812B Electrochemical Analyzer (CH Instrument, Inc.; Austin, TX), a silver/silver chloride (Ag|AgCl|KClsat) reference electrode, and a platinum wire as the counter electrode. Finally, a spectroscopic scan was performed to obtain the thickness of the protein layer after the adsorption assisted by potential. The procedure described above provided the data to calculate the thickness of the OTCE, the protein layer, and the adsorbed amount of the selected proteins on the thin carbon electrodes.
Molecular Dynamics Simulations
In order to understand the factors dominating the effect of the external electric field and protein molecules, molecular dynamics (MD) simulations were performed using NAMD 2.9 package37 with Amber force field 99SB.38 As representative examples of hard and soft proteins, lysozyme (LSZ) and myoglobin (Mb) were respectively selected. Although these proteins differ in thermal stability, they have similar molecular weights and dimensions (see Table 1). To feed the simulation, the protein coordinates were initially obtained from the Protein Data Bank (PDBID: 1LYZ and 1MBN, respectively). Next, the protein was solvated with water (TIP339) and placed in the center of a cubic box of 11.5 nm × 11.4 nm × 10.6 nm. Additionally, the system was neutralized by the addition of NaCl. The MD simulations were run at the respective IEP (see Table 1) by protonating or deprotonating stages using classical continuum electrostatics, available in H++.40 In all cases, the systems were minimized and stabilized until reaching the equilibrium state (~1 ns). Subsequently, the MD production was performed in the NVT ensemble at 298 K (maintained constant using Langevin dynamics, with a damping coefficient of 5 ps−1) using a time step of 2 fs, the SHAKE algorithm, and periodic boundary conditions. Also, the center of mass (COM) of the protein was fixed using the steered molecular dynamic (SMD) module to avoid displacement of the proteins during the simulation.41 In all cases, the external constant electric field was applied along the z direction for 40 ns using the following relationship:
| Equation 1 |
where Vm is the external electric potential and Lz is the length of the system in the z direction. For these experiments, three different electric fields were used: 0.075, 0.180, and 0.424 V·nm−1 (that correspond to 800, 2000, and 4500 mV, respectively). As pointed out in a recent paper by Wang,42 the simulation of the effect of electric potential in proteins (such as insulin) can be performed using larger electric fields, generating comparable results in shorter simulations. For this reason, we selected the electric field intensity of 0.424 V·nm−1 to understand the changes in the dipole moment and the solvent accessible surface area (SASA) in LSZ and Mb. The control simulations were carried out using identical conditions but without the application of the external electric field. As a final point, the simulation results were used to study the effect of the imposed electric field on the induced protein dipole moment and the SASA.
In order to complement these results and provide information related to the interaction between incoming proteins and proteins already in the surface, protein-protein interactions were also investigated by MD simulations. In this case, the simulations were configured using the same conditions described above for the single protein molecule excepting that a new box dimensions (15.0 nm × 14.5 nm × 25.0 nm) and only the highest electric field (0.424 V·nm−1) were used. To calculate the work required to form the Mb-Mb and LSZ-LSZ complexes, Steer Molecular Dynamic (SMD) simulations were used.43 At the beginning of the process, both proteins were set at a distance of 9 nm from each other. Next, one of them was pulled toward the z direction coordinate of COM of the second one (fixed COM simulating the protein previously adsorbed on the electrode). The optimal constant pulling velocity was found to be 0.2 nm·ns−1 whereas an appropriate stiff spring approximation43 was obtained using a spring force of 5 kcal·mol−1·nm2. The work was calculated as a function of time (t) using the following expression:
| Equation 2 |
where v is the constant pulling velocity, dt′ is the time step and f is the force done by the system.
Cross-Linking of BSA
Aiming to investigate the role of protein flexibility on the adsorption process under electrical stimulation, BSA molecules (a model soft protein) were cross-linked with suberic acid bis(N-hydroxy-succinimide ester).44 The cross-linker also known as disuccinimidyl subarate (DSS) is a homo-bifunctional cross-linker that reacts specifically with the primary amine of lysine residues to form stable amide bonds and that yields protein molecules with higher conformational rigidity. Because it is insoluble in water, 2 mg of DSS were first dissolved in 432 μL of DMF to obtain a concentration of 12.5 mmol·L−1. Then, the cross-linker solution was added to a BSA solution (5 mL) prepared by dissolving 25 mg of the protein in 10 mmol·L−1 phosphate buffer containing 150 mmol·L−1 of NaCl at pH = 7.8. After shaking at 80 rpm for 30 min, the reaction mixture was placed in a dialysis tube (12-14 kDa) and the ends were sealed using two tubing clips. Next, the sample was immersed in 150 mL of 10 mmol·L−1 citrate buffer at pH = 4.7 contained in a beaker and incubated in a cold room for 24 hs. In order to remove any unreacted DSS, and in agreement with the dialysis protocol, the buffer solution was replaced every 4 hs.
3. Results and Discussion
3.1 Effect of Applied Potential on the Adsorption of Hard Proteins
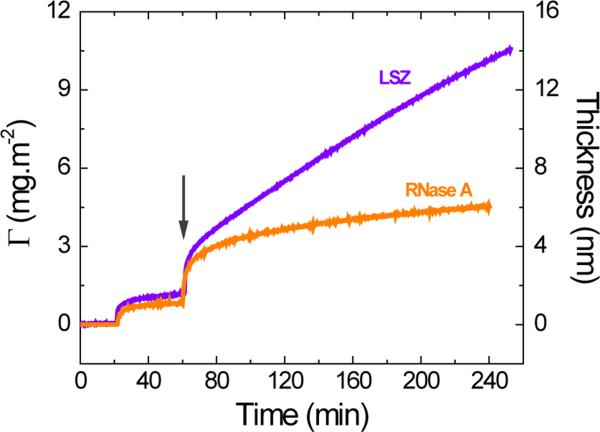
In order to compare the adsorption behavior of structure-stable proteins, two hard proteins16, 18 with similar molecular dimensions (LSZ and RNase A) were selected. In accordance with previous reports14,15 and after establishing the baseline, a solution containing the selected protein (0.1 mg·mL−1 dissolved in 10 mmol·L−1 phosphate buffer at the IEP of the selected protein) was introduced in the cell and the adsorption was recorded until a plateau was obtained (~40 min). As it can be observed in Figure 1, the adsorption at OCP rendered a layer with thickness values of 1.1 ± 0.1 nm and 1.6 ± 0.1 nm for RNase and LZS, respectively. Considering the molecular dimensions of these proteins, it is reasonable to consider that both proteins would be able to tumble upon the initial contact with the surface and adopt a side-on orientation with minimum structural rearrangements. In such case, the results shown in Figure 1 would indicate the formation of an incomplete layer of protein on the surface of the OTCE, with coverage of approximately 50% (at 60 min). Next, the selected potential (+800 mV) was applied to the electrode to induce a secondary adsorption process (marked with an arrow in Figure 1). Under these conditions, a rapid accumulation of proteins was observed immediately after the potential was applied yielding initial adsorption rates (dΓ/dt1) of (98 ± 7) × 10−2 mg·m−2·min−1 and (160 ± 2) × 10−2 mg·m−2·min−1 for RNase A and LSZ, respectively. It is important to point out that this process was attributed to the electrical polarization of the protein in the vicinity of the electrode surface and cannot be attributed to traditional electrostatic (experiments performed at the corresponding IEP) or hydrophobic interactions (equilibrated upon the formation of the first layer of protein). As the experiment progressed, the adsorption rate transitioned to a much slower process (dΓ/dt2, See Table 2), approximated to a first-order reaction, and that allowed the accumulation of significantly larger amounts of protein on the surface, again with respect to results obtained under OCP conditions.
Figure 1.
Effect of applied potential on the kinetic of 0.10 mg·mL−1 LSZ and RNase A at OCP and after the application of external potential to the OTCE. Experiments were performed in 10 mmol·L−1 buffer solution at the IEP of the respective protein and a flow rate of 1 mL·min-1. The arrow shows the time at which the external potential was applied.
Table 2.
Initial adsorption rate (obtained immediately after +800 mV were applied, dΓ/dt1) and linear approximation of the second adsorption processes (dΓ/dt2, calculated as the linear regression of the data points collected during the last 100 min of the experiment). Errors associated with the adsorbed amount correspond to the error propagation obtained from the thickness values from the ellipsometry experiment and modelled following the procedure described in the article. Errors in the adsorption rates correspond to the standard error in the linear regression calculation.
| Proteins | Γocp (mg·m−2) | dΓ/dt1 (× 10−2 mg-m−2-min−1) | dΓ/dt2 (× 10−2 mg-m−2-min−1) | Γ240 min (mg·m−2) |
|---|---|---|---|---|
| RNase A | 0.8 ± 0.2 | 98 ± 7 | 0.656 ± 0.004 | 4.5 ± 0.2 |
| LSZ | 1.2 ± 0.2 | 160 ± 2 | 3.67 ± 0.08 | 10.2 ± 0.2 |
| Mb | 0.9 ± 0.2 | 0.64 ± 0.02 | 0.391 ± 0.003 | 1.5 ± 0.2 |
| α-LAC | 1.5 ± 0.2 | 21 ± 1 | 0.313 ± 0.05 | 2.8 ± 0.2 |
| BSA | 2.4 ± 0.2 | 0.60 ± 0.08 | 0.258 ± 0.008 | 3.0 ± 0.2 |
| Cross-linked BSA | 2.7 ± 0.2 | 2.0 ± 0.1 | 1.019 ± 0.003 | 4.8 ± 0.2 |
| IgG | 5.0 ± 0.2 | 2.5 ± 0.2 | 1.033 ± 0.006 | 7.4 ± 0.2 |
| GOx | 2.4 ± 0.2 | 0.16 ± 0.04 | 0.080 ± 0.02 | 2.6 ± 0.2 |
After following the adsorption under the application of the external potential for 240 min, adsorbed layers of 13.6 ± 0.2 nm and 6.0 ± 0.2 nm were obtained for LSZ and RNAse A, respectively. Again, considering the molecular dimensions of the proteins and a side-on orientation, these results suggest the transition from an incomplete monolayer (obtained at OCP) into a multilayer consisted of approximately 4.5 and 2.7 molecules (in average) of LSZ and RNAse A, respectively. It is important to note that large values of dΓ/dt1 were obtained for both hard proteins indicating that both display similar ability to respond to the electric field (polarizability). However, the experimental data (dΓ/dt2, see Table 2) also showed that the capacity to bind additional protein molecules and further grow the layer was much higher for LSZ than for RNase A.
3.2 Effect of Applied Potential on the Adsorption of Soft Proteins
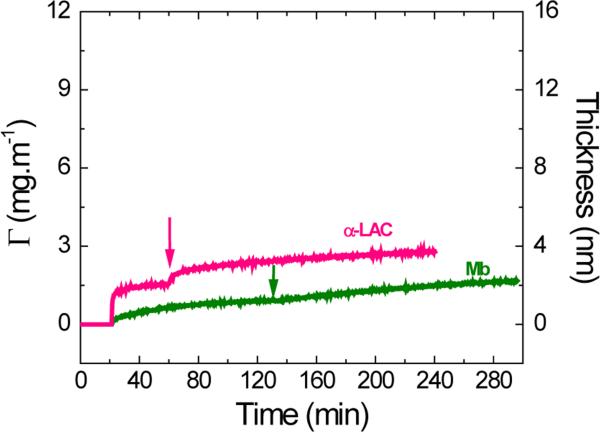
Along the same lines, the effect of the applied potential was evaluated on proteins with limited structural stability and that, as a consequence, have tendency to experience significant structural rearrangements upon adsorption. Owing to their similar molecular dimensions, α-LAC and Mb (considered model soft proteins16, 18) were selected for the initial set of experiments herein discussed. As previously described, the experiment started recording a baseline (impinging 10 mmol·L−1 buffer solution at IEP of the corresponding protein on the OTCE) for 20 min, while no potential was applied on the electrode surface. Then, a solution containing 0.10 mg·mL−1 of the selected protein (and at the corresponding IEP) was introduced in the cell at OCP with the purpose to coat the OTCE with a first layer of protein. As shown in Figure 2A, α-LAC was able to adsorb to the OTCE at OCP and form a monolayer in about 40 min, while Mb required approximately 110 min to reach a plateau in the signal (under the selected experimental conditions). Considering the thickness values obtained for each protein layer (dα-LAC = 2.0 ± 0.2 nm and dMb = 1.2 ± 0.2 nm) and their tendency to spread on the surface, the results suggest that while α-LAC is able to adsorb at a much higher rate and preserve much of its structural integrity, Mb undergoes significant spreading, increasing its footprint by a factor of approximately two. Next, the selected potential (+800 mV) was applied to the OTCE and maintained until the end of the experiment. As it can be also observed in Figure 2A, the potential only induced a limited accumulation of proteins yielding initial adsorption rates of (21 ± 1) × 10−2 mg·m−2·min−1 and (0.64 ± 0.02) × 10−2 mg·m−2·min−1 for α-LAC and Mb, respectively. In line with the results obtained with other proteins, the adsorption process gradually slowed down (dΓ/dt2) and allowed the accumulation of additional protein molecules on the surface. However, the obtained adsorbed amounts (Γ240min, see Table 2) indicate that the effect of external potential on the adsorption of soft proteins is less pronounced than on the adsorption of the hard counterparts.
Figure 2A.
Effect of applied potential on the adsorption kinetic of 0.10 mg·mL−1 α-LAC and Mb after adsorption of the respective protein monolayer onto OTCE at OCP. Experiments were performed in 10 mmol·L−1 buffer solution at IEP of the respective protein and a flow rate of 1 mL·min−1. The arrows show the time which the external potential was applied.
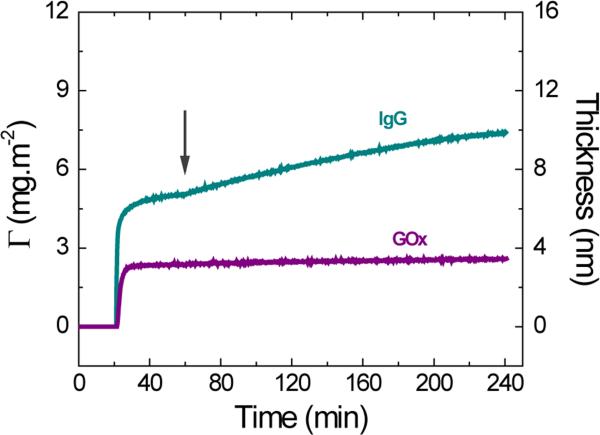
In addition to α-LAC and Mb, two soft proteins with similar molecular weights were selected to investigate the effect of molecular dimensions on the adsorption under electrical stimulation. For these experiments, GOx and IgG (Figure 2B) were selected. Again, a protein monolayer was allowed to adsorb on the OTCE at OCP (10 mmol·L−1 buffer solution containing 0.1 mg·mL−1 of protein at the IEP) after a baseline was collected injecting buffer solution for 20 min. Then, +800 mV were applied at t=60 min and hold during the remaining course of the experiment. Representative examples of the obtained dynamic adsorption curves are shown in Figure 2B. With pertinent differences, both GOx and IgG followed the pattern observed for other soft proteins. In this regard, the adsorption of the first layer of proteins onto the OTCE (at OCP) yielded thicknesses values of 3.1 ± 0.2 nm and 6.7 ± 0.4 nm for GOx and IgG, respectively. In agreement with previous reports,15 the corresponding thickness values are compatible with the formation of incomplete layers without a preferential side-on orientation. It is important to note that when the potential was applied, IgG exhibited a slightly higher initial adsorption rate ((2.5 ± 0.2) × 10−2 mg·m−2·min−1) than GOx ((0.16 ± 0.04) × 10−2 mg·m−2·min−1), suggesting that more compact proteins would be less susceptible to the potential-induced adsorption process. Although these values differ in almost an order of magnitude, they are still significantly smaller than those obtained from hard proteins and reveal the importance of the molecular stability on the potential-mediated adsorption process.
Figure 2B.
Effect of applied potential on the adsorption kinetic of 0.10 mg·mL−1 IgG and GOx after adsorption of the respective protein monolayer onto OTCE at OCP. Experiments were performed in 10 mmol·L−1 buffer solution at IEP of the respective protein and a flow rate of 1 mL·min−1. The arrows show the time which the external potential was applied.
3.3 Effect of Applied Potential on the Adsorption of BSA and Cross-linked BSA
It is then evident that the effect of the potential applied to the OTCE is more pronounced on the adsorption of hard proteins than on the adsorption of soft proteins. The data also indicate that the initial adsorption rate upon the application of the selected potential (dΓ/dt1, see Table 2) is most sensitive to the structural stability of the adsorbing protein. These findings are critical for the rational development of surfaces with enhanced biological activity (catalysis, biocompatibility, biorecognition, etc), for which high adsorption rates and thicker protein layers are preferred.
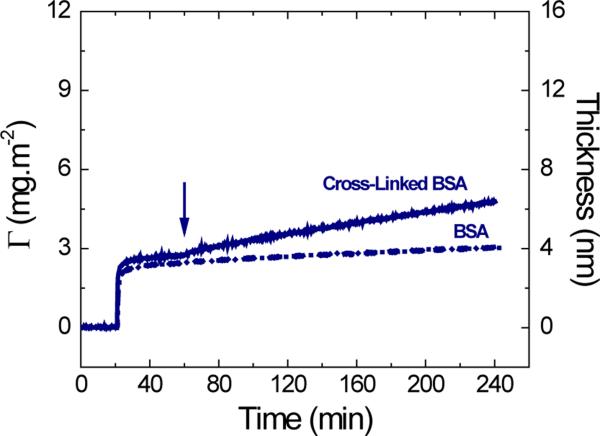
To verify this trend, the effect of the applied potential on the dynamic adsorption of BSA was compared to that of BSA cross-linked with DSS. DSS is a hydrophobic and lysine-specific cross-linker44 that can restrict the conformational rearrangements after the adsorption process, therefore decreasing the soft character of the original molecule. Again, the adsorption experiments were performed allowing the formation of a monolayer of either BSA or cross-linked BSA on the surface of the OTCE at OCP (10 mmol·L−1 citrate buffer containing 0.1 mg·mL−1 of protein at the IEP). As it can be observed in Figure 3, and in agreement with a previous report from our group,14 the protein monolayers adsorbed on the OTCE at OCP presented a slight difference in the thickness (dBSA = 3.3 ± 0.2 nm, dCL-BSA = 3.7 ± 0.3 nm). Such difference can be attributed to the restricted tendency of the BSA molecules to rearrange their molecular structure (spreading) upon the cross-linking step. Then, the potential was applied and maintained until the end of the dynamic adsorption experiment. It is important to point out that while the applied potential only induced a slight increase in the adsorbed amount of BSA, substantial variations were observed when the experiment was performed with the cross-linked version of the protein. While the trend observed is in line with that of other hard proteins, the adsorption rate and adsorbed amount are significantly lower than those observed for LSZ and RNase A. Although it may not be correct to consider the cross-linked BSA as part the group of hard proteins, it is evident that the internal flexibility of the protein has a mayor role on the capacity of proteins to respond to the electric field and can influence the adsorptive behavior upon the application of the external field.
Figure 3.
Effect of the applied potential on the adsorption kinetic of 0.10 mg·mL−1 BSA and BSA cross-linked with DSS after adsorption of the respective protein monolayer onto OTCE at OCP. Experiments were performed in 10 mmol·L−1 citrate at IEP and a flow rate of 1 mL·min−1. The arrow shows the time which the external potential was applied.
3.4. Molecular Modeling of LSZ and Mb
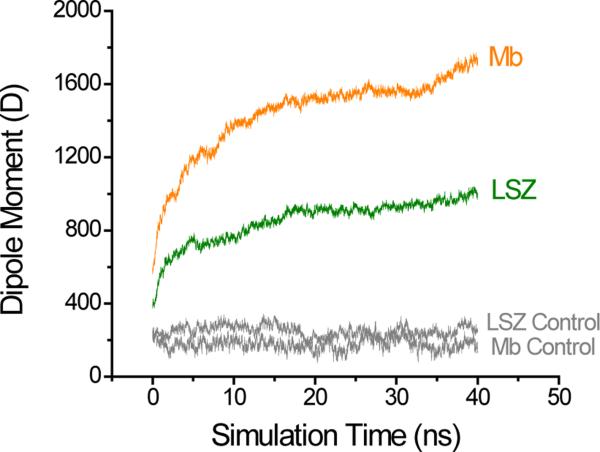
In order to provide an insight into the molecular mechanism that might be driving the differential adsorption behavior of proteins observed experimentally due to structural and dipole moment changes in proteins under the effect of applied electric potentials, MD simulations were performed using LSZ and Mb as representative examples of hard and soft proteins, respectively. In agreement with recent reports describing that the secondary adsorption process was promoted when higher potential values were applied to the substrate,42, 45 the simulations showed an increase in the dipole moment of both proteins, which depends on the intensity of the applied external electric field (see Supplementary Information). As representative examples, Figure 4 shows a summary of the calculated dipole moment as a function of simulation time for both proteins at 0.424 V·nm−1. As it can be observed, the potential applied was able to induce changes in the dipole moment of both proteins, but this change was larger for Mb than LSZ. More specifically, the application of the electric field induced an 8.0-fold change in the dipole moment of Mb, while the same conditions only induced a 5.5-fold change for LSZ (with respect to the simulations performed without electric field). While showing that the potential applied is able to induce larger conformational changes in flexible proteins (such as Mb or BSA46) yielding to more polarized molecules; the results of the simulations deviate from the trends observed in the previously described experiments and suggest that the polarizability of the protein may not be the only factor influencing the adsorption process of proteins upon the application of the external field.
Figure 4.
Simulation of the change in the dipole moment of LSZ and Mb induced by an electric field of 0.424 V·nm−1. To account for any other variation during the simulation, the control runs (gray) were performed under similar condition and with a zero electric field.
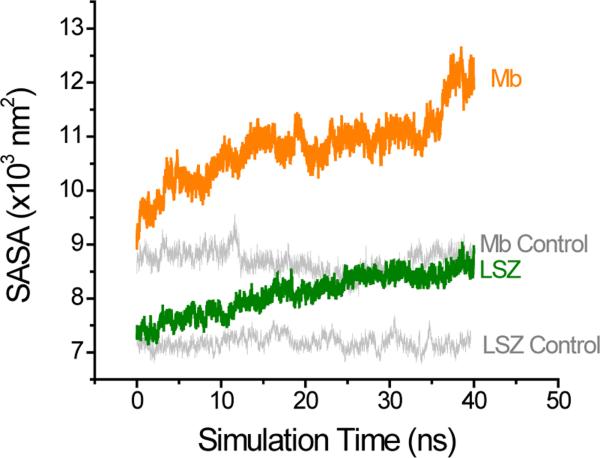
In order to explain the results observed it was required to consider the potential contribution of the solvent during the induction of the dipole moment in the selected proteins. This was accomplished by calculating the solvent accessible surface area (SASA). Changes in the SASA of proteins have been observed by different types of stress including electrical stress.47 Additionally, analysis of the SASA of proteins can be used to provide useful information of the protein potential ability to interact with the solvent and with others proteins.47,48 As shown in Figure 5, the presence of the external field induced not only changes in the dipole moment but also significant increases in the SASA. Also in line with the trend in the flexibility of the proteins, it was observed that the potential induced larger changes in the SASA for Mb than for LSZ. As a result, the simulations suggest that the applied potential induced significant changes in the protein structure (that resulted in an overall enlargement of the surface area) and the corresponding increases in hydration. In turn, this effect results in an increase in the solvation energy of the protein.
Figure 5.
Simulation of the SASA for LSZ and Mb induced by an electric field of 0.424 V·nm−1. To account for any other variation during the simulation, the control runs (gray) were performed under similar condition and with a zero electric field.
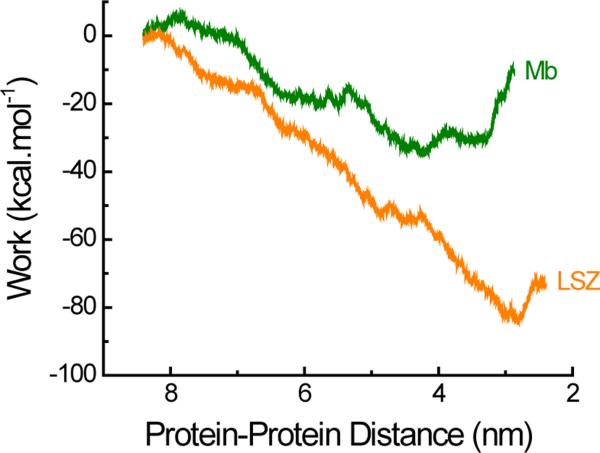
In light of these results, it is evident that both the protein dipole moment and the SASA should be considered together to identify the mechanisms driving the adsorption of protein assisted by potential. To address this need and provide a more realistic description of the protein accumulation phenomenon, the work function related to the interaction of two proteins under the effect of an external electric field was calculated. Figure 6 shows the results of such calculation considering that when the center of mass of the two proteins are (at least) 9 nm apart, they have no significant attraction. As it can be observed, the simulation results revealed that the work required to bring together two Mb molecules was much higher than that required to bring the two LSZ molecules together. In other words, LSZ showed stronger attractive forces than Mb under the influence of the applied potential. In fact, the adsorption rate of Mb was found to be considerably lower than that observed in the case of LSZ.
Figure 6.
Simulation of the work done during the Mb-Mb and LSZ-LSZ interactions induced by an electric field of 0.424 V·nm−1. The initial distance between the centers of mass of both proteins was 9 nm.
As a consequence, the results from the simulations (Figures 4-6) suggest that the secondary adsorption process, experimentally observed upon the application of the external field, could be attributed to the result of favorable electrostatic interactions (induced dipoles) and unfavorable increases in the SASA. Therefore, the high adsorption rate experimentally observed for LSZ upon the application of the external field could be explained by considering that the increase in the dipole moment is sufficiently large to overcome the moderate increase in the SASA. Although the intrinsic flexibility of Mb allows the development of a large dipole moment, this process is also accompanied by a significant increase in the SASA, which cannot compensate the electrostatic interaction and results in decreased protein-protein interactions.
4. Conclusions
The adsorption of hard and soft proteins was studied under the influence of an external potential using spectroscopic ellipsometry and complemented with MD simulations. According to the presented results, the potential applied to the electrode can affect the adsorption of all the selected proteins. However, “hard” proteins display a much stronger susceptibility to the applied potential than “soft” proteins, leading to the formation of ticker protein layers. While the induced changes in the dipolar moment of the adsorbing protein and the magnitude of applied potential are important factors, MD simulations were crucial to identify the contribution of the SASA, which seems to compete with the electrostatic interactions promoted by the induced dipoles. Although the results presented in this article cannot yet be extended to all proteins, the proposed approach could soon lead to the inclusion of other factors and provide insights on the molecular mechanism by which the adsorption of hard and soft proteins can be modulated by the application of an external electric potential.
Supplementary Material
Acknowledgements
Financial support for this project has been provided in part by the University of Texas at San Antonio and the National Institutes of Health through the National Institute of General Medical Sciences (2SC3GM081085) and the Research Centers at Minority Institutions (G12MD007591). The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing resources that have contributed to the research results reported within this paper.
Footnotes
The authors have declared no conflict of interests.
References
- 1.Norde W. Driving forces for protein adsorption at solid surfaces. Macromol. Symp. 1996;103(1):5–18. [Google Scholar]
- 2.Vermeer AWP, Norde W. CD Spectroscopy of Proteins Adsorbed at Flat Hydrophilic Quartz and Hydrophobic Teflon Surfaces. J. Colloid Interface Sci. 2000;225(2):394–397. doi: 10.1006/jcis.2000.6769. [DOI] [PubMed] [Google Scholar]
- 3.Norde W, Giacomelli CE. BSA structural changes during homomolecular exchange between the adsorbed and the dissolved states. J. Biotechnol. 2000;79(3):259–268. doi: 10.1016/s0168-1656(00)00242-x. [DOI] [PubMed] [Google Scholar]
- 4.Norde W. Colloids and Interfaces in Life Sciences and Bionanotechnology. CRC Press; 2011. [Google Scholar]
- 5.Norde W. My voyage of discovery to proteins in flatland ...and beyond. Colloids Surf. B. 2008;61(1):1–9. doi: 10.1016/j.colsurfb.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 6.Rabe M, Verdes D, Seeger S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011;162(1–2):87–106. doi: 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Vogler EA. Protein adsorption in three dimensions. Biomaterials. 2012;33(5):1201–1237. doi: 10.1016/j.biomaterials.2011.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alharthi SA, Benavidez TE, Garcia CD. Ultrathin Optically Transparent Carbon Electrodes Produced from Layers of Adsorbed Proteins. Langmuir. 2013;29(10):3320–3327. doi: 10.1021/la3049136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brusatori MA, Tie Y, Van Tassel PR. Protein Adsorption Kinetics under an Applied Electric Field: An Optical Waveguide Lightmode Spectroscopy Study. Langmuir. 2003;19(12):5089–5097. [Google Scholar]
- 10.Fraaije JG, Kleijn JM, van der Graaf M, Dijt JC. Orientation of adsorbed cytochrome c as a function of the electrical potential of the interface studied by total internal reflection fluorescence. Biophys. J. 1990;57(5):965–975. doi: 10.1016/S0006-3495(90)82616-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Y-Y, Li Y, Yang C, Xia X-H. Surface electric field manipulation of the adsorption kinetics and biocatalytic properties of cytochrome C on a 3D macroporous Au electrode. Anal. Bioanal. Chem. 2008;390(1):333–341. doi: 10.1007/s00216-007-1666-4. [DOI] [PubMed] [Google Scholar]
- 12.Bernabeu P, Caprani A. Influence of surface charge on adsorption of fibrinogen and/or albumin on a rotating disc electrode of platinum and carbon. Biomaterials. 1990;11(4):258–264. doi: 10.1016/0142-9612(90)90007-d. [DOI] [PubMed] [Google Scholar]
- 13.Benavidez TE, Garcia CD. Spectroscopic and electrochemical characterization of nanostructured optically transparent carbon electrodes. Electrophoresis. 2013;34(14):1998–2006. doi: 10.1002/elps.201300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benavidez TE, Garcia CD. Potential-Assisted Adsorption of Bovine Serum Albumin onto Optically Transparent Carbon Electrodes. Langmuir. 2013;29(46):14154–14162. doi: 10.1021/la4029657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benavidez TE, Torrente D, Marucho M, Garcia CD. Adsorption and catalytic activity of glucose oxidase accumulated on OTCE upon the application of external potential. J. Colloid Interface Sci. 2014;435(0):164–170. doi: 10.1016/j.jcis.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norde W, Lyklema J. Interfacial behaviour of proteins, with special reference to immunoglobulins. A physicochemical study. Adv. Colloid Interface Sci. 2012;179-182:5–13. doi: 10.1016/j.cis.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Claesson PM, Blomberg E, Fröberg JC, Nylander T, Arnebrant T. Protein interactions at solid surfaces. Adv. Colloid Interface Sci. 1995;57:161–227. [Google Scholar]
- 18.Bos MA, Shervani Z, Anusiem AC, Giesbers M, Norde W, Kleijn JM. Influence of the electric potential of the interface on the adsorption of proteins. Colloids Surf. B. 1994;3(1):91–100. [Google Scholar]
- 19.Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 1969;244(16):4406–4412. [PubMed] [Google Scholar]
- 20.Kendrew JC, Bodo G, Dintzis HM, Parrish R, Wyckoff H, Phillips D. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181(4610):662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 21.Malamud D, Drysdale JW. Isoelectric points of proteins: A table. Anal. Biochem. 1978;86(2):620–647. doi: 10.1016/0003-2697(78)90790-x. [DOI] [PubMed] [Google Scholar]
- 22.Squire PG, Moser P, O'Konski CT. Hydrodynamic properties of bovine serum albumin monomer and dimer. Biochem. 1968;7(12):4261–4272. doi: 10.1021/bi00852a018. [DOI] [PubMed] [Google Scholar]
- 23.Hecht HJ, Kalisz HM, Hendle J, Schmid RD, Schomburg D. Crystal Structure of Glucose Oxidase from Aspergillus niger Refined at 2·3 Å Reslution. J. Mol. Biol. 1993;229(1):153–172. doi: 10.1006/jmbi.1993.1015. [DOI] [PubMed] [Google Scholar]
- 24.Pazur JH, Kleppe K. The Oxidation of Glucose and Related Compounds by Glucose Oxidase from Aspergillus niger*. Biochemistry. 1964;3(4):578–583. doi: 10.1021/bi00892a018. [DOI] [PubMed] [Google Scholar]
- 25.Georganopoulou DG, Williams DE, Pereira CM, Silva F, Su T-J, Lu JR. Adsorption of Glucose Oxidase at Organic-Aqueous and Air-Aqueous Interfaces. Langmuir. 2003;19(12):4977–4984. [Google Scholar]
- 26.Hecht HJ, Schomburg D, Kalisz H, Schmid RD. The 3D structure of glucose oxidase from Aspergillus niger. Implications for the use of GOD as a biosensor enzyme. Biosens. Bioelectron. 1993;8(3–4):197–203. doi: 10.1016/0956-5663(93)85033-k. [DOI] [PubMed] [Google Scholar]
- 27.Noh H, Yohe ST, Vogler EA. Volumetric interpretation of protein adsorption: ion-exchange adsorbent capacity, protein pI, and interaction energetics. Biomaterials. 2008;29(13):2033–2048. doi: 10.1016/j.biomaterials.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lv Z, Wang J, Chen G, Deng L. Imaging recognition events between human IgG and rat anti-human IgG by atomic force microscopy. Int. J. Biol. Macromol. 2010;47(5):661–667. doi: 10.1016/j.ijbiomac.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Vega RA, Maspoch D, Shen CKF, Kakkassery JJ, Chen BJ, Lamb RA, Mirkin CA. Functional Antibody Arrays through Metal Ion-Affinity Templates. ChemBioChem. 2006;7(11):1653–1657. doi: 10.1002/cbic.200600271. [DOI] [PubMed] [Google Scholar]
- 30.Mora MF, Reza Nejadnik M, Baylon-Cardiel JL, Giacomelli CE, Garcia CD. Determination of a setup correction function to obtain adsorption kinetic data at stagnation point flow conditions. J. Colloid Interface Sci. 2010;346(1):208–215. doi: 10.1016/j.jcis.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nejadnik MR, Deepak FL, Garcia CD. Adsorption of Glucose Oxidase to 3-D Scaffolds of Carbon Nanotubes: Analytical Applications. Electroanalysis. 2011;23(6):1462–1469. doi: 10.1002/elan.201000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujiwara H. Spectroscopic ellipsometry. Principles and applications. J. Wiley & Sons; West Sussex, England: 2007. [Google Scholar]
- 33.Hampton MA, Nguyen AV. Nanobubbles and the nanobubble bridging capillary force. Adv. Colloid Interface Sci. 2010;154(1–2):30–55. doi: 10.1016/j.cis.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Tyrrell JWG, Attard P. Images of Nanobubbles on Hydrophobic Surfaces and Their Interactions. Phys. Rev. Lett. 2001;87(17):176104. doi: 10.1103/PhysRevLett.87.176104. [DOI] [PubMed] [Google Scholar]
- 35.Borkent BM, Dammer SM, Schönherr H, Vancso GJ, Lohse D. Superstability of Surface Nanobubbles. Phys. Rev. Lett. 2007;98(20):204502. doi: 10.1103/PhysRevLett.98.204502. [DOI] [PubMed] [Google Scholar]
- 36.Craig VSJ. Very small bubbles at surfaces-the nanobubble puzzle. Soft Matter. 2011;7(1):40–48. [Google Scholar]
- 37.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct., Funct., Bioinf. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahoney MW, Jorgensen WL. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000;112(20):8910–8922. [Google Scholar]
- 40.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33(suppl 2):W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comer J, Wells D, Aksimentiev A. Modeling Nanopores for Sequencing DNA. In: Zuccheri G, Samorì B, editors. DNA Nanotechnology. Vol. 749. Humana Press; 2011. pp. 317–358. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Li Y, He X, Chen S, Zhang JZH. Effect of Strong Electric Field on the Conformational Integrity of Insulin. J. Phys. Chem. A. 2014;118(39):8942–8952. doi: 10.1021/jp501051r. [DOI] [PubMed] [Google Scholar]
- 43.Park S, Khalili-Araghi F, Tajkhorshid E, Schulten K. Free energy calculation from steered molecular dynamics simulations using Jarzynski's equality. J. Chem. Phys. 2003;119(6):3559–3566. [Google Scholar]
- 44.Huang BX, Kim H-Y, Dass C. Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry. J. Am. Soc. Mass. 2004;15(8):1237–1247. doi: 10.1016/j.jasms.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Favi PM, Zhang Q, O'Neill H, Mamontov E, Diallo SO. Dynamics of lysozyme and its hydration water under an electric field. J. Biol. Phys. 2014;40(2):167–178. doi: 10.1007/s10867-014-9343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bekard I, Dunstan DE. Electric field induced changes in protein conformation. Soft Matter. 2014;10(3):431–437. doi: 10.1039/c3sm52653d. [DOI] [PubMed] [Google Scholar]
- 47.Budi A, Legge FS, Treutlein H, Yarovsky I. Comparative Study of Insulin Chain-B in Isolated and Monomeric Environments under External Stress. J. Phys. Chem. B. 2008;112(26):7916–7924. doi: 10.1021/jp800350v. [DOI] [PubMed] [Google Scholar]
- 48.Singh A, Munshi S, Raghavan V. Effect of External Electric Field Stress on Gliadin Protein Conformation. Proteomes. 2013;1(2):25–39. doi: 10.3390/proteomes1020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.