Abstract
The alarming increase in type 2 diabetes mellitus (T2DM) underscores the need for efficient screening and preventive strategies. Select protein biomarker profiles emerge over time during T2DM development. Periodic evaluation of these markers will increase the predictive ability of diabetes risk scores. Noninvasive methods for frequent measurements of biomarkers are increasingly being investigated. Application of salivary diagnostics has gained importance with the establishment of significant similarities between the salivary and serum proteomes. The objective of this study is to identify T2DM-specific salivary biomarkers by literature-based discovery. A serial interrogation of the PubMed database was performed using MeSH terms of specific T2DM pathological processes in primary and secondary iterations to compile cohorts of T2DM-specific serum markers. Subsequent search consisted of mining for the identified serum markers in human saliva. More than 60% of T2DM-associated serum proteins have been measured in saliva. Nearly half of these proteins have been reported in diabetic saliva. Measurements of salivary lipids and oxidative stress markers that can exhibit correlated saliva plasma ratio could constitute reliable factors for T2DM risk assessment. We conclude that a high percentage of T2DM-associated serum proteins can be measured in saliva, which offers an attractive and economical strategy for T2DM screening.
Keywords: diabetes mellitus, circulating biomarkers, salivary proteins, literature-based search
Introduction
The increasing prevalence of type 2 diabetes mellitus (T2DM) calls for developing concomitant screening strategies for early identification of high-risk individuals for the disease and/or its complications.1 The condition is comorbid with microvascular disorders such as retinopathy and neuropathy. While glucose and insulin are the most well-established biomarkers, advances in the “omics” technologies have identified a range of molecules including genetic variants, proteins, RNA transcripts, and small metabolites that could serve as predictors of diabetes risk.2
The natural history of T2DM suggests that the condition develops insidiously through periods of increased insulin secretion, insulin resistance, impaired glucose tolerance, and cell dysfunction. Several independent hypothesis-driven studies have investigated single or panels of biomarkers to establish T2DM risk scores.3,4 Long-term population-based studies have shown that biomarker trajectories along the course of T2DM development diverge over time. For example, decreased insulin sensitivity and increased interleukin-1 (IL-1) receptor antagonist in serum are evident as early as 13 and 6 years, respectively, prior to clinical diabetes.5,6 This suggests that repeated measures of diabetes-related variables will increase the predictive ability of diabetes risk scores.
Monitoring of serological parameters of diabetes typically involves invasive techniques with associated pain and distress. Hence, the development of noninvasive methods for frequent monitoring of biomarkers is a growing area of research. Some of the alternative methods evaluated include assessing skin auto-fluorescence for accumulation of advanced glycation end products and measuring the analytes in exhaled breath, urine, or saliva.7–10 Human saliva is a rich reservoir of analytes, comprising nearly 3,000 proteins and 12,000 peptides, and shares nearly 30% of proteins and 10% of peptides with the serum proteome and peptidome, respectively.11,12
Alterations in the salivary flow and composition in diabetes are well documented.8,13,14 Salivary glucose is increased in T2DM.14 Salivary immunoreactive insulin level has been correlated with plasma insulin level after a glucose load in individuals with normal glucose tolerance and in T2DM patients.15 Although the mechanism of manipulation of salivary content by systemic diseases is not fully understood, it is suggested that the circulating biomolecules can reach the saliva by active (eg, sIgA) or passive (eg, steroids) transportation, or ultrafiltration (eg, creatinine), or from the gingival crevicular fluid (GCF – a plasma filtrate arising from the junction between the sulcular epithelium and the tooth surface).16
In recent years, text mining methods are increasingly used to identify T2DM-specific risk factors from the vast clinical and experimental data in diabetes research.8,17 Based on the similarities between the serum and salivary proteomes, we hypothesized that the salivary proteome is likely to include a significant cohort of T2DM-specific serum proteins. We used the closed method of literature-based discovery to identify the presence in saliva of serum proteins that are correlated with the pathophysiological processes of T2DM.9,10,18 The ability to predict and identify disease occurrence/progression by biomarker panels is known to increase by including proteins of more than one pathological pathway.19 We evaluated select inflammatory mediators, adipokines, and ghrelin in diabetic saliva to validate our strategy for the identification of disease-specific salivary biomarkers.
Methods
Database mining
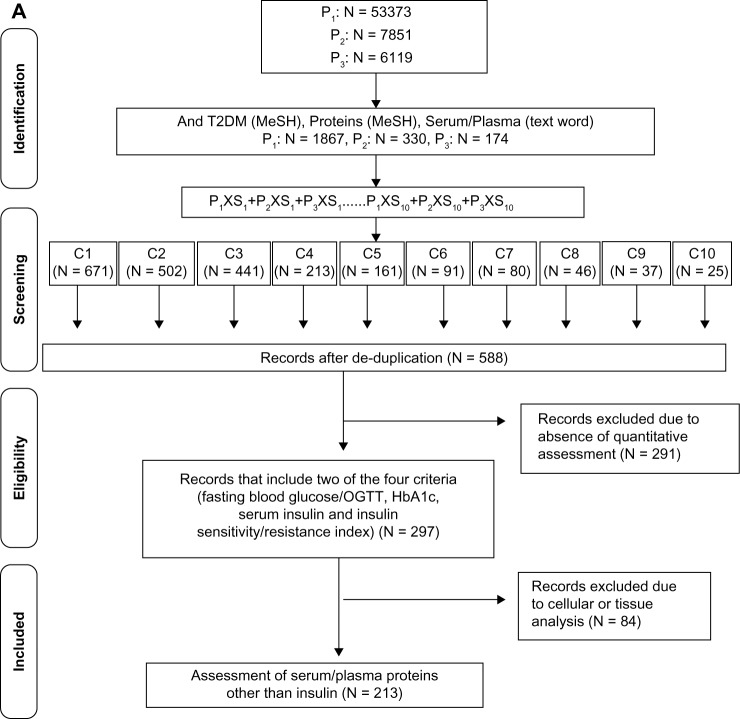
The PubMed database was serially interrogated using specific medical subject heading (MeSH) terms representing the primary sets of interest (P1, P2, and P3) and the secondary sets of interest (S1, S2, etc), resulting in the identification of common sets of interest (C1, C2, etc) (Fig. 1A). The MeSH includes terms that best reflect the article content as assigned by expert indexers at the National Library of Medicine (NLM).10 All English language publications in the PubMed database until January 30, 2014, was included. The MeSH terms insulin resistance, glucose intolerance, and insulin-secreting cells, representing critical processes/components in the pathogenesis of T2DM, constituted the P1, P2, and P3 primary sets, respectively. The three sets were then limited to diabetes mellitus, type 2 (MeSH), and proteins (MeSH), and filtered for humans. Each of the restricted primary set was then separately searched for the text word “serum” or “plasma”. The secondary sets consisted of factors and conditions integrally associated with the development of insulin resistance, glucose intolerance, or impaired β-cell function,9,20 and included the following MeSH terms: lipids/blood, inflammation mediators, adipokines, obesity, glycoproteins, oxidative stress, blood coagulation factors, iron-binding proteins, antibodies and globulins, and endothelium. Each secondary set was individually searched against each restricted primary set. S1–10 × (P1 + P2 + P3) constituted 10 common sets (C1–C10). Duplicate articles in the cumulative C1–C10 cohorts were removed. All retrospective, prospective, cross-sectional, and population studies were included for evaluation. The records so retrieved were screened for the inclusion criteria of measurements of 1) fasting blood glucose/OGTT, 2) glycosylated hemoglobin (HbA1c), 3) serum insulin, and 4) insulin sensitivity or insulin resistance index.3,6 All articles that satisfied three of the four criteria were then searched for names of protein markers and statistical significance. The resultant cohort obtained by this reductionist approach consisted of a total of 297 articles. Articles reporting cellular and/or tissue biomarkers alone and/or qualitative assessments of proteins were excluded. Each marker was counted once irrespective of the number of publications. The identified markers were then classified into six distinct functional groups: Group I: lipids and enzymes; Group II: inflammatory mediators; Group III: adipokines; Group IV: vascular and coagulation factors; Group V: gut hormones and glucose homeostasis; and Group VI: oxidative stress.
Figure 1.
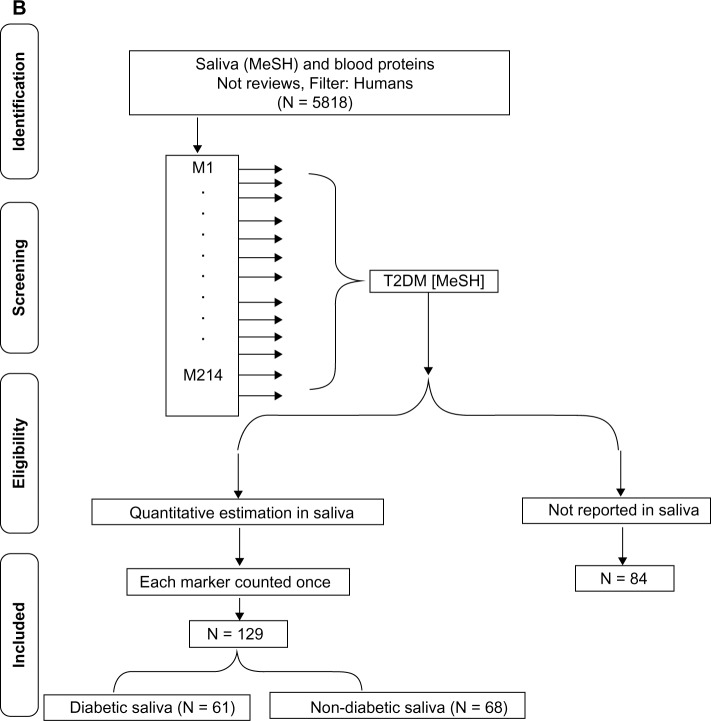
T2DM-specific circulating protein markers. (A) Literature-based search of the PubMed database for identifying T2DM-specific serum proteins using the MeSH terms insulin resistance (P1) or glucose intolerance (P2), or insulin secreting cells (P3) and each of the 10 secondary sets is shown. [S1: lipids/blood, S2: obesity, S3: adipokines, S4: antibodies and globulins, S5: glycoproteins, S6: blood, coagulation factors, S7: inflammation mediators, S8: oxidative stress, S9: endothelium, and S10: iron-binding proteins]. The S1–10 × (P1 + P2 + P3) constitute the 10 common sets C1–C10. (B) Flow diagram of the search strategy for identification of T2DM-associated circulating protein biomarkers reported in saliva. The PubMed database was searched for saliva (MeSH) and blood proteins, the cohort being filtered for humans and English language. The articles in the retrieved cohort were searched for circulating biomarkers identified in (A) and manually screened for quantitation in saliva. The publication cohort of T2DM-specific circulating protein biomarkers reported in saliva was further divided into two groups based on articles reporting quantitation in T2DM saliva (diabetic saliva) or other conditions (nondiabetic saliva).
Next we searched for reports of T2DM-associated serum proteins in saliva. The initial step consisted in retrieving articles reporting the presence of blood proteins in whole saliva using the MeSH term “saliva” and the text retrieval for “blood proteins”. The retrieved cohort was filtered for English language and humans. After excluding review articles, the cohort of articles reporting blood proteins in saliva was searched for the measurement of each of the identified T2DM-associated circulating protein biomarker. The retrieved articles were then manually screened for absolute measurement (not qualitative presence or relative/fold increase) of the markers and statistical significance. The publication cohort of T2DM-specific circulating proteins reported to be present in saliva was further divided into two groups based on measurement in T2DM saliva (diabetic saliva or DS) or other conditions (nondiabetic saliva or NDS). Each marker was counted once irrespective of the number of publications and the type of saliva (DS or NDS) (Fig. 1B). All searches and manual curation were performed independently by two faculty members and two student investigators. The retrieved information was assessed for similarity. In case of discrepancies, all four individuals repeated the search and consensus was reached after discussion.
Enzyme-linked immunosorbent assay (ELISA)
Unstimulated whole saliva (UWS) was collected into a prechilled centrifuge tube by the drooling method21,22 from 20 subjects with self-reported T2DM and HbA1c values (within the past 3 months) and 20 systemically healthy individuals attending the clinics of the Indiana University School of Dentistry (IUSD). Informed consent was obtained from all subjects in accordance with the Indiana University’s Institutional Review Board (IRB). Each sample was clarified by centrifuging at 4000g at 4 °C for 10 minutes and stored in the Complete Protease Inhibitor Cocktail (Roche) at –80 °C. All samples were depleted of amylase and immunoglobulins by incubating serially with antihuman amylase mAb (1:2500, cat. no. ab8944; Abcam) and protein G beads (Miltenyi Biotec Inc.) at 4 °C. Total protein of the precleaned saliva samples was determined by spectrophotometry. One microgram of protein from each precleaned UWS sample was assessed for the presence of IL-6/TNF-α (tumor necrosis factor α)/resistin/visfatin/ghrelin. The salivary resistin, visfatin, and ghrelin were determined using specific colorimetric kits following the manufacturer’s instructions (Cayman Chemical Company). IL-6 and TNF-α concentration in the saliva were determined using OptELISA kits (BD Biosciences).
Statistical analysis
The differences in the analyzed soluble proteins in UWS of normal and T2DM subjects were determined by Student’s t-test. P-Values <0.05 were considered significant.
Results
Compilation of serum proteins reported as markers for T2DM in PubMed
A total of 53,373, 7,851, and 6,119 articles were retrieved in the P1, P2, and P3 primary sets, respectively. Refining for T2DM and serum/plasma proteins and filtering for humans retrieved 1,867 (3.5%), 330 (4.2%), and 174 (2.8%) publications for each of the P1, P2, and P3 sets, respectively (Fig. 1A). When combined, the P1 × P2 cohort had 111 duplicate records, P1 × P3 cohort had 64 duplicates, and the P1 × P2 × P3 cohort had 7 duplicates. This suggested that >50% of articles were unique to each primary set and that the insulin resistance is a frequently used MeSH term in T2DM studies. Although the concepts of obesity, adipokines, and lipids/blood are highly related, each concept retrieved different sets of articles and shared only 8.9% of the articles. Therefore, to avoid missing information in the nonduplicate articles, the subsets were independently searched against each primary set. A total of 588 unique articles in the cumulative C1–C10 cohort were then manually curated for inclusion of well-accepted criteria of T2DM diagnosis, such as measures of fasting blood glucose/OGTT (oral glucose tolerance test), HbA1c, and insulin sensitivity/resistance. The resultant cohort of 297 articles (19.8%) was then screened for quantitative assessment of protein markers. A total of 214 proteins were identified, which were classified into six distinct functional groups as related to the processes of glucose and lipid metabolism, energy homeostasis, inflammation, endothelial perturbation, adipocyte activation, and oxidative stress. The number of circulating T2DM proteins ranged between 25 and 47 markers per functional group. Markers such as plasma insulin and glucose that belong to Group V have been evaluated in most studies as essential indicators of T2DM, while others such as vasculin or progranulin that belong to Group IV have been reported in isolated studies (Fig. 2A). Although the level of confidence was not determined, the inclusion of articles limited to those reporting statistical difference between the normal and T2DM samples supports the efficacy of association of the compiled markers with T2DM.
Figure 2.
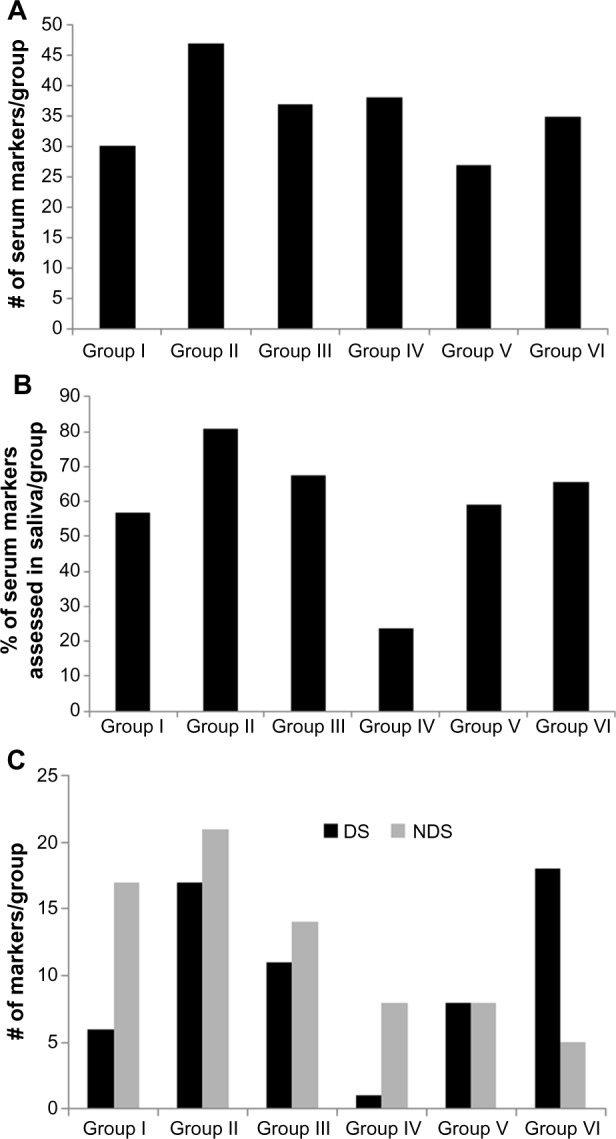
(A) Functional classification of the T2DM-associated circulating protein markers reported in the PubMed database. The T2DM-specific circulating proteins identified as in Figure 1A were classified into six functional groups. Group I: lipids and enzymes, Group II: inflammatory mediators, Group III: adipokines, Group IV: vascular and coagulation factors, Group V: gut hormones and glucose homeostasis, and Group VI: oxidative stress. (A) shows the total number of markers reported in each group. (B, C) Classification of T2DM specific circulating protein markers reported in saliva. The PubMed database was searched for reports of blood proteins (see text) in saliva (MeSH) and filtered for humans and English language. After excluding review articles, the retrieved cohort was screened for each of the 214 T2DM-associated circulating markers. Each marker was counted once irrespective of the number of publications. (B) shows the percentage of T2DM markers reported to be measured in human saliva in each group. The publication cohort of T2DM-specific serum proteins reported in saliva was further divided into two groups based on measurement in T2DM saliva (diabetic saliva or DS) or other condition (nondiabetic saliva or NDS). (C) shows the number of markers of each group reported to be measured in DS and NDS.
Identification of T2DM-associated serum proteins reported in saliva in PubMed
All identified serum/plasma T2DM markers were individually searched for quantitative assessment in saliva (Fig. 1B). Of the 214 circulating protein markers, 130 (60%) have been measured in saliva (Supplementary Table 1). While the inflammatory mediators (Group II), adipokines (Group III), and markers of oxidative stress (Group VI) represent the most investigated, vascular and coagulation factors (Group IV) represent the least evaluated group of T2DM serum proteins in human saliva (Fig. 2B). When screened for assessment in diabetes, it was observed that only 62 of the 130 salivary markers have been specifically measured in T2DM saliva, with the remaining being determined in other conditions including but not restricted to Sjogren’s syndrome or periodontitis. The number of markers assessed in diabetic and nondiabetic saliva per functional group is given in Figure 2C. It is important to note that, since most studies used control saliva samples, all 130 T2DM-associated protein markers with the exception of glutamic acid decarboxylase antibody can be measured in healthy saliva. Since the compiled cohort was restricted to quantitatively assessed markers, it is envisioned that, similar to circulating C-reactive protein and other serum markers for T2DM, differences in the concentration of select proteins between diabetic and nondiabetic saliva can be of diagnostic and/or prognostic significance.
Measurement of T2DM-associated serum protein markers in diabetic saliva
Assessment of multiple markers derived from different pathways of the disease pathogenesis is likely to exhibit greater potential for risk prediction of complex progressive diseases such as T2DM.19 The critical role of inflammation potentially initiated by changes in the adipose tissue is increasingly recognized as pivotal to the pathogenesis of T2DM.9,23 Gut hormones (Group IV) such as ghrelin regulate glucose homeostasis and exert antioxidant and anti-apoptotic effects, thereby playing a role in preventing diabetic complications such as nephropathy.24 Hence, as proof of concept of our data mining method and the information retrieved, we assessed representative inflammatory mediators from Group II (TNF-α and IL-6), adipokines from Group III (resistin and visfatin), and gut hormone from Group IV (acylated ghrelin and deacylated ghrelin) in diabetic saliva. We depleted the saliva samples of the abundant high-molecular-weight amylase and immunoglobulins to improve detection of the less abundant adipokines and ghrelin. While the concentration of TNF-α was significantly lower, that of IL-6 was significantly higher in T2DM saliva as compared to that in healthy saliva samples (Table 1). The salivary levels of acylated and deacylated ghrelin were slightly higher in T2DM saliva than in control saliva, although the difference was not significant (Table 1). Significantly decreased levels of ghrelin in the saliva of T2DM patients have been reported earlier.25 The differences in the salivary ghrelin levels in our study could be attributed to the nature of the sample (whole saliva vs precleaned) and the differences in the methods (ELISA vs radioimmunoassay/fast protein liquid chromatography) used. With respect to the adipokines, while the concentration of visfatin was significantly higher in T2DM saliva, that of resistin was equivalent to that in control samples (Table 1). The presence of resistin in saliva is at about half the concentration in serum and elevated levels in T2DM saliva has been reported earlier.26,27 The observed discrepancy could be due to the differences in the nature of the sample (whole saliva vs precleaned) and potentially the duration of diabetic status, since salivary resistin has been correlated with homeostatic model assessment (HOMA)-insulin resistance.27
Table 1.
Salivary markers in T2DM.
| MARKERS | T2DM | CONTROL | P VALUE |
|---|---|---|---|
| TNF-α | 19.1 ± 5.2 pg/ml | 28.6 ± 12.5 pg/ml | ≤0.015* |
| IL6 | 69.3 ± 18.6 pg/ml | 53 ± 20.3 pg/ml | ≤0.03* |
| Acylated ghrelin | 12.5 ± 0.5 pg/ml | 7.9 ± 1.8 pg/ml | ≤0.076 |
| Deacylated ghrelin | 10.7 ± 4.7 pg/ml | 8.1 ± 1.9 pg/ml | ≤0.061 |
| Resistin | 116.9 ± 98.6 pg/ml | 67.2 ± 27.1 pg/ml | ≤0.103 |
| Visfatin | 30.5 ± 5.12 pg/ml | 12.2 ± 5.6 pg/ml | ≤0.004* |
Notes: Unstimulated whole saliva (UWS) from 20 T2DM and 20 healthy saliva samples were assessed for the indicated marker by ELISA. Data given as mean ± SD.
P < 0.05.
Discussion
Discriminatory salivary biomarkers for systemic diseases such as breast, lung, and pancreatic cancers have been successfully identified and prevalidated.16,28 In this literature-based discovery, using T2DM serum proteome as the source, we identified a panel of 130 salivary proteins with potential for noninvasive risk assessment of T2DM. These include constituents that enter saliva from plasma and those that are secreted by the salivary glands. It has been suggested that the increased basement membrane permeability often associated with diabetes is a potential mechanism for increased passage of proteins and metabolites from the exocrine glands and for the enhanced leakage of serum-derived components into whole saliva.15
A significant concern regarding the use of salivary biomarkers for systemic diseases is the reliability for clinical application. While large proteins such as cytokines that reach saliva from crevicular fluid or via leaky passage from inflamed tissue are less likely to exhibit reliable saliva plasma ratio, small apolar molecules such as cortisol or low-density lipoproteins can diffuse efficiently and exhibit consistent saliva plasma ratio for reliable measurements.29 A few reports have correlated salivary markers with HbA1C levels – a measure of average serum glucose concentration – and hence indirectly with glycemic control. For example, salivary IL-6 or 1,5 anhydroglycitol has been positively correlated with HbA1c in T2DM.4,30 Salivary sex hormone binding globulin (SHBG) has been correlated negatively with HOMA-insulin resistance in nondiabetic, postmenopausal, obese women.7 Our comprehensive search of the published literature showed that, while 48% of T2DM-associated serum proteins have been assessed in diabetic saliva, 52% of the T2DM serum proteins were measured in nondiabetic saliva.
Our study has two limitations. First, the data are restricted to articles retrieved from the PubMed database, thereby potentially missing reports that might have been captured by including additional databases such as Embase and Science Citation Index. Second, the search included all articles reporting quantitation of proteins irrespective of the presence/absence of oral/systemic pathologies that might be related causally or otherwise to specific salivary protein(s). While this may be perceived as a potential drawback,31,32 since the search of salivary markers was restricted to serum proteins associated with T2DM pathophysiology, it is very likely that the identified salivary proteins will have considerable value as risk predictors of T2DM.
Evaluation of proteins from different functional classes is an acceptable alternative strategy to improve predictive ability. Assessment of select panels of serum biomarkers such as TNF-α, IL-6, E-selectin, soluble intercellular cell adhesion molecule-1 (sICAM-1), and soluble vascular cell adhesion molecule-1 (sVCAM-1) or adiponectin, leptin, monocyte chemoattractant protein-1, E-selectin, sICAM-1, and sVCAM-1 have been shown to significantly improve integrated discrimination and risk prediction of T2DM, albeit marginally over the classical risk factors.33–36 In the context of salivary markers for T2DM, the frequent prevalence of concomitant chronic periodontitis could potentially compromise the clinical significance.30–32 For example, elevated cytokines (TNF-α and IL-6) in saliva have been reported in chronic periodontitis with or without diabetes.30,31 Similarly, increased visfatin in the GCF has been reported in chronic periodontitis with or without diabetes.37 Increased resistin in the GCF has been observed in chronic periodontitis, more significantly in chronic periodontitis with T2DM.38 Thus, since chronic periodontitis represents a common confounding factor in diabetes, inclusion of markers that differentiate the cumulative effects of T2DM with periodontitis and T2DM alone either qualitatively or quantitatively will enhance the predictive power of a biomarker panel. In a recent review, Sattar suggested that the potential for clinical use of biomarkers is highest in diabetic complications.9 In this context, it is pertinent to note that many inflammatory and immune-associated proteins determined in saliva have been reported as potential biomarkers for diabetic retinopathy.9
In conclusion, we observed that, in addition to the previously reported 61 proteins, there are at least 84 distinct, quantifiable salivary proteins that have not been assessed in diabetic saliva. Since the risk factors are predictive over time, periodic assessment is of critical significance. Saliva represents a noninvasive biospecimen for frequent measurement of markers, facilitating early detection and improved strategies for diabetes prevention and management. In addition, the greater ease of testing special populations of patients (eg, confined, remote, pediatric) makes assessment of salivary biomarkers an attractive social and cost-effective strategy. However, despite the considerable excitement generated, caution must be exercised since the type of sample (stimulated/unstimulated; whole/glandular saliva), circadian variations, and susceptibility to preprocessing are some of the confounding parameters that should be addressed in biomarker selection and validation.39
Supplementary File
Supplementary Table 1. Information retrieved from PubMed database on T2DM serum markers reported in human saliva.
Acknowledgments
The authors acknowledge the contributions and publications by all the investigators whose works formed the basis of this literature-based study. The authors also thank Prof. Melinda Meadows and Prof. Lisa Maxwell for help with patient recruitment and saliva collection.
Footnotes
ACADEMIC EDITOR: Karen Pulford, Editor in Chief
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived the project, researched data, and wrote manuscript: MS. Conducted database search and compiled data: CB, MM, AV. Reviewed and critiqued manuscript: JB. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–53. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.Tuomilehto J, Lindstrom J, Hellmich M, et al. Development and validation of a risk-score model for subjects with impaired glucose tolerance for the assessment of the risk of type 2 diabetes mellitus-The STOP-NIDDM risk-score. Diabetes Res Clin Pract. 2010;87:267–74. doi: 10.1016/j.diabres.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Kolberg JA, Jorgensen T, Gerwien RW, et al. Development of a type 2 diabetes risk model from a panel of serum biomarkers from the Inter99 cohort. Diabetes Care. 2009;32:1207–12. doi: 10.2337/dc08-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mook-Kanamori DO, El-Din Selim MM, Takiddin AH, et al. 1,5-anhydrog-lucitol in saliva is a non-invasive marker of short-term glycemic control. J Clin Endocrinol Metab. 2014;99(3):E479–83. doi: 10.1210/jc.2013-3596. [DOI] [PubMed] [Google Scholar]
- 5.Carstensen M, Herder C, Kivimaki M, et al. Accelerated increase in serum interleukin-1 receptor antagonist starts 6 years before diagnosis of type 2 diabetes: Whitehall II prospective cohort study. Diabetes. 2010;59:1222–7. doi: 10.2337/db09-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabak AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimaki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373:2215–21. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akin F, Bastemir M, Alkis E, Kaptanoglu B. SHBG levels correlate with insulin resistance in postmenopausal women. Eur J Intern Med. 2009;20:162–7. doi: 10.1016/j.ejim.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Rao PV, Reddy AP, Lu X, et al. Proteomic identification of salivary biomarkers of type-2 diabetes. J Proteome Res. 2009;8:239–45. doi: 10.1021/pr8003776. [DOI] [PubMed] [Google Scholar]
- 9.Sattar N. Biomarkers for diabetes prediction, pathogenesis or pharmacotherapy guidance? Past, present and future possibilities. Diabet Med. 2012;29:5–13. doi: 10.1111/j.1464-5491.2011.03480.x. [DOI] [PubMed] [Google Scholar]
- 10.Vincent B, Vincent M, Ferreira CG. Making PubMed searching simple: learning to retrieve medical literature through interactive problem solving. Oncologist. 2006;11:243–51. doi: 10.1634/theoncologist.11-3-243. [DOI] [PubMed] [Google Scholar]
- 11.Loo JA, Yan W, Ramachandran P, Wong DT. Comparative human salivary and plasma proteomes. J Dent Res. 2010;89:1016–23. doi: 10.1177/0022034510380414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan W, Apweiler R, Balgley BM, et al. Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl. 2009;3:116–34. doi: 10.1002/prca.200800140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conner S, Iranpour B, Mills J. Alteration in parotid salivary flow in diabetes mellitus. Oral Surg Oral Med Oral Pathol. 1970;30:55–9. doi: 10.1016/0030-4220(70)90011-3. [DOI] [PubMed] [Google Scholar]
- 14.Jurysta C, Bulur N, Oguzhan B, et al. Salivary glucose concentration and excretion in normal and diabetic subjects. J Biomed Biotechnol. 2009;2009:430426. doi: 10.1155/2009/430426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchetti P, Benzi L, Masoni A, et al. Salivary insulin concentrations in type 2 (non-insulin-dependent) diabetic patients and obese non-diabetic subjects: relationship to changes in plasma insulin levels after an oral glucose load. Diabetologia. 1986;29:695–8. doi: 10.1007/BF00870278. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Xiao H, Karlan S, et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS One. 2010;5:e15573. doi: 10.1371/journal.pone.0015573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan AR, Awan FR. Mining of protein based biomarkers for type 2 diabetes mellitus. Pak J Pharm Sci. 2012;25:889–901. [PubMed] [Google Scholar]
- 18.Bhattacharya S, Ha-Thuc V, Srinivasan P. MeSH: a window into full text for document summarization. Bioinformatics. 2011;27:i120–8. doi: 10.1093/bioinformatics/btr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herder C, Karakas M, Koenig W. Biomarkers for the prediction of type 2 diabetes and cardiovascular disease. Clin Pharmacol Ther. 2011;90:52–66. doi: 10.1038/clpt.2011.93. [DOI] [PubMed] [Google Scholar]
- 20.Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363–74. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- 21.Srinivasan M, Kodumudi KN, Zunt SL. Soluble CD14 and toll-like receptor-2 are potential salivary biomarkers for oral lichen planus and burning mouth syndrome. Clin Immunol. 2008;126:31–7. doi: 10.1016/j.clim.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Zunt SL, Burton LV, Goldblatt LI, Dobbins EE, Srinivasan M. Soluble forms of Toll-like receptor 4 are present in human saliva and modulate tumour necrosis factor-alpha secretion by macrophage-like cells. Clin Exp Immunol. 2009;156:285–93. doi: 10.1111/j.1365-2249.2009.03854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bastard JP, Maachi M, Lagathu C, et al. Recent advances in the relationship between obesity, inflammation, and insulin resistance. Eur Cytokine Netw. 2006;17:4–12. [PubMed] [Google Scholar]
- 24.Yada T, Dezaki K, Sone H, et al. Ghrelin regulates insulin release and glycemia: physiological role and therapeutic potential. Curr Diabetes Rev. 2008;4:18–23. doi: 10.2174/157339908783502352. [DOI] [PubMed] [Google Scholar]
- 25.Aydin S. A comparison of ghrelin, glucose, alpha-amylase and protein levels in saliva from diabetics. J Biochem Mol Biol. 2007;40:29–35. doi: 10.5483/bmbrep.2007.40.1.029. [DOI] [PubMed] [Google Scholar]
- 26.Mamali I, Roupas ND, Armeni AK, Theodoropoulou A, Markou KB, Georgopoulos NA. Measurement of salivary resistin, visfatin and adiponectin levels. Peptides. 2012;33:120–4. doi: 10.1016/j.peptides.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Yin J, Gao H, Yang J, Xu L, Li M. Measurement of salivary resistin level in patients with type 2 diabetes. Int J Endocrinol. 2012;2012:359724. doi: 10.1155/2012/359724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao H, Zhang L, Zhou H, Lee JM, Garon EB, Wong DT. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol Cell Proteomics. 2012;11:M111.012112. doi: 10.1074/mcp.M111.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosch JA. The use of saliva markers in psychobiology: mechanisms and methods. Monogr Oral Sci. 2014;24:99–108. doi: 10.1159/000358864. [DOI] [PubMed] [Google Scholar]
- 30.Costa PP, Trevisan GL, Macedo GO, et al. Salivary interleukin-6, matrix metal-loproteinase-8, and osteoprotegerin in patients with periodontitis and diabetes. J Periodontol. 2010;81:384–91. doi: 10.1902/jop.2009.090510. [DOI] [PubMed] [Google Scholar]
- 31.Atieh MA, Faggion CM, Jr, Seymour GJ. Cytokines in patients with type 2 diabetes and chronic periodontitis: a systematic review and meta-analysis. Diabetes Res Clin Pract. 2014;104:e38–45. doi: 10.1016/j.diabres.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Choi YH, McKeown RE, Mayer-Davis EJ, Liese AD, Song KB, Merchant AT. Association between periodontitis and impaired fasting glucose and diabetes. Diabetes Care. 2011;34:381–6. doi: 10.2337/dc10-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao C, Song Y, Cook N, et al. The lack of utility of circulating biomarkers of inflammation and endothelial dysfunction for type 2 diabetes risk prediction among postmenopausal women: the Women’s Health Initiative Observational Study. Arch Intern Med. 2010;170:1557–65. doi: 10.1001/archinternmed.2010.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, Tinker L, Song Y, et al. A prospective study of inflammatory cytokines and diabetes mellitus in a multiethnic cohort of postmenopausal women. Arch Intern Med. 2007;167:1676–85. doi: 10.1001/archinte.167.15.1676. [DOI] [PubMed] [Google Scholar]
- 35.Mishra M, Kumar H, Bajpai S, Singh RK, Tripathi K. Level of serum IL-12 and its correlation with endothelial dysfunction, insulin resistance, proinflammatory cytokines and lipid profile in newly diagnosed type 2 diabetes. Diabetes Res Clin Pract. 2011;94:255–61. doi: 10.1016/j.diabres.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 36.Spranger J, Kroke A, Mohlig M, et al. Inflammatory cytokines and the risk to develop type 2 diabetes: results of the prospective population-based European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Diabetes. 2003;52:812–7. doi: 10.2337/diabetes.52.3.812. [DOI] [PubMed] [Google Scholar]
- 37.Pradeep AR, Raghavendra NM, Sharma A, et al. Association of serum and crev-icular visfatin levels in periodontal health and disease with type 2 diabetes mel-litus. J Periodontol. 2012;83:629–34. doi: 10.1902/jop.2011.110272. [DOI] [PubMed] [Google Scholar]
- 38.Gokhale NH, Acharya AB, Patil VS, Trivedi DJ, Setty S, Thakur SL. Resistin levels in gingival crevicular fluid of patients with chronic periodontitis and type 2 diabetes mellitus. J Periodontol. 2014;85:610–7. doi: 10.1902/jop.2013.130092. [DOI] [PubMed] [Google Scholar]
- 39.Esser D, Alvarez-Llamas G, de Vries MP, Weening D, Vonk RJ, Roelofsen H. Sample stability and protein composition of saliva: implications for its use as a diagnostic fluid. Biomark Insights. 2008;3:25–7. doi: 10.4137/bmi.s607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Information retrieved from PubMed database on T2DM serum markers reported in human saliva.