SUMMARY
In previous studies we observed that the helicase domain of Drosophila Dicer-2 (dmDcr-2) governs substrate recognition and cleavage efficiency, and that dsRNA termini are key to this discrimination. We now provide a mechanistic basis for these observations. We show that discrimination of termini occurs during initial binding. Without ATP, dmDcr-2 binds 3′ overhanging, but not blunt, termini. By contrast, with ATP, dmDcr-2 binds both types of termini, with highest affinity binding observed with blunt dsRNA. In the presence of ATP, binding, cleavage, and ATP hydrolysis are optimal with BLT termini compared to 3′ovr termini. Limited proteolysis experiments suggest the optimal reactivity of BLT dsRNA is mediated by a conformational change that is dependent on ATP and the helicase domain. We find that dmDcr-2’s partner protein, Loquacious-PD, alters termini dependence, enabling dmDcr-2 to cleave substrates normally refractory to cleavage, such as dsRNA with blocked, structured or frayed ends.
Graphical Abstract

INTRODUCTION
Dicer is an essential enzyme in all animals (Alvarez-Garcia and Miska, 2005), where it functions to process microRNAs (miRNAs) and small interfering RNAs (siRNAs). In both pathways Dicer cleaves double-stranded RNA (dsRNA) precursors to generate smaller RNA species (~20–30 nucleotides (nts)), which in concert with other factors, bind complementary mRNAs to downregulate their expression (Wilson and Doudna, 2013). In addition to this posttranscriptional regulation, in some organisms siRNAs produced by Dicer promote chromatin modifications that lead to transcriptional silencing (Sabin et al., 2013). Dicer-dependent small RNAs are required for proper expression of genes associated with many pathways, and Dicer mutations are implicated in numerous diseases (Foulkes et al., 2011).
In metazoa, Dicer is a large multi-domain enzyme (Figure 1A). Difficulties in overexpressing large amounts of the protein have hindered high-resolution structural studies. However, reconstructions from single particle electron micrographs (≥ 20Å) of Homo sapiens Dicer (hsDcr) define three conformational states (Lau et al., 2012; Taylor et al., 2013): an “L-shaped” molecule of hsDcr alone (apo; Figure 1B), a conformation favored by binding to a 37 basepair (bp) siRNA precursor, and a conformation favored by binding to an miRNA precursor. The interactions that promote different conformations are unclear. However, using domain-specific antibodies, tagged proteins, or proteins missing domains (Lau et al., 2012; Taylor et al., 2013), electron-microscopy (EM) studies also establish that the base of the L-shape is the helicase domain, and it is this domain that moves relative to the rest of the protein, or “core”, to yield different conformations (Taylor et al., 2013). While the helicase domain of hsDcr is conserved, all studies, from the first biochemical characterization (Zhang et al., 2002) until now, indicate the activity of hsDcr is independent of ATP. By contrast, Dicer of other organisms, including Caenorhabditis elegans DCR-1 and Drosophila melanogaster Dicer-2 (dmDcr-2), show an ATP dependence (Liu et al., 2003; Welker et al., 2011).
Figure 1. Domain organization and purification of dmDcr-2.
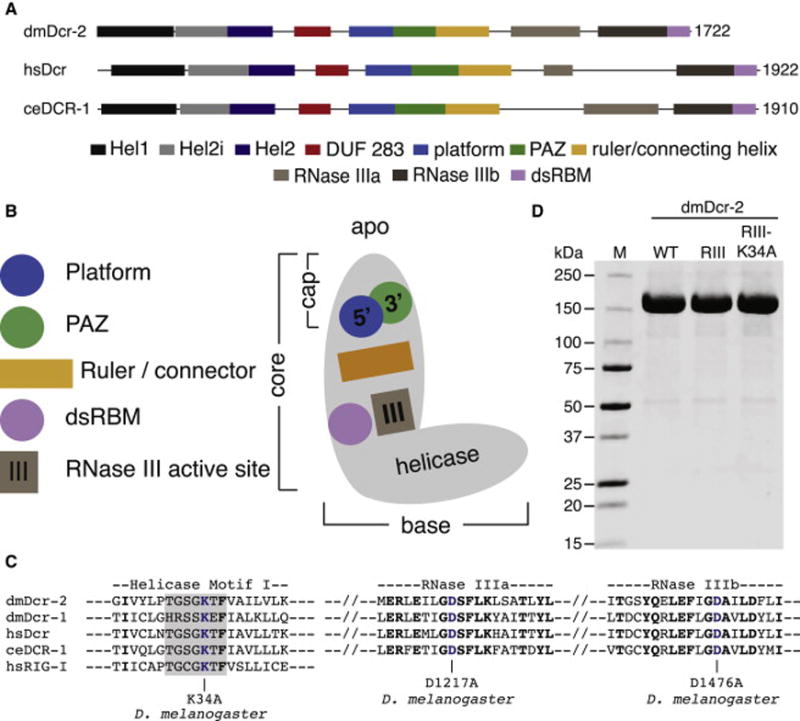
(A) Colored rectangles depict conserved domains. Information from Expresso structure-based alignments (Armougom et al., 2006), NCBI conserved domains (Marchler-Bauer et al., 2010), and crystallographic analyses (Kowalinski et al., 2011; Tian et al., 2014) was used in defining domain boundaries. Information is limited, so the ruler/connecting helix C-terminal boundary is arbitrary.
(B) Cartoon of Dicer domains in apo state (colored as in A) based on negative stain (Lau et al., 2012) and cryo-EM (Taylor et al., 2013) studies.
(C) Amino acids within Motif I (shaded) of Hel1, and the tandem RNase III domains (a and b), are shown with dmDcr-2 mutations (blue) used in this study. dmDcr-2RIII had a mutation in each of the RNase III domains, while dmDcr-2RIII-K34A had all three mutations shown. Studies of related helicases show that this mutation precludes ATP binding and hydrolysis (Linder and Jankowsky, 2011).
(D) Coomassie-stained SDS-PAGE of 10 μg of dmDcr-2 (WT), dmDcr-2RIII (RIII) or dmDcr-2RIII-K34A (RIII-K34A) after purification to homogeneity. M, marker proteins in kilodaltons (kDa).
At the top of Dicer’s core domain, or tip of the L (cap, Figure 1B), lies the PAZ domain (green circle) and adjacent platform domain (blue circle). An x-ray co-crystal structure of a hsDcr fragment containing these domains and part of the connector helix (orange rectangle) defines two pockets specific for dsRNA termini (Tian et al., 2014): the long recognized 3′ terminus binding pocket in the PAZ domain (MacRae et al., 2006), and the long-suspected 5′ terminus binding pocket in the platform domain (Park et al., 2011). By binding to dsRNA termini, these pockets position phosphodiester bonds ~20–30 nts distal for cleavage in the RNase III active site (MacRae et al., 2006).
In vitro, recombinant Dicer proteins cleave dsRNA in the absence of accessory dsRNA binding proteins (dsRBPs). However, in vivo, dsRBPs often facilitate Dicer cleavage. Adding TRBP to hsDcr allows it to process the suboptimal pre-siRNA 5 times faster (Chakravarthy et al., 2010) and promotes the optimal complex observed in cryo-EM analyses (Taylor et al., 2013). Similarly, Loquacious-PD (Loqs-PD) can facilitate cleavage by dmDcr-2, but only a subset of endogenous siRNA (endo-siRNA) precursors require Loqs-PD (Mirkovic-Hösle and Förstemann, 2014), and Loqs-PD is not required for producing exo-siRNAs from certain viruses (Marques et al., 2013). The selective requirement for Loqs-PD in siRNA precursor processing, and the mechanism by which Loqs-PD modulates dmDcr-2’s cleavage activity are unknown. Interestingly, both TRBP and Loqs-PD interact with Dicer’s helicase domain (Daniels et al., 2009; Hartig and Förstemann, 2011).
While Dicer is known to use dsRNA termini to position substrates in the RNase III active site ~22 nts away (MacRae et al., 2007), our previous studies indicate termini recognition plays a more active role in catalysis by the ATP-dependent Dicer enzymes, C. elegans DCR-1 and dmDcr-2 (Welker et al., 2011). With these enzymes, dsRNA with blunt (BLT) termini are processed more efficiently than those with 3′ overhanging (3′ovr) termini. Using dmDcr-2, here we show that dsRNA binding and ATP hydrolysis are also termini-dependent. Importantly, we provide evidence suggesting that the different cleavage efficiencies are accompanied by a termini-dependent, helicase-dependent and ATP-dependent conformational change of dmDcr-2. Further, we show that Loqs-PD modulates dmDcr-2’s termini dependence, allowing dmDcr-2 to cleave substrates with suboptimal termini that normally preclude cleavage.
RESULTS
A bottleneck to studies of Dicer, and mechanistic insight into the role of its helicase domain, is the difficulty in overexpressing large amounts of the enzyme (Ma et al., 2012; Nicholson, 2012). To enable such studies, we optimized overexpression and purification protocols for wildtype dmDcr-2, and two mutant variants (Figures 1C and 1D; Experimental Procedures). Our protocols allowed isolation of multimilligram quantities of highly purified protein per liter of insect cells infected with baculovirus, yields that far exceeded those of prior protocols.
dmDcr-2 shows different affinities and ATP requirements for binding to different termini
Towards the goal of understanding the mechanism by which dmDcr-2 discriminates termini, we considered the possibility that the enzyme had different binding affinities for different termini. Gel-shift analyses have been used to monitor binding of Dicer from various organisms to pre-miRNA (e.g. see (Feng et al., 2012)), but Dicer binding to siRNA precursors has only been monitored by filter binding and crosslinking (e.g. see (Fukunaga et al., 2014; Ma et al., 2012)). dsRBPs are not sequence-specific, and gel-shift assays are particularly informative in regard to how measured affinities correlate with the multiple, distinct complexes that can form with these proteins. We surveyed gel systems and found conditions that gave good separation of free dsRNA and dsRNA-protein complexes (Figure 2A); in these studies we used dmDcr-2RIII, which has a mutation in each RNase III domain to preclude cleavage (Figures 1C and 1D).
Figure 2. Binding affinity of dmDcr-2RIII for 106 BLT and 3′ovr dsRNA.

(A) Increasing dmDcr-2RIII was added to 20 pM dsRNA, labeled at the 5′ end of the sense strand (top in cartoons below lanes) with 32P, and incubated 30 min at 4°C, without (top panel) or with (middle panel) 5 mM ATP. Bottom panel assays were with 5 mM ATP and identical conditions but dsRNA was internally labeled (sense strand, 32P), and each strand had a 2′,3′-cyclic phosphate (open triangle). PhosphorImages show free dsRNA (fastest mobility) and complexes separated by 4% PAGE. (n=3).
(B) Radioactivity in gels as in (A) was quantified to generate binding isotherms. Radioactivity for dsRNAtotal and dsRNAfree was quantified to determine fraction bound. Fraction bound = 1 – (dsRNAfree/dsRNAtotal). All dsRNA of slower mobility than dsRNAfree was considered bound. Data points were fit using the Hill formalism, where fraction bound = 1/(1 + (Kdn/[P]n)). Error bars, standard deviation (STD); (n = 3).
We monitored binding in the absence or presence of ATP, comparing “106” dsRNAs, prepared from 106 nt sense and antisense RNAs, annealed to leave blunt (BLT) or 2-nt 3′overhanging (3′ovr) termini. For all conditions, except those with BLT dsRNA in the absence of ATP, the highest affinity complexes showed a doublet of bands (Figure 2A, compare top and middle panels). Since Dicer binds at the ends of a dsRNA (Zhang et al., 2002), it seemed likely that the highest mobility band of the doublet corresponded to dmDcr-2 binding at one end, and the slower mobility band of the doublet, one dmDcr-2 binding to each end. Indeed, the doublet disappeared in gel-shift assays performed with dsRNA containing terminal 2′,3′-cyclic phosphates to block dmDcr-2 binding to termini (Figure 2A, lower panel). The low affinity complexes observed with BLT dsRNA in the absence of ATP were similar to those observed with 2′,3′-cyclic phosphate substrates, raising the possibility that these complexes involved termini-independent interactions.
Multiple gel-shift assays were performed and quantified to estimate dissociation constants (Figure 2B; Table 1). The affinity of dmDcr-2 for 3′ovr termini in the absence of ATP (29 ± 2 nM) increased only slightly with ATP (18 ± 3 nM). The highest affinity binding occurred to termini of BLT dsRNA (7 ± 1 nM) in the presence of ATP. dmDcr-2 exhibited a distinctly different mode of binding with BLT dsRNA in the absence of ATP that was lower in affinity (56 ± 2 nM), and similar to the binding observed with 2′,3′-cyclic phosphate dsRNA. Together these data indicated that, in the absence of ATP, dmDcr-2 binds to 3′ovr, but not BLT, termini. By contrast, in the presence of ATP, dmDcr-2 binds to both BLT and 3′ovr termini, with the highest affinity binding observed with BLT dsRNA.
TABLE 1.
Summary of Kd, kobs and t1/2 valuesa
| Enzyme | dmDcr-2RIII | dmDcr-2WT | dmDcr-2WT + Loqs-PD | |||||
|---|---|---|---|---|---|---|---|---|
| Kd (nM) | kobs (min−1) (Cleavage) |
t1/2 (min) (Cleavage) |
kobs (min−1) (ATP hydrolysis) |
kobs (min−1) (Cleavage) |
t1/2 (min) (Cleavage) |
kobs (min−1) (ATP hydrolysis) |
||
| ATP | − | + | + | + | + | + | + | + |
| 106 BLT | 56 ± 2b | 7 ± 1 | 0.19 ± 0.01 | 3.7 | 0.04 ± 0.003 | 2.44 ± 0.1 | 0.3 | 0.05 ± 0.006 |
| 106 3′ovr | 29 ± 2 | 18 ± 3 | 0.01 ± 0.001 | 130.6 | 0.002 ± 0.002 | 0.84 ± 0.08 | 0.8 | 0.03 ± 0.001 |
mean ± STD for 3 independent experiments
distinct from termini binding (see text)
dmDcr-2 exhibits different efficiencies and ATP requirements for cleaving dsRNA with different termini
Our gel-shift studies showed that, in the absence of ATP, dmDcr-2 exhibits a low affinity interaction with BLT dsRNA that is distinct from the termini binding observed with 3′ovr dsRNA (Figure 2A). We wondered if the low affinity binding to BLT dsRNA in the absence of ATP could lead to cleavage. Using single-turnover cleavage assays (Figure 3A), we could not detect cleavage of BLT dsRNA in the absence of ATP even at long incubation times. By contrast, 3′ovr dsRNA was cleaved in the absence of ATP, albeit inefficiently (Figures 3A and 3B).
Figure 3. dsRNA termini dictate reactivity of dmDcr-2WT in an ATP-dependent manner.

(A) Single-turnover cleavage assays of 106 BLT and 3′ovr dsRNA (1 nM) with dmDcr-2WT (30 nM) at 25°C in cleavage assay buffer. Sense strand was 32P-internally labeled. Products were separated by 17% denaturing PAGE, and a representative PhosphorImage is shown. *, major siRNA product; ns, non-specific band; Left, marker nucleotide lengths; (n≥3).
(B) Quantification of single-turnover assays as in A, in the absence of ATP. Data points are mean ± STD (n=3). All dsRNA not corresponding to dsRNAuncleaved was considered cleaved. % 106 dsRNA cleaved versus time was fit to the pseudo-first-order equation: y = yo + A × (1−e−kt); where A = amplitude of rate curve, yo = baseline (~0), k = pseudo-first-order rate constant = kobs; t = time (Welker et al., 2011).
(C) Same as for (A) except with 5 mM ATP; (n≥3).
(D) Same as for (B) except with 5 mM ATP; (n=3).
(E) Multiple-turnover cleavage assays were with 8 nM dmDcr-2WT and 100 nM 32P-internally labeled 106 BLT or 3′ovr dsRNA with 5 mM ATP for times indicated. Products were separated by 12% denaturing PAGE, and a representative PhosphorImage is shown. *, major siRNA product; ns, non-specific band; (n≥3). Consistent with processivity, multiple siRNAs are produced per molecule of BLT dsRNA cleaved; at present it is unclear why the reaction plateaus at 40 min, although slow product release, or product inhibition, are possible.
(F) Quantification of multiple-turnover assays with 5 mM ATP as shown in (E) with methods as in (B). Data points are mean ± STD; (n=3).
(G) dmDcr-2WT (200 nM) was incubated with 106 BLT or 3′ovr dsRNA (600 nM) with 100 μM ATP in cleavage assay buffer at 25°C, and ATP hydrolysis monitored by thin-layer chromatography (TLC). PhosphorImage shows a representative TLC plate; (n≥3). Positions of origin, ATP and ADP are indicated.
(H) Quantification of ATP hydrolysis assays as in (G). Data points are mean ± STD (n=3). Data were fit to the pseudo-first-order equation: y = yo + A × (1−e−kt); where y = product formed (ADP in μM); A = amplitude of the rate curve, yo = baseline (~0), k = pseudo-first-order rate constant = kobs; t = time.
(see also Figures S1, S2 and S3).
In previous studies we observed that, in the presence of ATP, BLT dsRNA was cleaved more efficiently than 3′ovr dsRNA. The abundant protein provided by our optimized protocols allowed more quantitative studies, and confirmed that, with 5 mM ATP, single-turnover cleavage of 106 BLT dsRNA (kobs, 0.19 ± 0.01 min−1) was much more rapid than cleavage of 106 3′ovr dsRNA (kobs, 0.01 ± 0.001 min−1; Figures 3C and 3D; Table 1). The observed cleavage depended on termini and was not observed with dsRNA containing 2′,3′-cyclic phosphates (Figure S1). Further, experiments with 42 dsRNA of an independent sequence established that the termini-dependence of cleavage was not sequence-specific (kobs, 0.22 ± 0.02 min−1, BLT, 0.02 ± 0.004 min−1, 3′ovr; Figure S2). Cleavage assays with 106 dsRNA under multiple-turnover conditions also showed termini-dependent cleavage (Figures 3E and 3F). Finally, we observed that ATP hydrolysis was also termini-dependent. In the absence of dsRNA, dmDcr-2WT did not hydrolyze ATP (Figure S3A and S3B). However, under multiple-turnover conditions, dmDcr-2WT hydrolyzed ATP much more rapidly in the presence of BLT (kobs=0.04 ± 0.003 min−1) compared to 3′ovr dsRNA (kobs=0.002 ± 0.002 min−1) (Figures 3G and 3H; Table 1).
In addition to the primary siRNA product of 22 nts, dmDcr-2 generated smaller cleavage products (e.g., Figures 3C and 3E) that were observed regardless of the strand or sequence monitored (see Figure S2). In vitro, these heterogeneous cleavage products appeared at similar rates as primary siRNAs and resembled cleavage products of S. pombe Dcr1 in the presence of ATP (Colmenares et al., 2007). Dicer proteins without a helicase domain (Giardia Dicer) or with an ATP-independent helicase domain (hsDcr) generate uniform cleavage products (Colmenares et al., 2007), suggesting that product heterogeneity is coupled to Dicer’s ATPase activity (see below, Figure 5).
Figure 5. Characterization of cleavage events that require ATP hydrolysis.

(A) Single-turnover cleavage assays were with dmDcr-2WT (30 nM) and 106 BLT (lanes 1–6) or 3′ovr dsRNA (lanes 7–12) (1 nM) for 90 min at 25°C in cleavage assay buffer without (−) or with (+) 5 mM ATP, 10 mM glucose (Glu) and 1 unit hexokinase (HK). Sense strands were 5′32P-labeled (red dot) and blocked with 2′,3′-cyclic phosphate (open triangles). Hexokinase (1 unit) and glucose (10 mM; 0.2 micromoles) were incubated in reaction containing 5 mM ATP (20 min; 25°C) to deplete ATP before addition of dmDcr- 2. Products were resolved on a 17% polyacrylamide denaturing gel; a representative PhosphorImage is shown (n=2). *, 22 nt major siRNA product; **, 8 nt product. Marker nt lengths (left) with 10 nt positions based on 106-sense strand mapping (Welker et al., 2011); AH, alkaline hydrolysis.
(B) Same as for (A) except with 5 mM ATP analogs with or without hexokinase (HK) and glucose (Glu) to remove contaminating ATP (n=2).
Limited proteolysis assays suggest an ATP-, helicase- and termini-dependent conformational change in dmDcr-2
Our binding and kinetic assays indicated that dmDcr-2 binds and cleaves dsRNA with BLT termini differently than dsRNA with 3′ovr termini. Further, our previously reported trap experiments (Welker et al., 2011), and the multiple-turnover experiments reported here (Figures 3E, 3F, 3G and 3H), are consistent with processive cleavage of BLT dsRNA and a distributive cleavage of 3′ovr dsRNA. Because processive cleavage requires dmDcr-2 to produce multiple siRNAs without dissociating from dsRNA, it seemed possible that BLT termini triggered a conformational change in dmDcr-2 that led to a more stable complex.
To explore this possibility, we performed limited proteolysis experiments. After optimizing conditions with poly IC (Figures S4A), dmDcr-2RIII was incubated with trypsin for various times, with or without ATP, and with or without BLT or 3′ovr dsRNA (Figure 4A). A characteristic protease cleavage pattern was observed for dmDcr-2RIII alone (−), and this pattern did not change with the addition of ATP alone, or with either 106 BLT or 3′ovr dsRNA alone. However, with the addition of ATP and BLT dsRNA, a protease-resistant fragment was observed (*), consistent with a conformational change. While a fragment of similar size was stabilized with addition of ATP and 3′ovr dsRNA (arrow), the strong protease resistance was not observed.
Figure 4. dsRNA termini trigger alternate states of dmDcr-2 in an ATP and helicase-dependent manner.
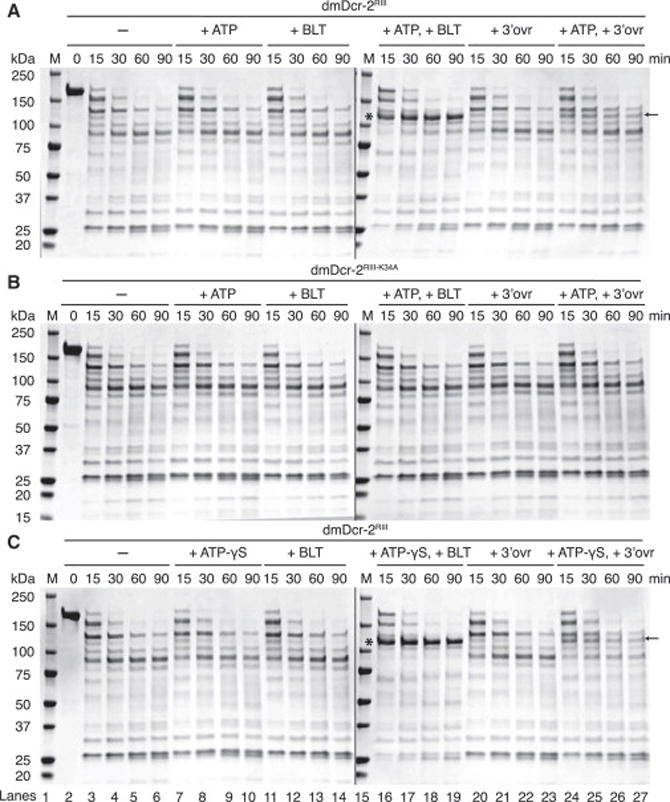
(A) 20 μg of dmDcr-2RIII (2 μM) was incubated with or without 10 μM 106 BLT or 3′ovr dsRNA, with or without 5 mM ATP in cleavage assay buffer (20 min; 25°C), before treating with trypsin (10 nM) for indicated times. At each time point, 4 μg of dmDcr-2 from the reaction mix was quenched with an equal volume of 2X SDS-PAGE loading buffer. Products were resolved by SDS-PAGE and visualized with Coomassie Brilliant Blue. *, protease-resistant fragment of ~116 kDa observed with BLT dsRNA and ATP and analyzed by mass spectrometry and Edman sequencing; ←, protease-resistant fragment observed with 3′ovr dsRNA and ATP; M, markers, kDa noted on left. Lanes 1–14 and lanes 15–27 (see bottom of Figure 4C) were on separate gels run concurrently for same time; (n≥3).
(B) Same as for (A) except with dmDcr-2RIII-K34A (2 μM); (n=3).
(C) Same as for (A) except with 5 mM ATP-γS; (n≥3).
(see also Figures S3, S4).
For both dsRNAs the protease-cleavage pattern observed with ATP was dependent on a functional helicase domain and was not observed with dmDcr-2RIII-K34A (Figure 4B). In addition, for both dsRNAs the protease-cleavage pattern observed with ATP was identical to that observed with ATP-γS (Figure 4C), an analog that cannot be hydrolyzed by dmDcr-2 (Figures S3C and S3D); further, the protease-resistant band (*) observed with ATP-γS was not due to contaminating ATP (Figure S4B). Adding ADP or AMP-PNP to BLT dsRNA gave lower levels of the protease-resistant fragment, similar to what was observed with ATP and 3′ovr dsRNA (Figure S4C; compare to Figure 4A). The results of Figure 4 suggest a model where dsRNA termini trigger alternate conformational states of dmDcr-2 that require a functional helicase domain and nucleotide binding, but not hydrolysis.
Characterization of cleavage events that require ATP hydrolysis
To gain information about which cleavage products required ATP hydrolysis, we monitored the initial cleavage from 106 BLT or 3′ovr dsRNA termini, using dsRNA where one strand had a 5′32P and a 2′,3′-cyclic phosphate to block cleavage from the other end (see cartoons below gel, Figure 5A). We compared single-turnover cleavage products with ATP, ATP-γS, ADP and AMP-PNP (Figures 5A and 5B). To control for artifacts from small amounts of ATP that might contaminate analogs, reactions were performed with or without treatment with the ATP-hydrolyzing enzyme, hexokinase, and its glucose substrate.
As expected, without ATP, or with hexokinase/glucose treatment to remove ATP, 3′ovr, but not BLT, dsRNA was cleaved to produce a 22 nt product from the 32P-labeled terminus (Figure 5A, compare lanes 7, 8, 12 with 1, 2, 6). Consistent with the idea that cleavage of 3′ovr dsRNA is not dependent on binding or hydrolysis of ATP, levels of 22 nt product were constant, regardless of reaction conditions or analog (Figure 5A, lanes 7–12; Figure 5B, lanes 5–6, 9–10,13–14). By contrast, with BLT dsRNA the 22 nt product was only observed under conditions that allowed hydrolysis; after treating ATP or analogs with hexokinase, cleavage products were essentially undetectable with BLT dsRNA (Figures 5A and 5B).
For both 106 BLT and 3′ovr dsRNA, the presence of ATP resulted in additional cleavage products, with the most abundant species being 8, 13–15, and 18 nts. Smaller species were also observed with some nucleotide analogs (Figure 5B), but disappeared after hexokinase/glucose treatment, indicating they resulted from contaminating ATP. Thus, nucleotide binding is sufficient to promote the protease-resistant fragment (Figure 4), but nucleotide hydrolysis is required to generate heterogeneous cleavage products from either dsRNA, and for the production of 22nt siRNAs from BLT, but not 3′ovr, dsRNA.
Loqs-PD modulates dmDcr-2’s termini dependence
dmDcr-2 requires Loqs-PD in vivo for processing certain endogenous substrates. Given our observation that dmDcr-2 was termini-dependent in vitro, and the diverse features of proposed endogenous precursors (Chung et al., 2008; Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008) we wondered if Loqs-PD might alter dmDcr-2’s termini dependence. We compared cleavage by dmDcr-2WT with or without Loqs-PD, using single-turnover cleavage assays. As in previous experiments (Figure 3), without Loqs-PD, dmDcr-2WT cleaved 106 BLT dsRNA more efficiently than 106 3′ovr dsRNA (Figure 6A, compare lanes 3–5 to 7–9), albeit cleavage of BLT, but not 3′ovr, dsRNA required ATP (compare lanes 6 and 10). Addition of Loqs-PD dramatically improved cleavage efficiency of 3′ovr dsRNA and allowed dmDcr-2WT to process BLT and 3′ovr dsRNAs with similar efficiencies (compare lanes 11–13 to 15–17; kobs, Table 1; Figures S5B and S5C). The increased cleavage efficiency with Loqs-PD required ATP; with Loqs-PD but without ATP, BLT dsRNA was not cleaved (lane 14) and 3′ovr dsRNA was cleaved minimally (lane 18). In multiple-turnover cleavage assays Loqs-PD also stimulated cleavage in the presence of ATP (Figures 6B and S5D), but again, the effect was more pronounced for 3′ovr dsRNA. Loqs-PD also promoted a dramatic increase (~15 fold) in rate of ATP hydrolysis by dmDcr-2WT with 3′ovr dsRNA, and a subtle increase with BLT dsRNA (Figures 6C and S5E; Table 1).
Figure 6. Loqs-PD modulates termini-dependent cleavage by dmDcr-2.
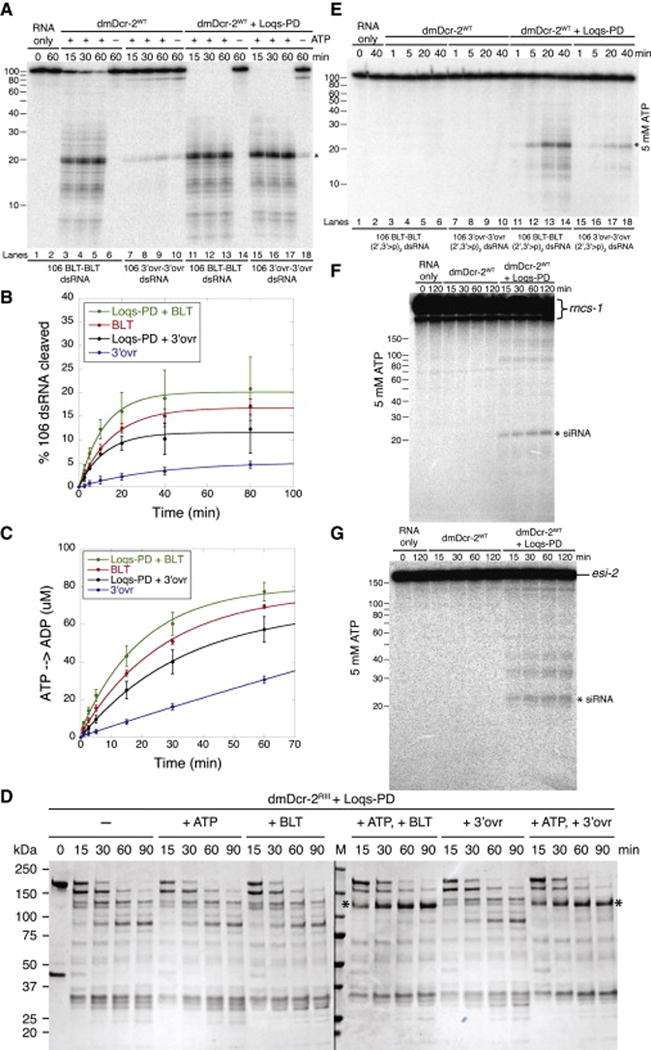
(A) Single-turnover cleavage assays were with dmDcr-2WT (30 nM) and 32P-internally labeled 106 BLT or 3′ovr dsRNA (1 nM), without or with Loqs-PD (30 nM) and −/+ 5 mM ATP, in cleavage assay buffer (25°C). Equimolar amounts of dmDcr-2WT and Loqs-PD were pre-incubated (5 min; ice) prior to adding dsRNA. Products were resolved on a 12% polyacrylamide denaturing gel and a representative PhosphorImage is shown. *, siRNA product; left, marked nt lengths; (n=3).
(B) Quantification of multiple-turnover cleavage assays with 8 nM dmDcr-2WT, 100 nM 32P-internally labeled 106 BLT or 3′ovr dsRNA, 8 nM Loqs-PD as indicated, and 5 mM ATP. Data points are mean ± STD; (n=2).
(C) Quantification of ATP hydrolysis assays with 200 nM dmDcr-2WT, 200 nM Loqs-PD and 600 nM 106 BLT or 3′ovr dsRNA with 100 μM ATP in cleavage assay buffer at 25°C. ATP hydrolysis was monitored by TLC; data points, mean ± STD (n=3).
(D) dmDcr-2RIII (2 μM, 20 μg) and Loqs-PD (2 μM, 4 μg) were incubated with or without 10 μM 106 BLT or 3′ovr dsRNA, −/+ 5 mM ATP in cleavage assay buffer (20 min; 25°C), before treating with trypsin (10 nM) for indicated times. Proteins were pre-incubated (5 min; ice) prior to adding dsRNA. At indicated times, 4 μg of dmDcr-2 and 0.8 μg of Loqs-PD were quenched with an equal volume of 2X SDS-PAGE loading buffer. Products were resolved by SDS-PAGE and visualized with Coomassie Brilliant Blue. *, protease resistant fragment of ~116 kDa observed with BLT and 3′ovr dsRNA with ATP and Loqs-PD. M, markers in kDa, indicated on left. Two separate gels were run concurrently for the same time; (n=3).
(E) Same as for (A) except with 32P-internally-labeled dsRNA with 2′,3′-cyclic phosphates (2′,3′>p) on each strand (n=3).
(F) Single-turnover cleavage reactions were incubated at 25°C in cleavage assay buffer for times indicated, and contained dmDcr-2WT (30 nM), 32P-internally labeled rncs-1 (1 nM), 5 mM ATP, and Loqs-PD when indicated (30 nM). Products were resolved on a 12% polyacrylamide denaturing gel; a representative PhosphorImage is shown. *, siRNA product; Marked nt lengths, left; (n=4).
(G) Same as for (F) except with esi-2; (n=3).
(see also Figures S5 and S6).
In these experiments Loqs-PD was allowing dmDcr-2 to react equally well with BLT and 3′ovr dsRNA. We wondered if addition of Loqs-PD would also minimize differences between BLT and 3′ovr dsRNA in limited proteolysis assays. Indeed, addition of Loqs-PD to reactions containing dmDcr-2RIII and ATP promoted indistinguishable patterns with 106 BLT or 3′ovr dsRNA, with similar amounts of the protease-resistant fragment (*, Figure 6D, compare to Figure 4A; see also Figure S5F and S5G).
Next, we tested whether Loqs-PD enabled dmDcr-2 to process dsRNA with 2′,3′-cyclic phosphates at both ends ((2′,3′>p)2 dsRNA). As shown previously (Figure S1), without Loqs-PD, dmDcr-2WT was unable to cleave BLT or 3′ovr (2′,3′>p)2 dsRNA (Figure 6E, lanes 3–10). However, with Loqs-PD and ATP, dmDcr-2WT cleaved both BLT and 3′ovr (2′,3′>p)2 dsRNA (Figure 6E, lanes 11–18); albeit the reaction was inefficient. Interestingly, BLT (2′,3′>p)2 dsRNA was cleaved faster than 3′ovr (2′,3′>p)2 dsRNA (Figure 6E, compare lanes 11–14 to 15–18), indicating termini continued to influence cleavage. When we monitored initial cleavage using 5′ end-labeled BLT and 3′ovr (2′,3′>p)2 dsRNA (Figure S6A), products were similar to those produced during initial cleavage of dsRNA without cyclic phosphates (Figure 5A). Thus, while Loqs-PD allowed dmDcr-2 to overcome the cyclic phosphate block, the reaction still appeared to initiate from the terminus.
We also tested effects of Loqs-PD on cleavage of substrates that mimic endogenous substrates. The C. elegans long non-coding RNA, rncs-1, folds to create a dsRNA of ~300 bp with terminal branches (Hellwig and Bass, 2008), while the Drosophila endo-siRNA precursor esi-2 is an extended hairpin with a terminal loop and frayed ends (Miyoshi et al., 2010)(Figure S6B). Without Loqs-PD, dmDcr-2WT was unable to cleave rncs-1 or esi-2 (Figure 6F and 6G). However, with Loqs-PD and ATP, dmDcr-2WT produced ~22 nt siRNAs from each RNA. We also analyzed rncs-1 reaction products with northern analyses, using probes to terminal structures that presumably preclude Dicer cleavage without Loqs-PD, or the extended dsRNA rod (Figure S6C). In the presence of Loqs-PD, termini probes mostly detected large fragments that might result from dmDcr-2 cleaving within the rod, while probes to the duplex rod detected only siRNA-sized products. Taken together, these results suggest that Loqs-PD enabled dmDcr-2 to bind and cleave dsRNA substrates internally.
DISCUSSION
Using highly purified, recombinant Drosophila Dicer-2, we performed biochemical assays to establish that the enzyme’s binding affinity, cleavage efficiency, and ATP hydrolysis efficiency, depend on the termini of its dsRNA substrates. In the presence of ATP, binding, cleavage, and ATP hydrolysis are optimal with BLT termini compared to 3′ovr termini. However, without ATP, dmDcr-2 can bind and cleave from 3′ovr, but not BLT, termini. Limited proteolysis experiments suggest the optimal reactivity of BLT dsRNA is mediated by a conformational change that is dependent on ATP and the helicase domain. We show that Loqs-PD alters the termini dependence of dmDcr-2. Our studies offer mechanistic insight into the role of dmDcr-2’s helicase domain and Loqs-PD in recognition and cleavage of endogenous substrates.
Model for differential substrate recognition and processing by dmDcr-2
Dicer’s helicase domain is a Super Family 2 (SF2) helicase and most similar to the RIG-I family of mammalian helicases (Fairman-Williams et al., 2010; Luo et al., 2013). Crystal structures of RIG-I have been reported (Civril et al., 2011; Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011; 2012), and the helicase domain of these structures fits well into EM maps of hsDcr (Lau et al., 2012; Taylor et al., 2013). Based on studies reported here, structural information for RIG-I, single particle electron micrographs of hsDcr (Lau et al., 2012; Taylor et al., 2013), and an x-ray co-crystal structure of dsRNA with a fragment of hsDcr containing the “cap” (Figure 1B; (Tian et al., 2014)) we propose a model by which dsRNA termini promote differential binding affinity, conformations, and reaction modes of dmDcr-2, in an ATP- and helicase-dependent manner (Figure 7). The model serves as a platform for summarizing our data, but further experimentation will be necessary to test its details.
Figure 7. Model for differential substrate recognition and processing by dmDcr-2.

dmDcr-2 (colored as in Figure 1) is shown interacting with BLT (left) and 3′ovr (right) dsRNA. Addition of ATP induces a conformational change that is favored to different degrees, as indicated by direction of red arrows. Loqs-PD (dsRBMs and isoform specific amino acids, orange spheres and tail, respectively) interacts with the helicase domain to stabilize the closed conformation. ATP hydrolysis is shown associated with the closed conformation. Additional details in text.
In our model we reorient the core domain (Figure 1B), containing the 3′ (green circle) and 5′ (blue circle) termini binding pockets (Tian et al., 2014), and draw the helicase domain with lobes to indicate sub-domains: Hel1, Hel2 and the Hel2i insertion characterized in Hef (Nishino et al., 2005). As cartooned (Figure 7), Hel2 (dark gray oval) is in a channel that continues into the RNase III active site, with Hel1 and Hel2i on either side (Lau et al., 2012). As in RIG-I co-crystals, and EM studies of hsDcr, in our model the apo enzyme can access multiple conformations upon binding dsRNA; we illustrate an intermediate closing of the helicase domain, as well as a clamping down of this domain around the dsRNA.
Consistent with our gel-shift analyses (Figure 2), in the absence of ATP, the model depicts 3′ovr, but not BLT, termini interacting with a termini binding pocket of dmDcr-2. At high dmDcr-2 concentrations, we observed low-affinity binding to BLT dsRNA in the absence of ATP, and this occurred at a dmDcr-2 concentration that also promoted binding to dsRNA blocked at termini with cyclic phosphates. In the model, dmDcr-2 is shown interacting with BLT dsRNA without engaging the termini binding pockets (-ATP); as yet it is unclear which domains of dmDcr-2 bind BLT dsRNA in the absence of ATP.
In the absence of ATP, dmDcr-2 can cleave 3′ovr, but not BLT, dsRNA; this cleavage is inefficient but precise, and always generates a product of 22 nts (e.g. Figure 5). Since dmDcr-2 binds 3′ovr termini in the absence of ATP (Figure 2A), this precise cleavage likely involves termini binding, presumably by the PAZ domain. Our model posits that 3′ovr dsRNA favors an intermediate closing of the helicase domain, which may allow a non-productive, cleavage-incompetent interaction where 3′ovr dsRNA angles away from Dicer’s core, as observed in certain EM images (Fig. 7; (Taylor et al., 2013)). The 3′ovr dsRNA would infrequently move close to the body of the enzyme allowing cleavage.
We find that ATP allows dmDcr-2 to bind to BLT termini, and also increases binding affinity for both BLT and 3′ovr termini (Figure 2A). Further, we observed a protease-resistant fragment that was dependent on ATP binding and dsRNA termini, occurring more readily in the presence of BLT, compared to 3′ovr, dsRNA (Figure 4). In our model the protease-resistant fragment reflects a closed conformation of the helicase domain that is favored in the presence of ATP and BLT dsRNA (Figure 7, bottom left). Although the proteolysis studies do not provide direct evidence of a conformational change, the observation of multiple conformations of hsDcr (Lau et al., 2012; Taylor et al., 2013), and the related RIG-I helicase (Kolakofsky et al., 2012; Luo et al., 2013) make this our preferred model. Our previous data (Welker et al., 2011) and the multiple-turnover data reported here (Figure 3) are consistent with a processive mode of cleavage with BLT dsRNA and a distributive mode of cleavage with 3′ovr dsRNA. In this light, a more closed conformation with BLT dsRNA would facilitate multiple cleavage events without dissociation.
We observed the protease resistant fragment in the presence of ATP as well as ATP-γS (Figure 4). RIG-I structures with nucleotide and BLT dsRNA show that ATP binding, without hydrolysis, enables compaction of the helicase domain (Kolakofsky et al., 2012; Luo et al., 2013). By analogy, our results indicate that ATP binding, without hydrolysis, is sufficient to promote the protease-resistant fragment that we propose signifies a closed conformation favored by BLT dsRNA (Figure 4). In contrast, generation of heterogeneous products with 3′ovr dsRNA is dependent on ATP hydrolysis (Figure 5), and hydrolysis is required for all products using a BLT dsRNA. By analogy to RIG-I, ATP hydrolysis may allow dmDcr-2 to translocate along dsRNA (Myong et al., 2009), possibly explaining the heterogeneous products. Further, we cannot exclude the possibility that heterogeneous products result from a threading of dsRNA into a closed helicase domain. ATP hydrolysis may also allow unwinding of BLT termini to facilitate interaction with termini binding pockets.
Addition of Loqs-PD allowed dmDcr-2 to cleave substrates that it could not cleave alone, such as those with 2′,3′-cyclic phosphates, long overhangs (esi-2), or terminal branched structures (rncs-1; Figure 6). Further, Loqs-PD promoted the protease resistant fragment (Figure 6D), allowing the 3′ovr cleavage pattern to look similar to that of BLT dsRNA. Loqs-PD interacts with dmDcr-2’s helicase domain using its second dsRNA-binding motif (dsRBM) and 22 amino acids unique to the PD isoform (Hartig and Förstemann, 2011; Miyoshi et al., 2010). In our model we show Loqs-PD interacting with the helicase domain to stabilize the closed conformation.
Implications for naturally occurring substrates
dsRBPs bind dsRNA of any sequence, and recognition of dsRNA termini, sometimes with protein domains different from those that recognize dsRNA, provides an alternate way to gain specificity. Both RIG-I (Hornung et al., 2006), and PKR (Nallagatla et al., 2007), dsRBPs involved in the mammalian innate immune response, recognize a 5′ triphosphate, presumably to distinguish viral transcripts with a 5′ triphosphate (“non-self”), from host transcripts (“self”), which are capped or processed to remove the 5′ triphosphate.
Our previous work (Welker et al., 2011), and that reported here, shows that both C. elegans DCR-1 and dmDcr-2 discriminate termini. However, except for miRNA precursors, the repertoire of termini found in natural Dicer substrates is unknown. miRNA precursors have 3′ovr termini and require only a single cleavage event to generate a mature miRNA; the less efficient distributive cleavage observed with 3′ovr dsRNA is suited to these substrates. In this study we focused on dmDcr-2, but D. melanogaster has a second Dicer, dmDcr-1, that lacks most of the helicase domain and is dedicated to miRNA processing (Lee et al., 2004).
dmDcr-2 is dedicated to siRNA processing, and the termini dependence we observe likely relates to these substrates. One type of siRNA, the endo-siRNAs, arise from genomic regions predicted to generate dsRNA, such as long hairpins, overlapping genes that give rise to sense-antisense pairs, repetitive elements and transposons (Okamura and Lai, 2008). These endo-siRNA precursors are inferred from deep-sequencing studies (Chung et al., 2008; Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008) and predicted to have termini features, such as long or frayed overhangs, that would prevent Dicer cleavage. Our studies suggest Loqs-PD can facilitate processing of these substrates.
A second type of siRNA arises from exogenous precursors, like viral dsRNA, and generates exo-siRNAs. Many organisms produce exo-siRNAs to mount an antiviral response (Ding and Voinnet, 2007; Marques and Carthew, 2007), including mammalian cells (Li et al., 2013; Maillard et al., 2013). For RNA viruses, viral replication intermediates consisting of genome-antigenome dsRNA are cleaved by Dicer to produce viral exo-siRNAs, and deep-sequencing shows an enrichment of siRNAs mapping to ends of viral genomes (Aliyari et al., 2008; Li et al., 2013; Maillard et al., 2013). In D. melanogaster the helicase activity of dmDcr-2 is required for an antiviral response, yet production of viral exo-siRNAs does not require Loqs-PD (Marques et al., 2013). Possibly, replication intermediates have termini that promote helicase-dependent cleavage, such as BLT termini.
EXPERIMENTAL PROCEDURES
Also see Supplemental Experimental Procedures.
Expression and purification of dmDcr-2 and Loqs-PD proteins
One-STrEP-FLAG (OSF)-tagged dmDcr-2WT, dmDcr-2RIII, and dmDcr-2RIII-K34A were expressed in Sf9 insect cells using the Bac-to-Bac Baculovirus Expression System (Invitrogen) with modifications, isolated from lysates with a StrepTrap HP column (GE Healthcare), dialyzed with PreScission Protease (GE Healthcare) to remove OSF-tag, purified using tandem ion-exchange chromatography and verified as monomeric by size exclusion chromatography. 6xHis-Loqs-PD was expressed in E. coli BL21-GOLD(DE3), isolated from lysates with Ni-NTA agarose (Qiagen), dialyzed with PreScission Protease to remove His-tag, and purified by cation-exchange chromatography.
dsRNA preparation
106 nt RNAs were prepared as described (Welker et al., 2011). Briefly, 106 nt sense, and BLT and 3′ovr antisense, strands were cloned into pss419 plasmid, which positions a hammerhead ribozyme at the 5′ end and a HDV ribozyme at the 3′ end. Ribozyme cleavage ensures precise and homogeneous termini of transcribed RNAs, leaving a 5′ hydroxyl and a 2′,3′-cyclic phosphate. 2′,3′-cyclic phosphates were removed by T4 PNK, which has a 2′,3′-cyclic phosphatase activity (Morse and Bass, 1997), except when 2′,3′-cyclic phosphates were desired. 106 nt single strands were gel purified after 8% denaturing PAGE. All denaturing gels contained 8 M urea unless otherwise specified. RNA strands were labeled as specified. 42 nt RNAs were chemically synthesized and gel purified after 8% denaturing PAGE. Equimolar single-strands were annealed, and dsRNAs gel purified after 8% native PAGE. rncs-1 and esi-2 were prepared as described (Hellwig and Bass, 2008; Miyoshi et al., 2010).
Gel mobility-shift assays
Gel-shift assays were with 20 pM dsRNA incubated (30 min, 4°C) with varying dmDcr-2 RIII, −/+ 5 mM ATP in binding buffer (25 mM TRIS pH 8.0, 100 mM KCl, 10 mM MgCl2, 10% (vol/vol) glycerol, 1 mM TCEP). Reactions were stopped by loading onto a 4% (19:1 acrylamide/bisacrylamide) native gel running at 150 V, 4°C in 0.5x Tris/borate/EDTA. Gels were electrophoresed for an additional 2.5 h, dried and autoradiographed.
Cleavage assays
Cleavage assays were at 25°C for times indicated, i n cleavage buffer (25 mM TRIS pH 8.0, 100 mM KCl, 10 mM MgCl2, 1 mM TCEP) with dmDcr-2WT and dsRNA as specified, −/+ 5 mM ATP. Reactions were started by adding dmDcr-2WT and stopped with 2 volumes of 2x formamide loading buffer (95% Formamide, 18 mM EDTA, 0.025% SDS, Xylene Cyanol, Bromophenol Blue). Multiple-turnover cleavage assays were as described above except reactions were stopped with 4 volumes 2x formamide loading buffer and loading 1/5th volume for denaturing PAGE. For reactions with Loqs-PD, equimolar dmDcr-2WT and Loqs-PD were incubated on ice (5 min), before addition to reaction. For reactions with hexokinase and glucose, hexokinase (1 unit, which phosphorylates 1 micromole of D-glucose/min at pH 7.6 at 25°C) and glucose (10 mM; 0.2 micromoles) were incubated in reaction mix containing 5 mM ATP (or analog) (20 min; 25°C) to deplete ATP (or contaminating ATP) before adding dmDcr-2. Products were separated by 12% or 17% denaturing PAGE as specified, visualized on a PhosphorImager screen (Molecular Dynamics) and quantified with ImageQuant software.
Limited proteolysis
Limited proteolysis was at 25°C in cleavage assay b uffer. In a 50 μl reaction, 20 μg of dmDcr-2RIII (2 μM) was incubated (20 min) with or without 10 μM dsRNA as specified, −/+ 5 mM ATP (or analogs), before treating with trypsin (10 nM) for times indicated; 4 μg of dmDcr-2RIII was then quenched at indicated times with an equal volume of 2x SDS-PAGE loading buffer. Hexokinase and glucose treatment was performed as described above with 7.5 μg of hexokinase in the reaction. Products were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue.
ATP hydrolysis assays
Reactions were at 25°C in cleavage buffer with 200 nM dmDcr-2WT, 600 nM dsRNA as specified, and 100 μM ATP. [α-32P] ATP (3000 Ci/mmol, 100 nM) was used to monitor hydrolysis. Reactions were started by adding dmDcr-2WT, quenched at indicated times in 100 mM EDTA, spotted onto PEI-cellulose plates, and chromatographed with 0.75 M KH2PO4 (adjusted to pH 3.3 with H3PO4) until solvent front reached top of plate. Plates were dried, visualized on a PhosphorImager screen (Molecular Dynamics) and quantified with ImageQuant software. For reactions with Loqs-PD, equimolar dmDcr-2WT and Loqs-PD were preincubated (ice, 5 min) before initiating reaction.
Supplementary Material
Highlights.
Drosophila Dicer-2 requires ATP to bind blunt, but not 3′ overhanging, dsRNA termini
Dicer-2 undergoes an ATP-, helicase-, and dsRNA termini- dependent conformational change
The proposed conformational change requires binding, but not hydrolysis, of ATP
Loquacious-PD modulates termini-dependent cleavage by Drosophila Dicer-2
Acknowledgments
We thank the Bass lab for invaluable feedback, V. Chandrasekaran, S. Alam, and C. Georgopoulos for advice on insect cell expression and protein purification; UU DNA/Peptide Core Facility for RNA synthesis (NCI CA04214); J. Schulze and D. Weber (Molecular Structure Facility, UC Davis) for assistance with Edman Sequencing and LC/MS/MS. This work was supported by the H.A. and Edna Benning Presidential Endowed Chair to BLB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
N.K.S., K.D.T., and B.L.B. designed experiments. N.K.S., and K.D.T. prepared samples. N.K.S., K.D.T., and P.J.A. conducted experiments. N.K.S., K.D.T., and B.L.B. analyzed data. B.L.B., N.K.S., and K.D.T. wrote the manuscript.
References
- Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Bennett M, Moldt M, Deimling T, Witte G, Schiesser S, Carell T, Hopfner KP. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakić M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007;27:449–461. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SM, Melendez-Peña CE, Scarborough RJ, Daher A, Christensen HS, El Far M, Purcell DFJ, Lainé S, Gatignol A. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol Biol. 2009;10:38. doi: 10.1186/1471-2199-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zhang X, Graves P, Zeng Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. Rna. 2012;18:2083–2092. doi: 10.1261/rna.033688.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Bahubeshi A, Hamel N, Pasini B, Asioli S, Baynam G, Choong CS, Charles A, Frieder RP, Dishop MK, et al. Extending the phenotypes associated with DICER1 mutations. Hum Mutat. 2011;32:1381–1384. doi: 10.1002/humu.21600. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Colpan C, Han BW, Zamore PD. Inorganic phosphate blocks binding of pre-miRNA to Dicer-2 via its PAZ domain. Embo J. 2014;33:371–384. doi: 10.1002/embj.201387176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Förstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011;39:3836–3851. doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig S, Bass BL. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci USA. 2008;105:12897–12902. doi: 10.1073/pnas.0805118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D, Kowalinski E, Cusack S. A structure-based model of RIG-I activation. Rna. 2012;18:2118–2127. doi: 10.1261/rna.035949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nat Struct Mol Biol. 2012;19:436–440. doi: 10.1038/nsmb.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping – the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Kohlway A, Pyle AM. Duplex RNA Activated ATPases (DRAs): Platforms for RNA sensing, signaling and processing. RNA Biol. 2013;10:111–120. doi: 10.4161/rna.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Kohlway A, Vela A, Pyle AM. Visualizing the determinants of viral RNA recognition by innate immune sensor RIG-I. Structure. 2012;20:1983–1988. doi: 10.1016/j.str.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Zhou K, Kidwell MA, Doudna JA. Coordinated activities of human Dicer domains in regulatory RNA processing. J Mol Biol. 2012;422:466–476. doi: 10.1016/j.jmb.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Carthew RW. A call to arms: coevolution of animal viruses and host innate immune responses. Trends Genet. 2007;23:359–364. doi: 10.1016/j.tig.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Marques JT, Wang JP, Wang X, de Oliveira KPV, Gao C, Aguiar ERGR, Jafari N, Carthew RW. Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog. 2013;9:e1003579. doi: 10.1371/journal.ppat.1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovic-Hösle M, Förstemann K. Transposon defense by endo-siRNAs, piRNAs and somatic pilRNAs in Drosophila: contributions of Loqs-PD and R2D2. PLoS ONE. 2014;9:e84994. doi: 10.1371/journal.pone.0084994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. Rna. 2010;16:506–515. doi: 10.1261/rna.1952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Bass BL. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry. 1997;36:8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- Nicholson AW. Dissecting Human Dicer: Some Assembly Required. J Mol Biol. 2012;422:464–465. doi: 10.1016/j.jmb.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin LR, Delás MJ, Hannon GJ. Dogma Derailed: The Many Influences of RNA on the Genome. Mol Cell. 2013;49:783–794. doi: 10.1016/j.molcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DW, Ma E, Shigematsu H, Cianfrocco MA, Noland CL, Nagayama K, Nogales E, Doudna JA, Wang HW. Substrate-specific structural rearrangements of human Dicer. Nat Struct Mol Biol. 2013;20:662–670. doi: 10.1038/nsmb.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ma JB, Park JE, Heo I, Kim VN, Patel DJ. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol Cell. 2014;53:606–616. doi: 10.1016/j.molcel.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell. 2011;41:589–599. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annual Review of Biophysics. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. Embo J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


