Antibiotic agents are among the most important contributors to the modernization of medicine, and it is difficult to imagine the continuation of advances of recent years without them. However, the emergence of antibiotic resistance threatens our ability to care for patients and is among the top public health threats of the 21st century. The Centers for Disease Control and Prevention conservatively estimates that at least 23,000 people die annually in the United States as a result of an infection with an antibiotic-resistant organism and that more than 2 million are sickened. From another perspective, the toll of antibiotic-resistant infections is much greater than the tolls of epidemic diseases such as human immunodeficiency virus infection or Ebola virus disease. According to a recent report from the United Kingdom,1 the human cost of the antibiotic-resistance crisis is estimated to be 300 million cumulative premature deaths by 2050, with a loss of up to $100 trillion (£64 trillion) to the global economy. This dire situation has been highlighted for years by the Infectious Diseases Society of America2 and is now a priority of the U.S. government.
Producing new antibiotics in the 21st century has been a daunting task. In the very successful era of the mid-20th century, antibiotic discovery typically consisted of screening cultures of soil-derived organisms such as streptomyces for activity against other microorganisms. This approach produced a plethora of “hits” between 1940 and 1960, from which multiple new antibiotics with expanded activity and potency were developed. However, by the 1970s, this golden era of antibiotic discovery started to fade, with the repeated identification of the same compounds. By the 1990s, with antibiotic resistance increasing, several antibiotic-discovery programs were launched that used genomics, high-tech chemical approaches, and high-throughput screening, but these proved expensive and inefficient. For example, the high-throughput screening approach of GlaxoSmithKline required 14 runs to discover one lead, at a cost of $1 million per campaign,3 and not one compound advanced to the final stage of clinical development; this has also been the experience of other pharmaceutical companies. As a result, and because of the poor economic return on investment of antibiotics,2 many companies halted their antibiotic research and development programs to focus on more economically favorable areas.
Against this bleak landscape, a recent report by Ling et al.4 brought a ray of light. The authors, using the isolation chip (iChip) that one of them had previously described,5 were able to culture microorganisms (in isolation from one another) from soil that had not been able to be cultured in vitro previously (estimated as approximately 99% of environmental bacteria). The myriad tiny agar-filled chambers of the iChip were first seeded with dilutions of soil containing approximately one bacterium per chamber and were then covered with a semipermeable membrane and placed back into the soil, permitting nutrients to diffuse into the chambers (Fig. 1).
Figure 1. Two Methods of Culturing Microorganisms from Soil.
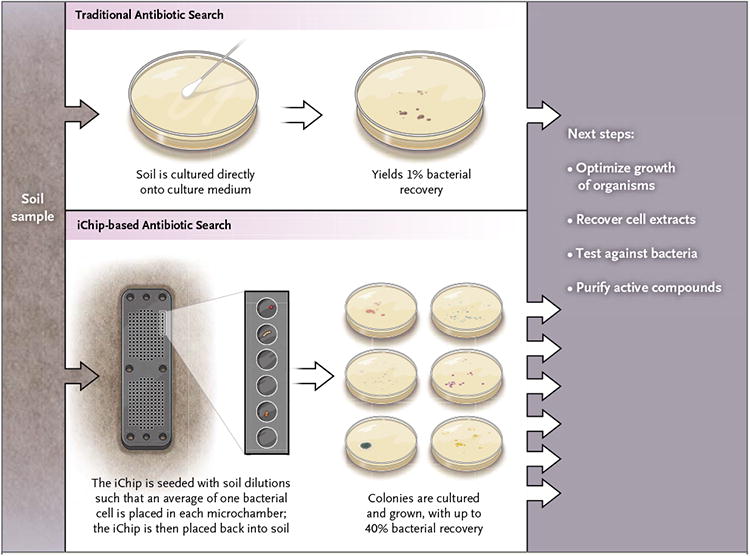
The traditional search for antibiotic agents involves culturing soil directly onto culture medium (e.g., an agar plate), which detects an estimated 1% of organisms present. Ling and colleagues4 used an isolation chip (iChip).5 After dilutions of soil are inoculated so that approximately one bacterial cell goes into each agar-filled chamber, the device is placed back in the soil. Many more bacteria survive and grow in the iChip than do on a traditional agar plate and, once established, are more likely to grow in vitro.
After prolonged incubation, many chambers contained bacterial colonies that now grew on enrichment medium, outside the soil. Further processing, extraction, and sophisticated chemical separation techniques identified an 11-amino-acid peptide antibiotic, designated teixobactin, which is produced by the provisionally named gram-negative bacterium Eleftheria terrae and inhibits the growth of Staphylococcus aureus. Teixobactin appears to act by forming a complex with precursors of peptidoglycan and teichoic acids of the cell wall of gram-positive bacteria. Teixobactin showed potent bactericidal activity against gram-positive pathogens, with minimal inhibitory concentrations of 0.5 μg per milliliter or less for staphylococci, streptococci (including pneumococci), Clostridium difficile, Bacillus anthracis, and enterococci, including multidrug-resistant strains. Teixobactin is similarly potent against Mycobacterium tuberculosis, a pathogen against which there is a current and urgent unmet medical need. In vivo studies in the mouse corroborated the activity of teixobactin against methicillinresistant S. aureus, and even after multiple rigorous attempts, the authors could not select teixobactinresistant mutants of S. aureus or M. tuberculosis.
This work represents a notable advance for the discovery of antibiotics that target gram-positive bacteria and M. tuberculosis. Gram-negative bacteria, like the producing species, are resistant to teixobactin because they lack one of the targets and because of the barrier effect of their outer membrane, which gram-positive bacteria do not have. The expectation is that tapping into a reservoir of microorganisms that is approximately 100 times as large as the reservoir that could be tested previously will provide fertile ground for the discovery of new compounds with activity against multidrug-resistant gram-negative bacteria like acinetobacter, pseudomonas, and carbapenemase-resistant Enterobacteriaceae. These pathogens are extremely urgent public health threats, and a new vein to fuel an antimicrobial-discovery pipeline is welcome. That said, it can take years for a discovery to yield an approved, commercial product.
If history has taught us any lesson about resistance, it is that the lack of selection of resistance to teixobactin in vitro should be viewed with great caution. Similar claims were made about vancomycin, because it targeted an essential component of bacterial cell walls thought to be irreplaceable. However, after large-scale use of vancomycin began in the 1980s, resistance soon emerged. Soil organisms have had millions of years to develop resistance to teixobactin, and it is possible that such resistance genes are already present in nature or that mutational resistance will arise in vivo after prolonged use. For now, though, we must take advantage of this expanded pool of testable organisms.
Acknowledgments
We thank William Miller, M.D., for scientific input during the writing of the manuscript.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org
References
- 1.Antimicrobial resistance: tackling a crisis for the health and wealth of nations. O'Neill J., chair 2014 Dec; http://amr-review.org/sites/default/files/AMR%20Review%20Paper%20 -%20Tackling%20a%20crisis%20for%20the%20health%20and %20wealth%20of%20nations_1.pdf.
- 2.Boucher HW, Talbot GH, Benjamin DK, Jr, et al. 10 × ′20 Progress — development of new drugs active against gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis. 2013;56:1685–94. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 4.Ling LL, Schneider T, Peoples AJ, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–9. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols D, Cahoon N, Trakhtenberg EM, et al. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol. 2010;76:2445–50. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


